Abstract
Puerto Ricans are a unique Hispanic population with European, Native American (Taino), and higher West African ancestral contributions than other non-Caribbean Hispanics. In admixed populations, such as Puerto Ricans, genetic variants can be found at different frequencies when compared to parental populations and uniquely combined and distributed. Therefore, in this review, we aimed to collect data from studies conducted in healthy Puerto Ricans and to report the frequencies of genetic polymorphisms with major relevance in drug response. Filtering for healthy volunteers or individuals, we performed a search of pharmacogenetic studies in academic literature databases without limiting the period of the results. The search was limited to Puerto Ricans living in the island, excluding those studies performed in mainland (United States). We found that the genetic markers impacting pharmacological therapy in the areas of cardiovascular, oncology, and neurology are the most frequently investigated. Coincidently, the top causes of mortality in the island are cardiovascular diseases, cancer, diabetes, Alzheimer’s disease, and stroke. In addition, polymorphisms in genes that encode for members of the CYP450 family (CYP2C9, CYP2C19, and CYP2D6) are also available due to their relevance in the metabolism of drugs. The complex genetic background of Puerto Ricans is responsible for the divergence in the reported allele frequencies when compared to parental populations (Africans, East Asians, and Europeans). The importance of reporting the findings of pharmacogenetic studies conducted in Puerto Ricans is to identify genetic variants with potential utility among this genetically complex population and eventually move forward the adoption of personalized medicine in the island.
Keywords: genetic polymorphisms, genotyping, healthy volunteers, personalized medicine, pharmacogenetics, Puerto Ricans
Introduction
Often, pharmacogenetic studies that include Hispanics or Latinos classify individuals from different ethnic groups as a single population. For the purpose of this review, Hispanics or Latinos are individuals from Spanish-speaking countries in the Americas (including the Caribbean), or are of Spanish culture or origin regardless of race, but with a presumed mixture of European, Native American, and African ancestries at varying degrees [1]. Bryc et al. found that Hispanic Latinos show a large variation in the ancestral contributions of each parental population (European, Native American, and African) among and within Hispanic ethnic groups [2]. For instance, although both Puerto Ricans and Mexicans are considered Hispanics or Latinos, Puerto Rican admixture is characterized by a large proportion of European ancestry but lower African and Native American ancestries. In contrast, Mexicans have higher contributions of Native American and European ancestries but minimal African ancestry [2, 3]. Caribbean Hispanics, including Puerto Ricans, have European, Native American (Taino), and higher West African contributions than other Hispanics, which have been not only historically recorded but also scientifically demonstrated [2]. Genome-wide allelic dissimilarity analysis showed that Puerto Ricans exhibit large heterogeneity when compared to Caucasians and African Americans from Kentucky in the United States [3].
A vast number of genetic polymorphisms involved in drug response have been identified without a clear perspective of the prevalence and implications of those variants in an admixed population. The top leading causes of death in the island as reported until 2010 were cardiovascular diseases, cancer, diabetes, Alzheimer’s disease, and stroke [4]. Although a vast number of the investigated genetic polymorphisms have involved commonly used drugs in the management of these diseases, their prevalence and implications of such variants have been poorly researched in admixed populations. Given the admixed nature of the Puerto Rican population and the genetic differences from other populations, our research group is focused on determining the prevalence of genetic variants with clinical relevance for Puerto Rico. The ultimate aim is to move forward the application of a personalized medicine paradigm in the island. This review summarizes all available pharmacogenetic studies performed in Puerto Rico, including data from the 1000 Genomes Project.
Methods
A literature search in the PubMed-based U.S. National Library of Medicine Medline, Scopus, and Google Scholar databases was performed from May to June 2015 following the same methodology used in previous studies [5, 6]. Because pharmacogenetics is a relatively new discipline in Puerto Rico, we decided not to filter or limit the period (dates) of this literature search. The articles were selected if the study fulfilled the following criteria: (i) reported genotype, allele, or metabolic phenotype frequencies; (ii) involved healthy volunteers (control groups from case-control studies were also included); and (iii) included Caribbean Hispanic Puerto Ricans from the Commonwealth of Puerto Rico (i.e. studies conducted in healthy Puerto Ricans residing outside of the island of Puerto Rico, mainly from mainland United States, were excluded). The search terms were “Puerto Rico,” “Puerto Ricans,” “Healthy,” “Pharmacogenetics,” “Allele,” “Genotypes,” “Frequency,” “Prevalence,” and a term for the “biomarker/probe drug” of clinical interest. A total of 104 previously identified genetic biomarkers were selected from those listed by the Food and Drug Administration [5, 7, 8] and the PharmGKB resource [5, 7, 8], as recommended by Cespedes-Garro et al. [5, 7, 8]. For the identification of phenotyping studies, we searched for commonly used probe drugs and included debrisoquine, sparteine, metoprolol, and dextromethorphan (CYP2D6), S-mephenytoin and omeprazole (CYP2C19), tolbutamide, losartan, and diclofenac (CYP2C9), isoniazid (NAT2), and caffeine (CYP1A2), as described earlier by others [5].
Additionally, a query of results from 55 Puerto Ricans in the 1000 Genomes Project database [9] and consultations with other colleagues in the field were made to enrich the review with updated information from other studies in Puerto Ricans, including unpublished data. The search was later divided into major categories based on the revised genetic biomarkers with at least one report in healthy Puerto Ricans.
β2 Adrenergic receptors (β2AR)
β2ARs are G-protein-coupled receptors expressed in cardiac myocytes and, vascular and bronchial smooth muscles, where they are involved in the opening of the airways. These receptors are the target of bronchodilators (i.e. albuterol), the rescue medicine for asthmatic patients. Among the most important identified variants in β2 AR are two missense mutations: Arg16Gly (rs1042713) and Glu27Gln (rs1042714). In vitro studies have shown that these mutations decrease the density of receptors through agonist-promoted down-regulation [10].
