SUMMARY
In the absence of pathogen infection, plant effector-triggered immune (ETI) receptors are maintained in a preactivation state by intermolecular interactions with other host proteins. Pathogen effector-induced alterations activate the receptor. In Arabidopsis, the ETI receptor RPM1 is activated via bacterial effector AvrB-induced phosphorylation of the RPM1-interacting protein RIN4 at Threonine 166. We find that RIN4 also interacts with the prolyl-peptidyl isomerase (PPIase) ROC1, which is reduced upon RIN4 Thr166 phosphorylation. ROC1 suppresses RPM1 immunity in a PPIase-dependent manner. Consistent with this, RIN4 Pro149 undergoes cis/trans isomerization in the presence of ROC1. While the RIN4P149V mutation abolishes RPM1 resistance, the deletion of Pro149 leads to RPM1 activation in the absence of RIN4 phosphorylation. These results support a model in which RPM1 directly senses conformational changes in RIN4 surrounding Pro149 that is controlled by ROC1. RIN4 Thr166 phosphorylation indirectly regulates RPM1 resistance by modulating the ROC1-mediated RIN4 isomerization.
INTRODUCTION
The NLR family immune receptors play a fundamental role in pathogen recognition in both plants and animals (Maekawa et al., 2011; Qi and Innes, 2013). NLRs contain a variable N-terminal domain; a nucleotide-binding, Apaf, Resistance protein, and CED-4 (ARC) domain in the middle; and a leucine-rich repeat domain in the C terminus. Animal NLRs are known to perceive conserved microbial molecular patterns, but a recent report showed that the human NLR protein NOD1 is capable of sensing the activity of Salmonella enterica effector SopE to activate proinflammatory responses (Keestra et al., 2013). All plant NLRs studied to date perceive variable pathogen effector proteins in a specific manner. The recognition of pathogen effectors triggers strong defenses in plants that are often associated with a form of programmed cell death at the site of infection termed the hypersensitive response (HR). How effectors activate plant NLRs is not well understood.
In the absence of pathogen infection, plant NLRs are kept in a preactivation state by intramolecular interactions between different NLR domains and intermolecular interactions with a second host protein (Maekawa et al., 2011; Hu et al., 2013). During infection, some of the NLR-interacting proteins are targeted by pathogen effectors. This is thought to cause a conformational change in NLRs and converts the latter into a postactivation state. In several cases, plant NLRs and their interacting proteins have been studied in detail. For instance, the tomato NLR protein Prf constitutively interacts with the Pto kinase (Mucyn et al., 2006). When infected with Pseudomonas syringae pv tomato (Pst), the effector proteins AvrPto and AvrPtoB interact with Pto to activate Prf-mediated resistance, likely by altering the conformation of the Pto-Prf complex (Tang et al., 1996; Scofield et al., 1996; Kim et al., 2002; Mucyn et al., 2009). Likewise, the Arabidopsis NLR RPS5 constitutively interacts with the PBS1 kinase (Ade et al., 2007). The P. syringae effector AvrPphB, a cysteine protease, specifically cleaves PBS1 to activate RPS5 resistance, likely by inducing a conformational change in the PBS1-RPS5 complex (Shao et al., 2003; Ade et al., 2007). The Arabidopsis NLRs RPM1 and RPS2 constitutively associate with the plasma-membrane-associated protein RIN4 (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003). Three P. syringae effectors, AvrRpt2, AvrB, and AvrRpm1, interact with RIN4 to activate RPS2 and RPM1 immunity. AvrRpt2 is a cysteine protease that cleaves RIN4 to activate RPS2 resistance (Axtell et al., 2003; Axtell and Staskawicz, 2003; Mackey et al., 2003; Day et al., 2005; Kim et al., 2005). The biochemical functions of AvrB and AvrRpm1 remain unknown, but they are known to induce RIN4 phosphorylation (Mackey et al., 2002; Desveaux et al., 2007). In particular, AvrB induces RIN4 phosphorylation at Thr166 through a host protein kinase RIPK, and this phosphorylation is sufficient for the activation of RPM1 (Chung et al., 2011; Liu et al., 2011).
Cyclophilins (CyPs) are a large family of proteins shared by prokaryotes and eukaryotes (Handschumacher et al., 1984; Stamnes et al., 1992). CyPs possess PPIase activity that catalyzes the isomerization between cis and trans isoforms of the X-prolyl peptide bond. Arabidopsis contains 29 CyPs (Romano et al., 2004) playing diverse roles including photo-damage protection (Dominguez-Solis et al., 2008), adaptation to abiotic stresses (Luan et al., 1994; Chen et al., 2007), and hormone signaling (Trupkin et al., 2012; Zhang et al., 2013). CyPs have also been found to play an important role in plant-pathogen recognition (Coaker et al., 2005). The maturation of the AvrRpt2 cysteine protease requires a eukaryotic CyP, and both the yeast CyP CPR1 and Arabidopsis CyP ROC1 (Rotamase CYP 1) can promote AvrRpt2 maturation in vitro (Coaker et al., 2005). Thus, Cyps such as ROC1 are proposed to positively regulate RPS2 resistance through its PPIase activity. A role of ROC1 in RPM1 resistance has not been determined. A previously identified gain-of-function Arabidopsis ROC1 mutant containing a Ser58Phe substitution in ROC1 (ROC1S58F) exhibits altered plant architecture (Ma et al., 2013). Here we show that ROC1S58F is enhanced in AvrRpt2 maturation and RIN4 cleavage. Surprisingly, ROC1S58F is not enhanced in RPS2 activation. Instead, it is compromised in RPS2 and RPM1 resistance. Detailed analyses showed that ROC1 inhibits RPM1 and RPS2 resistance through a direct interaction with RIN4. We further show that RIN4 Pro149 plays an essential role in the activation of RPM1 that can be uncoupled from RIN4 Thr166 phosphorylation and provide evidence that the RIN4 conformation defined by Pro149 mutations is subject to regulation by ROC1. In addition, RIN4 Thr166 phosphorylation reduces the ROC1-RIN4 interaction, suggesting that Thr166 phosphorylation is not directly sensed by RPM1. Instead, it indirectly activates RPM1 through reducing the ROC1 inhibition.
RESULTS
ROC1S58F Is Specifically Compromised in RPM1- and RPS2-Specified Immunity
Because ROC1 was previously shown to promote AvrRpt2 maturation (Coaker et al., 2005), we tested if ROC1S58F is affected in immunity specified by RPS2 and other NLRs. As shown in Figure 1A, virulent Pst grew to similar titers in wild-type (WT) (Col-0) and ROC1S58F plants, indicating that ROC1S58F was not affected in basal resistance. As expected, the rpm1, rps2, and rps5 plants were completely susceptible to Pst strains carrying avrB, avrRpt2, and avrPphB, respectively, whereas WT plants displayed full resistance to these strains (Figures 1B–1D). ROC1S58F exhibited partial susceptibility to Pst (avrB) or Pst (avrRpt2) but normal resistance to Pst (avrPphB), indicating that ROC1S58F is compromised in RPM1 and RPS2 resistance but retains full RPS5 resistance. This notion was further supported by a compromised HR in ROC1S58F to Pst (avrB) and Pst (avrRpt2) but normal HR to Pst (avrPphB) (Figure S1 available online). To further test the specificity of immune activation, the ROC1S58F mutant was crossed to the snc1 mutant, which carries a constitutive active NLR SNC1 and displays a dwarf phenotype indicative of autoimmune responses. All snc1 ROC1S58F double mutant plants were indistinguishable from snc1, indicating that the ROC1S58F mutation does not affect SNC1 immunity (Figure 1E). Together, these results suggest that ROC1S58F is specifically affected in immunity specified by RPM1 and RPS2 but not RPS5 and SNC1.
Figure 1. The ROC1S58F Mutant Is Specifically Compromised in RPS2- and RPM1-Specified Resistance.
