Abstract
Hybridization among sea turtle species has been widely reported in the Atlantic Ocean, but their detection in the Pacific Ocean is limited to just two individual hybrid turtles, in the northern hemisphere. Herein, we report, for the first time in the southeast Pacific, the presence of a sea turtle hybrid between the green turtle Chelonia mydas and the hawksbill turtle Eretmochelys imbricata. This juvenile sea turtle was captured in northern Peru (4°13′S; 81°10′W) on the 5th of January, 2014. The individual exhibited morphological characteristics of C. mydas such as dark green coloration, single pair of pre-frontal scales, four post-orbital scales, and mandibular median ridge, while the presence of two claws in each frontal flipper, and elongated snout resembled the features of E. imbricata. In addition to morphological evidence, we confirmed the hybrid status of this animal using genetic analysis of the mitochondrial gene cytochrome oxidase I, which revealed that the hybrid individual resulted from the cross between a female E. imbricata and a male C. mydas. Our report extends the geographical range of occurrence of hybrid sea turtles in the Pacific Ocean, and is a significant observation of interspecific breeding between one of the world’s most critically endangered populations of sea turtles, the east Pacific E. imbricata, and a relatively healthy population, the east Pacific C. mydas.
Keywords: Hybridization, Cheloniidae, Interspecific breeding, Cytochrome oxidase I, Female Eretmochelys imbricata
Introduction
Hybrids of sea turtles are uncommon in nature and they seem to occur only among species of the Cheloniidae family as no hybrids involving leatherback turtle (Dermochelys coriacea) have been found (Karl, Bowen & Avise, 1995). As far back as one hundred years ago (Wood, Wood & Critchley, 1983), hybrid individuals were observed to show intermediate morphological traits of species (Wood, Wood & Critchley, 1983). However, due to the high phenotypic plasticity inherent to species morphological traits, biochemical confirmation of the existence of sea turtles hybrids was needed to confirm the occurrence of hybridization. Protein electrophoresis was the earliest technique that proved the existence of hybrids Chelonia mydas × Eretmochelys imbricata and Caretta caretta × E. imbricata (Wood, Wood & Critchley, 1983; Conceição et al., 1990), but the use of advanced molecular and genetics assays reveal intercrossing among more species (Karl, Bowen & Avise, 1995). It is thought that sea turtle hybrids occur due to a disproportional abundance of species with similar distribution range and overlapping habitats such as nesting rookeries (Proietti et al., 2014). Also, unequal female:male ratio may also promote mating encounters between individuals of different species (Proietti et al., 2014). In any case, it is unclear whether this is a survival strategy in case of low population numbers or just a natural mechanism of evolution within these marine reptiles. Considering this, reporting the existence of hybrids is important for understanding the prevalence of and the patterns underlying hybridization among sea turtle species.
Although records of sea turtle hybrids exist from several places around the world (Karl, Bowen & Avise, 1995), there are only two reports of hybrids between C. mydas × E. imbricata, one in the Atlantic (Wood, Wood & Critchley, 1983) and the second one in the Pacific Ocean (Seminoff et al., 2003). In the Atlantic, several of these hybrids were found in a C. mydas nest in Suriname, likely the offspring of a female C. mydas crossed with a male E. imbricata; twenty years later, the Pacific record reported in Seminoff et al. (2003) highlighted that this hybridization is apparently not gender-biased, since theirs was the first observation of hybridization between a male C. mydas and a female E. imbricata. Herein, we report a new case of such hybridization between C. mydas and E. imbricata found in northern Peru. The finding adds support to the absence of gender bias between these species and constitutes the first report of a hybrid sea turtle in the Southeast Pacific.
