SUMMARY
Plasmodium falciparum parasites express and traffick numerous proteins into the red blood cell (RBC), where some associate specifically with the membrane skeleton. Importantly, these interactions underlie the major alterations to the modified structural and functional properties of the parasite-infected RBC. Plasmodium falciparum Erythrocyte Membrane Protein 3 (PfEMP3) is one such parasite protein that is found in association with the membrane skeleton. Using recombinant PfEMP3 proteins in vitro, we have identified the region of PfEMP3 that binds to the RBC membrane skeleton, specifically to spectrin and actin. Kinetic studies revealed that residues 38–97 of PfEMP3 bound to purified spectrin with moderately high affinity (KD(kin) = 8.5 × 10−8 M). Subsequent deletion mapping analysis further defined the binding domain to a 14 residue sequence (IFEIRLKRSLAQVL; KD(kin) = 3.8 × 10−7 M). Interestingly, this same domain also bound to F-actin in a specific and saturable manner. These interactions are of physiological relevance as evidenced by the binding of this region to the membrane skeleton of inside-out RBCs and when introduced into resealed RBCs. Identification of a 14 residue region of PfEMP3 that binds to both spectrin and actin provides insight into the potential function of PfEMP3 in P. falciparum-infected RBCs.
Keywords: Plasmodium falciparum, protein interactions, malaria, PfEMP3, spectrin, actin, red blood cell
INTRODUCTION
Infection with the Apicomplexan parasite Plasmodium falciparum causes the most severe form of human malaria, resulting in millions of deaths worldwide each year [1]. When compared to normal human red blood cells (RBCs), infected RBCs (IRBCs) have dramatically altered structural and functional properties that include decreased deformability and the appearance of electron dense protrusions, or knobs, at the RBC membrane that mediate binding to the vascular endothelium (see Cooke et al,. for review [2]). These parasite-induced modifications to the RBC are secondary to the synthesis and trafficking of a number of parasite-encoded proteins into the RBC. While some trafficked proteins remain soluble in the RBC cytosol, others are found in complexes with components of the RBC membrane skeleton or with other parasite-encoded proteins (see Cooke et al, for recent reviews [2, 3]).
One of the major protein components of the normal human RBC skeleton is spectrin. Spectrin is a flexible rod-like protein that is composed of two non-identical subunits, α and β [4], that associate in an anti-parallel manner to form αβ heterodimers. Heterodimers associate in a head to head fashion to form (αβ)2 tetramers, and these tetramers are in turn linked into a network by interactions with actin filaments, protein 4.1 and ankyrin (see Bennett and Gilligan for review [5]). The membrane skeleton is connected to the overlying cell membrane via interactions with protein 4.1 and glycophorins C and D (GPC and GPD, respectively), and between ankyrin and band 3 (see Lux and Palek for review [6]). Many P. falciparum proteins are trafficked to the RBC membrane, including the antigenically variable adherence ligand P. falciparum Erythrocyte Membrane Protein 1 (PfEMP1), Knob Associated Histidine Rich Protein (KAHRP), Mature parasite-infected Erythrocyte Surface Antigen (MESA) and Ring-infected Erythrocyte Surface Antigen (RESA; (see Cooke et al, for review [2]). Previously, we and others have performed detailed studies to identify the interactions of these proteins with components of the RBC skeleton and define the specific sub-domains involved. A 48 residue region of RESA that has been shown to bind to spectrin [7] resulted in increased resistance against heat shock of the IRBC [8, 9]. A 19 residue region of MESA binds to the 30 kDa region of protein 4.1 [10] in an interaction that displaces p55 from its normal host cell binding partner 4.1R, thereby potentially modulating the ternary 4.1R-GPC-p55 complex and altering the stability of the membrane skeleton in IRBCs [11]. KAHRP, which is necessary for the production of knobs [12, 13], binds to spectrin, actin and ankyrin in the membrane skeleton [14, 15] via a 72-residue region in the middle of KAHRP that binds to the repeat 4 region of α-spectrin (αR4) [16]. KAHRP also binds to the band 3-binding domain of ankyrin via the 5’ repeat region of the protein [15]. The cytoplasmic tail of PfEMP1 (VARC region) binds to both KAHRP [17] and to RBC spectrin and actin [14, 18]. The histidine-rich, 5’ and 3’ repeats regions of KAHRP bind to several different regions within the cytoplasmic tail of PfEMP1 [19]. Together these KAHRP-PfEMP1-spectrin-actin interactions have been proposed to cluster PfEMP1 at the knobs and anchor PfEMP1 in the IRBC membrane; an interaction that is essential for adhesion of IRBCs to endothelial cells under flow conditions [12].
Plasmodium falciparum Erythrocyte Membrane Protein 3 (PfEMP3) is an approximately 315 kDa protein that is synthesised by mature-stage parasites and exported to the RBC membrane [20–23]. The pfemp3 gene is located in the subtelomeric region of chromosome two, adjacent to kahrp [24–26]. pfemp3 comprises two exons; a structure typical for many parasite proteins that are trafficked to the RBC skeleton. It contains the VTS/PEXEL motif (RSLAQ; that signals export to the RBC; [27, 28]) and repetitive and non-repetitive highly charged sequences (Fig. 1A; [21]). Parasites generated by gene-specific targeted disruption of pfemp3 have demonstrated that PfEMP3 expression is not essential for the formation of knobs or for adhesion under flow conditions [29]. RBCs infected with genetically mutated parasites expressing truncated PfEMP3, however, do show decreased adherence, possibly resulting from the accumulation of PfEMP3 at the RBC membrane and reduced expression of PfEMP1 on the RBC membrane surface. In contrast, PfEMP3 null parasites show only a modest decrease in adhesion [29]. Taken together, these data suggest a role for PfEMP3 in transport and expression of PfEMP1 at the membrane surface [29]. To date, the specific interactions that localise PfEMP3 at the IRBC skeleton have not been characterised.
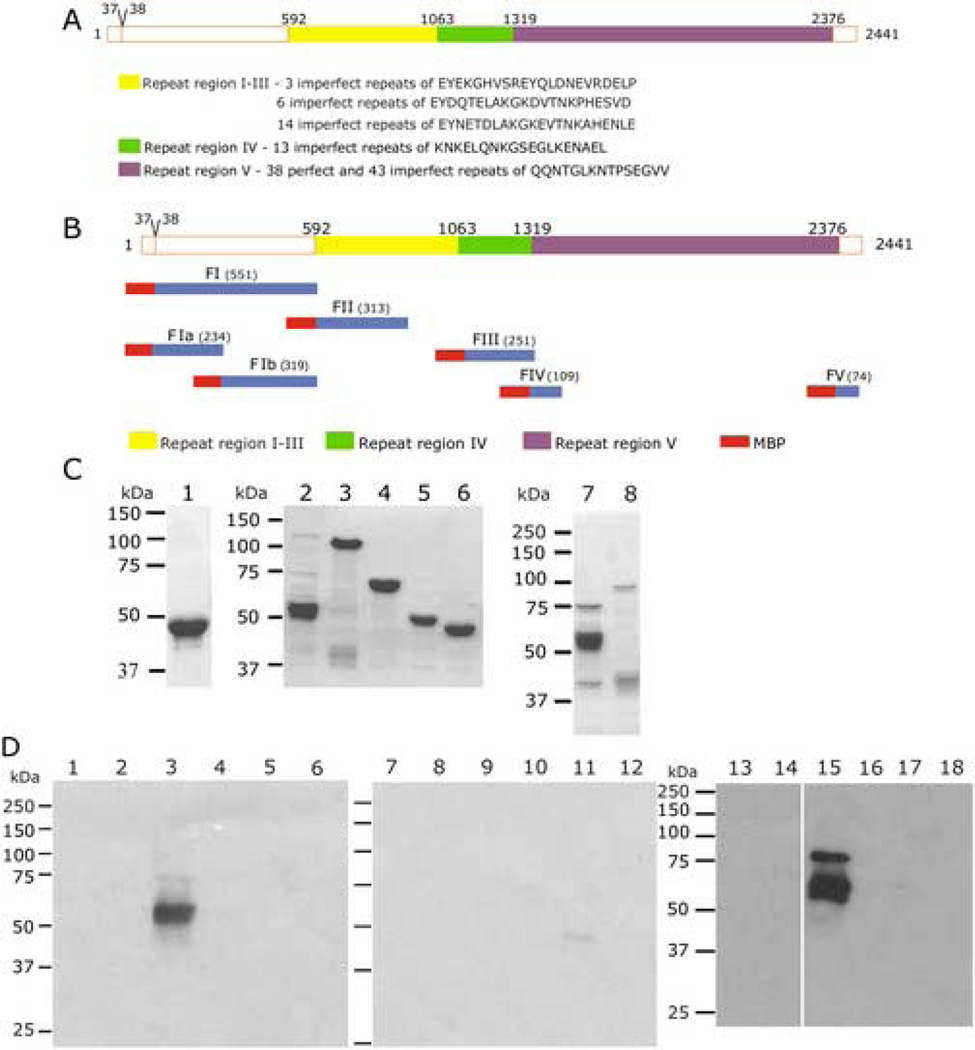
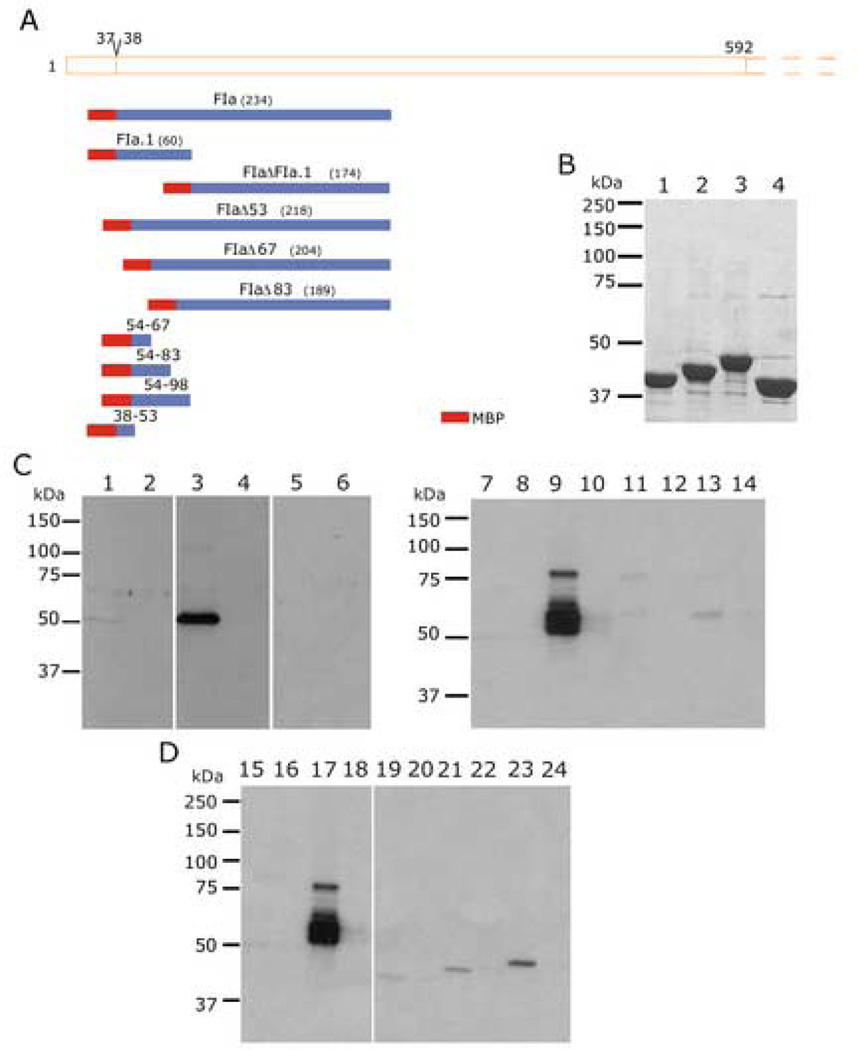
Fig. 1. Mapping the Region of PfEMP3 that Binds to the RBC Skeleton.