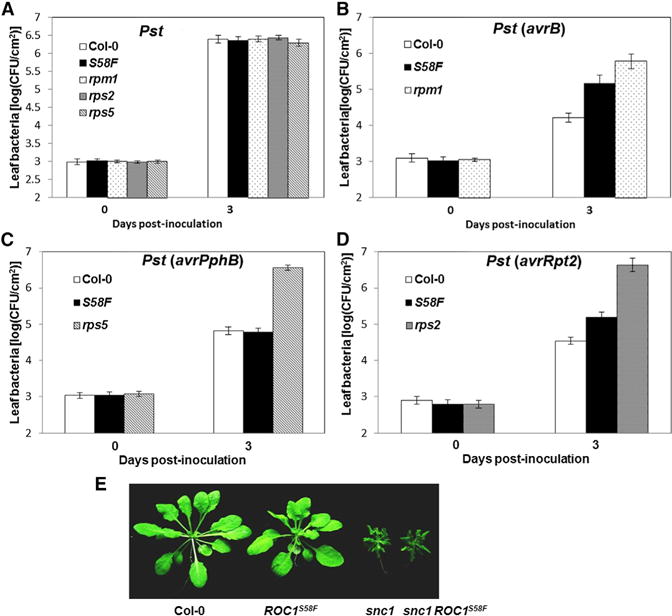
(A–D) Plants of the indicated genotypes were infiltrated with Pst (A), Pst (avrB) (B), Pst (avrPphB) (C), or Pst (avrRpt2) (D), and bacterial population in the leaf was determined at the indicated times. Error bars represent SD (n = 8, 4 biological repeats).
(E) Morphological phenotype of WT (Col-0), ROC1S58F, snc1, and snc1 ROC1S58F double mutant. Plants were photographed 5 weeks after germination. See also Figure S1.
The Diminished RPS2 Resistance in ROC1S58F Is Not Explained by AvrRpt2-Dependent RIN4 Cleavage
The results described above appear to be consistent with the possibility that ROC1S58F is less capable of promoting AvrRpt2 maturation, leading to reduced cleavage of RIN4 and incomplete release of RPS2 from RIN4 inhibition. We therefore determined maturation of the recombinant AvrRpt2, which leads to self-cleavage, in extracts from WT and ROC1S58F plants. As expected, the incubation with WT extracts promoted AvrRpt2 maturation, as indicated by the accumulation of self-cleavage product (Figure 2A). Surprisingly, the ROC1S58F extracts consistently showed more rapid AvrRpt2 cleavage than did the WT extracts. We further determined the RIN4 cleavage in plants inoculated with Pst (avrRpt2). The abundance of the intact RIN4 protein decreased in WT plants within 4 hr after inoculation, indicative of a cleavage by AvrRpt2 (Figures 2B and S2A). In ROC1S58F plants, the avrRpt2-induced RIN4 cleavage was more pronounced, and the amount of intact RIN4 in ROC1S58F was ~20% of that in WT plants 4 hr postinoculation. Thus, ROC1S58F appears to possess greater activity to promote AvrRpt2 maturation and RIN4 cleavage.
Figure 2. The ROC1S58F Mutation Enhances AvrRpt2 Maturation and RIN4 Cleavage.
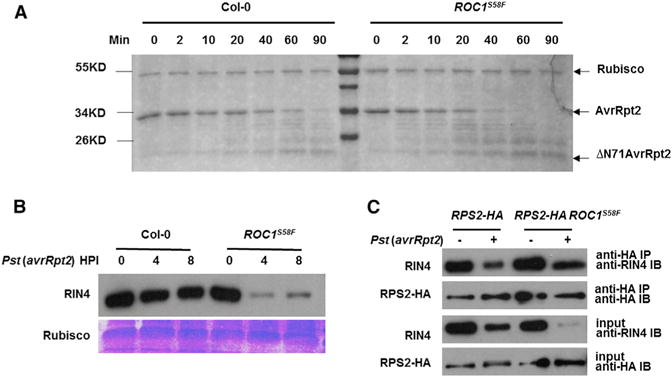
(A) ROC1S58F extract enhances AvrRpt2 autocleavage. Equal amounts of recombinant AvrRpt2 proteins were incubated with total protein extracts from WT (Col-0) or ROC1S58F plants for the indicated times, electrophoresed through SDS-PAGE, and AvrRpt2 self-cleavage was visualized by Coomassie brilliant blue (CBB) staining.
(B) The RIN4 cleavage by AvrRpt2 is more efficient in the ROC1S58F mutant. Plants of the indicated genotype were infiltrated with Pst (avrRpt2), and amounts of RIN4 was determined at the indicated hours postinoculation (HPI) by immuoblot with anti-RIN4 antibodies. Ponceau staining of Rubisco indicates equal loading of protein.
(C) The ROC1S58F mutation does not affect RPS2 accumulation and RPS2-RIN4 interaction. Plants carrying the RPS2-HA transgene under the control of RPS2 native promoter in the WT or ROC1S58F background were inoculated with Pst (avrRpt2) for 4 hr and examined for RPS2-HA accumulation by immunoblot and RPS2-RIN4 interaction by coIP. See also Figure S2.
The increased RIN4 cleavage but reduced RPS2 activation is puzzling, as this is not explained by the current model. We further examined RPS2-RIN4 interaction by coimmunoprecipitation (coIP) assay in WT and ROC1S58F plants carrying the same RPS2-HA transgene following inoculation with Pst (avrRpt2). Both plants accumulated similar levels of RPS2-HA (Figures 2C and S2B), indicating that ROC1S58F does not affect RPS2 abundance. As reported, a RIN4-RPS2-HA interaction was readily detected prior to bacterial infection in the WT RPS2-HA line. The inoculation with Pst (avrRpt2) reduced the amount of RIN4 associated with RPS2-HA concomitant with reduction of RIN4 abundance in total protein extract (Figure 2C). In the ROC1S58F background, although the total RIN4 protein was reduced to a much lower level compared to the WT RPS2-HA line upon Pst (avrRpt2) inoculation, the amount of RIN4 associated with RPS2-HA was comparable to that in the WT RPS2-HA line. Thus, the compromised RPS2 resistance in the ROC1S58F mutant was not caused by an increased RIN4 abundance or RIN4-RPS2 interaction. We thus reasoned that, in addition to AvrRpt2, ROC1 must modulate an additional protein (proteins) involved in RPS2 activation.
ROC1 Plays a Negative Role in RPM1 and RPS2 Immunity
We next used the Nicotiana benthamiana (Nb) transient expression system to test the role of ROC1 in RPM1- and RPS2-specified immune responses. Consistent with previous reports (Chung et al., 2011; Liu et al., 2011), the majority of leaves (15 out of 18) coexpressing RPM1-HA and a phospho-mimetic RIN4T166E mutant (pmRIN4) developed strong HR (Figure 3A). Only four out of 18 leaves coexpressing RPM1-HA, pmRIN4, and ROC1-FLAG developed HR (Figure 3A), indicating that overexpression of ROC1 significantly attenuated the HR triggered by RPM1 and pmRIN4. This HR was further attenuated upon the overexpression of ROC1S58F-FLAG. Likewise, HR triggered by RPS2-HA overexpression was attenuated by ROC1-FLAG overexpression (Figure 3B). Again, the ROC1S58F-FLAG overexpression further attenuated the RPS2 HR. An examination of the RPM1-HA, RPS2-HA, RIN4, and ROC1-FLAG proteins indicated that the differences in HR development were not caused by differential protein accumulation (Figures S3A and S3B).
Figure 3. ROC1 Negatively Regulates RPM1- and RPS2-Specified Immunity.