Methods
Sampling location
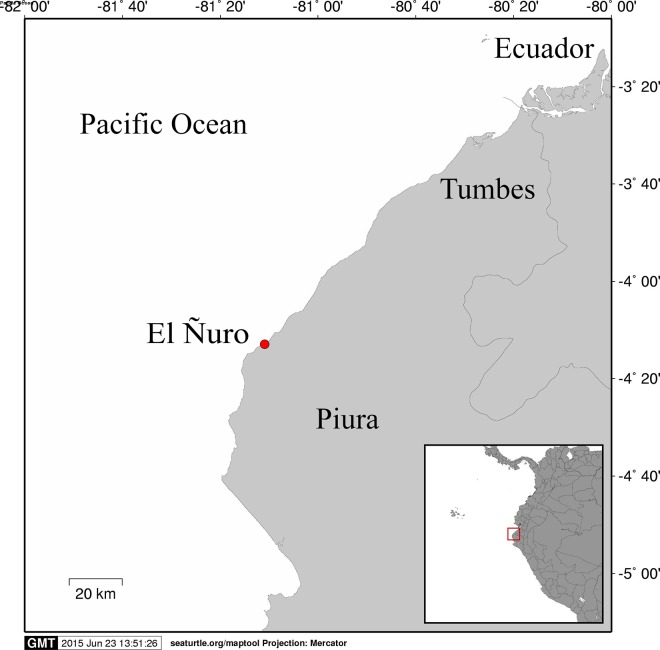
The individual was collected during a seasonal green turtle monitoring effort conducted at El Ñuro (4°13′S; 81°10′W, Fig. 1), a large sandy neritic area with rocky reefs in the coast of the department of Piura, northern Peru. The characteristics of the study site and details of the sampling strategy are available in Velez-Zuazo et al. (2014). Permits for the study were granted from the Dirección General Forestal y Fauna Silvestre: RD N°0383-2010-AG-DGFFS-DGEFFS and RD N°0606-2011-AG-DGFFS-DGEFFS.
Figure 1. Map of northern Peru showing the location where the hybrid sea turtle was found.
SEATURTLE.ORG Maptool. 2002. SEATURTLE.ORG, Inc. http://www.seaturtle.org/maptool/ (July 2015).
On the 5th of January, 2014, a small sea turtle was observed surfacing frequently during the first hour of the survey after deploying the entanglement net. At 9:50 AM the individual was caught in the net and was brought on board. During the first visual examination, it was evident that the sea turtle presented morphological characteristics of both Chelonia mydas and Eretmochelys imbricata (Fig. 2). The following body measurements were taken: notch to tip Curved Carapace Length (CCLn-t) and Curved Carapace Width (CCW), taken with a 100-cm soft measuring tape and 0.1 cm accuracy, notch to tip Straight Carapace Length (SCLn-t) and Straight Carapace Width (SCW) measured with a 100-cm Haglöf tree caliper, and weight, estimated with a 100 kg spring scale. Subsequently, a sample of tissue from the right shoulder area was obtained using a 4 mm-diameter biopsy punch (Acuderm) and stored in 90% ethanol at room temperature. Photographs of all characteristics were taken. Finally, inconel tags with unique identification codes were applied at both front flippers before releasing the individual into the water. The turtle was recaptured at 08:55 am on the following day, and released again after taken additional photographs.
Figure 2. Hybrid individual between Chelonia mydas and Eretmochelys imbricata captured at El Ñuro, northern Peru.
Morphological traits of Chelonia; (A) pair of pre-frontal scales, (B) four post-orbitales scales, (C) dark green coloration. Morphological traits of Eretmochelys; (C) peeling carapace, (D) pinkish plastron with epibionts, (E) two nails at the flipper.
