Abstract
Gene targeting technologies are essential for the analysis of gene functions. Knockout mouse generation via genetic modification of embryonic stem cells (ESCs) is the commonest example, but it is a time-consuming and labor-intensive procedure. Recently, a novel genome editing technology called CRISPR/Cas has enabled the direct production of knockout mice by non-homologous end joining (NHEJ)-mediated mutations. Unexpectedly, however, it generally exhibits a low efficiency in homologous recombination (HR) and is prone to high mosaicism. Meanwhile, gene targeting using ESCs is still being improved, as reported by Fukuda et al. in this issue. Here, we outline current gene targeting technologies with special emphasis on HR-mediated technologies, which are currently being performed using these two major strategies.
Keywords: Gene targeting, Genome editing, Homologous recombination, Knockin, Knockout
Forward genetics, also called classical genetics, aims to determine the genetic basis responsible for a phenotype (mutant phenotype → gene). By contrast, reverse genetics aims to understand the phenotypic effects of specific engineered gene sequences (gene → mutant phenotype). In 1981, embryonic stem cells (ESCs) were generated from mouse blastocysts [1]. The ESC can differentiate into all three germ layers, including gametes, in chimeras, and its genome can be transmitted to the next generation [2]. In addition, the ESC is suitable for gene modification using selective markers. Therefore, ESCs became a powerful tool for generation of gene-modified mice, resulting in the explosive development of reverse genetics after the 1980s.
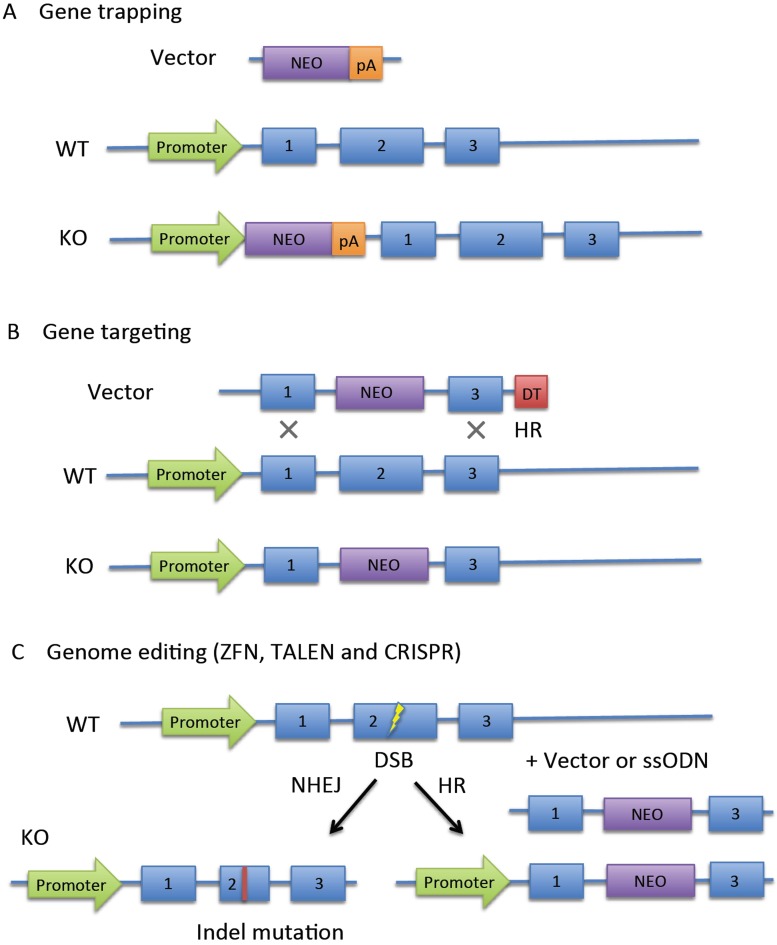
Knockout mice were generally produced by replacement or disruption of an existing gene with an artificial piece of DNA. Two popular methods, gene trapping and gene targeting, were usually applied to produce these mice. Gene trapping (Fig. 1A) is a high-throughput approach to introduce insertional mutations randomly in the genome. It is usually performed with gene trap vectors that consist of a promoterless reporter gene and/or selectable genetic marker and transcriptional termination sequence (polyadenylation sequence, pA). If the gene trap cassette is inserted into an intron of an expressed gene, the cassette is transcribed from the endogenous promoter, and downstream endogenous exons are ignored. Gene trapping is based on the random insertion of a gene trap cassette, whereas gene targeting targets a specific gene. In gene targeting, the targeting cassette has flanking homology regions (Fig. 1B), which make it possible to destroy the function of a desired gene. A targeting cassette usually contains a marker gene that provides resistance to a toxic agent (e.g., neomycin, NEO) and/or a reporter gene that produces an observable change (e.g., fluorescence). In addition, the cassette includes a negative selection marker (e.g., diphtheria toxin, DT) for complete selection. The Nobel Prize laureates in Physiology or Medicine in 2007 were Mario R. Capecchi, Martin J. Evans, and Oliver Smithies “for their discoveries of principles for introducing specific gene modifications in mice by the use of embryonic stem cells.” Although gene trapping and gene targeting are very popular today, low targeting efficiency and difficulty with germ-line transmission challenge research.