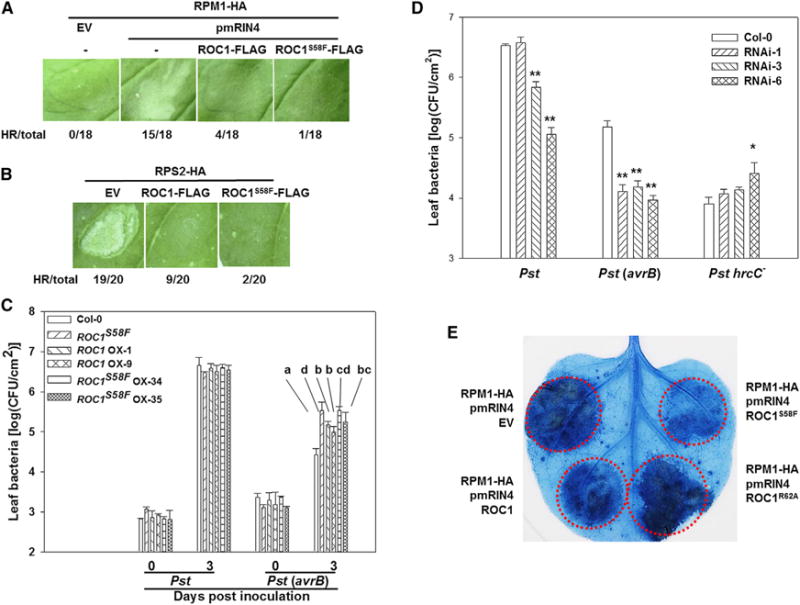
(A and B) Overexpression of ROC1-FLAG and ROC1S58F-FLAG inhibits RPM1 and RPS2 HR in Nb plants. Nb leaves were infiltrated with Agrobacterium containing the indicated constructs, and the development of HR was photographed 36 hr postinfiltration. pmRIN4, phosphomimetic RIN4 (RIN4T166E); EV, empty vector. The numbers under the photograph indicate ratios of infiltration showing HR to total number of infiltrations.
(C) Stable transgenic plants overexpressing ROC1-FLAG and ROC1S58F-FLAG are compromised in RPM1 resistance. Plants of the indicated genotypes were infiltrated with Pst or Pst (avrB), and bacterial population in the leaf was measured at the indicated times. Error bars represent SD. Different letters denote significant difference at p < 0.01 (Student’s t test, n = 8, 3 biological repeats).
(D) Silencing of ROC1 in Arabidopsis enhances disease resistance to Pst (avrB). WT (Col-0) and three independent ROC1 RNAi lines were infiltrated with the indicated bacterial strains, and the bacterial population was determined 3 days after inoculation. Error bars represent SD. * and ** denote significant difference at p < 0.05 and p < 0.01, respectively (Student’s t test, n = 8, 3 biological repeats). Similar results were obtained from two independent experiments.
(E) PPIase active site is required for ROC1 to inhibit RPM1 cell death. Nb leaves were infiltrated with Agrobacterium carrying the indicated constructs and stained with trypan blue 36 hr later. Fifteen leaves were tested with similar results. See also Figure S3.
To further substantiate these findings, we generated transgenic Arabidopsis lines overexpressing ROC1-FLAG and ROC1S58F-FLAG and inoculated these lines with Pst or Pst (avrB). The ROC1-FLAG and ROC1S58F-FLAG lines supported greater growth of Pst (avrB) but not Pst compared to Col-0 plants (Figures 3C), suggesting that ROC1 plays a negative role in RPM1 resistance. Overall, the ROC1S58F-FLAG lines reproducibly supported greater growth of Pst (avrB) than did the ROC1-FLAG lines. These lines expressed similar levels of ROC1 proteins (Figure S3C), indicating that ROC1S58F is a gain-of-function mutation inhibiting RPM1 resistance. This is consistent with the previous observation that ROC1S58F acts as a gain-of-function mutation in the modulation of plant architecture (Ma et al., 2013). To further determine the role of ROC1 in the regulation of RPM1 resistance, we inoculated ROC1 RNAi lines, which contain nearly nondetectable ROC1 transcripts (Ma et al., 2013), with various Pst strains. When inoculated with Pst (avrB), all RNAi lines showed reduced bacterial growth that was only ~10% of that in WT (Figures 3D and S3D). These results were obtained from plants grown under different growth conditions and were highly consistent, confirming a negative role of ROC1 in plant immunity. Some of the RNAi lines also displayed enhanced resistance to Pst (Figures 3D and S3D), suggesting that ROC1 also plays a role in basal resistance to the virulent bacteria. We further inoculated the RNAi lines with the Pst hrcC− mutant strain, which is unable to secrete type III effectors (Hauck et al., 2003) and considered only able to induce PAMP-triggered immunity (PTI) and not effector-triggered immunity (ETI). All RNAi lines supported similar or greater levels of Pst hrcC− growth compared to WT (Figure 3D), suggesting that the increased basal resistance to Pst was likely attributed to ETI. The normal Pst basal resistance in ROC1-FLAG overexpression plants (Figure 3C) and increased Pst basal resistance in ROC1 RNAi lines (Figure 3D) suggest that a minimal amount of ROC1 is sufficient to inhibit basal resistance to Pst. Together, these results support that ROC1 negatively regulates RPM1, RPS2, and potentially additional NLRs and that the ROC1S58F mutation enhances this negative regulation. Because the HR triggered by RPM1 and RPS2 in Nb plants are independent of bacterial effectors, the results additionally indicate that ROC1 modulates RPM1 and RPS2 activities through a host protein.
The PPIase Activity Is Required for ROC1 to Inhibit RPM1 and RPS2 Immunity
The enhanced AvrRpt2 maturation and stronger inhibition of RPM1 and RPS2 immunity in the ROC1S58F mutant plants suggested that ROC1S58F possesses greater PPIaase activity, which may be responsible for inhibiting RPM1 and RPS2 immunity. However, the recombinant ROC1S58F mutant protein expressed in E. coli was largely insoluble, preventing a direct measurement of the PPIase activity of ROC1S58F. Nonetheless, modeling of ROC1 to known CyP structures indicated that the Ser58Phe substitution resulted in new contacts between the Phe58 aromatic group with Asp73 and Phe74 and Lys59 with His54 (Arnold et al., 2006; Laskowski and Swindells, 2011) (Figure S3E). These new contacts occur in a loop containing several active site residues and may alter the ROC1 PPIase activity.
To test if the PPIase activity is required for ROC1 to inhibit RPM1 and RPS2 ETI, we introduced mutations into ROC1 that are known to disrupt PPIase activity. While transient expression of WT ROC1 and ROC1S58F in Nb plants partially inhibited the cell death triggered by pmRIN4 and RPM1, the ROC1R62A mutant, in which the invariant active site residue Arg62 was substituted with Ala, was completely unable to inhibit cell death (Figures 3E and S3F). In yeast, the CPR1H90Y substitution (corresponding to ROC1H99Y) is known to abolish the PPIase activity by perturbing the conformation of two invariant active site residues Trp121 and His126 (Cardenas et al., 1995). The ROC1H99Y mutant was largely incapable of inhibiting the RPS2-triggered HR in Nb plants (Figure S3F). Furthermore, introducing this mutation into the ROC1S58F mutant also abolished the ability of the latter to inhibit RPS2 HR (Figure S3G). Together, these results are consistent with a role of ROC1 PPIase activity in the inhibition of RPM1 and RPS2 immunity.