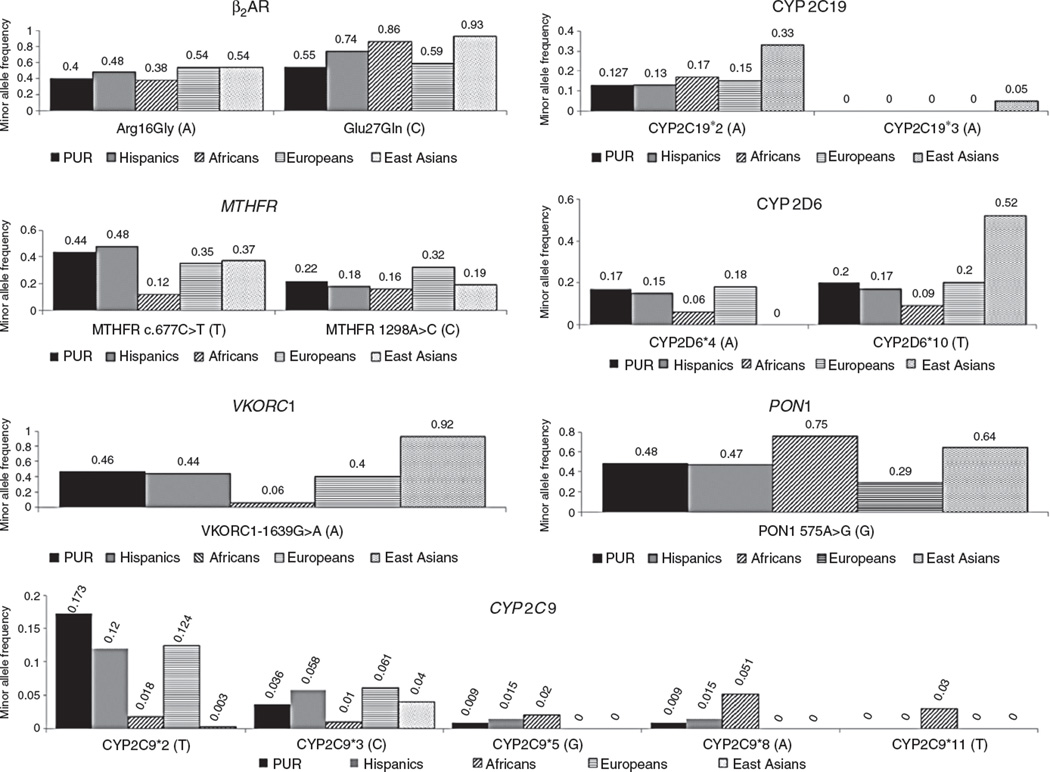
The Genetics of Asthma in Latino Americans study conducted in 667 asthmatic patients and parents of Mexican and Puerto Rican origins found that the presence of the ancestral allele of the genetic variant Arg-16Gly is associated with higher bronchodilator (albuterol) response in Puerto Ricans only. This study demonstrated ethnic-specific differences in individuals even when they belong to the same Hispanic or Latino population. Puerto Ricans have the highest prevalence, morbidity, and mortality of asthma [11]. Environmental factors and different patterns of linkage disequilibrium may explain these differences between Puerto Ricans and Mexicans. The ancestral allele frequency is 0.40 for the Arg16Gly variant in healthy Puerto Ricans (data from the 1000 Genomes Project) and ~0.56 for the Glu27Gln variant allele [9]. The frequencies of the ancestral allele for Arg16Gly and the Glu27Gln allele are more similar to those for Europeans (0.38 and 0.59, respectively; see Figure 1) than to other parental populations [9].
Figure 1.
Reported MAFs in Puerto Ricans, Hispanics, and parental populations (Africans, Europeans, and East Asians) for the most commonly studied genetic variants with pharmacogenetic relevance.
Data obtained from the 1000 Genomes Project.
Methylene-tetrahydrofolate reductase (MTHFR)
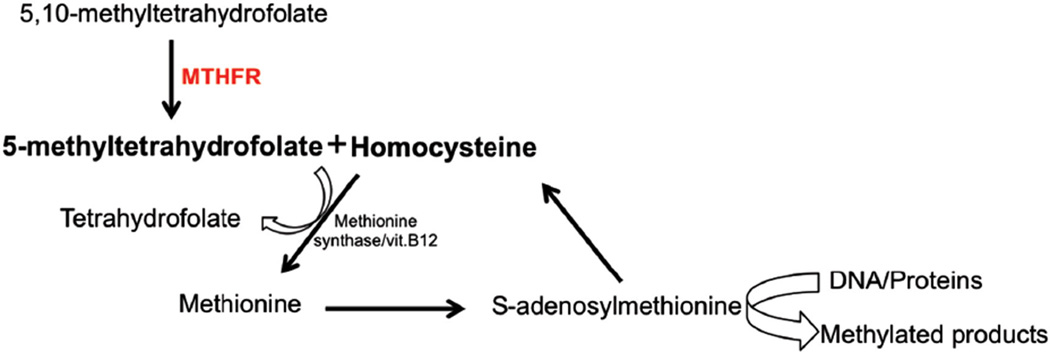
MTHFR is the enzyme responsible of the reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, a precursor of methionine synthesis (see Figure 2). The synthesis of methionine also requires homocysteine, a precursor whose accumulation in the plasma is associated with metabolic syndrome, cardiovascular events [12], neurologic disorders [13], and intestinal inflammation [14]. The most common MTHFR variants are two missense mutations that result in lower enzymatic activity [15]: c.677C>T (rs1801133; Ala222Val) and c.1298A>C (rs1801131; Glu429Ala). MTHFR deficiency due to c.677C>T and c.1298A>C has been associated with 5-fluorouracil-induced cytotoxicity in patients with colorectal cancer, but studies have been inconclusive [15, 16]. MTHFR c.677C>T has also been associated with methotrexate-induced toxicity manifested as liver, skin, and gastrointestinal toxicity, myelosuppression, and oral mucositis [16].
Figure 2.
Pathway of MTHFR and methionine synthesis.
Ayala-Rivera et al. determined the frequency of the common c.677C>T and c.1298A>C MTHFR gene mutations [17, 18], as well as the less common c.1317T>C variant [18], in healthy Puerto Ricans using 400 genomic DNA specimens from newborn dried blood samples. Genotyping tests were performed with restriction fragment length polymorphism (RFLP) assays. Summarizing, the authors reported a frequency of 0.14 homozygous for the c.677C>T variant and 0.03 homozygous for the c.1298A>C gene mutation. No individuals homozygous for the c.1317T>C variant were found in the study cohort. The overall minor allele frequencies (MAFs) for c.677C>T and c.1298A>C were 0.34 and 0.20, respectively [18].
Ayala-Rivera [18] also studied these loss-of-function single nucleotide polymorphisms (SNPs; c.677C>T, c.1298A>C, and c.1317T>C) in 68 samples collected from control families (i.e. 12 fathers, 30 pregnant mothers, and 26 babies) as part of a case-control study related to neural tube defects. They found that the frequencies of homozygotes for the c.677C>T (TT) and c.1298A>C (CC) mutations were 0.27 and 0.26, respectively. On the contrary, the frequency of heterozygotes for the c.1317T>C polymorphism was 0.038, but no homozygotes were reported. Finally, 0.13 was also found to be compound heterozygotes for the c.677C>T and c.1298A>C variants [18].
The frequency of the c.677C>T and c.1298A>C mutant alleles of MTHFR reported in healthy Puerto Ricans from the 1000 Genomes Project is 0.436 and 0.218, respectively (Figure 1). As can be seen, the frequencies of these two MTHFR mutant alleles in Puerto Ricans and Hispanics were closer to those earlier reported in East Asians (i.e. 0.37 and 0.19 for c.677C>T and c.1298A>C, respectively; see Figure 1) than to those in the other parental populations. However, the frequency of the 677T variant is higher among Puerto Ricans (0.436) and Hispanics (0.480) than in Europeans (0.35), Africans (0.12), and Asians (0.37).
