Summary
Carcinoma ex-pleomorphic adenoma (CA ex-PA) is a malignant salivary gland tumor that arises in association with pleomorphic adenoma (PA). Both PA and CA ex-PA have a broad spectrum of histology, and distinction from their histologic mimics may be difficult based on morphology alone. PLAG1 and HMGA2 abnormalities are the most common genetic events in both PA and CA ex-PA; however, the use of PLAG1 and HMGA2 as adjunct molecular tests has not been well established. Fluorescence in situ hybridization for PLAG1 and HMGA2 was performed on 22 CA ex-PA (10 myoepithelial carcinomas [MECAs], 10 salivary duct carcinomas [SDCs], 1 carcinoma with squamoglandular features, and 1 mixed MECA-adenocarcinoma not otherwise specified), 20 de novo carcinomas (11 MECAs and 9 SDCs), 16 PAs, and 11 PA-histologic mimics. All except 3 CAs ex-PA (86%) were positive for PLAG1 or HMGA2 rearrangements/amplifications. In contrast, 18 (90%) of 20 de novo carcinomas lacked abnormalities in PLAG1 or HMGA2 (P < .01). PLAG1 or HMGA2 rearrangements were identified in 6 (67%) of 9 hypocellular myxoid PAs and in 2 (29%) of 7 cellular PAs. Furthermore, all morphologic mimics of PA were negative for PLAG1 or HMGA2. PLAG1 and HMGA2 rearrangements are the most common genetic events in CA ex-PA regardless of the histologic subtype. Unlike CA ex-PA, de novo carcinomas were negative for PLAG1 and HMGA2. Interestingly, rearrangements of PLAG1/HMGA2 were identified in most hypocellular PAs but only in a small subset of cellular PAs. Fluorescence in situ hybridization for PLAG1 or HMGA2 can be used to distinguish between PA and CA ex-PA and their morphologic mimics.
Keywords: Carcinoma ex pleomorphic adenoma, Myoepithelial carcinoma, Salivary duct carcinoma, PLAG1, HMGA2
1. Introduction
Carcinoma ex-pleomorphic Adenoma (CA ex-PA) is a rare malignant neoplasm that arises in association with primary or recurrent pleomorphic adenoma (PA) [1,2]. Typically, they are high grade and show similar histology to their de novo counterparts. Salivary duct carcinoma (SDC) is the most common histologic subtype of CA ex-PA, followed by myoepithelial carcinoma (MECAs) [2,3]. However, any histologic subtype of salivary gland carcinoma can arise in association with PA [2,3]. CA ex-PA is classified based on the degree of tumor invasion through the preexistent PA capsule into the surrounding tissue as noninvasive, minimally invasive, and widely invasive [1]. The extent of invasion has been found to correlate with the clinical outcome in many studies [1,3,4]. Typically, no metastases or recurrences are found in patients with noninvasive/minimally invasive CA ex-PA in contrast to widely invasive tumors [1,4].
At the molecular level, rearrangements of PLAG1 gene (pleomorphic adenoma gene 1), a developmentally regulated zinc-finger proto-oncogene, located on 8q12 chromosomal region, are common abnormalities in PA and CA ex-PA [5-7]. The PLAG1 fusion partner genes include CTNNB1 (β-catenin), LIFR (leukemia inhibitory factor receptor), and FGFR1 (fibroblast growth factor receptor 1) [5,7-10]. Moreover, HMGA2 gene rearrangements with or without amplifications have been described in PA as well as in CA ex-PA [11]. Nevertheless, the diagnostic role of PLAG1 and HMGA2 abnormalities in salivary gland tumors has not been well established yet. The objectives of this study are to evaluate the morphologic spectrum of CA ex-PA in relation to its genetic features and examine the potential genetic link between CA ex-PA and its de novo counterparts. Furthermore, we sought to study PLAG1 and HMGA2 alterations in histologically different types of PAs and their morphologic mimics and investigate the presence of EWSR1 rearrangements in the latter.
2. Materials and methods
2.1. Tumors characteristics
A total of 66 salivary gland tumors (19 CA ex-PAs, 20 de novo carcinomas, 16 PAs, and 11 histologic mimics of PAs) were selected from the Department of Pathology files of Memorial Sloan-Kettering Cancer Center. Tumors were assessed for their anatomical location and size. The histologic type of the malignant component in CA ex-PA was defined according to the World Health Organization classification of salivary gland tumors [1]. The tumor was considered as myoepithelial if it was composed of at least 90% myoepithelial cells, displaying the typical histology of MECA, and/or showing immunohistochemical evidence of myoepithelial differentiation, that is, positivity for a keratin marker (Cam5.2, CK7, AE1/3) and at least one of the myoepithelial markers (S100, calponin, p63, or smooth muscle actin). CA ex-PAs were classified into noninvasive, minimally invasive, and widely invasive based on the World Health Organization criteria [1]. The percentage of the residual parent PA in CA ex-PA was estimated. Histologic analysis of CA ex-PAs was performed to include the following: mitotic rate, nuclear pleomorphism, tumor necrosis, and morphologic features of the PA component. In addition, PAs were assessed for cellularity, ductal, and myoepithelial differentiation; cell type; and matrix formation.