ROC1 Specifically Interacts with RIN4
The specific effect of ROC1S58F in RPM1 and RPS2 resistance but not RPS5 and SNC1 immune responses suggested an involvement of RIN4, which is required for the regulation of RPM1 and RPS2 but not RPS5 and SNC1. An alternative explanation is that ROC1 regulates the maturation of AvrRpt2 and AvrB but not AvrPphB. GST pull-down experiments were carried out to test these possibilities. Consistent with previous findings (Coaker et al., 2005), ROC1 interacted strongly with AvrRpt2 in vitro (Figure 4A). ROC1 also interacted strongly with RIN4 but not AvrB. The results support that ROC1 inhibits RPM1 and RPS2 immunity likely through RIN4. Both ROC1 and ROC1S58F interacted equally with RIN4 (Figure S4A), a result consistent with the possibility that the elevated PPIase activity, instead of strength of protein-protein interaction, is responsible for the reduction of RPM1 and RPS2 immunity in ROC1S58F plants. Luciferase complementation assays (Chen et al., 2008) indicated that ROC1-Nluc and Cluc-RIN4 strongly interacted in Nb plants (Figures 4B and S4B). As expected, RPM1-Nluc and RPS2-Nluc interacted with Cluc-RIN4. However, ROC1 failed to interact with RPM1 and RPS2, regardless of the orientation of the constructs used. These results indicated that, in plants, ROC1 can specifically interact with RIN4, but not RPM1 and RPS2. We further tested this possibility using coIP assay in Arabidopsis seedlings overexpressing ROC1-FLAG. Again, a clear ROC1-RIN4 interaction was detected (Figure 4C). RIN4 is a plasma-membrae-associated protein, whereas ROC1 is known to exist in multiple cellular compartments. Transient coexpression of ROC1-GFP and BFP-RIN4 in Nb leaves indicated that the two proteins colocalized in the cell (Figure 4D). Together, these results demonstrate that ROC1 is capable of interacting with RIN4 in the plant cell.
Figure 4. ROC1 Interacts with RIN4.
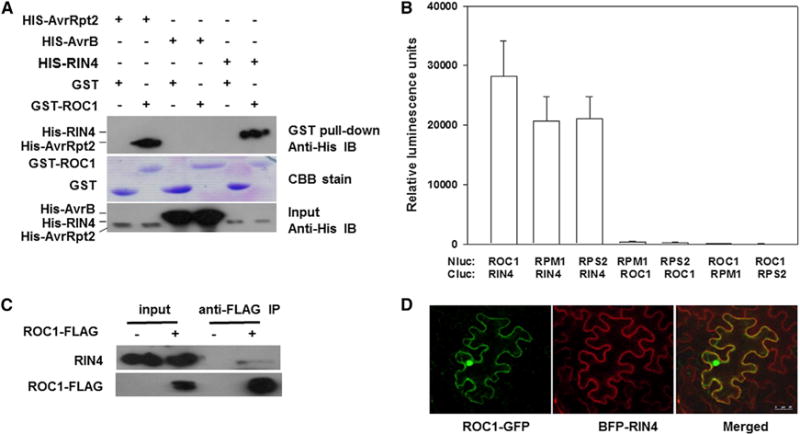
(A) ROC1and RIN4 interact in vitro. His-RIN4, His-AvrRpt2, or His-AvrB was incubated with GST, or GST-ROC1 recombinant protein for pull-down assay, and amounts of proteins in the blot were determined by immune blot or CBB staining.
(B) Luciferase complementation assay for ROC1-RIN4 interaction in Nb plants. Nb leaves were infiltrated with Agrobacterium carrying the indicated Nlu and Cluc constructs, and luminescence was measured 24 hr later. Error bars represent SD (n = 8, 3 biological repeats).
(C) CoIP assay for ROC1-RIN4 interaction in Arabidopsis seedlings expressing the ROC1-FLAG transgene. Immunoblots were detected with anti-RIN4 or anti-FLAG antibodies. The experiment was repeated twice with similar results.
(D) ROC1 and RIN4 colocalize at the plasma membrane in Nb plants. Agrobacteria containing the indicated constructs were infiltrated into Nb leaves for 24 hr, and fluorescence in the epidermal cell was visualized under a fluorescent microscope. See also Figure S4.
Previous reports showed that the C terminus of RIN4 (aa 142–210) is important for its function (Chung et al., 2011). GST pull-down assay indicated that RIN4 lacking aa 1–141 interacted normally with ROC1, indicating that the C terminus is sufficient for ROC1 interaction (Figure S5A).
RIN4 Thr166 Phosphorylation Reduces Its Interaction with ROC1
We examined if RIN4 Thr166 phosphorylation impacts ROC1 interaction. Interestingly, GST pull-down assays showed that pmRIN4 and the phospho-mimetic C-terminal fragment of RIN4 displayed much weaker interactions with ROC1 compared to the nonphosphorylated forms (Figures 5A and S5A). Luciferase complementation assays showed that the RIN4-ROC1 interaction in Nb plants was strongly reduced in the presence of AvrB-FLAG (Figures 5B and S5B), supporting that the AvrB-induced RIN4 phosphorylation leads to its dissociation from ROC1 in plants. We previously showed that the Xamthomonas campestris effector AvrAC, an uridylyl transferase, is capable of blocking RPM1 activation by inhibiting RIPK kinase activity (Feng et al., 2012). Coexpression of AvrAC-HA in Nb plants largely restored ROC1-RIN4 interaction even in the presence of AvrB (Figures 5B and S5B). In contrast, coexpression of a catalytic AvrACH469A mutant (mAvrAC) failed to restore ROC1-RIN4 interaction. These results further supported that the AvrB-induced phosphorylation of RIN4 via RIPK led to ROC1-RIN4 dissociation. Consistent with this notion, the RIN4 Thr166 phosphorylation has been shown to disrupt the RIN4-RIPK interaction (Liu et al., 2011).
Figure 5. RIN4 Thr166 Phosphorylation Weakens the ROC1-RIN4 Interaction.
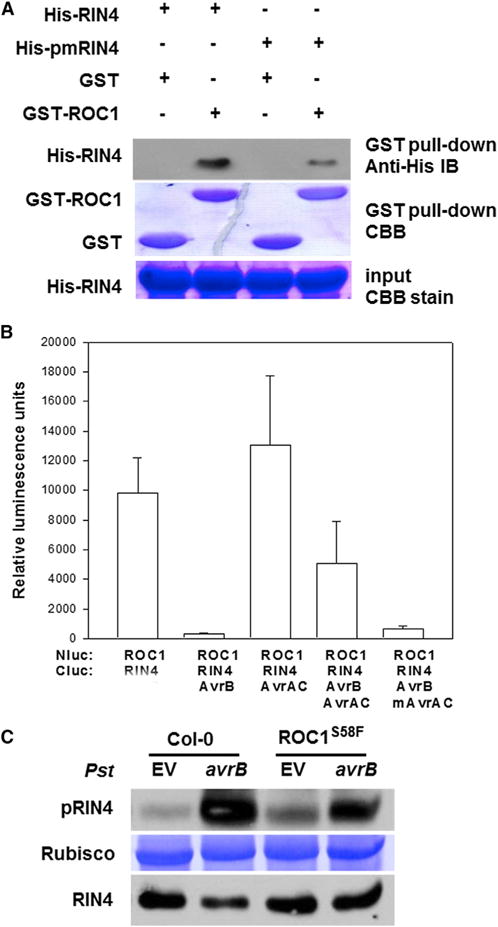
(A) The pmRIN4 mutation reduces the ROC1-RIN4 interaction in vitro. His-RIN4 or His-pmRIN4 was incubated with GST, or GST-ROC1 recombinant protein for pull-down assay, and amounts of proteins in the blot were determined by immune blot or CBB staining.
(B) AvrB diminishes the ROC1-RIN4 interaction in Nb plants, whereas AvrAC restores the interaction. The indicated Nluc and Cluc constructs along with AvrB-FLAG, AvrAC-HA, and mAvrAC-HA were transiently expressed in Nb plants for luciferase complementation assay. The experiment was performed twice with similar results. Error bars represent SD (n = 8; 2 biological repreats).
(C) The AvrB-induced RIN4 phosphorylation was diminished in ROC1S58F plants. WT (Col-0) and ROC1S58F plants were infiltrated with Pst containing an empty vector pVSP61 (EV) or avrB, and protein was extracted 2 hr after the onset of HR for immune blot analyses. CBB stain of Rubisco indicate equal loading of total protein. See also Figure S5.
