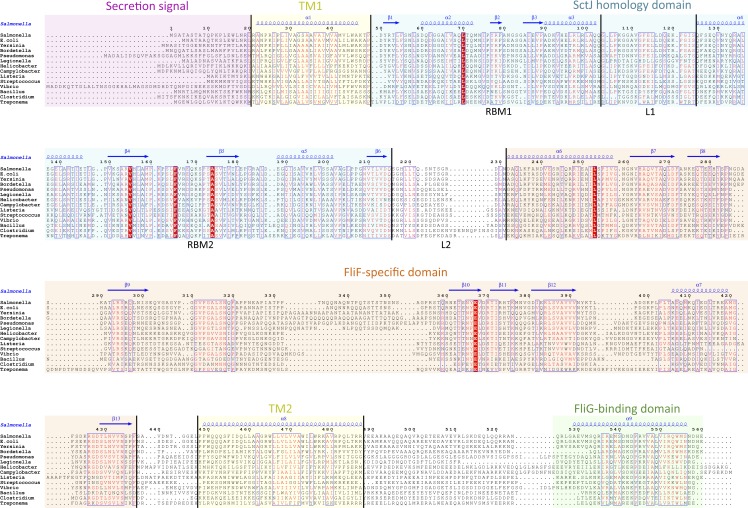
Figure 2. Domain organization of FliF.
Multiple sequence alignment of FliF sequences from various human pathogens (S. typhimurium, Escherichia coli, Yersinia pestis, Bordetella pertussis, Pseudomonas aeruginosa, Legionella pneumophilia, Helicobacter pylori, Campylobacter jejuni, Listeria monocytogenes, Streptococcus pneumonia, Vibrio cholerae, Bacillus subtilis, Clostridium difficile, Treponema palladium). Conserved residues are in red box, similar residues are in red characters. Identified domains are highlighted in colored boxes, with the TM helices in yellow, the FliG-binding domain in green, the signal sequence in purple, the SctJ homology domain in blue and the FliF-specific domain in orange. The predicted secondary structure elements for the S. typhimurium FliF are in blue at the top.