Fig. 1.
Summary of knockout strategies. A: Gene trapping depends on random insertion of a promoterless reporter gene, such as neomycin (NEO) and polyadenylation (pA) signals. The reporter is activated after insertion into introns of expressed genes in ESCs. B: Gene targeting relies on homologous recombination (HR) to introduce a reporter gene. In a random insertion event, a negative selection marker such as diphtheria toxin (DT) causes the death of a non-homologous recombinant. C: Genome editing technologies, including ZFN, TALEN, and CRISPR/Cas. Double-stranded breaks (DSB) generated by the artificial endonuclease are repaired by non-homologous end joining (NHEJ) or HR.
Recently, three representative genome editing technologies have been developed to target mutations to a specific location in the genome: zinc-finger nucleases (ZFNs) [3], transcription activator-like effector nucleases (TALENs) [4], and clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) nucleases [5]. These artificial nucleases enable genome editing by inducing double-strand breaks (DSBs) in targeted DNA that are repaired by non-homologous end joining (NHEJ) or homologous recombination (HR) [6,7,8,9]. NHEJ-mediated repair is error-prone and induces small insertions or deletions (indels) at the cleavage site, resulting in disruption of gene function by frameshift mutations or loss of key amino acid(s) (Fig. 1C left). In the presence of a single- or double-stranded DNA template (including vector) containing homology to the sequences flanking the DSB, mutant alleles with precise point mutations or DNA inserts can be produced by HR (Fig. 1C right). The efficiency of NHEJ-mediated mutation is so high that this technology is applied not only for gene knockout in ESCs but also for direct production of knockout mice via microinjection into zygotes. NHEJ occurs throughout the cell cycle, and is a dominant repair system in vertebrates [10]. By contrast, HR occurs only during the late S and G2 phase. Therefore, the efficiency of HR-mediated editing is generally low. In HR-mediated editing, the introduction of small epitope tags (V5, HA or Flag) or single loxP sites is relatively easy [11]; however, the introduction of long sequences encoding fluorescent proteins or exons and the simultaneous introduction of two loxP sites have been difficult, especially in the direct production of mice.
Today, many researchers aim to improve HR efficiency in gene targeting. For example, Hatada and colleagues reported that low-dose irradiation, with either γ-ray or x-ray exposure, increases HR efficiency in CRISPR/Cas genome editing in human pluripotent stem cells by activating the DNA repair/recombination machinery, including the ataxia-telangiectasia mutated (ATM), histone H2A.X and RAD51 proteins [12]. In another approach, Yu and colleagues developed a reporter-based screening approach for high-throughput identification of chemical compounds that increase HR efficiency [13]. They identified small molecules, L755507 and Brefeldin A, that increase HR efficiency 3-fold for large fragment insertions and 9-fold for point mutations. NHEJ inhibition is also effective for increasing HR efficiency. Recently, it was reported that SCR7 increased HR efficiency at the expense of NHEJ [14,15,16,17]. SCR7 is known as the inhibitor of DNA ligase IV, a key enzyme in the canonical NHEJ (C-NHEJ) pathway. Indeed, SCR7 increased the efficiency of HR-mediated genome editing up to 10–19-fold for at least five genes (Tex15, kell, Igkc, Os9 and Sgms2) in direct knockin mouse production [15, 16]. Gene silencing of the NHEJ key molecules KU70 and DNA ligase IV also promoted the efficiency of HR by 4–5-fold [17].
Knockdown of the bloom syndrome gene (Blm) also increases HR efficiency in various human cell lines [18]. Blm, which encodes a RecQ-type DNA helicase, is a suppressor of HR [19, 20]. In this issue, Fukuda and colleagues report that knockdown of Blm increases HR efficiency in ESCs [21]. Although the protocol for gene targeting in ESCs has already been well established, the HR efficiency is sometimes too low (< 1%). The authors report that blm knockdown enhances gene targeting efficiencies 2.3~5-fold for three gene loci (Prdm5, Prdm8 and Arl12ep). This is the first report showing the effectiveness of blm knockdown in producing gene-targeted mice via ESCs.