We next tested if ROC1 plays a role in AvrB-induced RIN4 phosphorylation by inoculating WT and ROC1S58F plants with Pst (avrB). While Pst (avrB) induced RIN4 Thr166 phosphorylation in both plants, the phosphorylation was notably less in ROC1S58F plants (Figure 5C). Together, these results suggest a mutual regulation between ROC1-RIN4 interaction and RIN4 phosphorylation.
RIN4 Pro149 Plays a Critical Role in RPM1 Activation
Because ROC1 interacts with the C terminus of RIN4, we speculated that RIN4 C terminus could be a substrate for the ROC1 PPIase. We therefore examined the importance of RIN4 C-terminal Pro residues in the regulation of RPM1 HR by individually substituting all four C-terminal Pros with Vals in pmRIN4. While the pmRIN4P159V, pmRIN4P189V, and pmRIN4P197V mutants still triggered normal HR in Nb plants when coexpressed with RPM1, the pmRIN4P149V mutant was completely unable to trigger HR (Figures 6A and S6A). We later found that RIN4T166D was more potent than RIN4T166E in triggering RPM1 HR. However, the RIN4P149V T166D mutant was completely unable to trigger RPM1 HR in Nb leaves (Figures 6B and S6B). The results demonstrated that Pro149 is specifically required for the phosphorylated RIN4 to trigger RPM1 HR. The results also indicated that secondary mutations within RIN4 can uncouple RIN4 Thr166 phosphorylation and RPM1 activation, suggesting that RIN4 Thr166 phosphorylation alone is insufficient to trigger RPM1 immune responses.
Figure 6. RIN4 Pro149 Is Subject to Isomerization by ROC1 and Plays a Critical Role in RPM1 Activation in Nb Plants.
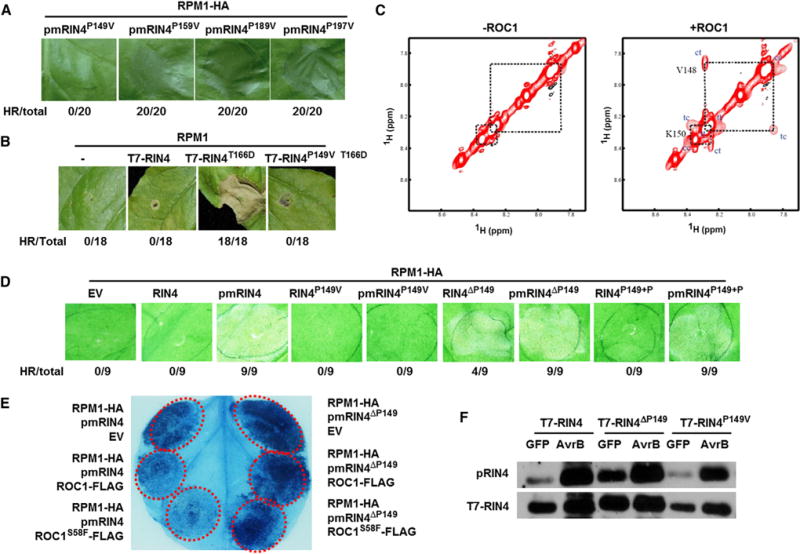
(A and B) Pro149Val substitution abolishes RPM1 HR triggered by pmRIN4 (A) and RIN4T166D (B) in Nb plants.
(C) ROC1 catalyzes cis/trans isomerization of RIN4 peptide. Shown are selected regions of ROESY spectra of the RIN4 peptide in the absence (−) or presence (+) of ROC1. Diagonal amide proton peaks of Val148 and Lys150 from cis and trans conformers are indicated by cc and tt, respectively, while ROE cross peaks due to conformational exchange resulted from ROC1-catalyzed isomerization are labeled by ct and tc. Both diagonal peaks and exchange cross peaks are negative and displayed in red.
(D) Deletion of Pro149 activates RPM1 HR in Nb plants. Numbers indicate number of leaves showing HR versus total number of infiltrated leaves.
(E) Cell death triggered by pmRIN4ΔP149 is insensitive to ROC1 inhibition. Nb leaves were infiltrated with Agrobacterium carrying the indicated constructs and photographed 72 hr later for HR ([A], [B], and [D]) or stained with trypan blue 36 hr later for cell death (E). The assays were repeated more than three times with consistent results.
(F) Effect of Pro149 mutations on AvrB-induced phosphorylation of Thr166. Nb leaves transiently expressing the indicated T7-RIN4 constructs were infiltrated with Agrobacterium carrying GFP or AvrB construct, and protein was extracted 16 hr later and subject to immunoblot analyses. The experiment was performed twice with similar results. See also Figure S6.
We then investigated if ROC1 can catalyze proline cis/trans isomerization of a RIN4 peptide spanning Pro149 (144-KVTVVPKFGDWD-155) using 2D 1H-1H rotating frame Overhauser effect spectroscopy (ROESY) NMR experiment, which is well documented for detecting conformational exchange in the μs to ms timescale. As expected, we observed strong rotating frame Overhauser effect (ROE) exchange cross peaks for amide protons of Val148 and Lys150 due to the cis/trans isomerization of Pro149 in the presence of ROC1, whereas no exchange cross peak was observed in the ROESY spectrum of the peptide without ROC1 (Figure 6C). This indicates that ROC1 does accelerate the cis/trans isomerization of Pro149, and thus, ROC1 can catalyze the conformational exchange of the RIN4.
To further examine the role of Pro149 in RPM1 immunity, we deleted this residue and tested its effect in the activation of RPM1. Surprisingly, RIN4ΔP149 can activate RPM1 HR in Nb plants without a phosphomimetic mutation, although to a lower level than the T166D/E mutation (Figures 6D and S6C). Luciferase complementation assays showed that RIN4 and RIN4ΔP149 interacted equally well with ROC1 (Figure S6D), indicating that the HR phenotype conferred by RIN4ΔP149 was likely caused by the conformation adopted by the mutant protein but not a defect in its interaction with ROC1. In this experiment we also tested the impact of ROC1H99Y and ROC1R62A mutations and observed a modest reduction in the ROC1-RIN4 interaction (Figure S6D), which does not appear to explain a complete lack of inhibition of RPM1 and RPS2 HR (Figures 3E and S3G). The contrasting phenotypes conferred by the RIN4P149V and RIN4ΔP149 mutants suggested that they adopt opposite conformations. We further tested if WT ROC1 and ROC1S58F were able to inhibit cell death in Nb leaves triggered by the pmRIN4ΔP149 mutant. While ROC1 and ROC1S58F clearly inhibited cell death triggered by pmRIN4 and RPM1 expression, they were unable to affect cell death triggered by pmRIN4ΔP149 and RPM1 (Figures 6E and S6E). These results further supported that the RIN4 Pro149-specified conformation is subject to regulation by ROC1.
We asked if pmRIN4 and RIN4ΔP149 similarly impact RPS2 function in Nb plants. While coexpression of WT RIN4 strongly inhibited cell death triggered by RPS2 overexpression, pmRIN4 and RIN4ΔP149 were less capable of inhibiting RPS2 cell death (Figures S6F and S6G), suggesting that the two RIN4 mutations may impact both RPM1 and RPS2 functions.
We next examined effect of RIN4ΔP149 mutation on the AvrB-induced RIN4 phosphorylation in Nb plants. Consistent with previous results (Liu et al., 2011), expression of AvrB strongly induced Thr166 phosphorylation of a T7-tagged RIN4 protein (Figure 6F). Unexpectedly, a constitutive Thr166 phosphorylation was observed when T7-RIN4ΔP149 was expressed in Nb plants. This result suggested that the Pro149-specified conformation also plays a role in RIN4 phosphorylation.