2.2. Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) on interphase nuclei from paraffin-embedded 4-μm sections was performed using custom probes of bacterial artificial chromosomes (BACs), flanking EWSR1 in 22q12, PLAG1 on 8q12, HMGA2 on 12q14, CTNNB1 on 3p21, LIFR on 5p13, and FGFR1 on 8p11 (Supplementary Table 1). BAC clones were chosen according to University of California, Santa Cruz genome browser (http://genome.uscs.edu). The BAC clones were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (Oakland, CA; http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described. Each BAC was validated for approximate cytogenetic location and specificity by hybridizing to normal human metaphases. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (ISIS) (Metasystems, Waltham, MA). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
3. Results
3.1. Clinicopathological characteristics
3.1.1. CA ex-PA cases
Table 1 shows the demographic and histologic features of CA ex-PA cases. The histologic types of the malignant component of the 22 CA ex-PA cases were MECA (10 cases), SDC (10 cases), carcinoma with squamoglandular features (1 case), and mixed MECA and adenocarcinoma not otherwise specified (NOS; 1 case). Detailed clinical and histologic data were available in all except 2 outside cases. All CA ex-PA tumors except 3 were located in the parotid gland. The nonparotid tumors were located in the base of tongue, the nasal cavity, and the lacrimal gland. In 2 cases, carcinoma was associated with a recurrent PA. The extent of invasion was classified as widely invasive in all cases except one, which was noninvasive. All SDCs were high grade showing increased mitotic activity and tumor necrosis (Fig. 1A). On the other hand, MECAs showed a wide range of mitotic activities ranging from 2 to 22 mitoses per 10 high-power fields (HPFs). Nuclear pleomorphism was noted in all CA ex-PA cases except 4. Tumor necrosis was identified in all MECAs (Fig. 1B), except for 2 cases, one of which showed low mitotic activity (2/10 HPFs) and bland nuclear features, whereas the second case had increased mitotic activity of 12/10 HPFs. The benign PA component was assessed in 19 cases, comprising a variable proportion of the entire neoplasm, ranging from less than 5% to 70% and being composed of benign epithelial and stromal/mesenchymal elements. The PA component was found either as a well-demarcated area (Fig. 1C) from the malignant component or intermixed with the malignant component. Extensive stromal hyalinization was noted in 61% (11/19) of the PA components. Among these, 3 were composed only of a discrete hyalinized stromal nodule without definite benign ductal elements (Fig. 1D).
Table 1.
Clinicopathological and demographic characteristics of CA ex-PAs
| No. | Age (y)/Sex | Location | Size (cm) | Subtype | Mitoses/10 HPFs | Necrosis | Nuclear pleomorphism | INV | PA% |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 47/F | Parotid | 1.3 | MECA | 2 | No | No | WI | 15 |
| 2 | 41/M | Parotid | 9.5 | MECA | 1 | Yes | No | WI | <5 |
| 3 | 67/F | Parotid | 1.1 | MECA | 12 | No | Yes | WI | 10 |
| 4 | 77/M | BOT | 2.1 | MECA | 22 | Yes | No | WI | 15 |
| 5 | 67/F | Parotid | 6 | MECA | 5 | Yes | Yes | WI | <5 |
| 6r | 63/F | Parotid | NA | MECA | NA | NA | NA | NA | NA a |
| 7 | 64/M | Parotid | 3 | MECA | 2 | Yes | Yes | WI | NA a |
| 8 | F | Parotid | NA | MECA | 6 | Yes | No | WI | 20 |
| 9 | 76/M | Parotid | 3.5 | MECA | 7 | Yes | Yes | WI | 30 |
| 10 | NA | NA | NA | MECA | NA | NA | NA | NA | NA a |
| 11 | 67/F | Parotid | 4.2 | SDC | 16 | Yes | Yes | WI | 25 |
| 12 | 63/M | Parotid | 1.4 | SDC | 8 | Yes | Yes | WI | 10 b |
| 13 | 75/M | Parotid | 2 | SDC | 8 | Yes | No | WI | 60 |
| 14 | 67/F | Parotid | 4.4 | SDC | 3 | Yes | Yes | WI | 10 |
| 15 | 70/M | Parotid | 1.2 | SDC | 6 | Yes | No | WI | 15 |
| 16 | 51/M | Parotid | 3.4 | SDC | 20 | Yes | Yes | WI | 30 |
| 17 | 87/F | Parotid | 2.4 | SDC | 9 | Yes | Yes | WI | 5 b |
| 18 | 43/M | Parotid | 1.5 | SDC | 4 | Yes | Yes | WI | 30 |
| 19 | 68/F | Parotid | 2 | SDC | 3 | Yes | Yes | WI | 70 |
| 20 | 98/M | Parotid | 2.7 | SDC | 10 | Yes | Yes | WI | 5 b |
| 21 | 74/F | Lacrimal gland | 4.2 | CA-SG | 16 | Yes | Yes | WI | 25 |
| 22 | 35/M | Parotid | 5.8 | MECA and ACA NOS | 6 | Yes | Yes | NI | 40 |
Abbreviations: INV, invasion; BOT, base of tongue; NA, not available; PA, pleomorphic adenoma; MECA, myoepithelial carcinoma; SDC, salivary duct carcinoma; F, female; M, male; WI, widely invasive; BOT, base of tongue; NA: not applicable; CA-SG, carcinoma with squamoglandular features; ACA NOS, adenocarcinoma not otherwise specified; WI, widely invasive; NI, noninvasive.