VKORC1
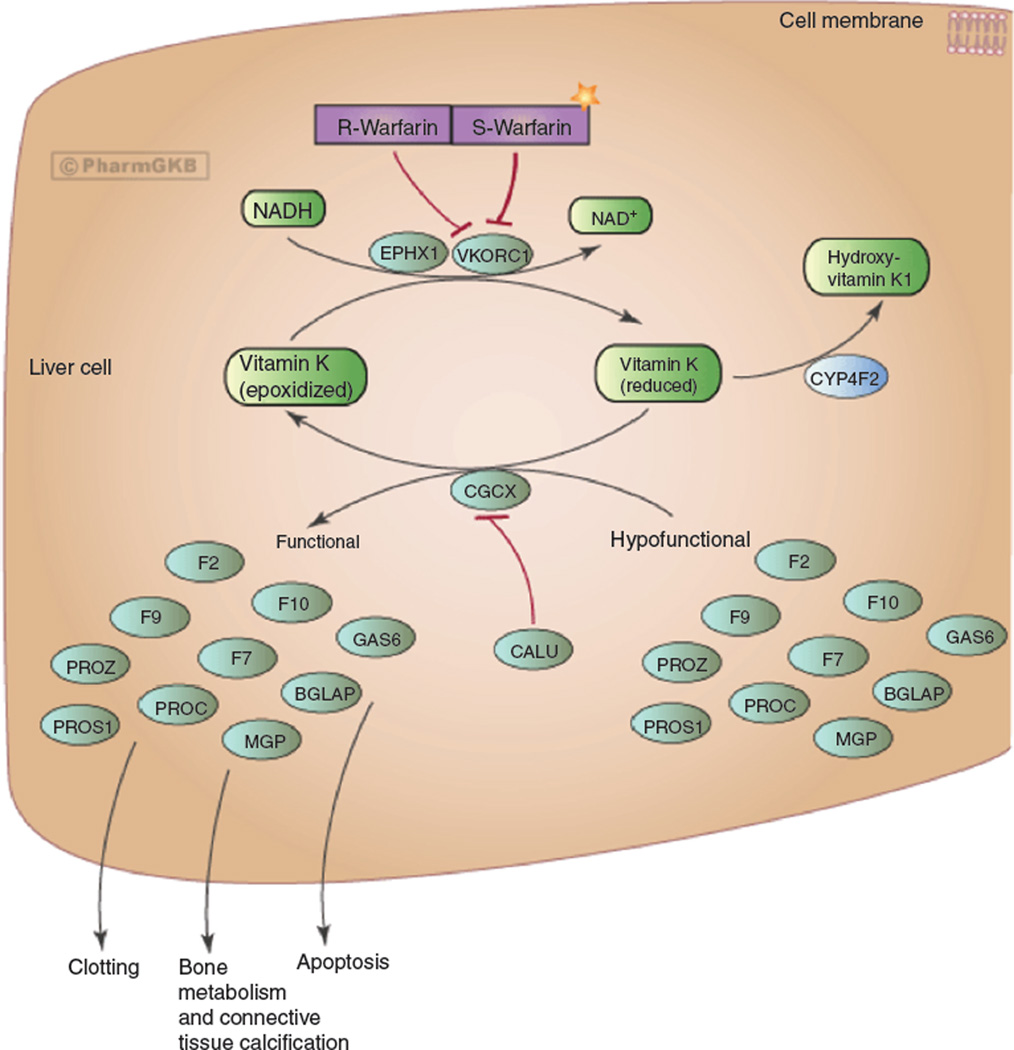
VKORC1 is a small integral protein targeted by the oral anticoagulant warfarin. Warfarin inhibits the recycling of vitamin K by VKORC1, which is essential for the activation of coagulation factors during the vitamin K cycle (see Figure 3) [19]. Several genetic polymorphisms and haplotypes in VKORC1 have been associated with variations in warfarin requirements resulting in warfarin sensitivity or resistance. One of the most important genetic polymorphisms, −1639G>A, is a change of guanine to adenine (G>A) in the promoter region of the gene, which affects the transcription factor binding site and decreases reporter gene activity (when compared to the ancestral allele), suggesting a decrease in enzyme expression and lower warfarin requirements [20]. VKORC1 genotypes are considered the biggest predictors of warfarin dose, accounting for approximately 25% of the variability in dose requirements [21].
Figure 3.
Mechanism of action of warfarin.
From Whirl-Carrillo et al. [19]. Used with permission from PharmGKB and Stanford University.
A previous noninterventional, cross-sectional study by Duconge et al. [22] determined the frequency of the −1639G>A allele. This study was conducted in 92 specimens of Puerto Rican newborns, representing all geographic regions of the island. The observed frequency for the VKORC1 −1639G>A variant allele was 0.29 [22]. The −1639G>A allele frequency in Puerto Ricans participating at the 1000 Genomes Project is 0.464, comparable to that found among Europeans (see Figure 1) [9].
Cytochrome P450s
Cytochrome P450 enzymes catalyze the oxidation, reduction, and hydrolysis (phase I metabolism) of drugs. They also have a function in the synthesis of steroid hormones, and their highest expression occurs in the liver. Only a few CYP450s are important in the metabolism of many drugs at pharmacologically relevant concentrations. The most studied cytochromes in pharmacogenetics are CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP3A5. For a broader review of CYP450s, the reader should refer to Zanger and Schwab [23].
CYP2C9
CYP2C9 is involved in the metabolism of S-warfarin, phenytoin, glyburide, losartan, and nonsteroidal anti-inflammatory drugs. The most studied variants in Caucasians are CYP2C9*2 (rs1799853; 430C>T) and *3 (rs1057910; 1075A>C), both associated with decreased enzyme activity and lower warfarin requirements [24–27]. Other variants, such as CYP2C9*5 (rs28371686; 1080C>G), *6 (rs9332131; 818delA), *8 (rs7900194; 449G>A), and *11 (rs28371685; 1003C>T), are more frequent among African descendants and are also associated with lower dose requirements of warfarin [28, 29].
In a previous study by Duconge et al. [22], the observed frequencies in 92 neonates were 0.065 and 0.054 for CYP2C9*2 and *3, respectively. Due to the known relevance of CYP2C9 and VKORC1 genotypes in warfarin dose requirements, the prevalence of allelic variants were calculated individually and as a composite. The prevalence of combinatorial genotypes of CYP2C9 and VKORC1 was as follows: 0.16 were carriers of CYP2C9 and VKORC1 combined variants, 0.09 were carriers of CYP2C9 polymorphisms, 0.35 were carriers of the VKORC1 haplotype A (identified by −1639G>A), and the remaining 0.40 were wild-types. Triple carriers (i.e. CYP2C9*1/*2 or *1/*3 plus VKORC1 AA) accounted for 0.03 of the study cohort. No statistically significant deviations from Hardy-Weinberg equilibrium (HWE) of genotype proportions in these loci were found according to the goodness-of-fit χ2-test [22]. Notably, one individual carried the rare allele CYP2C9*6 (818delA, frameshift), which has been associated with decreased enzyme activity in people of African descent [22, 30]. In summary, among the Puerto Rican population, 0.60 of the study cohort carried at least one SNP predicting deficient warfarin metabolism or responsiveness and another 0.13 were double carriers with allelic variants in both genes [22].