To substantiate the results obtained from the transient expression experiments, we generated stable transgenic Arabidopsis plants carrying various RIN4 mutant forms in the rps2 rin4 double mutant and rpm1 rps2 rin4 triple mutant background. As expected, pmRIN4 T1 transgenic plants of the rps2 rin4 background were often dwarfed or even died at early stages of development, whereas those of the rpm1 rps2 rin4 background were completely normal (Figures 7A and S7A), a phenotype indicative of constitutive RPM1 activation. The phenotype correlated with the amount of mutant RIN4 protein in these plants (Figure S7A). In contrast, rps2 rin4 pmRIN4P149V plants were completely normal, indicating that Pro149 is essential for pmRIN4 to activate RPM1. rps2 rin4 RIN4ΔP149 transgenic plants also developed dwarf or lethal phenotype, whereas rpm1 rps2 rin4 RIN4ΔP149 plants were completely normal, indicating that RIN4ΔP149 constitutively activates immunity in an RPM1-dependent manner. To determine if the autoimmune phenotype of RIN4ΔP149 was caused by a constitutive Thr166 phosphorylation of this mutant protein, we generated transgenic plants in which RIN4 Thr166 was substituted with Ala. The rps2 rin4 RIN4T166A ΔP149 plants showed similar phenotypes, as did rps2 rin4 RIN4ΔP149 plants (Figures 7A and S7A), indicating that RIN4ΔP149 activates RPM1 in the absence of Thr166 phosphorylation.
Figure 7. RIN4 Pro149 Plays a Critical Role in RPM1 Activation in Arabidopsis Plants.
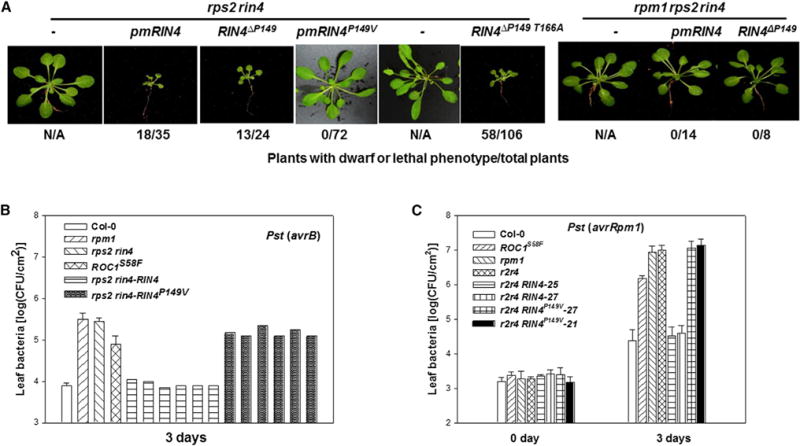
(A) The RIN4ΔP149 mutation triggers RPM1-dependent dwarfism independent of RIN4 phosphorylation. rps2 rin4 double mutant and rpm1 rps2 rin4 triple mutant plants were transformed with the indicated RIN4 constructs under the control of the native RIN4 promoter, and the morphological phenotype of representative T1 plants were photographed 5 weeks after germination. Numbers under the photograph indicate ratios of plants with dwarf or lethal phenotype to normal plants. (B and C) RIN4 Pro149 is essential for Arabidopsis resistance to Pst (avrB) and Pst (avrRpm1). Plants of the indicated genotypes were infiltrated with Pst (avrB) (B) or Pst (avrRpm1) (C), and bacterial growth in the leaf was determined 3 days postinoculation. In (B), six individual T1 transgenic plants were tested for each construct. In (C), two independent T2 transgenic lines were tested. Error bars indicate SD (n = 8). See also Figure S7.
We further examined RPM1 disease resistance in T1 transgenic plants carrying WT RIN4 and RIN4P149V (Figure 7B). The nontransgenic rps2 rin4 and rpm1 plants were fully susceptible to Pst (avrB) and supported a high level of bacterial growth. The RPM1 resistance was fully restored in rps2 rin4 RIN4 plants. In contrast, rps2 rin4 RIN4P149V plants were fully susceptible to Pst (avrB). All plants showed identical susceptibility to Pst (Figure S7C). Inoculation of selected T2 plants with Pst (avrRpm1) further showed that the rps2 rin4 RIN4P149V transgenic plants were completely abolished in RPM1 resistance (Figure 7C). Luciferase complementation assay indicated that both RIN4 and RIN4P149V interacted equally well with RPM1 (Figure S7D), further supporting that the Pro149-specified conformation, but not a lack of RPM1-interaction, is responsible for the phenotype.
DISCUSSION
In this study, our in-depth analyses revealed a negative regulation of RPM1 and RPS2 immunity by ROC1. ROC1 exerts its regulation through a direct interaction with RIN4. Its positive role in AvrRpt2 maturation and negative role in RPM1 and RPS2 activation suggest a previously unknown mechanism of ROC1 in immune regulation. We further identified a conformation switch specified by RIN4 Pro149 for RPM1 and RPS2 regulation and showed that this switch is regulated by ROC1.
In addition to ROC1, three other immunophilin genes have been shown to affect plant susceptibility to P. syringae (Pogorelko et al., 2014), highlighting the importance of this family during immune signaling. It is intriguing that ROC1 is exploited by P. syringae for AvrRpt2 maturation (Coaker et al., 2005). This may reflect a tight association of AvrRpt2 with host immune system during host-pathogen coevolution. Alternatively, AvrRpt2 may target a host substrate of ROC1 for virulence.
ROC1S58F enables more efficient RIN4 cleavage by AvrRpt2, and its extract allows more rapid autoprocessing of AvrRpt2, suggesting that ROC1S58F possesses greater PPIase activity. Several lines of evidence indicate that ROC1 plays a negative role in RPM1 and RPS2 immunity. Overexpression of WT ROC1 and ROC1S58F inhibited the RPS2 and RPM1 HR in Nb plants. Likewise, the ROC1S58F mutant and stable transgenic plants overexpressing ROC1 and ROC1S58F were compromised in resistance to Pst (avrB). Overexpression of ROC1S58F inhibited RPM1 and RPS2 immunity more strongly than did the overexpression of WT ROC1, suggesting that the ROC1 PPIase activity is positively correlated with its ability to inhibit immunity. Indeed, ROC1 mutations known to impair PPIase activity largely abolished its ability to inhibit RPS2 and RPM1 HR. Most importantly, silencing of ROC1 in Arabidopsis enhanced disease resistance to Pst (avrB).
The ability of ROC1 and ROC1S58F to inhibit RPS2- and RPM1-triggered HR in Nb plants independent of effectors indicates that ROC1 regulates RPS2 and RPM1 resistance through a host protein. Indeed, GST pull-down, luciferase complementation, coIP, and colocalization experiments showed a specific ROC1-RIN4 interaction. The ROC1S58F mutation does not affect this interaction, a result consistent with an elevated ROC1 PPIase activity being responsible for the inhibition of RPM1 HR. These led to the hypothesis that RIN4 is a substrate of ROC1 PPIase. Indeed, we found that ROC1 catalyzed the isomerization of a RIN4 peptide spanning Pro149. The pmRIN4-triggered RPM1 HR in Nb plants was specifically abolished by a RIN4 Pro149 to Val substitution, but not other Pro to Val substitutions, in the RIN4 C terminus. Furthermore, the Arabidopsis RIN4P149V mutant was completely unable to activate RPM1 resistance to Pst (avrB). In contrast, the RIN4ΔP149 mutation constitutively activates RPM1 HR in Nb plants and causes dwarfism in Arabidopsis plants. The RIN4P149V and RIN4ΔP149 mutant proteins likely adopt opposite conformations. Whereas the conformation of RIN4P149V is inhibitory to RPM1, the RIN4ΔP149 conformation favors RPM1 activation. Interestingly, the RPM1 HR triggered by the pmRIN4ΔP149 mutant is no longer inhibited by ROC1 or ROC1S58F, whereas the HR triggered by pmRIN4 is sensitive to ROC1 inhibition. These results strongly support the possibility that ROC1 maintains RIN4 in a conformation that is inhibitory to RPM1 and RPS2 activation. A regulatory role of ROC1 in NLR activation may not be limited to RPM1 and RPS2, as multiple pathogen effectors have been shown to interact with RIN4 (Luo et al., 2009; Wilton et al., 2010). Indeed, ROC1 RNAi lines displayed elevated resistance to Pst, but not Pst hrcC− mutant bacteria, which is consistent with a role of ROC1 in inhibiting ETI. The results suggest the presence of additional NLRs that weakly recognize effectors in Pst.