The prior PA at the same site was not available for review.
Hyalinized nodule.
Fig. 1.
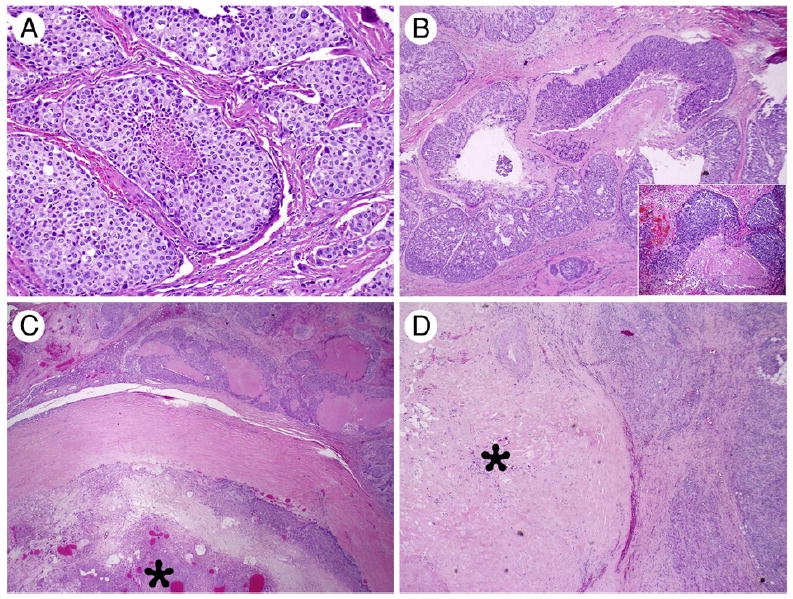
Morphologic spectrum of CA ex-PA. A, High-grade SDC type: the tumor cells show abundant eosinophilic cytoplasm and prominent nuclei, arranging in a solid growth pattern with central necrosis (×200). B, MECA with multinodular growth pattern and tumor necrosis (×40) with an inset (×400) showing high-power view of the tumor with hyalinized necrotic stroma and pseudoglandular spaces. C, CA ex-PA associated with a well-demarcated PA component (star; ×20). D, CA ex-PA in which only a discrete hyalinized stromal nodule (star) is noted, most likely representing a burned-out PA (×20).
3.1.2. De novo carcinomas
The de novo carcinoma group included 11 MECAs and 9 SDCs. There was no statistically significant difference in the frequency of nuclear pleomorphism, mitotic activity, and presence/absence of tumor necrosis between CA ex-PA and de novo carcinoma groups.
3.1.3. Pleomorphic adenomas
Typical PA morphology with hypocellular myxoid matrix and ductal/tubular differentiation was identified in 9 cases (Fig. 2A). Seven PAs were cellular composed of monomorphic myoepithelial/mesenchymal cells with occasional ductal structures and focal stromal matrix (Fig. 2B).
Fig. 2.
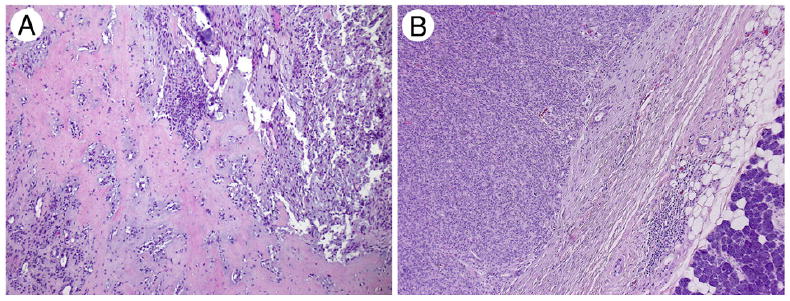
PA phenotype. A, Typical PA showing hypocellular proliferation of ductal and myoepithelial cells with a hyalinized/myxoid stroma (×10). B, Cellular PA with no stromal matrix (×40).
3.2. Immunohistochemical studies
Review of the immunohistochemical studies were performed on 18 of 21 MECAs (CA ex-PA and de novo), and the carcinomas were positive for a cytokeratin and at least one of the myoepithelial markers (S100, calponin, p63, and smooth muscle actin). The other 3 cases showed the typical morphologic appearance of MECA.