Although genome editing technologies enable us direct production of mutant mice by microinjection into zygotes, these founder (F0) mice show mosaicism in some cases [22]. Therefore, it takes about several months or a year to obtain a homozygotic knockout line after all in such cases. In addition, direct production of knockin mice with long exogenous sequence has still been difficult at present except for limited laboratories. By contrast, knockin ESC lines are obtained efficiently by combination with CRISPR/Cas and classic selection [23] which cannot be applied to genome editing using zygotes. These ESCs can be applied to phenotypic analyses both in vitro and in vivo: in vitro differentiation of ESCs and production of ESC-derived mice via the conventional chimera method or the tetraploid complementation method [24]. And it is also important to know that use of ESCs reduces the number of mice required and is good for animal welfare. Thus, ESCs still have several merits especially in kncokin of mice.
Genome engineering, including genome editing, is making rapid progress. Molecules and systems that increase HR efficiency will make it easy to produce knockin mice, including conditional knockout, introduction of reporter genes and precise point mutations, in the near future.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981; 292: 154–156. [DOI] [PubMed] [Google Scholar]
- 2.Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 1984; 309: 255–256. [DOI] [PubMed] [Google Scholar]
- 3.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA 1996; 93: 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 2011; 29: 143–148. [DOI] [PubMed] [Google Scholar]
- 5.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes DE. Non-homologous end joining as a mechanism of DNA repair. Curr Biol 2001; 11: R455–R457. [DOI] [PubMed] [Google Scholar]
- 7.van den Bosch M, Lohman PH, Pastink A. DNA double-strand break repair by homologous recombination. Biol Chem 2002; 383: 873–892. [DOI] [PubMed] [Google Scholar]
- 8.Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet 2006; 40: 363–383. [DOI] [PubMed] [Google Scholar]
- 9.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010; 79: 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006; 5: 1021–1029. [DOI] [PubMed] [Google Scholar]
- 11.Yang H, Wang H, Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 2014; 9: 1956–1968. [DOI] [PubMed] [Google Scholar]
- 12.Hatada S, Subramanian A, Mandefro B, Ren S, Kim HW, Tang J, Funari V, Baloh RH, Sareen D, Arumugaswami V, Svendsen CN. Low-Dose Irradiation Enhances Gene Targeting in Human Pluripotent Stem Cells. Stem Cells Transl Med 2015; 4: 998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, Qi LS. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 2015; 16: 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, Kumar S, Pandey M, Singh RK, Ray P, Natarajan R, Kelkar M, De A, Choudhary B, Raghavan SC. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 2012; 151: 1474–1487. [DOI] [PubMed] [Google Scholar]
- 15.Singh P, Schimenti JC, Bolcun-Filas E. A mouse geneticist’s practical guide to CRISPR applications. Genetics 2015; 199: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 2015; 33: 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 2015; 33: 543–548. [DOI] [PubMed] [Google Scholar]
- 18.So S, Nomura Y, Adachi N, Kobayashi Y, Hori T, Kurihara Y, Koyama H. Enhanced gene targeting efficiency by siRNA that silences the expression of the Bloom syndrome gene in human cells. Genes Cells 2006; 11: 363–371. [DOI] [PubMed] [Google Scholar]
- 19.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 1995; 83: 655–666. [DOI] [PubMed] [Google Scholar]
- 20.Larsen NB, Hickson ID. RecQ Helicases: Conserved Guardians of Genomic Integrity. Adv Exp Med Biol 2013; 767: 161–184. [DOI] [PubMed] [Google Scholar]
- 21.Fukuda M, Inoue M, Muramatsu D, Miyachi H, Shinkai Y. Knockout mouse production assisted by Blm knockdown. J Reprod Dev 2015; 62:DOI . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yen ST, Zhang M, Deng JM, Usman SJ, Smith CN, Parker-Thornburg J, Swinton PG, Martin JF, Behringer RR. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev Biol 2014; 393: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gennequin B, Otte DM, Zimmer A. CRISPR/Cas-induced double-strand breaks boost the frequency of gene replacements for humanizing the mouse Cnr2 gene. Biochem Biophys Res Commun 2013; 441: 815–819. [DOI] [PubMed] [Google Scholar]
- 24.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA 1993; 90: 8424–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]