Abstract
Inhibitor-induced conformational ensemble shifts in a multi-drug resistant HIV-1 protease variant, MDR769, are characterized by site-directed spin labeling (SDSL) double electron-electron resonance (DEER) spectroscopy. For MDR769 compared to native enzyme, changes in inhibitor IC50 values are related to a parameter defined as |ΔC|, which is the relative change in the inhibitor-induced shift to the closed state. Specifically, a linear correlation is found between |ΔC| and the fold-change in IC50, provided that inhibitor-binding is not too weak. Moreover, inhibitors that exhibit MDR769 resistance no longer induce a strong shift to a closed conformational ensemble as seen previously in native enzyme.
HIV-1 protease (HIV-1 PR) is a 99-amino acid C2-symmetric homodimer responsible for HIV viral maturation (1), and this protein is the target of therapies utilizing protease inhibitors (PIs). Residues 46 to 56 in each subunit of HIV-1 PR comprise a β-hairpin known as the flap, which together regulate access to the active-site (2). Cross-resistance to several inhibitors can arise from primary and compensatory amino acid changes (3), which occurs because of natural polymorphisms (4), drug-pressure selected mutations (5, 6), or combinations of both. These substitutions have been shown to alter HIV-1 PR flap conformational heterogeneity (7–9) and dynamics (10) in the free protease.
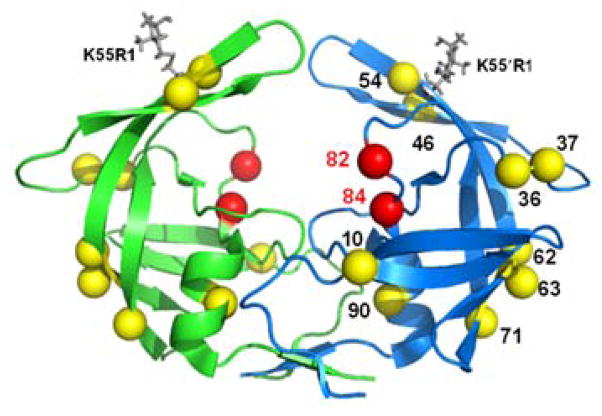
The failure of Highly Active Antiretroviral Therapy (HAART) in HIV/AIDS treatment is attributed to the emergence of drug-pressure selected mutations in the HIV genome after exposure to one or several inhibitors (5, 6). One particular multi-drug resistant HIV-1 PR variant is the clinical isolate MDR769 (Figure 1). MDR769 is resistant to various inhibitors (11) and exhibits higher level of resistance to most FDA-approved PIs, with the exception of darunavir (DRV), tipranavir (TPV) and lopinavir (LPV) (12). Crystal structures of this variant reveal an expanded active site pocket in the apo, substrate-bound and inhibitor-bound forms (12–15). Fewer H-bonding and van der Waals interactions result between PIs and the binding cleft, contributing to drug resistance in MDR769 (13).
Figure 1.
Ribbon diagram of MDR769 (PDB file 1TW7) protease colored by subunit, rendered in PyMol 1.3. Primary mutations relative to wild-type (LAI) are shown as red spheres, while compensatory mutation sites are rendered as yellow spheres. MTSL spin probes (K55R1) are incorporated in silico via MMM 2011.1 (16) and shown as gray capped sticks.
Previous studies have shown that flap conformational sampling can be altered by drug-pressure selected mutations (9), as well as sequence variations among subtypes (7). For subtype B HIV-1 PR, we also demonstrated that PIs and the substrate-mimic CA-p2 induce shifts in the conformational sampling ensemble towards the closed state (17).
In this study, the relationship between changes in flap closure and drug potency were investigated by defining a parameter |ΔC|, which is the magnitude of the difference in inhibitor-induced conformational shift to the closed state between two HIV-1 protease variants. For instance, if a given protease inhibitor induced X% and Y% of the closed population (%closed) in variants A and B, respectively, |ΔC| can be calculated by using Eq 1.
| (Eq 1) |
This parameter is used to calculate the inhibitor-induced percentage change of flap closure between subtype B and MDR769. The values of |ΔC| are then compared to drug potency using previously reported half maximal inhibitory concentration (IC50) measurements.