3.3. FISH results
3.3.1. Carcinoma ex-pleomorphic adenoma
Most CA ex-PA cases (19/22; 86%) showed abnormalities of either PLAG1 or HMGA2 by FISH analysis (Table 2). Most tumors showed greater than 50% break-apart signals (range, 30%-85%). The 3 negative CA ex-PAs were of SDC subtype and showed a clear-cut benign PA component. Sixteen (73%) CA ex-PA tumors showed PLAG1 gene rearrangements with or without amplifications (Fig. 3A). Among the 16 PLAG1-rearranged CA ex-PA cases, 12 showed an unbalanced pattern, being associated with deletion of telomeric parts (n = 5), centromeric amplification (n = 3), centromeric amplification and deletion of telomeric parts (n = 1), amplifications of both centromeric and telomeric ends (n = 1), and deletion of centromeric parts (n = 2). HMGA2 rearrangements were identified in 3 of 8 CA ex-PA tumors (Fig. 3B), all of which were associated with an unbalanced pattern, showing in one case each telomeric amplification, centromeric amplification, and amplification of both telomeric and centromeric regions. The 3 CA ex-PA tumors associated with a burned-out hyalinized nodule were positive for PLAG1 rearrangements. In 2 cases, the carcinoma and the coexisting PA components were analyzed separately showing the following: one showed PLAG1 rearrangements in both benign and malignant components, whereas the other showed HMGA2 rearrangements and amplification only in the carcinoma component.
Table 2.
Molecular characteristics of CA ex-PAs
| No. | Subtype | PLAG1 | HMGA2 | FGFR1 | LIFR | CTNNB1 | EWSR1 |
|---|---|---|---|---|---|---|---|
| 1 | MECA | Rearranged | Neg | Neg | Neg | Neg | Neg |
| 2 | MECA | Rearranged and C′ amp | NP | Rearranged and C′ amp | NP | NP | Neg |
| 3 | MECA | Rearranged and C′ amp | NP | Rearranged and C′ amp and T′ del | NP | NP | NP |
| 4 | MECA | Neg | Rearranged and T′ amp | NP | NP | NP | Neg |
| 5 | MECA | Neg a | Rearranged and C′T′ amp b | NP | NP | NP | Neg |
| 6 | MECA | Rearranged and C′ amp | Neg | Rearranged and C′ amp | Neg | Neg | Neg |
| 7 | MECA | Rearranged and C′/T′ amp | NP | Rearranged and C′ amp | Neg | Neg | Neg |
| 8 | MECA | Rearranged and T′ del and C′ amp NP | Rearranged and C′ amp | NP | NP | NP | |
| 9 | MECA | Rearranged a | NP | NP | NP | NP | NP |
| 10 | MECA | Neg | Rearranged and C′ amp | NP | NP | NP | Neg |
| 11 | SDC | Rearranged and T′ del | NP | Neg | NP | Rearranged | NP |
| 12 | SDC | Rearranged and T′ del | NP | Neg | NP | Rearranged | NP |
| 13 | SDC | Rearranged | NP | Neg | Neg | Neg | NP |
| 14 | SDC | Neg | Neg | NP | NP | NP | NP |
| 15 | SDC | Neg | Neg | NP | NP | NP | NP |
| 16 | SDC | Rearranged and T′ del | NP | NP | NP | Neg | NP |
| 17 | SDC | Rearranged and C′ del | NP | NP | NP | NP | NP |
| 18 | SDC | Rearranged and T′ del | NP | NP | NP | Neg | NP |
| 19 | SDC | Neg | Neg | NP | NP | NP | NP |
| 20 | SDC | Rearranged | NP | NP | NP | Neg | NP |
| 21 | CA-SG | Rearranged and C′ del | NP | NP | NP | Neg | NP |
| 22 | MECA and ACA | Rearranged and T′ del | NP | NP | NP | Neg | NP |
Abbreviations: Neg, negative; C′, centromeric; amp, amplified; NP, not performed; T’, telomeric; del, deletion; MECA, myoepithelial carcinoma; SDC, salivary duct carcinoma; CA-SG, carcinoma with squamoglandular features; ACA, adenocarcinoma.
Present in both carcinoma and PA.
Present in carcinoma but not in PA component.
Fig. 3.