Recently, Renta et al. analyzed 104 specimens from dried blood samples of healthy Puerto Ricans (unpublished results) to identify two rare variants (i.e. c.−1766T>C and c.−1188T>C) that are in linkage disequilibrium with the CYP2C9*8, as earlier described by others. The rare c.−1188T>C (rs4918758) SNP was detected in the study cohort with an MAF of 0.26. The frequency of the heterozygous genotype (C/T) was 0.43 and the homozygous genotype (C/C or mutant homozygous) was 0.06 (unpublished results). The observed frequency of 0.26 for c.−1188T>C in this cohort resembles that early reported in the HapMap Yoruba population from Nigeria (MAF=0.273, n=88, p=0.84) and was not significantly different from another cohort of Puerto Ricans in the 1000 Genomes Project (MAF=0.37, n=55, p=0.23) [9].
Only two heterozygous for the rare c.−1766T>C polymorphism (rs933209) were identified (MAF=0.01 overall). The observed allele frequency was comparable to those reported in both the HapMap Yoruba population from Nigeria (MAF=0.014, n=88, p=0.74) and the Puerto Rican population in the 1000 Genomes Project (MAF=0.009, n=55, p=0.60) [9].
CYP2C19
CYP2C19 metabolizes the oral antiplatelet, clopidogrel, omeprazole (proton pump inhibitor), and drugs that work in the central nervous system, such as amitriptyline (antidepressant) and carbamazepine (antiepileptic) [23]. From 16 identified genetic variants, seven have been associated with an inactive enzyme [31]. The most commonly studied variants are CYP2C19*2 (rs4244285; c.681G>A; results in a splicing defect) and CYP2C19*3 (rs4986893; c.636G>A; encodes for a stop codon), both producing an inactive enzyme (http://www.cypalleles.ki.se/cyp2c19. htm). CYP2C19*2 and *3 have been associated with a lower response to clopidogrel, which is activated by CYP2C19 metabolism [23]. Individuals with reduced enzyme activity are categorized as poor metabolizers.
In a study by Orengo-Mercado et al. [32], the frequency distribution of CYP2C19*2 and *3 were ascertained in unrelated samples from healthy Puerto Ricans using TaqMan®genotyping assays. The prevalence of the CYP2C19*2 was 0.09, from which 15 carriers were heterozygous and 1 individual was homozygous (corresponding to 0.152 heterozygous and 0.01 homozygous among the population, respectively). CYP2C19*3 was not found in any of the samples tested [32].
Duconge et al. [33] interrogated the CYP2C19 locus in 122 Puerto Ricans by applying a RFLP technique and confirmed the results with the Illumina Physiogenomic Bead- Array™. They found a CYP2C19*2 allele frequency of 0.139 with the RFLP method and a similar frequency with the Illumina assay (0.141). The frequency of heterozygotes and homozygotes was 0.1229 and 0.0164, respectively. None of the individuals were carriers of the CYP2C19*3 [33]. Another study conducted by Duconge et al. [34] found an MAF of 0.148 for CYP2C19*2 in 98 examined samples, whereas the CYP2C9*3 allele had a frequency of 0.035 in 95 individuals [34]. These two studies were conducted using dried blood samples from the Puerto Rico Newborn Screening Program. To date, the 1000 Genomes Project reported frequencies of 0.127 and 0.000 for CYP2C19*2 and *3, respectively, for Puerto Ricans [9].
The CYP2C19*2 frequency (0.141) in Puerto Ricans is similar to that reported in Hispanics (MAF=0.13) and Europeans (MAF=0.15; see Figure 1). The CYP2C19*3 is considered rare in non-Asian descendants; therefore, it is not surprising that the CYP2C19*3 allele was absent from the studies by Duconge et al. [33] and Orengo-Mercado et al. [32]. Interestingly, Duconge et al. reported five individuals who were heterozygous for the CYP2C19*3 variant after performing SNP imputation [34]. Given the expected alterations in the pattern of linkage disequilibrium due to significant admixture in Puerto Ricans, this finding should be taken with caution. The 1000 Genomes Project (phase I) reports an MAF of 0.05 for the CYP2C9*3 in the East Asian population, but it has not been found in the rest of the populations tested until the date [9].
CYP2D6
CYP2D6 is the only gene that encodes for a protein from the CYP2D subfamily [23]. CYP2D6 metabolizes between 15% and 25% of prescription drugs, including tamoxifen (anticancer drug), codeine (opiate), debrisoquine (antihypertensive), and psychotropic drugs, such as paroxetine (antidepressant), aripiprazole, and risperidone (both atypical antipsychotics) [3, 23]. CYP2D6 is a highly polymorphic gene. The CYP2D6*4 allele (rs3892097; 1846G>A) encodes for a nonfunctional enzyme [3]. The presence of CYP2D6*4 has been associated with breast cancer mortality and relapse due to decreased CYP2D6 activity and consequent inability to metabolize tamoxifen into its active compound [35]. The CYP2D6*10 (rs1065852; 100C>T) genetic variant is associated with decreased enzyme activity resulting from a missense mutation. CYP2D6 is also subjected to structural variants, such as deletions and amplifications, where the latter results in higher than normal enzymatic activity. CYP2D6 gene amplifications are associated with ultra-rapid metabolism of codeine resulting in morphine-induced intoxication in children [36].
The frequency of the nonfunctional CYP2D6*4 (g.1934G>A transition) SNP was also assessed by RFLP in 67 genomic DNA specimens randomly selected from the existing repository of the Puerto Rico Newborn Screening Program (unpublished results). The MAF of the CYP2D6*4 variant in this healthy cohort of Puerto Ricans was found to be 0.15 [95% confidence interval (95% CI): 9.8–22], with 16 heterozygous (G/A) (corresponding to a frequency of 0.239; 95% CI: 15–36) and two homozygous (A/A) for the mutant allele (corresponding to a frequency of 0.03; 95% CI: 0.21–11). The CYP2D6*4 mutant allele was found with an MAF of 0.17 of the Puerto Rican cohort participating of the 1000 Genomes Project, very similar to the frequency mentioned above for the same population [9]. Although slightly lower, the observed prevalence is quite similar to that reported among Caucasians (MAF=0.18; see Figure 1) [9, 37]. Individuals who are heterozygous for the CYP2D6*4 variant are expected to show a reduced metabolic capacity for this major drug clearance pathway [38]. The observed genotype proportions for this SNP were in HWE (χ2=0.24, p<0.05).