A. Schematic representation of PfEMP3 and representative sequences located in each distinct peptide repeat region. Repeat regions are shown in solid, shaded boxes, whereas non-repeat regions are shown in open boxes. Residue numbers are indicated above the schematic. The intron splice site, located between the first and second exon of pfemp3 is indicated by the division at residues 37–38 on the schematic.
B. Schematic representation of PfEMP3 and the initial sub-fragments used to analyse binding to the RBC skeleton. Residue numbers and fragment lengths are indicated above the schematic and adjacent to the fragment name (shown in brackets), respectively.
C. Purified MBP-PfEMP3 fusion proteins. 2µg (total protein) of each purified protein was resolved by SDS-PAGE in 10% polyacrylamide gels, prior to visualisation with Coomassie Brilliant Blue. The protein samples are: MBP (lane 1), -FI (lane 2), -FII (lane 3), -FIII (lane 4), -FIV (lane 5), -FV (lane 6), -FIa (lane 7) and -FIb (lane 8).
D. IOV binding assay immunoblots. IOVs (lanes 1, 3, 5, 7, 9, 11, 13, 15 and 17) and BSA (lanes 2, 4, 6, 8, 10, 12, 14, 16 and 18) were coated on the wells of the 96 well plate prior to adding MBP-PfEMP3 fusion proteins or MBP control protein: MBP (lanes 1, 2, 13 and 14), MBP-PfEMP3-FI (lanes 3 and 4), -FII (lanes 5 and 6), -FIII (lanes 7 and 8), FIV (lanes 9 and 10), -FV (lanes 11 and 12), -FIa (lanes 15 and 16) and -Flb (lanes 17 and 18) . -FI, and its sub-fragment –FIa bound to IOVs ( lanes 3 and 15) but not to BSA (lanes 4 and 16). Residual levels of –FV bound to IOVs (lane 11). MBP did not bind to either IOVs (lanes 1 and 13) or BSA (lanes 2 and 14).
Here, we have examined the association of PfEMP3 with the RBC membrane skeleton. Using a series of recombinant PfEMP3 fragments in vitro with inside-out vesicles (IOVs), formed from normal human RBCs and purified spectrin and actin, we have defined the binding region of PfEMP3 to a 14 residue sequence. Biosensor interaction assays yielded kinetic data indicative of a moderate affinity interaction between PfEMP3 fusion proteins and spectrin (KD(kin) of at least 3.8 × 10−7 M), and F-actin co-sedimentation interaction assays demonstrated that PfEMP3 binds F-actin in a specific and saturable manner. Continued elucidation of PfEMP3 and other interactions will facilitate construction of a detailed model of the protein interactions occurring at the membrane skeleton of IRBCs, and allow a better understanding of the complex structural modifications of IRBCs that ultimately result in severe P. falciparum malaria.
MATERIALS AND METHODS
Construction of pMAL-c2 Clones, and Expression and Purification of Recombinant Proteins
Specific oligonucleotide primers (Table 1) designed using the pfemp3 nucleotide sequence [25], were used to PCR amplify regions of pfemp3 from P. falciparum 3D7 genomic DNA. PCR products were cloned into the pMAL-c2 Maltose-Binding Protein (MBP) expression plasmid (New England Biolabs, Beverly MA, USA) and the nucleotide sequence and codon reading frame confirmed by automated sequencing. MBP-PfEMP3 fusion proteins were expressed and purified from appropriate E coli BL21(DE3) (Novagen Inc., Milwaukee, WI, USA) strains using standard techniques [30]. Purified proteins were dialed overnight against phosphate-buffered saline (PBS; 10 mM Na2HPO4/NaH2PO4, pH 7.4, containing 0.15 M NaCl) at 4 °C, with one buffer change before proteins were concentrated by centrifugal filtration. Total protein concentrations were determined by Bio-Rad Protein Assays (Bio-Lab Laboratories, Hercules, CA, USA).
Table 1. Oligonucleotide primer sequences.
Oligonucleotide primers sequences used in the construction of the pfemp3 DNA fragments.
| Primer | Fragment a | Sequence (5 ’ → 3 ’)b |
|---|---|---|
| p766 | FI (+), FIa (+), FIa.1 (+ ) | ccgagatctTTTACGGTTGTTAAGAAT |
| p767 | FI (−), FIb (−) | cgagaattcttaTGGTTCATGTTCTAAT |
| p768 | FII (+) | cgcggatccCCAACCAAATTACCTGAA |
| p769 | FII (−) | cgagaattcttaTCTAGATTTTCGCGTGC |
| p773 | FIII (−) | cgcgaattcttaCATGTTTCCTATACTT |
| p774 | FIV (+) | cgaggatccATAGGAAACATGGAACAA |
| p775 | FIV (−) | ccggaattcttaTCCTTCACTAGGTGTAT |
| p776 | FIII (+) | cgaggattcTCTGATGGATTAAAAGAA |
| p797 | FV (−) | cgcgaattcttaATTTTTTTTTCCTCTCAA |
| p799 | FV (+) | cgaggatccGATTTAAAGAATAAAGCTA |
| p942 | FIa (−), FIa.4 (−), FIaΔFIa.1 (−) | ccggaattcttaTAGTTCATTTATAGCAC |
| p943 | FIb (+) | caatggatccAATGAACTAAAAGAAAGG |
| p994 | FIa.1 (−) | ccggaattcttaCTCTTTTCTTTTATGATCCCTG |
| p995 | FIa.2 (+), FIaΔFIa.1 (+) | cgcggattcGCTTTAAAACAGAAAACT |
| p996 | FIa.2 (−) | ccggaattcttaCTTTAATGCTTCTCCATTTA |
| p997 | FIa.3 (+) | cgcggatccGAAAAAGAAAATAAAGAAACA |
| p998 | FIa.3 (−) | ccggaattcttaTTTCAAATCCTTTTCTAC |
| p999 | FIa.4 (+) | cgcggatccGAAATGGAATTGAAAGAGAAG |
| p1107 | FIaΔ5 3 (+) | ggggacaagtttgtacaaaaaagcaggcttaATATTTGAAATAAGACTTAAAAG |
| p1108 | FIaΔ6 7 (+) | ggggacaagtttgtacaaaaaagcaggcttaGGGAATACAAGGTTAAGC |
| p1109 | FIaΔ8 3 (+) | ggggacaagtttgtacaaaaaagcaggcttaGAGGCATTAAAAGAAAAGC |
| p1110 | FIaA53 (−), FIaA67 (−), FIaA83 (−) | ggggaccactttgtacaagaaagctgggtcTAGTTCATTTATAGCAC |
| p1236 | 54–83 (+), 54–98 (+) | ccggaattcATATTTGAAATAAGACTTAAAAG |
| p1237 | 54–83 (−) | cccaagcttttaCTTAGTTCTAGGATCCCTTAC |
| p1238 | 54–98 (−) | cccaagcttttaCTCTTTTCTTTTATGATCCCTG |
| p1212 | 54–67 (+) | aattcATATTTGAAATAAGACTTAAAAGATCATTAGCCCAGGTTTTGtaag |
| p1211 | 54–67 (−) | gatccttaCAAAACCTGGGCTAATGATCTTTTAAGTCTTATTTCAAATATg |
| p1245 | 38–53 (+) | ccggaattcTTTACGGTTGTTAAGAAT |
| p1246 | 38–53 (−) | cccaagcttttaATTATACACATTGACA |
(+) and (−) refer to coding (forward) and non-coding (reverse) DNA sequences, respectively.
Gene specific sequence shown in upper case; non-complimentary sequence shown in lower case and enzyme restriction sites are underlined.