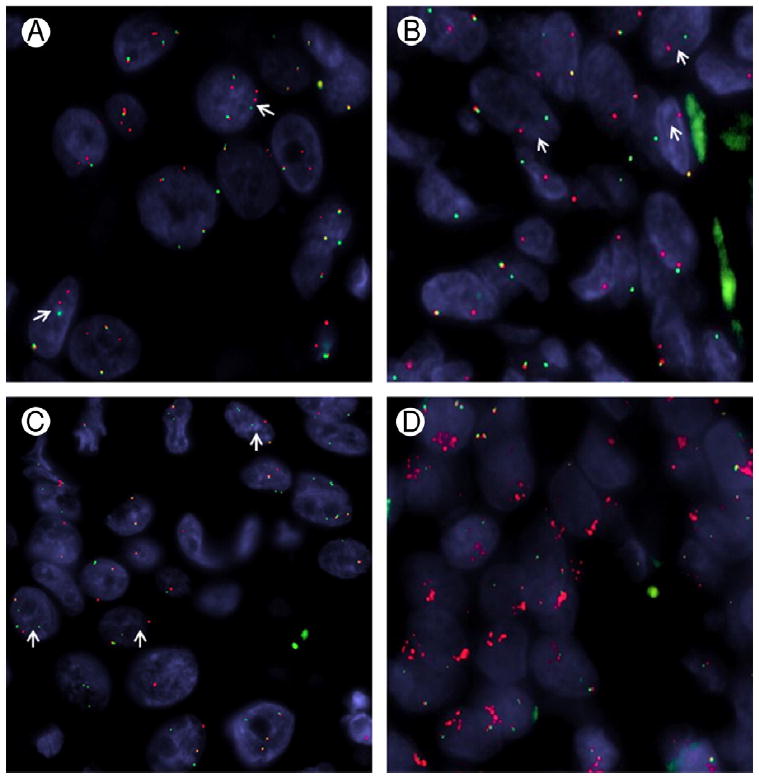
PLAG1, HMGA2, CTNNB1, and FGFR1 gene abnormalities by FISH. A, PLAG1 gene rearrangement (arrows) in an SDC CA ex-PA (CA ex-PA #12). B, HMGA2 break-apart (arrows) in a typical PA with hypocellular matrix (PA #5). C, SDC-type CA ex-PA showing a CTNNB1 gene rearrangement (arrows; CA ex-PA #12). D, Unbalanced FGFR1 rearrangement pattern associated with telomeric deletion (green signal) and subsequent centromeric amplification (red signal), MECA ex-PA (CA ex-PA #2).
In addition, FISH was performed on 14 of 16 PLAG1-rearranged carcinomas to identify potential fusion partners, including FGFR1, CTNNB1, and LIFR. Two of the 11 tested tumors showed CTNNB1 rearrangements with an unbalanced telomeric loss (Fig. 3C). FGFR1 gene rearrangements were identified in 5 of 6 CAs ex-PA of MECA type (Fig. 3D), but in none of the 3 SDC ex-PA cases tested. All the rearranged FGFR1 tumors showed an unbalanced pattern, which correlated with the unbalanced PLAG1 rearrangements. The 4 examined CA ex-PA tumors for LIFR abnormalities were negative.
3.3.2. De novo carcinoma
All except 2 (90%) de novo carcinomas (10 MECAs and 8 SDCs) lacked abnormalities in PLAG1 and HMGA2 genes (P < .01; Table 3). One de novo carcinoma showed PLAG1 rearrangement and 1 showed HMGA2 rearrangement, and both carcinomas were salivary duct type. FGFR1 gene rearrangements were investigated in 3 de novo MECAs and 1 myoepithelioma, but no gene abnormalities were found. Furthermore, none of the tested MECAs (8 CA ex-PAs and 4 de novo carcinomas) showed EWSR1 gene rearrangements.
Table 3.
Molecular characteristics of de novo MECA and SDC
| No. | Histology | PLAG1 | HMGA2 | EWSR1 |
|---|---|---|---|---|
| 1 | MECA | Neg | Neg | Neg |
| 2 | MECA | Neg | Neg | Neg |
| 3 | MECA | Neg | Neg | Neg |
| 4 | MECA | Neg | Neg | Neg |
| 5 | MECA | Neg | Neg | NP |
| 6 | MECA | Neg | Neg | NP |
| 7 | MECA | Neg | Neg | NP |
| 8 | MECA | Neg | Neg | NP |
| 9 | MECA | Neg | Neg | NP |
| 10 | MECA | Neg | Rearranged | NP |
| 11 | MECA | NA | Neg | NP |
| 12 | SDC | Neg | Neg | NP |
| 13 | SDC | Neg | Neg | NP |
| 14 | SDC | Rearranged and C′ del | NP | NP |
| 15 | SDC | Neg | Neg | NP |
| 16 | SDC | Neg | Neg | NP |
| 17 | SDC | Neg | Neg | NP |
| 18 | SDC | Neg | Neg | NP |
| 19 | SDC | Neg | Neg | NP |
| 20 | SDC | Neg | Neg | NP |
Abbreviations: MECA, myoepithelial carcinoma; SDC, salivary duct carcinoma; Neg, negative; NP, not performed; NA, not applicable; C′ del, deletion of centromeric part.