Orengo-Mercado et al. found an MAF of 0.09 for the CYP2D6*10 (c.100C>T) variant in a cohort of 100 unrelated healthy Puerto Ricans. From this sample, 0.14 were heterozygous for the allelic variant (C/T), whereas 0.02 were homozygous (T/T), with no deviations from HWE [32]. CYP2D6*10 is a major variant allele in people of Asian heritage. The mutant (T) allele of the CYP2D6*10 is present with a frequency of ~0.50 in East Asians [39, 40], 0.20 in Caucasians, and 0.09 in Africans (see Figure 1) [9, 39, 41, 42]. The observed MAF of 0.09 in Puerto Ricans was lower than that previously reported in Europeans (MAF=0.20) [9, 32], which led Orengo-Mercado et al. to suggest that the African ancestry contribution to the genetic make-up of Puerto Ricans may explain this relative lower frequency of the CYP2D6*10 mutant allele [32]. However, data from the 1000 Genomes Project show a similar frequency of the CYP2D6*10 polymorphism among Puerto Ricans (MAF=0.20) and Europeans (MAF=0.20) [9]. The observed differences in CYP2D6*10 allele frequencies reported by Orengo-Mercado et al. [32] and the 1000 Genomes Project [9] may be due to the sampling methods employed in both studies.
CYP2D6*31 (c.4042G>A, p.R440H) is a rare allele that has been previously associated with severely reduced functional activity in Spaniards and Puerto Ricans [43]. The corresponding estimates of MAF and genotype frequencies for the CYP2D6*31 variant were determined among Puerto Ricans also using a TaqMan®custom SNP assay. In a screening of 100 specimens from a cohort of healthy Puerto Ricans, the calculated MAF was 0.01 (95% CI: 0.0004–0.038), with two heterozygotes (0.02; 95% CI: 0.001–0.074) and no homozygotes found for this rare mutation (HWE: χ2=0.01, p<0.05; unpublished results). These results were confirmed by RFLP and were not significantly different from prior reports by González-Tejera et al. [44] and Gaedigk et al. [43] in ~50 Puerto Rican psychiatric patients (MAF=0.02, 2 heterozygous: 4%).
PON1
Paraoxonase I is encoded by PON1. The enzyme is an esterase with recognized roles in the hydrolysis of organophosphates and in the prevention of atherosclerosis [45]. Bouman et al. suggested that the reaction catalyzed by PON1 was a rate-limiting step in the conversion of clopidogrel into its active metabolite [46]. Their conclusion was based on the finding that individuals with the Glu192Arg (rs662; 575A>G) polymorphism had lower PON1 activity, lower concentration of the active metabolite of clopidogrel in plasma, lower platelet inhibition, and therefore higher risk of stent-thrombosis [46].
Using the Illumina Physiogenomic BeadArray™ technology, Duconge et al. screened the PON1 polymorphism (rs662) in 71 healthy Puerto Ricans and reported an allele frequency of 0.45 (i.e. 34 heterozygous and 15 homozygous for the variant allele) [34]. Orengo-Mercado et al. also interrogated this locus in 99 healthy individuals and found an MAF of 0.50 for this SNP [32]. The MAF reported by the 1000 Genomes Project in Puerto Ricans is 0.482, showing a similar prevalence among Puerto Ricans when compared to reports by Duconge et al. and Orengo-Mercado et al. [9, 32, 34]. This variant is more prevalent among African descendants (MAF=0.75) and East Asians (0.64). The PON1 variant allele is more frequent in Puerto Ricans and Hispanics (0.47) than among Europeans (0.29), evidencing the genetic contribution of Africans and Asians to these populations (see Figure 1) [9].
Conclusions
As expected, the most frequently investigated genetic polymorphisms are those concerning to the areas of cardiovascular (i.e. VKORC1, CYP2C9, and CYP2C19) and neuropsychiatry (i.e. CYP2D6) as well as oncology (i.e. MTHFR and CYP2D6) to a less extent. These results are in good agreement with the current statistics of top leading causes of death and morbidities in the island as well as the global trend of research in this field [4, 47]. Concerning the revised genetic biomarkers, studies predominantly targeted a member of the highly polymorphic superfamily of CYP450 genes (i.e. CYP2C9, CYP2C19, and CYP2D6). This seems to be a consequence of logical expectations about the potential clinical utility of genotyping patients for these loci when making treatment decision with drugs for which these enzymes represent a major metabolic pathway (pharmacokinetics).
Collected reports also show that, of the entire Caribbean region, Puerto Rico is one of the territories with highest development and investment in scientific activities, including the evolving field of pharmacogenomics and personalized medicine. This review combines the results from seven studies conducted in healthy Puerto Rican volunteers living in the island and data from the 1000 Genomes Project (n=55 Puerto Ricans). We found two major limitations of pharmacogenetic studies in Puerto Ricans: (a) the relatively small sample size, that is, most of the mentioned studies included only 100 individuals or less (see Table 1), and (b) Puerto Ricans are often deemed as merely a member of the Hispanic population, although there are evidences supporting the substantial genetic diversity within Hispanics [2]. In this regard, results from studies in other Hispanic groups (e.g. Mexican Americans) are often extrapolated to the Puerto Rican population without any validation of such findings in the population of the Island. Other limitations are (i) limited resources and funding availability, (ii) lack of a local network and incentive to establish strong ties or collaborative agreements to conduct studies island-wide, (iii) skepticism about the cost-effectiveness of genotyping, and (iv) insufficient support from the local government and institutions.
Table 1.
Prevalence of genetic variants with pharmacogenetic relevance among Puerto Ricans.
| Gene | Allele | Rs number | DNA change | Effect | MAF | Na | References |
|---|---|---|---|---|---|---|---|
| B2AR | Arg16Gly | rs1042713 | β2AR down-regulation | 0.400 | 55 | [8] | |
| Glu27Gln | rs1042714 | β2AR down-regulation | 0.560 | 55 | [8] | ||
| MTHFR | rs181133 | c.677C>T | Reduced enzyme activity | 0.340 | 400 | [16] | |
| 0.436 | 55 | [8] | |||||
| rs1801131 | c.1298A>C | Reduced enzyme activity | 0.200 | 400 | [16] | ||
| 0.218 | 55 | [8] | |||||
| VKORC1 | rs9923231 | −1639G>A | Reduced enzyme expression | 0.290 | 92 | [20] | |
| 0.464 | 55 | [8] | |||||
| CYP2C9 | *2 | rs1799853 | 3608C>T | Reduced enzyme activity | 0.065 | 92 | [20] |
| 55 | [8] | ||||||
| *3 | rs1057910 | 1075A>C | Reduced enzyme activity | 0.054 | 92 | [20] | |
| CYP2C19 | *2 | rs4244285 | 681G>A | Inactive enzyme | 0.090 | 99 | [48] |
| 0.141 | 122 | [31] | |||||
| 0.148 | 98 | [32] | |||||
| 0.127 | 55 | [8] | |||||
| *3 | rs4986893 | 636G>A | Inactive enzyme | 0.00 | 96 | [48] | |
| 0.000 | 72 | [31] | |||||
| 0.035 | 95 | [32] | |||||
| 0.000 | 55 | [8] | |||||
| CYP2D6 | *4 | rs3892097 | 1846G>A | Inactive enzyme | 0.150 | 67 | Unpublished |
| 0.170 | 55 | [8] | |||||
| *10 | rs1065852 | 100C>T | Reduced enzyme activity | 0.090 | 100 | [48] | |
| 0.200 | 55 | [8] | |||||
| *31 | 4042G>A | Inactive enzyme | 0.010 | 100 | Unpublished | ||
| PON1 | rs662 | 575A>G | Reduced enzyme activity | 0.450 | 71 | [32] | |
| 0.482 | 55 | [8] | |||||
| 0.500 | 99 | [48] |
N indicates the number of individuals tested in each study.