In Vitro Binding Assays using IOVs
Inside out vesicles (IOVs) were prepared from normal human RBCs [10] and resuspended in IOV Incubation Buffer (IB; 138 mM NaCl, 5 mM KCl, 6 mM Na2HPO4, 5 mM glucose, pH9.0). IOV binding assays were performed using previously described methods [17]. Briefly, IOVs diluted in IB were used to coat wells of 96 well microtiter plates (Dynatech Laboratories, Inc., Chantilly VA, USA) overnight at 4 °C. Wells were then blocked overnight at 4 °C with 5% (w/v) BSA in IB, before washing twice with IB. Then, 0.5 −4 µg of total recombinant MBP-PfEMP3 fusion proteins diluted in IB containing 0.05% (v/v) Tween 20 were added to each well and incubated overnight at 4 °C. Wells were then washed 5 times with IB before proteins were stripped from the wells using heated (70 °C) reducing sample buffer. Proteins were resolved by SDS-PAGE and transferred to Polyscreen® polyvinylidene difluoride (PVDF) membrane (NEN® Life Science Products Inc., Boston MA, USA). Interacting proteins were detected via immunoblotting using a primary polyclonal rabbit anti-MBP antiserum (J. Bettadapura, Monash University) and secondary anti-rabbit Ig-horseradish peroxidase conjugate (Silenus Labs Pty. Ltd., Melbourne VIC, Australia). Autoradiographic images were obtained using NEN Renaissance™ Western Blot chemiluminescence reagent (NEN Life Science Products Inc., Boston MA, USA).
Triton X-100 Detergent Solubility
Triton X-100 (TX100) detergent solubilities were performed on lysed and resealed RBCs pre-incubated with MBP-PfEMP3 fusion proteins. Lysis and resealing was performed as previously described [31]. Briefly, 1ml of 75% haematocrit RBCs was dialysed against cold hypotonic buffer (5mM KPO4 pH7.4, 20 mM KCl) for 80 min at 4 °C, in the presence or absence of desired MBP fusion proteins. RBCs were then resealed by dialysing against pre-warmed isotonic buffer (5mM KPO4 pH7.4, 160 mM KCl, 5 mM glucose) for 60 min at 37 °C, washed extensively with PBS before extraction with PBS containing 1.0% (v/v) TX100 on ice for 20 mins in the presence of protease inhibitors (Roche Diagnostics, Castle Hill NSW, Australia). Samples were centrifuged at 13,000 rpm for 5 min at 4 °C before removal of the supernatant. The TX100 insoluble pellet was then washed in PBS/TX100/protease inhibitor solution before resuspension in PBS containing 2% (w/v) SDS and incubated at room temperature for 20 mins in the presence of protease inhibitors. Samples were then centrifuged as before and the pellet and supernatant fractions collected. Samples were resolved in 12% (w/v) polyacrylamide gels, before western blot detection was performed (as above) using polyclonal rabbit-anti-MBP antiserum.
In Vitro Binding Assays using Spectrin
Spectrin binding assays were performed in a similar manner to the IOV binding assays, with some modifications. Briefly, PBS was used instead of IB and 100 ng of purified human spectrin (Sigma, Castle Hill NSW, Australia) was used to coat the wells of a 96 well microtitre plate (Dynatech Laboratories Inc., Chantilly VA, USA). Assays were then processed as described above.
Binding Assays using Resonant Mirror Detection
Protein interactions were studied using the resonant mirror detection method [32–34] of the IAsys™ (Affinity Sensors, Cambridge, UK), using previously described methods [17]. Spectrin or BSA were immobilised on aminosilane cuvettes, and the PfEMP3 fusion proteins added in aqueous solution.
Actin Co-sedimentation Assays
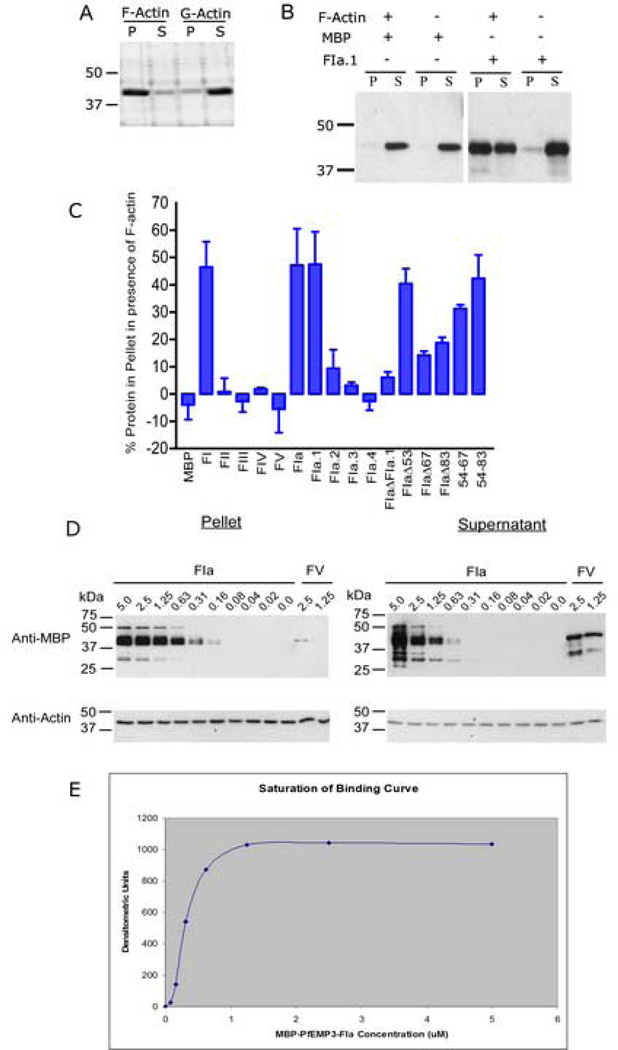
Monomeric G-actin (Cytoskeleton, Denver CO, USA; ≥ 95% pure (by SDS-PAGE analysis) was polymerised to 0.4 mg/ml in the presence of 50 mM KCl, 2 mM MgCl2, 1 mM ATP for 1 hour at room temperature. F-actin (5 µM) and recombinant MBP-PfEMP3 fusion proteins (5 µM) were incubated in PBS for 30 min at room temperature and then centrifuged at 25 °C for 10 min at 85,000 rpm (313,000 × g) in a Beckman Optima™ TLX ultracentrifuge using a TLA 120.2 rotor. The pellet was reconstituted into twice the original sample volume and equal volumes of pellet and supernatant were resolved by SDS-PAGE in 10% (w/v) polyacrylamide gels. Gels were stained with Coomassie Brilliant Blue or transferred to PVDF for immunoblot detection using polyclonal anti-MBP antiserum. Immunoblots were performed as detailed above, and the resulting autoradiographic data analysed by densitometry using the public domain NIH Image program (http://rsb.info.nih.gov/nih-image/). The percentage protein in the pellet was then calculated in both the presence and absence of F-actin, before subtraction of the % protein in the pellet in the absence of F-actin. The graphed data was obtained from two independent assays (mean ± standard deviation).
Actin co-sedimentation assays were also used to demonstrate specificity and saturability of the interaction between PfEMP3 and F-actin. Briefly, PfEMP3-FIa was titrated from 5 µM to 0.0 µM, alongside the negative interaction control protein -FV (2.5 µM and 1.25 µM). Pellet and supernatant fractions were processed for immunoblot detection as described above. A saturation of binding curve was constructed from the data obtained from a representative co-sedimentation experiment by densitometric analysis and subsequent subtraction of the signal obtained for the presence of non-binding –FV in the pellet. Equal loading of pellet and supernatant samples was demonstrated by probing duplicate blots with monoclonal anti-actin antibodies (Sigma, Castle Hill NSW, Australia).
RESULTS
Cloning, Protein Expression and Purification
The P. falciparum pfemp3 gene possesses a typical two exon gene structure and VTS/PEXEL motif (RSLAQ) that are common to many genes that encode proteins that are trafficked from the intracellular parasite into the RBC. pfemp3 possesses a first exon of 108 bp separated from a downstream second 7.2 kb exon by an intronic sequence (Fig. 1A). Lack of PCR amplification of the full length 7.2 kb second exon P. falciparum 3D7 genomic DNA precluded the expression and use of full length PfEMP3 proteins during the course of this investigation. Therefore, a fragment-based approach was used, in which defined fragments that are representative of the highly repetitive protein sequences encoded by the second exon of pfemp3 were PCR amplified, cloned into the pMAL-c2 expression vector, sequence confirmed and used to express and purify the encoded fusion proteins (Fig. 1B). Longer DNA fragments encoding PfEMP3’s highly repetitive regions ( Fig. 1A) could also not be cloned in full length, therefore representative fragments of these repeat regions were cloned and used for protein expression. The resulting purified MBP-PfEMP3 fusion proteins used in the interaction assays are shown in Figs. 1C, Fig. 2B and Fig. 4B. Immunoblot detection using polyclonal anti-MBP sera was performed on all purified MBP fusion proteins to demonstrate presence of the MBP tag and confirm the identity of each protein (data not shown).