Pulsed electron paramagnetic resonance (EPR) spectroscopy, specifically site-directed spin labeling (SDSL) double electron-electron resonance (DEER) is a powerful tool to monitor the conformational change in free and inhibitor-bound HIV-1 PR variants (7, 9, 17, 18). For DEER distance measurements in HIV-1 PR, nitroxide radical labels are attached to K55C/K55C′ (termed K55R1 after labeling) on solvent-exposed flap sites. Because HIV-1 PR is a homodimer, two labels are incorporated into the protease for a single cysteine substitution, and the dipolar interaction between these probes provides detailed distance and population distribution profiles that reflect various flap conformational ensembles, namely the curled/tucked, closed, semi-open and wide-open conformational states (7, 9).
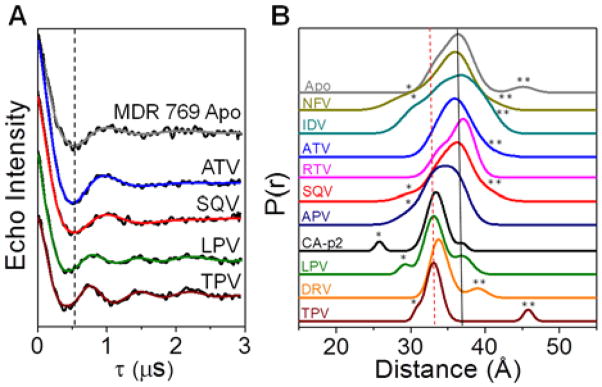
Ligand-induced conformational shifts for MDR769 were acquired from DEER measurements, with methanethiosulfonate (MTSL) spin probes incorporated at sites K55R1/K55′R1. Figure 2 shows select DEER data and distance profiles (complete data in Supporting Information). The effects of inhibitors on the average flap distance can clearly be seen in the DEER echo curves in Figure 2A. LPV and TPV shift the frequency with less dampening of the oscillations; generating distance profiles with most probable distances of 33 Å and narrower breadths. In Figure 2B, the solid line indicates a distance of ~37 Å, which coincides with that expected for the semi-open conformational state, where the distance profile for apo MDR769 indicates a longer most probable flap distance (37 Å versus 36 Å) relative to subtype B, as seen previously (9). The dashed line at 33 Å marks the distance expected for the inhibitor-induced closed conformation (9, 17).
Figure 2.
(A) Background-subtracted DEER dipolar evolution curves (black) with fits from Tikhonov regularization (TKR) analysis for MDR769. Vertical dashed line marks the local minimum for Apo. (B) Stack plot of distance profiles for free MDR769 and with FDA-approved inhibitors or substrate-mimic, CA-p2. Semi-open and closed populations have flap distances of ~37 Å (black solid line) and ~33 Å (red dashed line), respectively. The minor population at ~26 – 30Å corresponds to the curled/tucked flap conformation (asterisk), whereas those at ~40 – 45 Å are assigned to the wide-open populations (double asterisk). Full details of data analyses are given in Supporting Information.
In our method to ensure proper background subtraction, the Tikhonov regularization (TKR) distance profiles from DeerAnalysis2008 are deconstructed to a linear combination of Gaussian populations. Nominally, four Gaussian populations are required for sufficient regeneration of the TKR data, and these are assigned to curled/tucked, closed, semi-open, and wide-open HIV-1 PR conformational states (7, 9). Population assignments are based on assessing MTSL distances in HIV-1 PR models from molecular dynamics simulation and X-ray studies (19–22). Minor populations located at 26–30 Å and 40–45 Å are assigned to the curled or tucked (23) and wide-open states (20); respectively.
For apo subtype B HIV-1 PR, previous DEER studies reveal a distance profile containing a predominant semi-open conformation, where inhibitor binding shifts the conformational ensemble to increasing populations of the closed state (17). For subtype B, the fractional occupancy of the closed state was >60% for 7 out of 10 inhibitor, with a concomitant shift in the most probable distance from 36 Å to ~33 Å. These PIs are ritonavir (RTV), saquinavir (SQV), amprenavir (APV), lopinavir (LPV), darunavir (DRV) tipranavir (TPV), and Ca-P2 substrate mimic. For the remaining three inhibitors, atazanavir (ATV) has ~40% closed population, whereas only <15% closed population is seen for nelfinavir (NFV) and indinavir (IDV).