We conclude that the substantial admixture among Puerto Ricans seems to be responsible for the divergences of the observed allele frequencies on these relevant pharmacogenes compared to other reference populations in the HapMap and the 1000 Genomes Project. Indeed, admixture seems to enhance the amount of combinatorial genotypes across the Puerto Rican genomes, resulting in enriched genetic mosaics with a subsequent reduction of the relative frequencies of major alleles [32]. The combination of genetic markers previously associated with drug response and diseases may interplay differently from other populations and their validity must be carefully studied given the genetic heterogeneity of Puerto Ricans. Other environmental, socioeconomic, and cultural factors need to be considered in association studies of different populations.
Phenotyping studies of pharmacogenetically relevant biomarkers in healthy Puerto Ricans were not mentioned in this review because they were either conducted outside of the Commonwealth of Puerto Rico or not fully published in peer-reviewed journals. In addition, there are other pharmacogenes with known clinical implementation (i.e. Clinical Pharmacogenetics Implementation Consortium guidelines) whose frequencies among Puerto Ricans are not yet known. Pharmacogenes with clinical implementation considered as having PharmGKB high level of evidence and with either recommended or actionable pharmacogenetics are CFTR, CYP2C9, CYP2C19, CYP2D6, G6PD, DPYD, HLA-B, IFNL3, TPMT, UGT1A1, and VKORC1 (https://www.pharmgkb.org/cpic/pairs) [8, 19]. Noteworthy, only four of these pharmacogenes have been studied in Puerto Ricans, as mentioned in this review. Therefore, it is necessary to conduct additional studies to expand our current knowledge of these valid genetic markers in Puerto Ricans, so that we can ultimately bring the anticipated benefits of pharmacogenetics to this population. Currently, we are performing a more comprehensive pharmacogenetic ascertainment of Puerto Ricans by interrogating 231 pharmacogenes and ~1936 markers of pharmacokinetics and pharmacodynamic relevance, using the DMET Plus array, which includes the majority of these recommended genes as well as the 32 “Core ADME” genes.
Other studies on nonpharmacogenetic biomarkers, but somehow with certain pharmacological implications (i.e. PCFT-SLC46A1, OCA1, HPS, and HIV serotypes), and that also included healthy subjects, have been conducted in Puerto Ricans within the island [49–52]. Reports of the frequencies of genetic biomarkers and phenotyping of drug metabolism or valid biomarkers in the Puerto Rican population are necessary to support efforts towards the adoption of a personalized medicine paradigm in the island. The availability of these reports will also help promulgate a public policy that seeks to reduce current health-care disparities among individuals of Hispanic heritage within the United States and its territories. Accordingly, additional pharmacogenetic studies of valid genetic biomarkers, including phenotyping, within the unique population of Puerto Ricans are warranted.
Acknowledgments
We thank the Laboratory of Personalized Medicine, Hartford, CT, and the UPR-MSC Research Center in Minority Institutions Center for Genomics in Health Disparities and Rare Disorders for their support and for providing the resources and facilities for performing genetic assays. The authors also want to thank Mohan Kocherla, MSc; Krystyna Gorowski, MSc; Myrna E. Casillas, PharmD; Sonia E. Rodríguez, PharmD; and Jennifer Serrano, PharmD, for their help in the genotyping tests and data analysis.
Research funding: This review was supported in part by the National Heart, Lung, and Blood Institute grant number SC1 HL123911, the Research Center in Minority Institutions award number 8G12 MD007600 from the National Institute on Minority Health and Health Disparities, and the Minority Biomedical Research Support-Research Initiative for Scientific Enhancement at the University of Puerto Rico, Medical Sciences Campus, grant number R25GM061838. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the United States Government. No writing assistance was utilized in the production of this manuscript.
Footnotes
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Authors have no relevant affiliation or financial involvement with any organization or entity with a financial interest in or conflicts of interest with the subject matter or materials discussed in the article that need to be disclosed.
Employment or leadership: None declared.
Honorarium: None declared.
Competing interests: The funding organization(s) played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
Contributor Information
Karla Claudio-Campos, Department of Pharmacology and Toxicology, School of Medicine, University of Puerto Rico, Medical Sciences Campus (UPR-MSC), San Juan, Puerto Rico.
Carmelo Orengo-Mercado, Molecular Genetics Lab, Department of Biochemistry, School of Medicine, University of Puerto Rico, Medical Sciences Campus (UPR-MSC), San Juan, Puerto Rico.
Jessicca Y. Renta, Molecular Genetics Lab, Department of Biochemistry, School of Medicine, University of Puerto Rico, Medical Sciences Campus (UPR-MSC), San Juan, Puerto Rico
Muriel Peguero, Pharmaceutical Sciences Department, School of Pharmacy, University of Puerto Rico, Medical Sciences Campus (UPR-MSC), San Juan, Puerto Rico.
Ricardo García, Pharmaceutical Sciences Department, School of Pharmacy, University of Puerto Rico, Medical Sciences Campus (UPR-MSC), San Juan, Puerto Rico.
Gabriel Hernández, Pharmaceutical Sciences Department, School of Pharmacy, University of Puerto Rico, Medical Sciences Campus (UPR-MSC), San Juan, Puerto Rico.
Susan Corey, Department of Pharmacology and Toxicology, School of Medicine, University of Puerto Rico, Medical Sciences Campus (UPR-MSC), San Juan, Puerto Rico.
Carmen L. Cadilla, Molecular Genetics Lab, Department of Biochemistry, School of Medicine, University of Puerto Rico, Medical Sciences Campus (UPR-MSC), San Juan, Puerto Rico
References
- 1.United States Census Bureau. About Hispanic Origin. Hispanic Origin. [accessed 2015 Sep 20];2013 [Internet]. Available at: http://www.census.gov/topics/population/hispanic-origin/about.html.