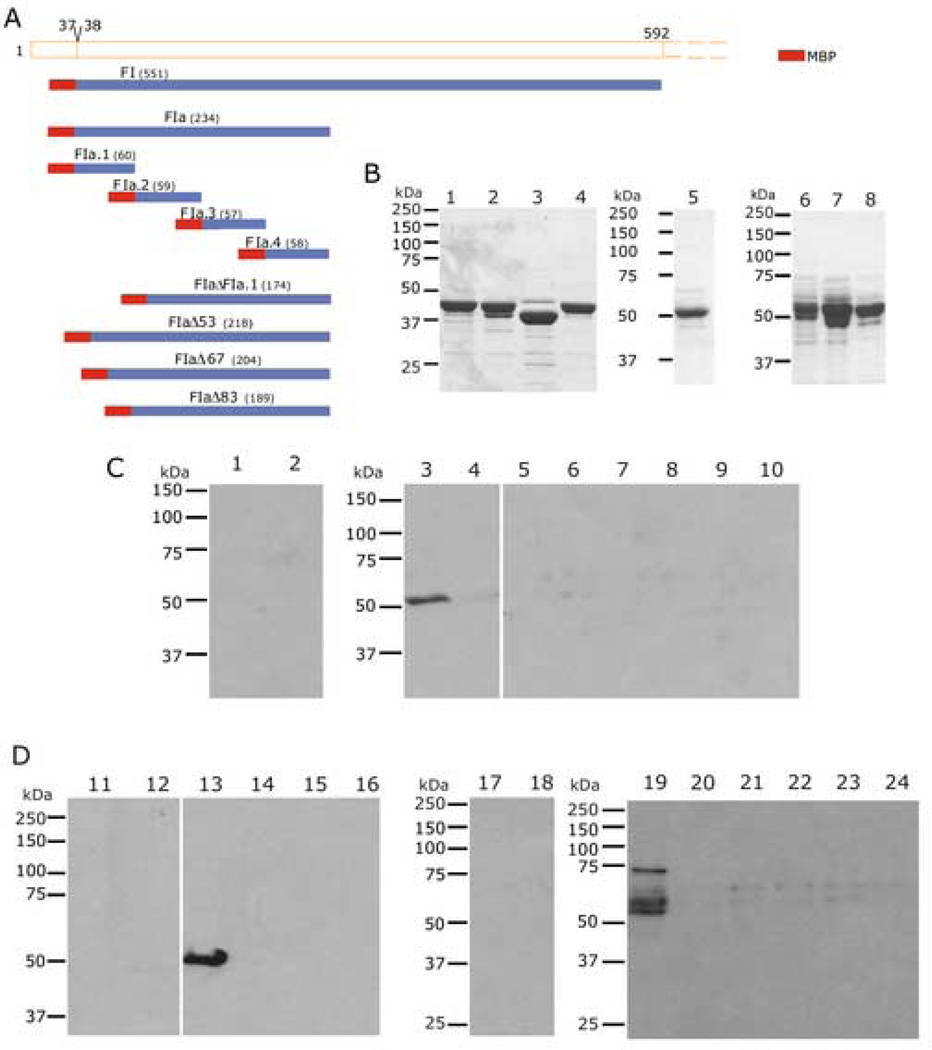
Fig. 2. Finer Mapping of the Region of PfEMP3 that Binds to the Skeleton.
A. Schematic representation of PfEMP3-FI region (residues 38–592) and the smaller PfEMP3 sub-fragments and deletion mutant proteins.
B. Purified MBP-PfEMP3 fusion proteins. 2µg (total protein) of each purified protein was resolved by SDS-PAGE, prior to visualisation with Coomassie Brilliant Blue. The protein samples are: -FIa.1 (lane 1), -FIa.2 (lane 3), -FIa.3 (lane 3), -FIa.4 (lane 4), -FIaΔFIa.1 (lane 5), -FIaΔ53 (lane 6), -FIaΔ67 (lane 7) and -FIaΔ83 (lane 8).
C and D. IOV binding assay immunoblots. IOVs (lanes 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21 and 23) and BSA (lanes 2, 4, 6, 8, 10, 12, 14, 16,18, 20, 22 and 24) were coated on the wells of the 96 well plate prior to adding MBP-PfEMP3 fusion proteins or MBP control protein: MBP (lanes 1, 2, 11, 12, 17 and 18), -FIa.1 (lanes 3 and 4), -FIa.2 (lanes 5 and 6), -FIa.3 (lanes 7 and 8), -FIa.4 (lanes 9 and 10), -FIa (lanes 13 and 14), -FIaΔFIa.1 (lanes 15 and 16), -FIaΔ53 (lanes 19 and 20), -FIaΔ67 (lanes 21 and 22) and –FIaΔ83 (lanes 23 and 24). The sub-fragment of –Fla, FIa.1 bound to IOVs (lane 3) and not to BSA (lane 4), thereby mapping the membrane skeleton binding region of PfEMP3 to a 60 residue region. Deletion of these 60 residues from the binding fragment FIa, resulting in the generation –FIaΔFIa.1, abolished binding to IOVs (lane 15). Finer deletion analysis was performed to dissect the 60 residue region, using -FIaΔ53, -FIaΔ67 and -FIaΔ83, showed binding of -FIaΔ53 to IOVs (lane 19) but not to BSA (lane 20). Both -FIaΔ67 and -FIaΔ83 did not bind to IOVs (lanes 21 and 23, respectively) or BSA (lanes 22 and 24, respectively).
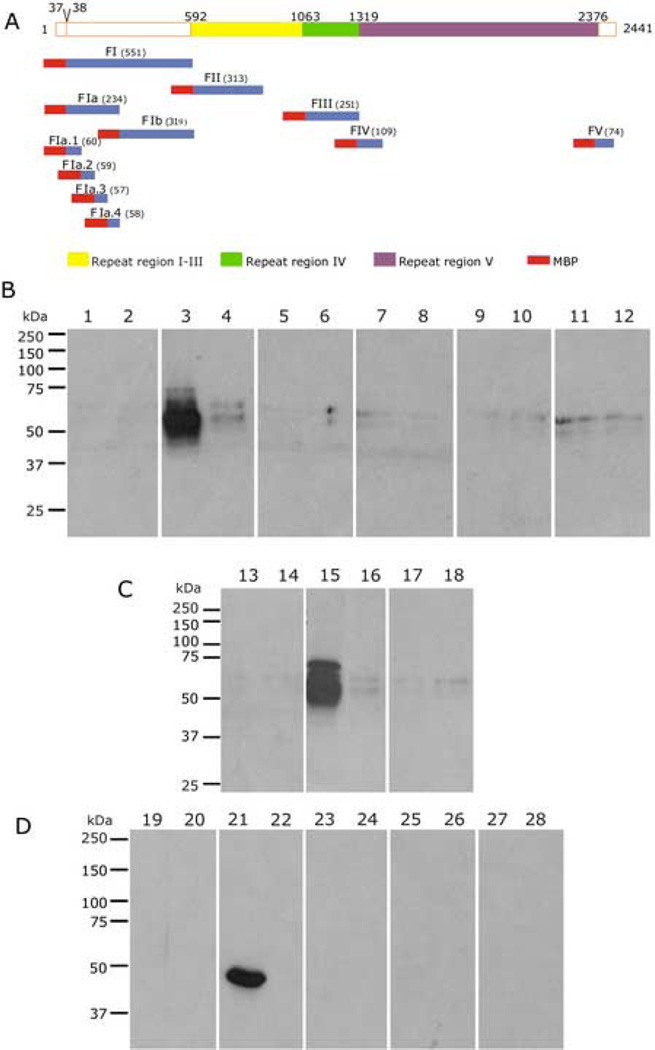
Fig. 4. Mapping the Region of PfEMP3 that Binds to Spectrin.
A. Schematic representation of PfEMP3 and its sub-fragment proteins.
B, C and D. Spectrin binding assay immunoblots. Spectrin (lanes 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25 and 27) and BSA (lanes 2, 4, 6, 8, 10, 12, 14, 16,18, 20, 22, 24, 26 and 28) were coated on the wells of the 96 well plate prior to adding MBP-PfEMP3 fusion proteins or MBP control protein: MBP (lanes 1, 2, 13, 14, 19 and 20), -FI (lanes 3 and 4), -FII (lanes 5 and 6), -FIII (lanes 7 and 8), -FIV (lanes 9 and 10), -FV (lanes 11 and 12), -FIa (lanes 15 and 16), -FIb (lanes 17 and 18), -FIa.1 (lanes 21 and 22), –FIa.2 (lanes 23 and 24), -Fla.3 (lanes 25 and 26) and – Fla.4 (lanes 27 and 28). Fragment –FI and its successively smaller sub-fragments – FIa and –Fla.1 all bound to IOVs (lanes 3, 15 and 21, respectively) but not to BSA (lanes 4, 16 and 22, respectively). These data map the region of PfEMP3 that binds to spectrin to the same 60 residues that bind to IOVs.
Mapping the PfEMP3 RBC Membrane Skeleton Binding Domain
IOVs prepared from normal human RBCs using the methodology described possess spectrin and actin in a properly linked and conformed network, in addition to the many other membrane skeleton proteins (for review, see Fairbanks et al., [35]). A sample of prepared IOVs was resolved by SDS-PAGE and stained with Coomassie Brilliant Blue to confirm the presence of the major RBC membrane skeleton proteins, including spectrin and actin (data not shown).
An initial series of IOV interaction assays were performed using the MBP-PfEMP3 fusion proteins –FI (residues 38–587 of full length PfEMP3), -FII (residues 588–901), -FIII (residues 1069–1320), -FIV (residues 1319–1428) and –FV (residues 2368–2441) to broadly map the region of PfEMP3 that binds to the RBC skeleton. A representative immunoblot is shown in Figure 1D. PfEMP3-FI bound to IOVs (lane 3) but not to BSA (lane 4), whereas the -FII, -FIII, -FIV and -FV fragments and MBP did not bind to either IOVs or BSA (lanes 5 through 12, respectively). These initial interaction assays indicated that residues 38–587 of PfEMP3 contain the binding domain for RBC membrane skeleton protein(s). Subsequent division of –FI into –FIa (residues 38–271) and –FIb (residues 269–587) protein sub-fragments further refined the binding domain to a 234 residue region, as evidenced by the binding of –FIa to IOVs (lane 15), but not to BSA (lane 16).
To further define the PfEMP3 skeleton-binding domain, smaller protein fragments were generated. Representative immunoblots of binding assays using these proteins (Figs. 2C and D) show that -FIa.1 (residues 38–97), a 60 residue fragment (FTVVKNYNKIDNVYNIFEIRLKRSLAQVLGNTRLSSRGVRDPRTKEALKEKQFRDH KRKE) of -FI, binds to IOVs (Fig. 2C, lane 3), but not to BSA (lane 4). Other sub-regions (-FIa.2, -FIa.3 and -FIa.4, corresponding to residues 98–156, 157–214 and 215–271, respectively) were also examined for binding ability. None of these bound to IOVs or to BSA (lanes 5 through 10, respectively). Based on these data, several deletion mutant PfEMP3 protein sub-fragments were generated. PfEMP3-FIaΔFIa.1, in which residues 38–97 had been deleted did not bind to IOVs or BSA (Fig. 2D, lanes 15 and 16, respectively). The deletion mutant proteins -FIaΔ53 bound to IOVs but not to BSA (lanes 19 and 20), whereas -FIaΔ67 and -FIaΔ83 did not bind to either IOVs (lanes 21 and 23, respectively) or BSA (lanes 22 and 24). Taken together, these data map the region of PfEMP3 that binds to the RBC membrane skeleton to residues 54–67 (14 residues in total; IFEIRLKRSLAQVL).