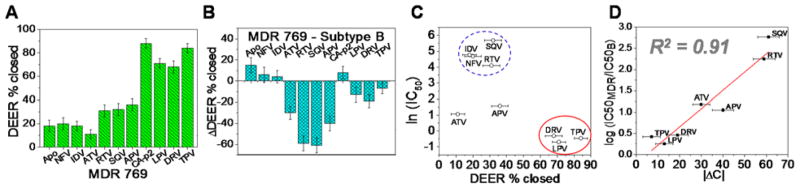
For MDR769, only 3 out of 10 inhibitors, LPV, DRV, and TPV, shift the population to >60% closed. Six inhibitors retain the most probable distance at ~35–37 Å (Figure 2B). For APV, RTV or SQV, only 31–36% closed population is observed (Figure 3A), which is 40–60% less than that seen previously for subtype B (Figure 3B). Likewise, ATV exhibited a 30% decrease in the shift to the closed population. Because MDR769 represents a clinical isolate from a patient failing extensive antiretroviral therapy, it is not surprising that many of the inhibitors lose the ability to induce flap closure. Within error, IDV and NFV induced similar degrees of flap closure in subtype B and MDR769 (≤20%). CA-p2, a substrate mimic that serves as positive control, was found to have comparable flap closure to subtype B.
Figure 3.
(A) DEER percentage closed (% closed) for MDR769 protease. (B) DEER % difference between MDR769 and subtype B (17). (C) Logarithmic plot of half maximal inhibitory concentration (IC50) (12) against DEER % closed for MDR769. Inhibitors clustered into two distinct groups but ATV and APV appear to be outliers. (D) Plot of the log IC50 fold-change (12) versus the magnitude of % difference in DEER closed population (|ΔC|) between MDR769 and subtype B reveals a linear correlation. Inhibitors that exhibited low % closed population in both MDR769 and subtype B PR (IDV and NFV) are excluded from analysis.
The relationship between induced conformational shifts detected with DEER spectroscopy and underlying biological implications were examined by plotting previously reported half-maximal inhibitory concentrations (IC50) for MDR769 (12) in logarithmic scale, against the percentage of closed state (Figure 3C). The correlation between induced conformational shifts and IC50 measurements agrees with X-ray models (12, 13), where drugs that bind the HIV-1 PR active site pocket and inhibit viral propagation have multiple interactions with HIV-1 PR that stabilize the closed conformation. Results show that inhibitors with percentage closed (%closed) ≥68%, namely LPV, DRV and TPV, have excellent drug potency (IC50 ≤ 0.7 nM) to the HIV virus. DEER populations indicated that the distance distribution widths are also narrower for these PIs, suggesting conformationally rigid flaps that are locked-in onto the PI in the active site pocket, consistent with inhibitor-bound MDR769 PR crystal structures that reveal multiple inhibitor-flap interactions (12, 13). In contrast, most PIs with %closed ≤32% have IC50 in the 60–300 nM range. Currently, there is no available crystal structure for MDR769 bound to inhibitors NFV, IDV, RTV, SQV, APV or ATV; interestingly the DEER data for these inhibitors reveal conformational flexibility, which may complicate crystallographic resolution or even interfere with crystallization.
Even though APV and ATV have IC50 of 3–5 nM, DEER data suggest that these inhibitors may bind to MDR769 without promoting substantial flap closure. The presence of outliers, APV and ATV, in Figure 3C may arise because of the active site mutation, D25N that was introduced in the DEER constructs. This substitution is often incorporated in spectroscopic investigations (24, 25) because it imparts sample stability and homogeneity. Without inhibitor-bound crystal structures, it is difficult to understand why APV and ATV are affected by the D25N substitution, but we speculate that the mode of binding may be altered by this substitution.
Nevertheless, when the logarithmic fold-change of IC50 (MDR769 relative to wild-type)—a measure of drug resistance—is plotted against the magnitude of |ΔC|, defined earlier as % change in flap closure, ATV and APV now fit within a linear trend (Figure 3D) with a coefficient of determination (R2) equal to 0.91. Here, however, NFV and IDV were excluded because of their weak ability to induce flap closure in both subtype B and MDR769. The weak effects of these two inhibitors is not surprising, given they have the largest wild-type Kd values, ~60–67 times the Kd of DRV (10 pM) (17). Finally, the linear trend in Figure 3D suggests that aside from IC50, the parameter |ΔC| can be used to evaluate inhibitor effectiveness against emerging drug resistant constructs or the effectiveness of novel inhibitors for drug potency against various HIV-1 PR variants. Other drug-resistant variants or inhibitors can be used to further test and validate the proposed method.
Supplementary Material
Acknowledgments
Funding Sources
This work is supported by NSF MBC-0746533 (GEF), UF Center for AIDS Research, and NHMFL-IHRP.
We thank Dr. Alexander Angerhofer for the helpful discussions and Dr. Maria Cristina Dancel for her assistance with mass spectrometry experiments.