Molecular analysis
To identify the maternal lineage of our individual and the likely origin of the mother we conducted a phylogenetic reconstruction analyzing the nucleotide variation in the gene cytochrome oxidase I (cox1) from the mitochondrial DNA (mtDNA). Whole genomic DNA was isolated using a Qiagen DNeasy blood and tissue kit according to manufacturer’s instructions and eluted in 50 μl of buffer AE (Qiagen, Valencia, CA, USA). Approximately 679 base-pairs of cox1 were targeted and amplified through Polymerase Chain Reaction (PCR), using specific primers (M13-tailed cocktail primers Fish-F1t1and Fish-R1t1, Ivanova et al., 2007). PCR conditions for an 8 μl amplification product were as follow: 1 μl of genomic DNA at a concentration of ∼20 ng/μl, 5 μl of Taq Master Mix (Qiagen, Valencia, CA, USA), 0.5 μl of each 10 uM primer cocktail and ultrapure water. Cycling conditions were an initial denaturing step (94 °C for 2 min) followed by 35 cycles of 30 s at 94 °C, 40 s at 52 °C, and 1 min at 72 °C, and a final extension of 10 min at 72 °C. The amplification product was purified using a phosphatase and exonuclease and sequencing of both strands was conducted using an automated station ABI 3130xl sequencer (Applied Biosystems, Foster City, CA, USA). Both forward and reverse sequences were edited using Sequencher 4.8 (Gene Codes) and aligned with sea turtle species baseline sequences downloaded from the Barcode of Life Project (BOLD, www.boldsystems.org).
To conduct the phylogenetic reconstruction we used two approaches. First, we used BOLD Identification System to compare our sequence to the species level barcode records. Identification using this method relies in a Neighbor-Joining analysis. Second, we downloaded barcode sequences for all extant species of sea turtle and from different regions of the world (See Supplementary Information). All sequences, including the sequence from our specimen, were aligned using ClustalW, with default parameters, as implemented in Mega 5.05 (Tamura et al., 2011). A Maximum Likelihood (ML) algorithm implemented in RAxML (Stamatakis, 2014) using the GTR model of nucleotide substitution with GAMMA correction and a rapid Bootstrap analysis using the auto MRE bootstopping criterion was used. We set the leatherback sea turtle (Dermochelys coriacea) sequence as outgroup for the phylogenetic inference. Figtree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/) to draw the consensus tree and for including values of branch support for all analyses.
Results
Morphological characteristics
The hybrid sea turtle measured 47.3 cm CCLn-t, 42.3 cm CCW, 44.6 cm SCLn-t, 34.1 cm SCW, and weighted 11 kg, and it was categorized as a juvenile since Chelonia mydas and Eretmochelys imbricata reach sexual maturity at >85 and 78.8 cm CCL respectively (Liles et al., 2011; Zárate et al., 2013). Most of the external morphological characteristics resemble those of C. mydas such as dark green coloration of the carapace and skin, non-serrated marginal carapace, pair of pre-frontal scales, four post-orbital scales (Fig. 2 and Table 1), and the presence of a median ridge in the lower jaw. Visible E. imbricata characteristics were a pair of claws on each front flipper, long beak, non-imbricated peeling scutes, plastron with pink coloration, and with high presence of epibionts. High density of epibionts on the plastron were previously observed in E. imbricata captured at El Ñuro, but such densities of barnacles are uncommon on C. mydas at this site (Kelez et al., 2010–2014, personal observations).
Table 1. Comparison of morphological traits among hawksbill, green and hybrid turtles including the individual found at El Ñuro.