Confirmatory evidence for the association of PfEMP3 with the membrane skeleton was obtained from TX100 extraction of lysed and resealed RBCs that had been incubated in the presence of 5 µM MBP-PfEMP3-FI or –FV fusion proteins. MBP-PfEMP3-FI was detected in both the TX100 soluble and insoluble fractions (Fig. 3, lanes 1 and 2, respectively), whereas –FV was detected only in the TX100 soluble fraction (lane 3). Similarly extracted lysed RBC controls are also shown (lanes 5 and 6). Collectively, these data demonstrate the classical detergent solubility profiles characteristic of skeleton-associated proteins and define this membrane skeleton binding region of PfEMP3 to the 14 residue region IFEIRLKRSLAQVL.
Fig. 3. Membrane Skeleton Association of PfEMP3 FI.
TX100 detergent extractions were performed on lysed and resealed RBCs that had been incubated with 5 µM purified MBP-PfEMP3-FI or –FV. TX100 soluble and insoluble fractions were resolved by SDS-PAGE and immuno-blotted with anti-MBP antiserum. MBP-PfEMP3-FI was detected in both the TX100 soluble and insoluble fractions (lanes 1 and 2, respectively). –FV, a region of PfEMP3 that does not bind to the membrane skeleton, was detected only in the TX100 soluble fraction (lane 3) but not the TX100 insoluble fraction (lane 4). The control TX100 soluble and insoluble fractions of normal lysed and resealed (no MBP fusion protein) RBCs are shown in lanes 5 and 6, respectively.
PfEMP3 binds to Spectrin
To determine if PfEMP3 and spectrin are binding partners, we performed binding assays between fragments of PfEMP3 and purified spectrin (Figs. 4 and. 5). Similar to the above IOV binding assays, PfEMP3-FI and its sub-fragments -FIa and - FIa.1 bound to spectrin (Fig. 4B, C and D, lanes 3, 15 and 21, respectively), but not to BSA (Fig. 4B, C and D, lanes 4, 16 and 22, respectively). These assays defined the region of PfEMP3 that binds to spectrin to the same 60 residue region that bound IOVs in earlier experiments. Further assays used the smaller PfEMP3 deletion and sub-fragment proteins (Fig. 5A). The –FIa fragment bound to spectrin (Fig. 5C, lane 3) but its deletion mutant -FIaΔFIa.1, which lacks residues 38–97, failed to bind to spectrin ( lane 5). The deletion mutant – FlaΔ53 bound to spectrin (lane 9 and 17), whereas –FlaΔ67 and –FlaΔ83 failed to bind (lanes 11 and 13, respectively). Use of smaller fragments spanning the 60 residue region showed that −54–67 bound to spectrin at low levels (lane 19), with increasing binding levels observed for −54–83 (lane 21) and −54–98 (lane 23). Taken together, these data define the minimal binding region of PfEMP3 for spectrin to the same 14 residue region (IFEIRLKRSLAQVL) that binds IOVs. These data also indicate the presence of sequences downstream of the 14 residue region that act to increase the intensity of binding to spectrin conferred by the 14 residue region.
Fig. 5. Deletion Mapping of PfEMP3 Binding to Spectrin.
A. Schematic representation of PfEMP3-FI region (residues 38–592) and the smaller PfEMP3 sub-fragments and deletion fragments used to map the spectrin binding domain.
B. Additional MBP-PfEMP3 fusion proteins that were used in these analyses are shown. 2µg (total protein) of each purified protein was resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. The protein samples are: −5438–592) and the smaller PfEMP3 sub-fragments and deletion fragments used to map 67 (lane 1), −5438–592) and the smaller PfEMP3 sub-fragments and deletion fragments used to map 83 (lane 2), −5438–592) and the smaller PfEMP3 sub-fragments and deletion fragments used to map 98 (lane 3) and −38–53 (lane 4).
C and D. Spectrin binding assay immunoblots. Spectrin (lanes 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21 and 23) and BSA (lanes 2, 4, 6, 8, 10, 12, 14, 16,18, 20, 22 and 24) were coated on the wells of the 96 well plate prior to adding MBP-PfEMP3 fusion proteins or MBP control protein: MBP (lanes 1, 2, 7, 8, 15 and 16), MBP-PfEMP3-FIa (lanes 3 and 4), -FIaΔFIa.1 (lanes 5 and 6), -FIaΔ53 (lanes 9 and 10), -FIaΔ67 (lanes 11 and 12), -FIaΔ83 (lanes 13 and 14), -FIaΔ53 (lanes 17 and 18), −54–67 (lanes 19 and 20), −54–83 (lanes 21 and 22) and –54–98 (lanes 23 and 24). PfEMP3-FIa (lane 3) and its deletion mutant -FIaΔ53 (lane 9 and 17) and −54–67, −54–83 and −54–98 (lanes 19, 21 and 23, respectively) each bound to spectrin only. Taken together, these data map the minimal binding region of PfEMP3 to spectrin to the 14 residue region represented on the protein fragment −54–67.
Kinetic Analysis of PfEMP3 – Spectrin Interactions
An IAsys™ system was used to determine the kinetics of the observed interactions. Biophysical data was obtained using aminosaline cuvettes coated with spectrin or BSA and PfEMP3 proteins were applied in aqueous solution. The data obtained (ka, kd and subsequent KD(kin) and KD(Scat)) are shown in Table 2. These data are in accordance with the microtitre plate interaction data and show strong, saturable interactions between spectrin and PfEMP3 fragments -FI (KD(kin) = 7.0 × 10−8 M) , - FIa ( KD(kin) = 7.5 × 10−8 M) and -FIa.1 (KD(kin) = 8.5 × 10−8 M) and no interaction with any of the other fragments. No interaction was observed between -FIaΔFIa.1 and spectrin, again confirming the binding region as the 60 residues encoded by -FIa.1. KD(kin) data was also obtained using the PfEMP3 deletion mutants proteins. PfEMP3-FIaΔ53 bound strongly to spectrin coated cuvettes (KD(kin) = 2.75 × 10−7 M) , but -FIaΔ67 did not. Kinetic analyses using protein −38–53 showed no binding, but strong binding of protein −54–67 to spectrin (KD(kin) = 3.80 × 10−7 M), respectively. These data further confirm the region of PfEMP3 that binds to spectrin is encoded in the 14 residue sequence (IFEIRLKRSLAQVL) contained within PfEMP3 sub-fragment −54–67.
Table 2. Quantitation of the interaction between PfEMP3 and Spectrin.
Purified MBP-PfEMP3 fusion proteins and purified RBC spectrin were used in interaction assays using an IAsys™ resonant mirror biosensor (Affinity Sensors, Cambridge, UK). The binding assays were performed as previously described [17] in PBS containing 0.05% (v/v) Tween 20. From the binding curves obtained using the resonant mirror detection method, ka, kd, KD(kin) and KD(Scat) were determined using the software package FASTFIT™.
| PfEMP3 Fragments |
ka (M−1s−1) |
kd (s−1) |
KD(kin) (µM) |
KD(Scat) (µM) |
|---|---|---|---|---|
| FI | 3.0 ± 0.1×104 | 2.1 ± 0 .1× 10−3 | 0.07 | 0.059 |
| FII, FIII, FIV, FV | No binding | No binding | No binding | No binding |
| FIa | 8.4 ± 0.1×104 | 6.3 ± 0 .2× 10−3 | 0.075 | 0.067 |
| Fib | No binding | No binding | No binding | No binding |
| FIa.1 | 7.9 ± 0.2×104 | 6.7 ± 0 .1× 10−3 | 0.085 | 0.080 |
| FIa.2, FIa.3, FIa.4 | No binding | No binding | No binding | No binding |
| FIaΔFIa.1 | No binding | No binding | No binding | No binding |
| FIaΔ53 | 2.4 ± 0.2×103 | 6.6 ± 0 .2× 10−4 | 0.275 | 0.236 |
| FIaΔs67 | No binding | No binding | No binding | No binding |
| 38–53 | No binding | No binding | No binding | No binding |
| 54–67 | 15 ± 0.2×103 | 5.7 ± 0 .2× 10−4 | 0.380 | 0.300 |
| MBP | No binding | No binding | No binding | No binding |
PfEMP3 Binds to F-actin
To detect interactions between polymerised F-actin and regions of PfEMP3, co-sedimentation assays were used. Under the sedimentation conditions used, F-actin was found predominantly in the pellet, whereas monomeric G-actin was found mostly in the supernatant fraction (Fig. 6A). Subsequently, eqi-molar concentrations of F-actin and recombinant MBP-PfEMP3 fusion proteins were interacted and co-sedimented. Resultant supernatant and pellet fractions were analysed by immunoblot (Fig. 6B) and show that the MBP fusion tag alone is unable to bind F-actin, whereas the MBP-PfEMP3-FIa.1 fusion protein co-sedimented with F-actin, (although a proportion of the recombinant protein also remained in solution). In further immunoblots, densitometry was used to determine the percentage of recombinant protein in the pellet, corrected by subtraction of the % of recombinant protein that pelleted in the absence of F-actin, and the resultant data graphed (Fig. 6C). Interestingly the MBP-PfEMP3 fusion proteins that were found to pellet, and therefore to bind F-actin, were consistent with those that were also shown to bind spectrin. Fragments -FI, -FIa and -FIa.1 bind to F-actin, whereas the other fragments show no binding (Fig. 6C). Deletion analysis confirmed the same 60 residue region as the binding domain, as evidenced by the inability of -FIaΔFIa.1 to bind F-actin. Further deletion analysis showed that residues 38–53 were not essential for binding, however deletion of further residues within this larger region resulted in only a partial loss of binding. This was evidenced by binding of -FIaΔ53, but only partial binding of -FIaΔ67 and -FIaΔ83. These data again show that residues 54–67 are sufficient to confer almost all binding observed by the 60 residue region encoded by -FIa.1, however the presence of other residues downstream of the 14 residue region appear to increase the intensity of interaction, almost to the level detected for –FIa.1 (Fig. 6C).