- 2.Bryc K, Velez C, Karafet T, Moreno-Estrada A, Reynolds A, Auton A, et al. Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc Natl Acad Sci USA. 2010;107(Suppl 2):8954–8961. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claudio-Campos K, Duconge J, Cadilla CL, Ruaño G. Pharmacogenetics of drug metabolizing enzymes in U.S. Hispanics. Drug Metabol Drug Interact. 2015;30:87–105. doi: 10.1515/dmdi-2014-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Division de Prevencion y Control de Enfermedades Cronicas. Secretaria Auxiliar para la Promocion de la Salud. Plan de accion de enfermedades cronicas para Puerto Rico 2014–2020. 2014 [Google Scholar]
- 5.Cespedes-Garro C, Naranjo M, Ramirez R, Serrano V, Fariñas H, Barrantes H, et al. Pharmacogenetics in Central American healthy volunteers: interethnic variability. Drug Metab Perspect Ther. 2015;30:19–31. doi: 10.1515/dmdi-2014-0025. [DOI] [PubMed] [Google Scholar]
- 6.Moher D, Liberati A, Tetzlaff J, Altman DG, Altman D, Antes G, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement (Chinese edition) J Chin Integr Med. 2009;7:889–896. [Google Scholar]
- 7.Food and Drug Administration. Drugs Table of Pharmacogenomic Biomarkers in Drug Labeling. 2014 [Internet]. Available at: http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm.
- 8.PharmGKB. CPIC Gene-Drug Pairs [Internet] [accessed 2015 Jun 7]; Available at: https://www.pharmgkb.org/page/cpic. [Google Scholar]
- 9.Amigo J, Salas A, Phillips C, Carracedo A. SPSmart: adapting population based SNP genotype databases for fast and comprehensive web access. BMC Bioinform. 2008;9:428. doi: 10.1186/1471-2105-9-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33:9414–9419. doi: 10.1021/bi00198a006. [DOI] [PubMed] [Google Scholar]
- 11.Choudhry S, Ung N, Avila PC, Ziv E, Nazario S, Casal J, et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am J Respir Crit Care Med. 2005;171:563–570. doi: 10.1164/rccm.200409-1286OC. [DOI] [PubMed] [Google Scholar]
- 12.Catena C, Colussi G, Nait F, Capobianco F, Sechi L. Elevated homocysteine levels are associated with the metabolic syndrome and cardiovascular events in hypertensive patients. Am J Hypertens. 2015;28:943–950. doi: 10.1093/ajh/hpu248. [DOI] [PubMed] [Google Scholar]
- 13.Ansari R, Mahta A, Mallack E, Luoa JJ. Hyperhomocysteinemia and neurologic disorders: a review. J Clin Neurol. 2014;10:281–288. doi: 10.3988/jcn.2014.10.4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu S, Li J, Bing Y, Yan W, Zhu Y, Xia B, et al. Diet induced hyperhomocysteinemia increases intestinal inflammation in an animal model of colitis. J Crohns Colitis. 2015;9:708–719. doi: 10.1093/ecco-jcc/jjv094. [DOI] [PubMed] [Google Scholar]
- 15.Etienne-Grimaldi M-C, Francoual M, Formento J-L, Milano G. Methylenetetrahydrofolate reductase (MTHFR) variants and fluorouracil-based treatments in colorectal cancer. Pharmacogenomics. 2007;8:1561–1566. doi: 10.2217/14622416.8.11.1561. [DOI] [PubMed] [Google Scholar]
- 16.Yang L, Hu X, Xu L. Impact of methylenetetrahydrofolate reductase (MTHFR) polymorphisms on methotrexate-induced toxicities in acute lymphoblastic leukemia: a meta-analysis. Tumor Biol. 2012;33:1445–1454. doi: 10.1007/s13277-012-0395-2. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Fragoso L, Garcia-Garcia I, Leavitt G, Renta JY, Ayala M, Cadilla CL. MTHFR polymorphisms in Puerto Rican children with isolated congenital heart disease and their mothers. Int J Genet Mol Biol. 2010;2:43–47. [PMC free article] [PubMed] [Google Scholar]
- 18.Ayala-Rivera M. Methylenetetrahydrofolate reductase, MTHFR, gene mutations in the Puerto Rican population. Ph.D. thesis. 1999 Jun;1:26–43. Retrieved from the University of Puerto Rico Medical Sciences Campus library. [Google Scholar]
- 19.Whirl-Carrillo M, McDonagh E, Hebert J, Gong L, Sangkuhl K, Thorn CF, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan H-Y, Chen J-J, Lee MT, Wung J-C, Chen Y-F, Charng M-J, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and interethnic differences in warfarin sensitivity. Hum Mol Genet. 2005;14:1745–1751. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz UI, Stein CM. Genetic determinants of dose and clinical outcomes in patients receiving oral anticoagulants. Clin Pharmacol Ther. 2006;80:7–12. doi: 10.1016/j.clpt.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Duconge J, Cadilla C, Windemuth A, Kocherla M, Gorowski K, Seip RL, et al. Prevalence of combinatorial CYP2C9 and VKORC1 genotypes in Puerto Ricans: implications for warfarin management in Hispanics. Ethn Dis. 2009;19:390–395. [PMC free article] [PubMed] [Google Scholar]
- 23.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Crespi C, Miller V. The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH: cytochrome P450 oxidoreductase. Pharmacogenetics. 1997;7:203–210. doi: 10.1097/00008571-199706000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Human Cytochrome P450 (CYP) Allele Nomenclature Committee. The human cytochrome P450 (CYP) allele nomenclature database [Internet] Available at: http://www.cypalleles.ki.se/index.htm. [Google Scholar]
- 26.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharma. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 27.Steward DJ, Haining RL, Henne KR, Davis G, Rushmore TH, Trager WF, et al. Genetic association between sensitivity to warfarin and expression of CYP2C9*3. Pharmacogenetics. 1997;7:361–367. doi: 10.1097/00008571-199710000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. CYP2C9*8 is prevalent among African Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10:1243–1255. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie H-G, Prasad HC, Kim RB, Stein CM. CYP2C9 allelic variants: ethnic distribution and functional significance. Adv Drug Deliv Rev. 2002;54:1257–1270. doi: 10.1016/s0169-409x(02)00076-5. [DOI] [PubMed] [Google Scholar]
- 30.Kidd R, Curry T, Gallagher S. Identification of a null allele of CYP2C9 in an African-American exhibiting toxicity to phenytoin. Pharmacogenetics. 2001;11:803–808. doi: 10.1097/00008571-200112000-00008. [DOI] [PubMed] [Google Scholar]