| Hawksbill | Hybrids | Green | |||
|---|---|---|---|---|---|
| Characteristics | Suriname1 | Mexico2 | Peru3 | ||
| Imbricated scutes | Yes | Yes | Yes | No | No |
| Elongated snout | Yes | Yes | Yes | Yes | No |
| Second claw | Yes | Yes | No | Yes | No |
| Marginal scute serration | Yes | Yes | Yes | No | No |
| Pre-frontal scales | Four | Variable | Two | Two | Two |
| Post-orbital scales | Three | Not reported | Three | Four | Four |
| Mandibular middle ridge | No | Yes | Yes | Yes | Yes |
Maternal lineage identification and origin
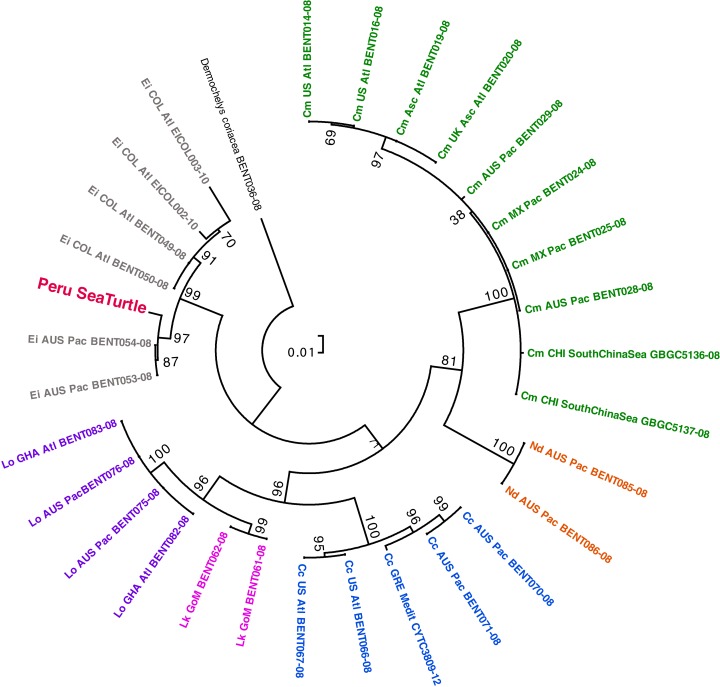
We obtained a 717bp sequence of mtDNA cox1 that we used for phylogenetic tree reconstruction (GenBank accession number KU254594). Both, the NJ phylogenetic used by BOLD and the ML phylogenetic tree we reconstructed placed our specimen within the Eretmochelys imbricata clade. The results provided by BOLD species identification, indicated that the sequence of our individual matched to E. imbricata with a probability interval of 93.22–99.57% at the species level, and 100% at the genus level. Similarly, the ML phylogenetic reconstruction placed our specimen within the E. imbricata clade (99% node support for ML consensus tree), specifically within the clade composed of individuals from populations from the Pacific Ocean basin (97% node support for ML consensus tree) (Fig. 3).
Figure 3. Phylogenetic placement of the hybrid specimen as suggested after a Maximum Likelihood analysis.
Species were color-labeled by genus and are Chelonia mydas (green), Natator depressus (orange), Caretta caretta (blue), Lepidochelys kempii (fucsia), Lepidochelys olivacea (purple), and Eretmochelys imbricata (gray). The specimen under study is indicated with a dark pink label.
Discussion
To the best of our knowledge, this is the first time a hybrid sea turtle has been reported in the southeast Pacific. Furthermore, it represents the second case of a crossing between a female Eretmochelys imbricata and a male Chelonia mydas in the entire Pacific Ocean, and only the third case in all oceans. Morphologically, the appearance of the hybrid individual mostly resembled that of C. mydas; however, the comparison with the rest of E. imbricata and C. mydas hybrids suggest that only the mandibular middle ridge from C. mydas and the elongated snout from E. imbricata is present in hybrids, while other morphological traits are somehow variable (Table 1). Because the individual generally resembled a C. mydas, it may be possible that the existence of more hybrids is going unnoticed without careful morphological evaluation. In addition, we suggest that whenever possible, skin samples for molecular analyses should be taken from individuals with indistinct morphological characteristics.