Footnotes
Supporting Information Available
Experimental details, sample preparation, time-domain echo curves and data analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Huff JR. J Med Chem. 1991;34:2305–2314. doi: 10.1021/jm00112a001. [DOI] [PubMed] [Google Scholar]
- 2.Shao W, Everitt L, Manchester M, Loeb DD, Hutchison CA, 3rd, Swanstrom R. Proc Natl Acad Sci U S A. 1997;94:2243–2248. doi: 10.1073/pnas.94.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee SY, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia P, Ruane P, Hellinger J, Shirvani V, Zolopa A, Shafer RW. Antimicrob Agents Chemother. 2010;54:4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandaranayake RM, Kolli M, King NM, Nalivaika EA, Heroux A, Kakizawa J, Sugiura W, Schiffer CA. J Virol. 2010;84:9995–10003. doi: 10.1128/JVI.00505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemente JC, Hemrajani R, Blum LE, Goodenow MM, Dunn BM. Biochemistry. 2003;42:15029–15035. doi: 10.1021/bi035701y. [DOI] [PubMed] [Google Scholar]
- 6.Weber IT, Agniswamy J. Viruses. 2009;1:1110–1136. doi: 10.3390/v1031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kear JL, Blackburn ME, Veloro AM, Dunn BM, Fanucci GE. J Am Chem Soc. 2009;131:14650–14651. doi: 10.1021/ja907088a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torbeev VY, Raghuraman H, Mandal K, Senapati S, Perozo E, Kent SB. J Am Chem Soc. 2009;131:884–885. doi: 10.1021/ja806526z. [DOI] [PubMed] [Google Scholar]
- 9.Galiano L, Ding F, Veloro AM, Blackburn ME, Simmerling C, Fanucci GE. J Am Chem Soc. 2009;131:430–431. doi: 10.1021/ja807531v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piana S, Carloni P, Rothlisberger U. Protein Sci. 2002;11:2393–2402. doi: 10.1110/ps.0206702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer S, Shafer RW, Merigan TC. Aids. 1999;13:661–667. doi: 10.1097/00002030-199904160-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Liu Z, Brunzelle JS, Kovari IA, Dewdney TG, Reiter SJ, Kovari LC. Biochem Biophys Res Commun. 2011;412:737–742. doi: 10.1016/j.bbrc.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logsdon BC, Vickrey JF, Martin P, Proteasa G, Koepke JI, Terlecky SR, Winters MA, Merigan TC, Kovari LC. J Virol. 2004;78:3123–3132. doi: 10.1128/JVI.78.6.3123-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin P, Vickrey JF, Proteasa G, Jimenez YL, Wawrzak Z, Winters MA, Merigan TC, Kovari LC. Structure. 2005;13:1887–1895. doi: 10.1016/j.str.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, Wang Y, Brunzelle J, Kovari IA, Kovari LC. Protein J. 2011;30:173–183. doi: 10.1007/s10930-011-9316-2. [DOI] [PubMed] [Google Scholar]
- 16.Polyhach Y, Bordignon E, Jeschke G. Phys Chem Chem Phys. 13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 17.Blackburn ME, Veloro AM, Fanucci GE. Biochemistry. 2009;48:8765–8767. doi: 10.1021/bi901201q. [DOI] [PubMed] [Google Scholar]
- 18.Fanucci GE, Cafiso DS. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Ding F, Layten M, Simmerling C. J Am Chem Soc. 2008;130:7184–7185. doi: 10.1021/ja800893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornak V, Okur A, Rizzo RC, Simmerling C. Proc Natl Acad Sci U S A. 2006;103:915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rick SW, Erickson JW, Burt SK. Proteins. 1998;32:7–16. doi: 10.1002/(sici)1097-0134(19980701)32:1<7::aid-prot3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 22.Spinelli S, Liu QZ, Alzari PM, Hirel PH, Poljak RJ. Biochimie. 1991;73:1391–1396. doi: 10.1016/0300-9084(91)90169-2. [DOI] [PubMed] [Google Scholar]
- 23.Scott WR, Schiffer CA. Structure. 2000;8:1259–1265. doi: 10.1016/s0969-2126(00)00537-2. [DOI] [PubMed] [Google Scholar]
- 24.Sayer JM, Liu F, Ishima R, Weber IT, Louis JM. J Biol Chem. 2008;283:13459–13470. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louis JM, Ishima R, Torchia DA, Weber IT. Adv Pharmacol. 2007;55:261–298. doi: 10.1016/S1054-3589(07)55008-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.