3.3.3. PA and histologic mimics
PLAG1 or HMGA2 rearrangements were identified in 6 (67%) of 9 typical PAs showing myxoid matrix and in 2 (29%) of 7 cellular PAs (Table 4). Eight (50%) of 16 PAs showed PLAG1 or HMGA2 gene rearrangements by FISH analysis (Table 4). All tumors showed a balanced gene rearrangement, except for 2 cases, which showed PLAG1 rearrangement with associated centromeric amplification in one case, whereas the other showed HMGA2 gene rearrangements with deletion of the telomeric parts. All tumors in the control group including 2 epithelial MECAs (EMCs), 2 basal cell adenocarcinomas, and 7 polymorphous low-grade adenocarcinomas (PLGAs) were negative for PLAG1 and HMGA2 abnormalities (Table 5). In addition, no EWSR1 rearrangements were detected in 1 myoepithelioma and 6 PLGAs tested.
Table 4.
Histologic and molecular characteristics of PAs
| No. | Cellular a | PLGA1 | HMGA2 | CTNNB1 | FGFR1 |
|---|---|---|---|---|---|
| 1 | No | Rearranged | NP | Rearranged | Neg |
| 2 | No | Neg | Neg | NP | NP |
| 3 | No | Neg | Neg | NP | NP |
| 4 | No | Neg | Rearranged | NP | NP |
| 5 | No | Neg | Rearranged | NP | NP |
| 6 | No | Neg | Neg | NP | NP |
| 7 | No | Rearranged | NP | NP | NP |
| 8 | No | Rearranged | NP | Neg | NP |
| 9 | No | Rearranged | NP | Rearranged | NP |
| 10 | Yes | Rearranged | NP | NP | NP |
| 11 | Yes | Neg | Neg | NP | NP |
| 12 | Yes | Rearranged/C′ amp | NP | NP | Rearranged/T del |
| 13 | Yes | Neg | Neg | NP | NP |
| 14 | Yes | Neg | Neg | NP | NP |
| 15 | Yes | Neg | Neg | NP | NP |
| 16 | Yes | Neg | Neg | NP | NP |
Abbreviations: NP, not performed; Neg, negative; C′, centromeric; amp: amplified; T del, deletion of telomeric part.
Cellular PAs are predominantly monomorphic with a predominant 1 cell type with focal stromal matrix.
Table 5.
Molecular features of control group
| DX | n | PLAG1 | HMGA2 | MYB |
|---|---|---|---|---|
| EMC | 2 | Neg | Neg | NP |
| PLGA | 7 | Neg | Neg | Neg |
| BACA | 2 | Neg | Neg | NP |
Abbreviations: DX, diagnosis; EMC, epithelial-myoepithelial carcinoma; PLGA, polymorphous low-grade adenocarcinoma; BACA, basal cell adenocarcinoma; Neg, negative; NP, not performed; BACA, basal cell adenocarcinoma.
4. Discussion
In this study, we examined PLAG1 and HMGA2 gene alternations in different histologic subtypes of CA ex-PA, their de novo counterparts, and the entities that enter in their differential diagnosis. CA ex-PA is a carcinoma that arises in association with a coexisting PA component or a prior excised PA at the same site [1,2]. Occasionally, the carcinoma component might completely efface the preexistent benign PA. In some cases, only a hyalinized nodule without epithelial elements might be seen. This raises the possibility of a preexisting PA component, but pathologists are often reluctant to accept as enough evidence for a diagnosis of CA ex-PA in these cases. CA ex-PA is typically aggressive and might have worse prognosis compared with its de novo counterparts, especially in cases of MECA [12]. Moreover, CA ex-PA and, to a higher extent, PA have a broad spectrum of histologic features, and distinction from their histologic mimics such as EMC, PLGA, and basal cell adenoma/adenocarcinoma may be difficult based on morphology alone. Therefore, evaluation of genetic alterations by FISH ancillary test could represent a valuable tool in the diagnostically challenging cases.
Genetic alterations of PLAG1 and HMGA2 genes, including gene rearrangements and amplifications, have been previously described in PA [11,13,14]. These genetic abnormalities have been also identified in histologically similar soft tissue tumors [15,16] and CA ex-PA [6,17]. In the study by Bahrami et al [15], 12 (63%) of 19 CAs ex-PAs were positive for PLAG1 rearrangements and 1 of 3 cases were positive for HMGA2 rearrangements by FISH analysis. In accordance with the study by Bahrami et al, we found PLAG1 and/or HMGA2 rearrangements in 19 (86%) of 22 CAs ex-PAs. However, in contrast with the study by Bahrami et al, which covered a wider histologic spectrum of CA ex-PA types (adenocarcinoma NOS, SDC, MECA, EMC, squamous cell carcinoma, and carcinosarcoma), our work focused mainly on the 2 most common histologic types of CA ex-PA (SDC and MECA) [2,3,15]. The 3 PLAG1/ HMGA2-negative CA ex-PA cases in our study were of SDC type showing an overt benign PA component, suggesting either undetectable PLAG1 abnormalities by FISH resolution or alternative oncogenic events. Interestingly, in 2 cases analyzed, PLAG1 gene rearrangements were present in both the benign and malignant components in one case, whereas in the other, HMGA2 gene rearrangement and amplification were present only in the malignant component. Similar to the results in Bahrami et al [18], PLAG1 and/or HMGA2 alterations were found in 4 (50%) of 8 CA ex-PAs with SDC morphology. Furthermore, PLAG1 or HMGA2 FISH abnormalities were detected in 8 (80%) of 10 SDCs containing a hyalinized nodule rather than a recognizable PA, in keeping with a preexisting, but burned-out, PA [18]. Our findings further support this assumption, with all 3 CA ex-PA tumors associated with a hyalinized nodule being positive for PLAG1 rearrangements. This result has a diagnostic impact in the surgical pathology of salivary gland as indicated earlier. As reported previously, we found CTNNB1 rearrangements in 2 SDC ex-PA cases, in keeping with a t(3;8)(p21;q12) [5,7,9,10]. In addition, the presence of t(8;8)(q12.1;p12), resulting in FGFR1-PLAG1 fusion has been identified in a subgroup of PA [19]. Of interest and in keeping with our prior data, FGFR1 rearrangements (often associated with telomeric deletion and centromeric amplifications) were identified in 5 of 6 PLAG1-rearranged MECAs ex-PA, but not in de novo myoepithelial tumors [16].