Difference in abundance between species has been proposed as a condition that may promote hybridization. Nesting numbers in C. mydas rookeries had been recovering over the last 30 years in the eastern Pacific, and are significantly higher when compared to the E. imbricata eastern Pacific nesting regions (Seminoff et al., 2012). In fact, due to its small size and the numerous threats it faces, the eastern Pacific E. imbricata is one of the eleven most endangered sea turtle populations in the world (Wallace et al., 2011). This contrast between populations is evidenced in foraging aggregations of sea turtles in northern Peru. From 2010 to 2014, during seasonal (n = 11) in-water surveys at El Ñuro, a total of 186 different individual C. mydas were captured but only two E. imbricata were observed (Velez-Zuazo et al., 2014). Also, for both species, foraging and nesting habitat overlap occurs to some degree in the eastern Pacific. Both species are distributed throughout the same coastal line and can be found in algal seabed, rocky reefs and especially in mangrove habitats, as recently reported in Gaos et al. (2012). However, there are no rookeries in Peru and also adult E. imbricata are uncommon in Peruvian waters (Hays-Brown & Brown, 1982; Kelez, Velez-Zuazo & Manrique, 2003; Alfaro-Shigueto et al., 2010; Rosales, Vera & Llanos, 2010; Quiñones et al., 2011); all but one of the E. imbricata reported in Peru have been juveniles (Forsberg, 2008). Therefore, it is likely that mating of this hybrid’s progenitors occurred in waters to the north of Peru where E. imbricata rookeries are located (i.e., Ecuador, Costa Rica, Nicaragua, El Salvador, Mexico). Moreover, given that both E. imbricata × C. mydas hybrids reported so far in the eastern Pacific resulted from a crossing between a male C. mydas and a female E. imbricata, it is possible that male E. imbricata are available in insufficient numbers to breed during reproductive seasons due to the reduced population size of the eastern Pacific E. imbricata. For these reasons, we hypothesize that, the current population status and habitat overlap may be the reason for the existence of hybrids of these two species in the southeast Pacific Ocean.
Taxonomically, C. mydas and E. imbricata belong to the Chelonini and Carettini tribes, respectively, and were phylogenetically separated more than 50 millions of years ago (Karl, Bowen & Avise, 1995). Therefore, this hybridization case clearly shows interbreeding between ancient sea turtles linages, an event reported for other populations. For example, at the Bahia rookery (Brazil, Southwest Atlantic) 42% of sampled females identified as hawksbills were actually hybrids between Caretta caretta and E. imbricata; these hybrids are fertile and reproductively successful (Lara-Ruiz et al., 2006). This brings over the table a previous discussion about what is the most appropriate scheme for taxonomic classification of species (Avise & Johns, 1999). Although we were unable to ascertain the generation and fertility of the hybrid captured in northern Peru, we cannot overrule that reproduction between hybrids could be occurring. We recommend that additional research be conducted to further identify hybrid individuals in the eastern Pacific, as well as estimate their abundance, fertility and the ecological role they are playing in marine ecosystems.
Supplemental Information
Supplementary Data. Sequences used for phylogenetic analyses. All sequences were downloaded from the Barcode of Life Database System, except for the sequence obtained in this study.
Acknowledgments
We thank the support of El Ñuro fishermen community, L. Klinge and all the volunteers during in-water surveys. We thank J.A. Seminoff, B.J. Wallace, M.C. Proietti and two anonymous reviewers for revising this manuscript. Thanks to SEATURTLE.ORG Maptool (2002), http://www.seaturtle.org/maptool/ for helping to create Fig. 1.
Funding Statement
Duke Marine Laboratory, Fund for Nature–Japan, Principia College, Krilca Gifts, CNPC-Peru and ecOceanica turtle adoption program provided funds for our research in northern Peru. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
There are no competing interests.
Author Contributions
Shaleyla Kelez conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Ximena Velez-Zuazo conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper, genetic Analysis.
Aldo S. Pacheco conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
We were granted permits from the Direccion Gerenal Forestal y Fauna Silvestre: RD N°0383-2010-AG-DGFFS-DGEFFS and RD N°0606-2011-AG-DGFFS-DGEFFS.
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
We were granted permits from the Direccion Gerenal Forestal y Fauna Silvestre: RD N°0383-2010-AG-DGFFS-DGEFFS and RD N°0606-2011-AG-DGFFS-DGEFFS.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
GenBank Accession Number KU254594.