- 31.Luo H-R, Poland RE, Lin K-M, Wan Y-JY. Genetic polymorphism of cytochrome P450 2C19 in Mexican Americans: a cross-ethnic comparative study. Clin Pharmacol Ther. 2006;80:33–40. doi: 10.1016/j.clpt.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Orengo-Mercado C, Nieves B, López L, Vallés-Ortiz N, Renta JY, Santiago-Borrero PJ, et al. Frequencies of functional polymorphisms in three pharmacokinetic genes of clinical interest within the admixed Puerto Rican population. J Pharmacogenomics Pharmacoproteomics. 2013;4:1–6. doi: 10.4172/2153-0645.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duconge J, Cadilla CL, Renta JY, Silen-Rivera P, Piovanetti P, Garcia-Berdecia R, et al. Prevalence of CYP2C19 gene polymorphisms in the Puerto Rican population: a preliminary report. P R Health Sci J. 2008;27:2–3. [PMC free article] [PubMed] [Google Scholar]
- 34.Duconge J, Escalera O, Korchela M, Ruaño G. Clinical implications of genetic admixture in Hispanic Puerto Ricans: impact on the pharmacogenetics of CYP2C19 and PON1. In: Sanodou D, editor. Clinical Applications of Pharmacogenetics. Rijeka: InTech; 2012. pp. 151–163. [Google Scholar]
- 35.Bijl MJ, van Schaik RH, Lammers L, Hofman A, Vulto AG, van Gelder T, et al. The CYP2D6*4 polymorphism affects breast cancer survival in tamoxifen users. Breast Cancer Res Treat. 2009;118:125–130. doi: 10.1007/s10549-008-0272-2. [DOI] [PubMed] [Google Scholar]
- 36.Kelly LE, Rieder M, van den Anker J, Malkin B, Ross C, Neely MN, et al. More codeine fatalities after tonsillectomy in North American children. Pediatrics. 2012;129:e1343–e1347. doi: 10.1542/peds.2011-2538. [DOI] [PubMed] [Google Scholar]
- 37.Sachse C, Brockmoller J, Bauer S, Roots I. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet. 1997;60:284–295. [PMC free article] [PubMed] [Google Scholar]
- 38.Leathart JB, London SJ, Steward A, Adams JD, Idle JR, Daly AK. CYP2D6 phenotype-genotype relationships in African-Americans and Caucasians in Los Angeles. Pharmacogenetics. 1998;8:529–541. doi: 10.1097/00008571-199812000-00010. [DOI] [PubMed] [Google Scholar]
- 39.Roh HK, Dahl ML, Johansson I, Ingelman-Sundberg M, Cha YN, Bertilsson L. Debrisoquine and S-mephenytoin hydroxylation phenotypes and genotypes in a Korean population. Pharmacogenetics. 1996;6:441–447. doi: 10.1097/00008571-199610000-00008. [DOI] [PubMed] [Google Scholar]
- 40.Yoon YR, Cha IJ, Shon JH, Kim KA, Cha YN, Jang IJ, et al. Relationship of paroxetine disposition to metoprolol metabolic ratio and CYP2D6*10 genotype of Korean subjects. Clin Pharmacol Ther. 2000;67:567–576. doi: 10.1067/mcp.2000.106128. [DOI] [PubMed] [Google Scholar]
- 41.Gaedigk A, Gotschall RR, Forbes NS, Simon SD, Kearns GL, Leeder JS. Optimization of cytochrome P4502D6 (CYP2D6) phenotype assignment using a genotyping algorithm based on allele frequency data. Pharmacogenetics. 1999;9:669–682. doi: 10.1097/01213011-199912000-00002. [DOI] [PubMed] [Google Scholar]
- 42.Johansson I, Oscarson M, Yue Q. Genetic analysis of the Chinese cytochrome P4502D locus: characterization of variant CYP2D6 genes present in subjects with diminished capacity for debrisoquine hydroxylation. Mol Pharmacol. 1994;46:452–459. [PubMed] [Google Scholar]
- 43.Gaedigk A, Isidoro-García M, Pearce RE, Sánchez S, García-Solaesa V, Lorenzo-Romo C, et al. Discovery of the nonfunctional CYP2D6 31 allele in Spanish, Puerto Rican, and US Hispanic populations. Eur J Clin Pharmacol. 2010;66:859–864. doi: 10.1007/s00228-010-0831-4. [DOI] [PubMed] [Google Scholar]
- 44.González-Tejera G, Gaedigk A, Corey S. Genetic variants of the drug-metabolizing enzyme CYP2D6 in Puerto Rican psychiatry patients: a preliminary report and potential implications for breast cancer patients. P R Health Sci J. 2010;29:299–304. [PubMed] [Google Scholar]
- 45.Mackness M, Mackness B. Paraoxonase 1 and atherosclerosis: is the gene or the protein more important? Free Radic Biol Med. 2004;37:1317–1323. doi: 10.1016/j.freeradbiomed.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 46.Bouman H, Schömig E, van Werkum JW, Velder J, Hackeng CM, Hirschhäuser C, et al. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat Med. 2011;17:110–116. doi: 10.1038/nm.2281. [DOI] [PubMed] [Google Scholar]
- 47.Holmes MV, Shah T, Vickery C, Smeeth L, Hingorani AD, Casas JP. Fulfilling the promise of personalized medicine? Systematic review and field synopsis of pharmacogenetic studies. PLoS One. 2009;4:e7960. doi: 10.1371/journal.pone.0007960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavallari LH, Vaynshteyn D, Freeman K, Wang D, Perera M, Takahashi H, et al. CYP2C9 promoter region single-nucleotide polymorphisms linked to the R150H polymorphism are functional suggesting their role in CYP2C9*8-mediated effects. Pharmacogenet Genom. 2013;23:228–231. doi: 10.1097/FPC.0b013e32835e95c7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renta JY, Cadilla CL, Vega ME, Hillyer GV, Estrada C, Jiménez E, et al. Longitudinal studies on maternal HIV-1 variants by biological phenotyping, sequence analysis and viral load. Cell Mol Biol (Noisy-le-grand) 1997;43:1097–1114. [PubMed] [Google Scholar]
- 50.Mahadeo KM, Diop-Bove N, Ramirez SI, Cadilla CL, Rivera E, Martin M, et al. Prevalence of a loss-of-function mutation in the proton-coupled folate transporter gene (PCFT-SLC46A1) causing hereditary folate malabsorption in Puerto Rico. J Pediatr. 2011;159:623–627. doi: 10.1016/j.jpeds.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santiago Borrero PJ, Rodríguez-Pérez Y, Renta JY, Izquierdo NJ, Del Fierro L, Muñoz D, et al. Genetic testing for oculocutaneous albinism type 1 and 2 and Hermansky-Pudlak syndrome type 1 and 3 mutations in Puerto Rico. J Invest Dermatol. 2006;126:85–90. doi: 10.1038/sj.jid.5700034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cordova A, Barrios NJ, Ortiz I, Rivera E, Cadilla C, Santiago-Borrero PJ. Poor response to desmopressin acetate (DDAVP) in children with Hermansky-Pudlak syndrome. Pediatr Blood Cancer. 2005;44:51–54. doi: 10.1002/pbc.20210. [DOI] [PubMed] [Google Scholar]