The AvrB-induced phosphorylation of RIN4 Thr166 plays a crucial role in RPM1 activation (Chung et al., 2011; Liu et al., 2011). Our analyses showed that RIN4ΔP149 is constitutively phosphorylated in plants, supporting a role of RIN4 conformation in the regulation of phosphorylation. At the first glance, these results were consistent with the current model in which RIN4 Thr166 phosphorylation is directly sensed by RPM1. However, pmRIN4P149V is unable to trigger RPM1 immunity, indicating that phosphorylation can be uncoupled from activation of RPM1 in the context of P149V. Furthermore, the RIN4T166AΔP149 mutant still caused dwarfism in Arabidopsis plants in a RPM1-dependent manner, indicating that RIN4ΔP149 constitutively activates immune responses in the absence of Thr166 phosphorylation. Thus, the conformation adopted by the RIN4ΔP149 mutant protein can uncouple the requirement of Thr166 phosphorylation for RPM1 activation.
NLR proteins have been shown to recognize a change in the general fold of an effector target. For example, the RPS5 NLR indirectly recognizes the P. syringae AvrPphB effector, which acts as a protease and cleaves the PBS1 kinase (Shao et al., 2003). Recent experiments indicated that the requirement of PBS1 cleavage for activating RPS5 can be bypassed, as a 5 aa insertion in PBS1 surrounding the cleavage site was sufficient to trigger RPS5 activation (DeYoung et al., 2012). In light of our findings, it is possible that RPM1 senses conformational changes in RIN4 in the region surrounding aa 149–166 and that both Thr166 phosphorylation and ΔPro149 mutation render sufficient changes in RIN4 conformation to activate RPM1. However, this model does not take into account the ROC1-RIN4 dissociation after RIN4 Thr166 phosphorylation. A more plausible explanation is that the Thr166 phosphorylation is indirectly sensed by RPM1, likely by impeding RIN4-ROC1 interaction. It should be noted, however, that the remaining RIN4-ROC1 interaction after RIN4 phosphorylation still allows ROC1S58F or the overexpressed ROC1 to dampen RPM1-specified defenses. Taken together, we suggest a model for RPM1 activation by AvrB (Figure S7E). The ROC1 PPIase activity maintains RIN4 in a configuration resembling RIN4P149V that is unable to activate RPM1. The AvrB-induced RIN4 Thr166 phosphorylation interferes with RIN4 Pro149 isomerization by ROC1, allowing the accumulation of a pool of RIN4 with a conformation similar to that of RIN4ΔP149, triggering RPM1 immunity.
EXPERIMENTAL PROCEDURES
Plant Materials
Arabidopsis thaliana plants used include WT (Col-0), rps2-101C (Mindrinos et al., 1994), rin4 rps2 double mutant and rpm1 rps2 rin4 triple mutant (Kim et al., 2005), nproRPS2::RPS2-HA (Axtell and Staskawicz), rpm1 (formerly described as rps3-1; Bisgrove et al., 1994), rps5-2 (Warren et al., 1998), and ROC1S58F and transgenic lines silenced for ROC1 (Ma et al., 2013). Arabidopsis plants were grown in a growth room at 20°C (night) and 24°C (day) with a 10 hr light/14 hr dark photoperiod or otherwise indicated. N. benthamiana plants were grown in a growth room at 24°C with a 10 hr light/14 hr dark photoperiod.
Bacterial Strains, Bacterial Growth, and HR Assay in Arabidopsis Plants
Pst strains used include DC3000, DC3000 (avrRpt2), DC3000 (avrB), DC3000 (avrRpm1), DC3000 (avrPphB), and DC3000 hrcC− mutant. For bacterial growth assay, bacteria were inoculated at a concentration of 1 × 106 CFU/ml with a needleless syringe. For HR assays in Arabidopsis, bacteria were infiltrated at a concentration of 5 × 107 CFU/ml.
Stable Transgenic Plants, Agrobacterium-Mediated Transient Expression, HR, Luciferase Complementation, and Protein Colocalization in N. benthamiana
The 35S::ROC1-FLAG and 35S-ROC1S58F-FLAG constructs were introduced into Arabidopsis Col-0, and WT RIN4, RIN4T166E, RIN4P149V, RIN4ΔP149, and RIN4 T166A,ΔP149 constructs under the native RIN4 promoter were introduced into rps2 rin4 double mutant or rpm1 rps2 rin4 triple mutant plants through Agrobacterium-mediated transformation.
For HR and cell death assay in N. benthamiana plants, Agrobacterium strains carrying the desired constructs were grown on LB plates for 30 hr, then cultured at 28°C in LB media for 12 hr. For ROC1 inhibition of RPS2- and RPM1-dependent HR, Agrobacterium was infiltrated at 4 × 108 CFU/ml (ROC1 constructs) and 7.5 × 107 CFU/ml (RPM1, RPS2, and RIN4 constructs). For RPM1-dependent HR triggered by RIN4 and its derivatives, 2 × 108 CFU/ml bacteria were infiltrated for each construct. HR development was documented by directly visualizing leaf collapse or staining with trypan blue at the indicated times.
For luciferase complementation assays, Agrobacterium containing the desired constructs was infiltrated into Nb leaves at the following concentrations: 4 × 108 CFU/ml for RIN4 constructs, 8 × 108 CFU/ml for RPM1 and RPS2 constructs, and 2 × 108 CFU/ml for ROC1 constructs. Leaf discs were taken 24 hr later, incubated with 1 mM luciferin in a 96-well plate, and luminescence was recorded with the GLOMAX 96 microplate luminometer (Promega). Each data point consisted of at least eight replicates.
For protein colocalization experiments, Nb leaves transiently expressing the desired constructs were mounted in water, and images were captured under a confocal microscope (Leica TCS SP5).
Plant Protein Extraction, CoIP, and Immunoblotting
Total protein was extracted from plants by grinding tissues in 100 μl grinding buffer (25 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM DTT, and plant protease inhibitor cocktail). Debris was removed by centrifugation at 14,000 rpm for 10 min. Samples were separated on SDS-PAGE, and immunoblotting was performed by using standard protocols with anti-RIN4, anti-RIN4 pT166, anti-T7, anti-HSP90, anti-FLAG, anti-Cluc, or anti-HA antibodies.
For coIP assays, total protein extracts from 10-day-old Col-0 or 35S::ROC1-FLAG transgenic seedlings were subject to anti-FLAG IP. Total protein was incubated with agarose-conjugated anti-FLAG antibody for 4 hr. Immunoprecipitates were washed five times with a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1mM DTT, 0.1% Triton X-100, and protease inhibitor cocktail. The resulting protein was separated by a 15% NuPAGE gel (Invitrogen), and the presence of RIN4, ROC1-FLAG was detected by immunoblot.
For the AvrRpt2-induced disappearance of RIN4, Pst (avrRpt2) was infiltrated into Col-0 leaves at a concentration of 5 × 107 CFU/ml for 4 hr, and total protein was isolated for immunoblotting.
For Pst (avrB)-induced RIN4 phosphorylation in Arabidopsis plants, 5-week-old plants grown under 11/13 hr light-dark cycle were syringe-infiltrated with Pst (avrB) or empty vector (pVSP61). Leaf samples were harvested at 2 hr after the onset of HR for protein extraction. For AvrB-induced RIN4 phosphorylation in Nb plants, Agrobacterium containing T7-tagged RIN4 constructs were syringe infiltrated in Nb plants. Agrobacterium carrying the GFP or AvrB construct was then syringe infiltrated into the same leaf areas 16 hr after the first infiltration. Leaf samples were harvested at 24 hr after the second infiltration. Protein samples were subject to immunoblot analyses as described (Liu et al., 2011).