References
- Alfaro-Shigueto et al. (2010).Alfaro-Shigueto J, Mangel JC, Caceres C, Seminoff JA, Gaos A, Yanez I. Hawksbill turtles in Peruvian coastal fisheries. Marine Turtle Newsletter. 2010;129:19–21. [Google Scholar]
- Avise & Johns (1999).Avise JC, Johns GC. Proposal for a standardized temporal scheme of biological classification for extant species. Proceedings of the National Academy of Sciences. 1999;96(13):7358–7363. doi: 10.1073/pnas.96.13.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceição et al. (1990).Conceição MB, Levy JA, Marins LF, Marcovaldi MA. Eletrophoretic characterization of a hybrid between Eretmochelys imbricata and Caretta caretta (Cheloniidae) Comparative Biochemistry and Physiology–Part B. 1990;97(2):275–278. doi: 10.1016/0305-0491(90)90280-7. [DOI] [Google Scholar]
- Forsberg (2008).Forsberg K. Proyecto tortugas marinas: iniciativas y esfuerzos para la conservacion de las tortugas marinas en Tumbes. In: Kelez S, van Oordt F, de Paz N, Forsberg K, editors. Book of abstracts, II Simposio de tortugas marinas en el Pacifico Sur Oriental. Peru: Lima; 2008. pp. 69–70. Available at http://media.wix.com/ugd/413492_1483a24c9a914fc0bac8302e360f914a.pdf . [Google Scholar]
- Gaos et al. (2012).Gaos AR, Lewison RL, Yañez IL, Wallace BP, Liles MJ, Nichols WJ, Baquero A, Hasbún CR, Vasquez M, Urteaga J. Shifting the life-history paradigm: discovery of novel habitat use by hawksbill turtles. Biology Letters. 2012;8(1):54–56. doi: 10.1098/rsbl.2011.0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays-Brown & Brown (1982).Hays-Brown C, Brown WM. Status of sea turtles in the Southeastern Pacific: emphasis on Peru. In: Bjorndal KA, editor. Biology and conservation of Sea Turtles. Washington, D.C.: Smithsonian Institution Press; 1982. pp. 235–240. Available at http://igalsela.tripod.com/English/Sela-Turtles-1981.pdf . [Google Scholar]
- Ivanova et al., (2007).Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes. 2007;7:544–548. doi: 10.1111/j.1471-8286.2007.01748.x. [DOI] [Google Scholar]
- Karl, Bowen & Avise (1995).Karl SA, Bowen BW, Avise JC. Hybridization among the ancient mariners: characterization of marine turtle hybrids with molecular genetic assays. Journal of Heredity. 1995;86:262–268. doi: 10.1093/oxfordjournals.jhered.a111579. [DOI] [PubMed] [Google Scholar]
- Kelez, Velez-Zuazo & Manrique (2003).Kelez S, Velez-Zuazo X, Manrique C. Current status of sea turtles along the northern coast of Peru: preliminary results. In: Seminoff JA, editor. Proceedings of the Twenty-Second Annual Symposium on Sea Turtle Biology and Conservation, NOAA Technical Memorandum NMFS-SEFSC-503; Washington, D.C.: NOAA; 2003. pp. 264–265. Available at www.nmfs.noaa.gov/pr/pdfs/species/turtlesymposium2002.pdf . [Google Scholar]
- Lara-Ruiz et al. (2006).Lara-Ruiz P, Lopez G, Santos F, Soares L. Extensive hybridization in hawksbill turtles (Eretmochelys imbricata) nesting in Brazil revealed by mtDNA analyses. Conservation Genetics. 2006;7(5):773–781. doi: 10.1007/s10592-005-9102-9. [DOI] [Google Scholar]
- Liles et al. (2011).Liles MJ, Jandres MV, López WA, Mariona GI, Hasbún CR, Seminoff JA. Hawksbill turtles Eretmochelys imbricata in El Salvador: nesting distribution and mortality at the largest remaining nesting aggregation in the eastern Pacific Ocean. Endangered Species Research. 2011;14(1):23–30. doi: 10.3354/esr00338. [DOI] [Google Scholar]