In this study, we investigated the potential genetic link between CA ex-PAs and their de novo counterparts. Also, in contrast to CA ex-PA, all but 2 de novo carcinomas (90%) lacked abnormalities in PLAG1 or HMGA2 genes (P < .01). These 2 latter cases might have had a PA component that was not sampled or was completely obscured by the malignant component. Recently, we found that myoepithelial CA ex-PA correlates with a worse clinical behavior compared with de novo MECA [12]. This highlights the valuable role of this FISH assay in separating CA ex-PA from de novo counterparts and therefore improves tumor stratification.
PA has a broad spectrum of morphologic features, and distinction from its histologic mimics may be challenging. Furthermore, a subset of PA, which is usually cellular and often showing one predominant uniform cell type, can morphologically resemble other salivary gland tumors, in particular PLGA. In this current study, we selected 9 cases of typical PA showing hypocellular myxoid stroma with ductal and myoepithelial elements and 7 cases of cellular PA with a dominant 1-cell type. Interestingly, PLAG1 or HMGA2 rearrangements were identified in most (78%) hypocellular myxoid PAs, but only in one-third (29%) of cellular PAs. This implies that cellular PA might represent a discrete genetic subgroup; however, additional molecular and immunohistochemical studies are required to characterize this further. In contrast, all morphologic mimics of PA and CA ex-PA, including PLGA, EMC, and basal cell adenoma/adenocarcinoma, were negative for PLAG1 or HMGA2 abnormalities. Given the positive predictive value, this assay can therefore be used to distinguish PA from its morphologic mimics.
In our previous study, we have found PLAG1 gene rearrangements in a significant proportion of cutaneous and benign soft tissue myoepithelial tumors that are associated with tubuloductal differentiation and morphologically resemble salivary PA [16]. In addition to the shared PLAG1 and HMGA2 gene rearrangements between soft tissue myoepithelial tumors and PA of salivary gland, EWSR1 gene rearrangements have been reported as a common event in myoepithelial tumors arising outside salivary glands and in clear cell hyalinizing carcinoma of salivary gland [20,21]. In this study, none of the tested salivary myoepithelial tumors (8 CA ex-PAs, 4 de novo carcinomas, and 1 myoepithelioma) showed EWSR1 gene abnormalities, confirming previously reported findings [8,21].
Given the morphologic similarities between PLGA and myoepithelial tumors, we also examined EWSR1 gene alterations by FISH in 6 PLGAs, but none were positive.
In summary, PLAG1 and HMGA2 gene abnormalities are the most common genetic events in different histologic subtypes of CA ex-PA. PLAG1/HMGA2 rearrangements were identified in most hypocellular PAs, but only in a small subset of cellular PAs. In contrast to CA ex-PA, most de novo SDC and MECA were negative for PLAG1 and HMGA2 gene abnormalities. FISH ancillary tests for PLAG1 or HMGA2 gene alterations can be used to distinguish between CA ex-PA and its de novo counterparts as well as separate PA from its morphologic mimics.
Supplementary Material
Footnotes
Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.humpath.2014.08.017.
Competing interests: None.
Contributor Information
Nora Katabi, Email: katabin@makcc.org.
Cristina R. Antonescu, Email: antonesc@mskcc.org.