AvrRpt2 Cleavage and GST Pull-Down Assays
For AvrRpt2 autocleavage, 20 μg AvrRpt2-His was incubated with 10 μg of Arabidopsis crude extract in a total volume of 100 μl in a buffer containing 100 mM Tris-HCl (pH 7.5) and 10% glycerol at 20°C. Aliquots (10 μl) of the reaction mixture were withdrawn at various time points and examined by 15% SDS-PAGE and Coomassie brilliant blue staining.
For GST pull-down assays, soluble GST-fusion protein was immobilized on G beads (GE), incubated with His-tagged proteins, and extensively washed before the bound protein was eluted for immunoblot analysis (Cui et al., 2010).
NMR Analysis of Proline Isomerization
All NMR samples contained 1.2 mM RIN4 peptide spanning Pro149 144-KVTVVPKFGDWD-155 in 20 mM sodium phosphate, 50 mM NaCl (pH 6.5) with 90% H2O/10% D2O and 0.01% DSS. 2D 1H-1H total correlation spectroscopy (TOCSY) experiment (mixing time of 75 ms), and 2D 1H-1H ROESY experiment (mixing time of 200 ms) data were collected for the peptide NMR samples with 48 μM or without GST-ROC1 on Bruker Avance 500 or 700 MHz spectrometers at 293K. Partial 1H signals assignments of the peptide were obtained based on TOCSY and ROESY spectra. Two distinct sets of 1H NMR signals were observed for some residues in the TOCSY and ROESY spectra due to the existence of both cis and trans conformations for the proline.
Supplementary Material
Acknowledgments
The authors would like to thank Fred Ausubel and David Mackey for sharing plant materials and Brian Staskawicz for helpful discussions. J.M.Z. was funded by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant number XDB11020200), Chinese Ministry of Science and Technology (2011CB100700), and Chinese Natural Science Foundation (31230007). D.L. was funded by Chinese Natural Science Foundation (31370290) and Chinese Ministry of Agriculture (2014ZX0800932B). G.C., Y.H.C., and K.Y. were funded by a grant from the National Institutes of Health (RO1GM092772) and the UC Davis Research Investments in Science and Engineering (RISE RI-091). All NMR experiments were carried out at Beijing Nuclear Magnetic Resonance Center.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2014.09.007.
References
- Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol. 2003;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- Bisgrove SR, Simonich MT, Smith NM, Sattler A, Innes RW. A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell. 1994;6:927–933. doi: 10.1105/tpc.6.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas ME, Lim E, Heitman J. Mutations that perturb cyclophilin A ligand binding pocket confer cyclosporin A resistance in Saccharomyces cerevisiae. J Biol Chem. 1995;270:20997–21002. doi: 10.1074/jbc.270.36.20997. [DOI] [PubMed] [Google Scholar]
- Chen AP, Wang GL, Qu ZL, Lu CX, Liu N, Wang F, Xia GX. Ectopic expression of ThCYP1, a stress-responsive cyclophilin gene from Thellungiella halophila, confers salt tolerance in fission yeast and tobacco cells. Plant Cell Rep. 2007;26:237–245. doi: 10.1007/s00299-006-0238-y. [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe. 2011;9:125–136. doi: 10.1016/j.chom.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coaker G, Falick A, Staskawicz B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science. 2005;308:548–550. doi: 10.1126/science.1108633. [DOI] [PubMed] [Google Scholar]
- Cui H, Wang Y, Xue L, Chu J, Yan C, Fu J, Chen M, Innes RW, Zhou JM. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe. 2010;7:164–175. doi: 10.1016/j.chom.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Huang J, Chisholm ST, Li D, Staskawicz BJ. Molecular basis for the RIN4 negative regulation of RPS2d-isease resistance. Plant Cell. 2005;17:1292–1305. doi: 10.1105/tpc.104.030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Singer AU, Wu AJ, McNulty BC, Musselwhite L, Nimchuk Z, Sondek J, Dangl JL. Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog. 2007;3:e48. doi: 10.1371/journal.ppat.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Qi D, Kim SH, Burke TP, Innes RW. Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol. 2012;14:1071–1084. doi: 10.1111/j.1462-5822.2012.01779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Solis JR, He Z, Lima A, Ting JL, Buchanan BB, Luan S. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc Natl Acad Sci USA. 2008;105:16386–16391. doi: 10.1073/pnas.0808204105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou JM. A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature. 2012;485:114–118. doi: 10.1038/nature10962. [DOI] [PubMed] [Google Scholar]
- Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- Keestra AM, Winter MG, Auburger JJ, Frässle SP, Xavier MN, Winter SE, Kim A, Poon V, Ravesloot MM, Waldenmaier JF, et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature. 2013;496:233–237. doi: 10.1038/nature12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA. 2005;102:6496–6501. [Google Scholar]
- Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJ, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe. 2011;9:137–146. doi: 10.1016/j.chom.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Lane WS, Schreiber SL. pCyP B: a chloroplast-localized, heat shock-responsive cyclophilin from fava bean. Plant Cell. 1994;6:885–892. doi: 10.1105/tpc.6.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Caldwell KS, Wroblewski T, Wright ME, Michelmore RW. Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell. 2009;21:2458–2472. doi: 10.1105/tpc.107.056044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Song L, Yang Y, Liu D. A gain-of-function mutation in the ROC1 gene alters plant architecture in Arabidopsis. New Phytol. 2013;197:751–762. doi: 10.1111/nph.12056. [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol. 2011;12:817–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucyn TS, Wu AJ, Balmuth AL, Arasteh JM, Rathjen JP. Regulation of tomato Prf by Pto-like protein kinases. Mol Plant Microbe Interact. 2009;22:391–401. doi: 10.1094/MPMI-22-4-0391. [DOI] [PubMed] [Google Scholar]
- Pogorelko GV, Mokryakova M, Fursova OV, Abdeeva I, Piruzian ES, Bruskin SA. Characterization of three Arabidopsis thaliana immunophilin genes involved in the plant defense response against Pseudomonas syringae. Gene. 2014;538:12–22. doi: 10.1016/j.gene.2014.01.029. [DOI] [PubMed] [Google Scholar]
- Qi D, Innes RW. Recent advances in plant NLR structure, function, localization, and signaling. Front Immunol. 2013;4:348. doi: 10.3389/fimmu.2013.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano PGN, Horton P, Gray JE. The Arabidopsis cyclophilin gene family. Plant Physiol. 2004;134:1268–1282. doi: 10.1104/pp.103.022160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- Stamnes MA, Rutherford SL, Zuker CS. Cyclophilins: a new family of proteins involved in intracellular folding. Trends Cell Biol. 1992;2:272–276. doi: 10.1016/0962-8924(92)90200-7. [DOI] [PubMed] [Google Scholar]
- Tang X, Frederick RD, Zhou JM, Halterman DA, Jia Y, Martin GB. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- Trupkin SA, Mora-García S, Casal JJ. The cyclophilin ROC1 links phytochrome and cryptochrome to brassinosteroid sensitivity. Plant J. 2012;71:712–723. doi: 10.1111/j.1365-313X.2012.05013.x. [DOI] [PubMed] [Google Scholar]
- Warren RF, Henk A, Mowery P, Holub E, Innes RW. A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell. 1998;10:1439–1452. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M, Subramaniam R, Elmore J, Felsensteiner C, Coaker G, Desveaux D. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc Natl Acad Sci USA. 2010;107:2349–2354. doi: 10.1073/pnas.0904739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li B, Xu Y, Li H, Li S, Zhang D, Mao Z, Guo S, Yang C, Weng Y, Chong K. The cyclophilin CYP20-2 modulates the conformation of BRASSINAZOLE-RESISTANT1, which binds the promoter of FLOWERING LOCUS D to regulate flowering in Arabidopsis. Plant Cell. 2013;25:2504–2521. doi: 10.1105/tpc.113.110296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