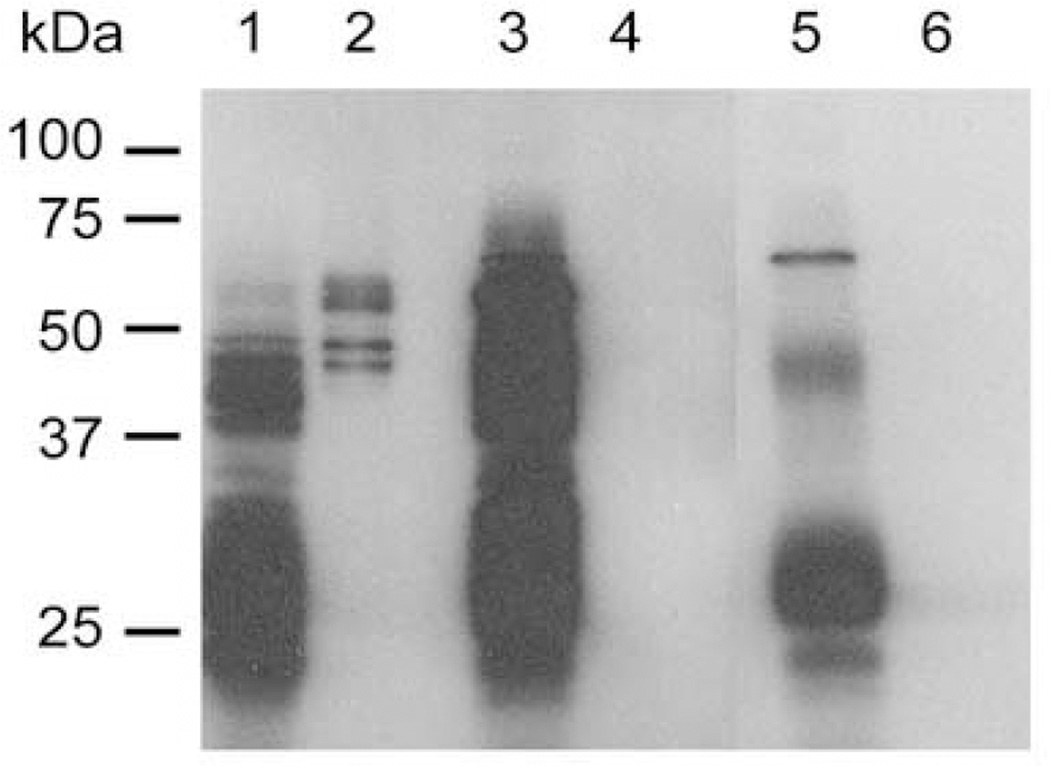
Fig. 6. The Region of PfEMP3 that Binds to Actin.
A. Preliminary co-sedimentation assays were performed using polymerised F-actin and monomeric G-actin. Samples of F-actin and G-actin were then centrifuged at 25 °C for 10 min at 85,000 rpm (313,000 × g). Pellet and supernatant samples from each reaction were then resolved by SDS-PAGE in 10 % (w/v) polyacrylamide gels and stained with Coomassie Brilliant Blue. Nearly all the polymerised F-actin was found in the pellet fraction (P), whereas most of the monomeric G-actin was found in the supernatant fraction (S). Only residual levels of actin were observed in the other fractions.
B. Eqi-molar solutions (5 µM) of F-actin and recombinant MBP-PfEMP3 fusion proteins were interacted and co-sedimented. Immunoblot analysis of resultant supernatant and pellet fractions was performed. Representative immunoblots show MBP alone is unable to bind F-actin, whereas the MBP-PfEMP3-F1a.1 fusion protein co-sedimented in the presence of F-actin, (although a proportion of the recombinant protein also remained in solution).
C. Eqi-molar solutions (5 µM) of F-actin and purified MBP-PfEMP3 fusion proteins were interacted and then co-sedimented. The pellet and supernatant fractions were resolved by SDS-PAGE and the immunoblot data analysed by densitometry. The graphed data show pelleting of F-actin with -FI, and its successively smaller sub-fragments, -FIa, -FIa.1. Deletion mutagenesis further defined the binding region to 14 residues (residues 54–67 of PfEMP3), as evidenced by -FIaΔ53 pelleting with F-actin, whereas –FIaΔ67 did not. The presence of only these 14 residues in the −54–67 fragment also resulted in pelleting with F-actin.
D. Binding of F-actin to -FIa was demonstrated to be specific and saturable. Constant 5 µM actin was incubated with titrated serial dilutions of –FIa and centrifuged. Pellet and supernatant fractions were analysed by immunoblot, using anti-MBP and anti-actin antibodies and subsequent densitometry.
E. Saturation of binding curve constructed from densitometric analyses of titrated – FIa co-sedimented with constant 5 µM actin (Fig. 5D). The graphed data are from a representative assay (mean ± standard deviation).
Further, co-sedimentation assays demonstrated that the interaction between F-actin and PfEMP3-FIa.1 is specific and saturable (Fig. 6D). Constant 5 µM F-actin was interacted with titrated serial dilutions of PfEMP3-FIa (from 5 µM to 0.0 µM) and the negative control protein PfEMP3-FV (1.25 µM and 2.5 µM). Immunoblots of a representative experiment are shown (Fig. 6D), and the corresponding densitometric data corrected by subtraction of the relative percentage amount of PfEMP3-FV pelleted in the absence of F-actin. Graphical representation of this saturation of binding curve is shown in Fig. 6E.
DISCUSSION
Here, we have characterised the interactions that bind PfEMP3 to the membrane skeleton of IRBCs. We have used a recombinant protein approach and two independent in vitro assays to identify spectrin as a binding partner for PfEMP3 and to map the minimal binding domain a 14 residue sequence, IFEIRLKRSLAQVL, corresponding to res idues 54–67 of PfEMP3. Additionally, biosensor assays determined moderately high affinity interactions between various fragments of PfEMP3 and spectrin (Table 2; –FIa (KD(kin) = 7.0 × 10−8 M) and its 60 residue sub-fragment –FIa.1 (KD(kin) = 8.5 × 10−8 M)). Deletion of these 60 residues (38–97) in – FIaΔFIa.1 abolished binding, whereas deletion of residues 38–53 of this region in -FIaΔ53 retained binding ability (KD(kin) = 2.75 × 10−7 M), but deletion of residues 38–67 in –FIaΔ67 abolished binding (Table 2). Positive confirmation of these 14 residues conferring the binding domain of PfEMP3 to spectrin was obtained using the protein −54–67 (KD(kin) = 3.8 × 10−7 M). A low level of −54–67 binding to spectrin was also detected (Fig. 5), with binding levels showing modest increases when residues down stream of the 54–67 residue region were included in the proteins −54–83 and −54–98. Interestingly, the same phenomenon was observed in the actin co-sedimentation assays (Fig. 6C), where residues downstream of the 14 residue region appeared to increase the intensity of the interaction. Such enhancement of interactions by adjacent residues has previously been reported in the binding of the parasite protein MESA to its binding partner protein 4.1R [36]. The observation that the PfEMP3-membrane skeleton interaction occurs in vitro to IOVs, where the supramolecular structure of the spectrin-actin network and the conformation of the individual proteins is conserved, as well as in vivo provides confidence that this is a physiologically relevant interaction.
In a previous study using various truncated constructs of PfEMP3 fused to green fluorescent protein, Knuepfer and colleagues demonstrated peripheral localization of a transgene product containing the first 500 residues of PfEMP3, but not of products composed of only the first 66 or 88 PfEMP3 residues when those constructs were transfected into P. falciparum parasites. Products of 120 residues fused to GFP showed some weak peripheral association [23]. The authors concluded that a region between residues 120–500 is responsible for both the tight association they described with Maurer’s clefts and the membrane skeleton. These results are in apparent contradiction to those reported here, where we define the membrane skeleton-binding domain to sequences in the first 97 residues of PfEMP3. These results may be reconciled if we propose that only proteins that are trafficked via the Maurer’s clefts are in the correct conformation and localization to bind to spectrin and actin at the membrane skeleton. The shorter constructs would fail to bind, either because of steric hindrance due to the presence of GFP as has pre viously been reported [37] or because of failure to properly expose binding domains on either the host or parasite proteins. This proposition could be tested by tracking of a chimeric PfEMP3 construct lacking the first 67 residues of PfEMP3. At any rate, using the techniques described in this paper, we were unable to detect any other membrane skeleton binding domains within residues 120–500 of PfEMP3 and conclude that efficient association of this protein with the membrane skeleton requires trafficking in the protein aggregates associated with Maurer’s clefts [23]. Interestingly, the 14 residue region identified here as the spectrin and actin binding domain of PfEMP3 (IFEIRLKRSLAQVL) also contains the putative VTS/PEXEL motif RSLAQ. This motif has recently been described as a common sequence that results in the trafficking of parasite proteins across the parasitophorous vacuole and into the red cell cytosol [27, 28]. Many of the 300–400 proteins that have this sequence do not interact with the red cell membrane skeleton. In the case of PfEMP3, the region of sequence common to recombinant proteins that bind to spectrin is a 14 residue sequence that includes the VTS/PEXEL. Whether any residues that contribute to membrane skeleton binding are part of the VTS/PEXEL is not known, however, the lack of membrane skeleton association of many VTS/PEXEL proteins leads us to conclude that it is the flanking residues in this 14 residue region that are most likely to be responsible for the association with spectrin.