- Quiñones et al. (2011).Quiñones J, Zeballos J, Quispe S, Delgado L. Southernmost records of hawksbill turtles along the east Pacific coast of South America. Marine Turtle Newsletter. 2011;130:16–19. [Google Scholar]
- Proietti et al. (2014).Proietti MC, Reisser J, Marins LF, Marcovaldi MA, Soares LS, Monteiro DS, Wijeratne S, Pattiaratchi C, Secchi ER. Hawksbill × loggerhead sea turtle hybrids at Bahia, Brazil: where do their offspring go? PeerJ. 2014;2:e1712. doi: 10.7717/peerj.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales, Vera & Llanos (2010).Rosales CA, Vera M, Llanos J. Varamientos y captura incidental de tortugas marinas en el litoral de Tumbes, Perú. Revista Peruana de Biologia. 2010;17(3):293–301. doi: 10.15381/rpb.v17i3.4. [DOI] [Google Scholar]
- Seminoff et al. (2012).Seminoff JA, Alfaro-Shigueto J, Amorocho D, Arauz R, Baquero Gallegos A, Chacon Chaverri D, Gaos A, Kelez S, Mangel JC, Urteaga J, Wallace B. Biology and conservation of sea turtles in the eastern Pacific Ocean, a general overview. In: Seminoff JA, Wallace B, editors. Sea Turtles of the Eastern Pacific, Advances in Research and Conservation. Tucson: University of Arizona Press; 2012. pp. 11–38. [Google Scholar]
- Seminoff et al. (2003).Seminoff JA, Karl SA, Schwartz T, Resendiz A. Hybridization of the green turtle (Chelonia mydas) and hawksbill turtle (Eretmochelys imbricata) in the Pacific Ocean: indication of an absence of gender bias in the directionality of crosses. Bulletin of Marine Science. 2003;73:643–652. [Google Scholar]
- Stamatakis (2014).Stamatakis A. A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (RAxML version 8) 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura et al. (2011).Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez-Zuazo et al. (2014).Velez-Zuazo X, Quiñones J, Pacheco AS, Klinge L, Paredes E, Quispe S, Kelez S. Fast growing, healthy and resident green turtles (Chelonia mydas) at two neritic sites in the central and northern coast of Peru: implications for conservation. PLoS ONE. 2014;9(11):e1712. doi: 10.1371/journal.pone.0113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace et al. (2011).Wallace BP, DiMatteo AD, Bolten AB, Chaloupka MY, Hutchinson BJ, Abreu-Grobois FA, Mortimer JA, Seminoff JA, Amorocho D, Bjorndal KA, Bourjea J, Bowen BW, Dueñas RB, Casale P, Choudhury BC, Costa A, Dutton PH, Fallabrino A, Finkbeiner EM, Girard A, Girondot M, Hamann M, Hurley BJ, López-Mendilaharsu M, Marcovaldi MA, Musick JA, Nel R, Pilcher NJ, Troëng S, Witherington B, Mast RB. Global conservation priorities for marine turtles. PLoS ONE. 2011;6(9):e1712. doi: 10.1371/journal.pone.0024510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, Wood & Critchley (1983).Wood JR, Wood FE, Critchley K. Hybridization of Chelonia mydas and Eretmochelys imbricata. Copeia. 1983;1983(3):839–842. doi: 10.2307/1444361. [DOI] [Google Scholar]
- Zárate et al. (2013).Zárate P, Bjorndal KA, Parra M, Dutton PH, Seminoff JA, Bolten AB. Hatching and emergence success in green turtle Chelonia mydas nests in the Galápagos Islands. Aquatic Biology. 2013;19(3):217–229. doi: 10.3354/ab00534. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data. Sequences used for phylogenetic analyses. All sequences were downloaded from the Barcode of Life Database System, except for the sequence obtained in this study.