References
- 1.Barnes LEJ, Reichart P, Sidransky D. Classification of tumours Pathology and genetics of head and neck tumours. Lyon: World Health Organization; IARC; 2005. [Google Scholar]
- 2.Ellis GL, Auclair PL. Tumors of the salivary glands 3rd series. Washington DC: Armed Forces Institute of Pathology; 1996. [Google Scholar]
- 3.Katabi N, Gomez D, Klimstra DS, Carlson DL, Lee N, Ghossein R. Prognostic factors of recurrence in salivary carcinoma ex pleomorphic adenoma, with emphasis on the carcinoma histologic subtype: a clinicopathologic study of 43 cases. Hum Pathol. 2010;41:927–34. doi: 10.1016/j.humpath.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Brandwein M, Huvos AG, Dardick I, Thomas MJ, Theise ND. Noninvasive and minimally invasive carcinoma ex mixed tumor: a clinicopathologic and ploidy study of 12 patients with major salivary tumors of low (or no?) malignant potential. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1996;81:655–64. doi: 10.1016/s1079-2104(96)80071-0. [DOI] [PubMed] [Google Scholar]
- 5.Kas K, Voz ML, Roijer E, et al. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet. 1997;15:170–4. doi: 10.1038/ng0297-170. [DOI] [PubMed] [Google Scholar]
- 6.Martins C, Fonseca I, Roque L, et al. PLAG1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: a combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol. 2005;18:1048–55. doi: 10.1038/modpathol.3800386. [DOI] [PubMed] [Google Scholar]
- 7.Voz ML, Agten NS, Van de Ven WJ, Kas K. PLAG1, the main translocation target in pleomorphic adenoma of the salivary glands, is a positive regulator of IGF-II. Cancer Res. 2000;60:106–13. [PubMed] [Google Scholar]
- 8.Andre F, Bachelot T, Campone M, et al. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res. 2013;19:3693–702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 9.Astrom AK, Voz ML, Kas K, et al. Conserved mechanism of PLAG1 activation in salivary gland tumors with and without chromosome 8q12 abnormalities: identification of SII as a new fusion partner gene. Cancer Res. 1999;59:918–23. [PubMed] [Google Scholar]
- 10.Debiec-Rychter M, Van Valckenborgh I, Van den Broeck C, et al. Histologic localization of PLAG1 (pleomorphic adenoma gene 1) in pleomorphic adenoma of the salivary gland: cytogenetic evidence of common origin of phenotypically diverse cells. Lab Invest. 2001;8:1289–97. doi: 10.1038/labinvest.3780342. [DOI] [PubMed] [Google Scholar]
- 11.Persson F, Andren Y, Winnes M, et al. High-resolution genomic profiling of adenomas and carcinomas of the salivary glands reveals amplification, rearrangement, and fusion of HMGA2. Genes Chromosomes Cancer. 2009;48:69–82. doi: 10.1002/gcc.20619. [DOI] [PubMed] [Google Scholar]
- 12.Kong DE, Drill E, Gonen M, Klimstra DS, Ghossein R, Katabi N. Proliferative grading predicts outcome in myoepithelial carcinoma of salivary glands: a clinicopathologic study of 49 cases. Mod Pathol. 2014;27:320. doi: 10.1097/PAS.0000000000000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geurts JM, Schoenmakers EF, Roijer E, Astrom AK, Stenman G, van de Ven WJ. Identification of NFIB as recurrent translocation partner gene of HMGIC in pleomorphic adenomas. Oncogene. 1998;16:865–72. doi: 10.1038/sj.onc.1201609. [DOI] [PubMed] [Google Scholar]
- 14.Geurts JM, Schoenmakers EF, Roijer E, Stenman G, Van de Ven WJ. Expression of reciprocal hybrid transcripts of HMGIC and FHIT in a pleomorphic adenoma of the parotid gland. Cancer Res. 1997;57:13–7. [PubMed] [Google Scholar]
- 15.Bahrami A, Dalton JD, Krane JF, Fletcher CD. A subset of cutaneous and soft tissue mixed tumors are genetically linked to their salivary gland counterpart. Genes Chromosomes Cancer. 2012;51:140–8. doi: 10.1002/gcc.20938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antonescu CR, Zhang L, Shao SY, et al. Frequent PLAG1 gene rearrangements in skin and soft tissue myoepithelioma with ductal differentiation. Genes Chromosomes Cancer. 2013;52:675–82. doi: 10.1002/gcc.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahrami A, Dalton JD, Shivakumar B, Krane JF. PLAG1 alteration in carcinoma ex pleomorphic adenoma: immunohistochemical and fluorescence in situ hybridization studies of 22 cases. Head Neck Pathol. 2012;6:328–35. doi: 10.1007/s12105-012-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahrami A, Perez-Ordonez B, Dalton JD, Weinreb I. An analysis of PLAG1 and HMGA2 rearrangements in salivary duct carcinoma and examination of the role of precursor lesions. Histopathology. 2013;63:250–62. doi: 10.1111/his.12152. [DOI] [PubMed] [Google Scholar]
- 19.Persson F, Winnes M, Andren Y, et al. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene. 2008;27:3072–80. doi: 10.1038/sj.onc.1210961. [DOI] [PubMed] [Google Scholar]
- 20.Antonescu CR, Katabi N, Zhang L, et al. EWSR1-ATF1 fusion is a novel and consistent finding in hyalinizing clear-cell carcinoma of salivary gland. Genes Chromosomes Cancer. 2011;50:559–70. doi: 10.1002/gcc.20881. [DOI] [PubMed] [Google Scholar]
- 21.Antonescu CR, Zhang L, Chang NE, et al. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–24. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