The affinity of the interaction between the 14 residue PfEMP3 region encoded by recombinant PfEMP3 protein -FIa.1 (residues 38–98) and spectrin (KD(kin) = 8.5 × 10−8 M) is of similar magnitude to the affinity of other protein-proteins interactions occurring at the RBC membrane skeleton. Thus, in uninfected RBCs, protein 4.1 binds to p55 with moderate affinity (Kd = 2.5 × 10−9 M; [38]), protein 4.2 binds to band 3 with a Kd = 2 – 8 × 10−7 M; [39], and protein 4.2 binds to ankyrin with Kd = 1 – 3.5 × 10−7 M [39]. This level of binding is also typical of that seen for malaria proteins binding at the membrane skeleton to host or malaria proteins. These include protein 4.1 and MESA (KD(kin) = 1.3 × 10−7 M; [11]) and between KAHRP and PfEMP1 (KD(kin) = 1 × 10−7 M; [17]).
We have identified the same 14 residue region of PfEMP3 (IFEIRLKRSLAQVL) as the binding region for both spectrin and actin. The capacity of a single domain to interact with two distinct host proteins has not previously been reported for other well-characterised malaria protein interactions at the RBC membrane. Although KAHRP has been shown to be capable of binding to PfEMP1, spectrin, ankyrin and actin, different regions of KAHRP were responsible for binding to each protein [15–18]. Since it is not known precisely which domain on spectrin is bound by PfEMP3, it is unclear whether a single molecule of PfEMP3 could simultaneously bind molecules of both spectrin and actin. Binding may be a stochastic process with PfEMP3 molecules binding one or other potential binding partner at random. This would lead to a somewhat random localisation with respect to knobs, which is indeed what has been observed [21]. Perhaps also, this dual binding capability has evolved to provide a large number of potential binding sites to anchor this protein. As KAHRP is also capable of binding actin, these two malaria proteins may be expected to compete for actin binding sites at the membrane skeleton. Previous rheological studies have already shown increased rigidification of the RBC membrane in parasites that express and localise PfEMP3 at the RBC skeleton [40] and this probably requires a sufficient number of the highly charged PfEMP3 molecules to be bound at the membrane, which the availability of additional binding sites would help ensure. Alternatively, it may be that PfEMP3 binding distorts the membrane skeleton in a way that brings together the binding domains on spectrin and actin and leads to membrane rigidification. This might also be responsible for the distorted appearance of the RBC that is almost always seen in scanning electron micrographs of IRBCs.
Previous rheological studies have also demonstrated a lower extent of rigidification of the RBC membrane skeleton in RBCs infected with a specific pfemp3 gene knocko ut par asite [40]. This observation, in conjunction with the data presented in this study, supports our working hypothesis that PfEMP3 may rigidify the RBC membrane skeleton by initiating novel protein interactions (PfEMP3-spectrin and PfEMP3–actin interactions) or by altering the affinity of other protein interactions at the IRBC skeleton. As such, PfEMP3 may act in an analogous fashion to protein 4.1, which strengthens the low affinity ( Ka = 5 × 10−3 M−1) spectrin-actin interaction by several orders of magnitude (spectrin-actin-protein 4.1; Ka = 1 × 10−12 M−2) [41]. Alternatively, the presence of PfEMP3 at the RBC skeleton may have a competitive effect on other established interactions spectrin and actin interactions including spectrin-actin-protein 4.1 [41], spectrin-ankyrin [42], spectrin-actin-adducin [43], actin-dematin [44], actin-tropomyosin [44] and actin-tropomodulin [45]. In order to clarify the likely effect PfEMP3 has on other protein interactions in the RBC membrane skeleton, further mapping studies would be required to define the domain in spectrin and actin that binds PfEMP3, and to determine if these domain(s) overlap with previously described binding domains in spectrin. The N-terminal region of β-spectrin contains the binding domain for actin and protein 4.1, whereas the ankyrin binding site has been mapped to a region near the C-terminal region of β-spectrin [46–48]. It is also possible that PfEMP3 may act in collaboration with other parasite-encoded proteins, such as PfEMP1 and KAHRP, both of which have been shown to interact with spectrin and actin [14, 18]. Further mapping of the protein interacting domains of PfEMP3 and other parasites proteins with the RBC membrane skeleton will help to elucidate this complicated set of interactions and facilitate our understanding of the changes induced in the architecture of the membrane skeleton of P. falciparum-IRBCs and their subsequent impact on the pathophysiology of malaria.
Acknowledgments
We thank Dr. Jayaram Bettadapura and Vicki Gatzigiannis for providing the pMAL-c2 constructs of PfEMP3-FI - FV. LMS was supported by a PhD Scholarship from Biota Holdings Limited. KLW is supported by an Australian NHMRC Howard Florey Centenary Research Fellowship. BMC is a NHMRC Senior Research Fellow. This research was supported by grants from the NHMRC and the NIH (grant #DK32094).
Abbreviations
- RBC
Red Blood Cell
- IRBC
infected Red Blood Cell
- PfEMP3
Plasmodium falciparum Erythrocyte Membrane Protein 3
- KD(kin) and KD(Scat)
dissociation constants determined by kinetic and Scatchard Analysis, respectively
- GPC and GPD
Glycophorin C and D, respectively
- PfEMP1
Plasmodium falciparum Erythrocyte Membrane Protein 1
- KAHRP
Knob Associated Histidine Rich Protein
- MESA
Mature parasite-infected Erythrocyte Surface Antigen
- RESA
Ring-infected Erythrocyte Surface Antigen
- αR4
α-spectrin repeat region 4
- IOV
Inside-out Vesicle
- PCR
Polymerase Chain Reaction
- MBP
Maltose Binding Protein
- PBS
Phosphate-buffered Saline
- IB
IOV Incubation Buffer
- BSA
Bovine Serum Albumin
- PVDF
Polyscreen®polyvinylidene difluoride
- TX100
Triton X-100
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooke BM, Mohandas N, Coppel RL. The malaria-infected red blood cell: structural and functional changes. Adv Parasitol. 2001;50:1–86. doi: 10.1016/S0065-308X(01)50029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooke BM, Mohandas N, Coppel RL. Malaria and the red blood cell membrane. Semin Hematol. 2004;41:173–188. doi: 10.1053/j.seminhematol.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Speicher DW, Morrow JS, Knowles WJ, Marchesi VT. Identification of proteolytically resistant domains of human erythrocyte spectrin. Proc Natl Acad Sci U S A. 1980;77:5673–5677. doi: 10.1073/pnas.77.10.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett V, Gilligan DM. The spectrin-based membrane skeleton and micron-scale organization of the plasma membrane. Annual Review of Cell Biology. 1993;9:27–66. doi: 10.1146/annurev.cb.09.110193.000331. [DOI] [PubMed] [Google Scholar]
- 6.Lux SE, Palek J. In: Blood: Principles and Practice of Hematology. Handin RI, Lux SE, Stossel TP, editors. Philidelphia: JB Lippincott Co; 1995. pp. 1701–1818. [Google Scholar]
- 7.Foley M, Corcoran L, Tilley L, Anders R. Plasmodium falciparum: mapping the membrane-binding domain in the ring-infected erythrocyte surface antigen. Exp Parasitol. 1994;79:340–350. doi: 10.1006/expr.1994.1096. [DOI] [PubMed] [Google Scholar]
- 8.Da Silva E, Foley M, Dluzewski AR, Murray LJ, Anders RF, Tilley L. The Plasmodium falciparum protein RESA interacts with the erythrocyte cytoskeleton and modifies erythrocyte thermal stability. Mol Biochem Parasitol. 1994;66:59–69. doi: 10.1016/0166-6851(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 9.Silva MD, Cooke BM, Guillotte M, Buckingham DW, Sauzet JP, Le Scanf C, Contamin H, David P, Mercereau-Puijalon O, Bonnefoy S. A role for the Plasmodium falciparum RESA protein in resistance against heat shock demonstrated using gene disruption. Mol Microbiol. 2005;56:990–1003. doi: 10.1111/j.1365-2958.2005.04603.x. [DOI] [PubMed] [Google Scholar]
- 10.Bennett BJ, Mohandas N, Coppel RL. Defining the Minimal Domain of the Plasmodium falciparum Protein MESA Involved in the Interaction with the Red Cell Membrane Skeletal Protein 4.1. The Journal of Biological Chemistry. 1997;272:15299–15306. doi: 10.1074/jbc.272.24.15299. [DOI] [PubMed] [Google Scholar]
- 11.Waller KL, Nunomura W, An X, Cooke BM, Mohandas N, Coppel RL. Mature parasite-infected erythrocyte surface antigen (MESA) of Plasmodium falciparum binds to the 30-kDa domain of protein 4.1 in malaria-infected red blood cells. Blood. 2003;102:1911–1914. doi: 10.1182/blood-2002-11-3513. [DOI] [PubMed] [Google Scholar]
- 12.Crabb B, Cooke BM, Reeder JC, Waller RF, Caruana SR, Davern KM, Wickham ME, Brown GV, Coppel RL, Cowman AF. Targeted Gene Disruption Shows That Knobs Enable Malaria-Infected Red Cells to Cytoadhere under Physiological Shear Stress. Cell. 1997;89:287–296. doi: 10.1016/s0092-8674(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 13.Rug M, Prescott SW, Fernandez KM, Cooke BM, Cowman AF. The role of KAHRP domains in knob formation and cytoadherence of P falciparum-infected human erythrocytes. Blood. 2006;108:370–378. doi: 10.1182/blood-2005-11-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilejian A, Rashid MA, Aikawa M, Aji T, Yang YF. Selective association of a fragment of the knob protein with spectrin, actin and the red cell membrane. Molecular and Biochemical Parasitology. 1991;44:175–182. doi: 10.1016/0166-6851(91)90003-o. [DOI] [PubMed] [Google Scholar]
- 15.Magowan C, Nunomura W, Waller KL, Yeung J, Liang J, Van Dort H, Low PS, Coppel RL, Mohandas N. Plasmodium falciparum histidine-rich protein 1 associates with the band 3 binding domain of ankyrin in the infected red cell membrane. Biochim Biophys Acta. 2000;1502:461–470. doi: 10.1016/s0925-4439(00)00069-7. [DOI] [PubMed] [Google Scholar]
- 16.Pei X, An X, Guo X, Tarnawski M, Coppel R, Mohandas N. Structural and functional studies of interaction between Plasmodium falciparum knob-associated histidine-rich protein (KAHRP) and erythrocyte spectrin. J Biol Chem. 2005;280:31166–31171. doi: 10.1074/jbc.M505298200. [DOI] [PubMed] [Google Scholar]
- 17.Waller KL, Cooke BM, Nunomura W, Mohandas N, Coppel RL. Mapping the binding domains involved in the interaction between the Plasmodium falciparum knob-associated histidine-rich protein (KAHRP) and the cytoadherence ligand P. falciparum erythrocyte membrane protein 1 (PfEMP1) The Journal of Biological Chemistry. 1999;274:23808–23813. doi: 10.1074/jbc.274.34.23808. [DOI] [PubMed] [Google Scholar]
- 18.Oh SS, Voigt S, Fisher D, Yi SJ, LeRoy PJ, Derick LH, Liu S, Chishti AH. Plasmodium falciparum erythrocyte membrane protein 1 is anchored to the actin-spectrin junction and knob-associated his tidine-rich protein in the erythrocyte skeleton. Mol Biochem Parasitol. 2000;108:237–247. doi: 10.1016/s0166-6851(00)00227-9. [DOI] [PubMed] [Google Scholar]
- 19.Waller KL, Nunomura W, Cooke BM, Mohandas N, Coppel RL. Mapping the domains of the cytoadherence ligand Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) that bind to the knob- associated histidine-rich protein (KAHRP) Mol Biochem Parasitol. 2002;119:125–129. doi: 10.1016/s0166-6851(01)00395-4. [DOI] [PubMed] [Google Scholar]
- 20.Trelka DP, Schneider TG, Reeder JC, Taraschi TF. Evidence for vesicle-mediated trafficking of parasite proteins to the host cell cytosol and erythrocyte surface membrane in Plasmodium falciparum infected erythrocytes. Mol Biochem Parasitol. 2000;106:131–145. doi: 10.1016/s0166-6851(99)00207-8. [DOI] [PubMed] [Google Scholar]
- 21.Pasloske BL, Baruch DI, van Schravendijk MR, Handunnetti SM, Aikawa M, Fujioka H, Taraschi TF, Gormley JA, Howard RJ. Cloning and characterization of a Plasmodium falciparum gene encoding a novel high-molecular weight host membrane-associated protein, PfEMP3. Mol Biochem Parasitol. 1993;59:59–72. doi: 10.1016/0166-6851(93)90007-k. [DOI] [PubMed] [Google Scholar]
- 22.Van Schravendijk MR, Pasloske BL, Baruch DI, Handunnetti SM, Howard RJ. Immunochemical characterization and differentiation of two approximately 300-kD erythrocyte membrane-associated proteins of Plasmodium falciparum, PfEMP1 and PfEMP3. Am J Trop Med Hyg. 1993;49:552–565. doi: 10.4269/ajtmh.1993.49.552. [DOI] [PubMed] [Google Scholar]
- 23.Knuepfer E, Rug M, Klonis N, Tilley L, Cowman AF. Trafficking determinants for PfEMP3 export and assembly under the Plasmodium falciparum-infected red blood cell membrane. Mol Micro biol. 2005;58:1039–1053. doi: 10.1111/j.1365-2958.2005.04895.x. [DOI] [PubMed] [Google Scholar]
- 24.Lanzer M, de Bruin D, Wertheimer SP, Ravetch JV. Transcriptional and nucleosomal characterization of a subtelomeric gene cluster flanking a site of chromosomal rearrangements in Plasmodium falciparum. Nucleic Acids Res. 1994;22:4176–4182. doi: 10.1093/nar/22.20.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner MJ, Tettelin H, Carucci DJ, Cummings LM, Aravind L, Koonin EV, Shallom S, Mason T, Yu K, Fujii C, Pederson J, Shen K, Jing J, Aston C, Lai Z, Schwartz DC, Pertea M, Salzberg S, Zhou L, Sutton GG, Clayton R, White O, Smith HO, Fraser CM, Adams MD, Venter JC, Hoffman SL. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- 26.Pasloske BL, Baruch DI, Ma C, Taraschi TF, Gormley JA, Howard RJ. PfEMP3 and HRP1: co-expressed genes localized to chromosome 2 of Plasmodium falciparum. Gene. 1994;144:131–136. doi: 10.1016/0378-1119(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 27.Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, Lopez-Estrano C, Haldar K. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 28.Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 29.Waterkeyn JG, Wickham ME, Davern KM, Cooke BM, Coppel RL, Reeder JC, Culvenor JG, Waller RF, Cowman AF. Targeted mutagenesis of Plasmodium falciparum erythrocyte membrane protein 3 (PfEMP3) disrupts cytoadherence of malaria-infected red blood cells. Embo J. 2000;19:2813–2823. doi: 10.1093/emboj/19.12.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ausubel FM, Brend R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. 1, 2 and 3. New York: Greene Publishing Associate and Willey Interscience; 1994. [Google Scholar]
- 31.Dluzewski AR, Rangachari K, Wilson RJ, Gratzer WB. Properties of red cell ghost preparations susceptible to invasion by malaria parasites. Parasitology. 1983;87(Pt 3):429–438. doi: 10.1017/s0031182000082950. [DOI] [PubMed] [Google Scholar]
- 32.Cush R, Cronin JM, Stewart WJ, Maule CH, Molloy J, Goddard NJ. The resonant mirror: A novel optical biosensor for direct sensing of biomolecular interactions. Part I: Principle of operation and associated instrumentation. Biosensors and Bioelectronics. 1993;8:347–353. [Google Scholar]
- 33.George AJ, French RR, Glennie MJ. Measurement of kinetic binding constants of a panel of anti-saporin antibodies using a resonant mirror biosensor. J Immunol Methods. 1995;183:51–63. doi: 10.1016/0022-1759(95)00031-5. [DOI] [PubMed] [Google Scholar]
- 34.Watts HJ, Lowe CR, Pollard-Knight DV. Analytical Chemistry. 1994;66:2465–2470. doi: 10.1021/ac00087a010. [DOI] [PubMed] [Google Scholar]
- 35.Fairbanks G, Steck TL, Wallach DF. Electrophoretic analys is of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971;10:2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- 36.Kun JFJ, Waller KL, Coppel RL. Plasmodium falciparum: Structural and Functional Domains of the Mature-Parasite-Infected Erythrocyte Surface Antigen. Experimental Parasitology. 1999;91:258–267. doi: 10.1006/expr.1998.4374. [DOI] [PubMed] [Google Scholar]
- 37.Knuepfer E, Rug M, Cowman AF. Function of the plasmodium export element can be blocked by green fluorescent protein. Mol Biochem Parasitol. 2005;142:258–262. doi: 10.1016/j.molbiopara.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Marfatia SM, Lue RA, Branton D, Chishti AH. In vitro binding studies suggest a membrane-associated complex between erythroid p55, protein 4.1, and glycophorin C. J Biol Chem. 1994;269:8631–8634. [PubMed] [Google Scholar]
- 39.Korsgren C, Cohen CM. Associations of Human Erythrocyte Band 4.2. Binding to ankyrin and to the cytoplasmic domain of band 3. The Journal of Biological Chemistry. 1988;263:10212–12028. [PubMed] [Google Scholar]
- 40.Glenister FK, Coppel RL, Cowman AF, Mohandas N, Cooke BM. Contribution of parasite proteins to altered mechanical properties of malaria-infected red blood cells. Blood. 2002;99:1060–1063. doi: 10.1182/blood.v99.3.1060. [DOI] [PubMed] [Google Scholar]
- 41.Ohanian V, Wolfe LC, John KM, Pinder JC, Lux SE, Gratzer WB. Analysis of the ternary interaction of the red cell membrane skeletal proteins spectrin, actin, and 4.1. Biochemistry. 1984;23:4416–4420. doi: 10.1021/bi00314a027. [DOI] [PubMed] [Google Scholar]
- 42.Tyler JM, Reinhardt BN, Branton D. Associations of Erythrocyte Membrane Proteins. Binding of purified bands 2.1 and 4.1 to spectrin. The Journal Of Biological Chemistry. 1980;255:7034–7039. [PubMed] [Google Scholar]
- 43.Hughes CA, Bennett V. Adducin: a physical model with implications for function in assembly of spectrin-actin complexes. J Biol Chem. 1995;270:18990–18996. doi: 10.1074/jbc.270.32.18990. [DOI] [PubMed] [Google Scholar]
- 44.Siegel DL, Branton D. Partial Purification and Characterization of an Actin-bundling Prote in, Band 4.9, from Human Erythrocytes. The Journal of Cell Biology. 1985;100:775–785. doi: 10.1083/jcb.100.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mak AS, Roseborough G, Baker H. Tropomyosin from human erythrocyte membrane polymerizes poorly but binds F-actin effectively in the presence and absence of spectrin. Biochimica et Biophysica Acta. 1987;912:157–166. doi: 10.1016/0167-4838(87)90084-7. [DOI] [PubMed] [Google Scholar]
- 46.Karinch AM, Zimmer WE, Goodman SR. The Identification and Sequence of the Actin-binding Domain of Human Red Blood Cell b-Spectrin. The Journal of Biological Chemistry. 1990;265:11833–11840. [PubMed] [Google Scholar]
- 47.Becker PS, Schwartz MA, Morrow JS, Lux SE. Radiolabel-transfer cross-linking demonstrates that protein 4.1 binds to the N-terminal region of beta spectrin and to actin in binary interactions. European Journal of Biochemistry. 1990;193:827–836. doi: 10.1111/j.1432-1033.1990.tb19406.x. [DOI] [PubMed] [Google Scholar]
- 48.Kennedy SP, Warren SL, Forget BG, Morrow JS. Ankyrin binds to the 15th repetitive unit of erythroid and nonerythroid beta-spectrin. J Cell Biol. 1991;115:267–277. doi: 10.1083/jcb.115.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]