Abstract
The coiled-coil is one of the most ubiquitous and well studied protein structural motifs. Significant effort has been devoted to dissecting subtle variations of the typical heptad repeat sequence pattern that can designate larger topological features such as relative α-helical orientation and oligomer size. Here we report the X-ray structure of a model coiled-coil peptide, HA2-Del-L2seM, which forms an unanticipated core antiparallel dimer with potential sites for discrete higher-order multimerization (trimer or tetramer). In the X-ray structure, a third, partially-ordered α-helix is weakly associated with the antiparallel dimer and analytical ultracentrifugation experiments indicate the peptide forms a well-defined tetramer in solution. The HA2-Del-L2seM sequence is closely related to a parent model peptide, HA2-Del, which we previously reported adopts a parallel trimer; HA2-Del-L2seM differs by only hydrophobic leucine to selenomethione mutations and thus this subtle difference is sufficient to switch both relative α-helical topology and number of α-helices participating in the coiled-coil. Comparison of the X-ray structures of HA2-Del-L2seM (reported here) with the HA2-Del parent (reported previously) reveals novel interactions involving the selenomethionine residues that promote antiparallel coiled-coil configuration and preclude parallel trimer formation. These novel atomic insights are instructive for understanding subtle features that can affect coiled-coil topology and provide additional information for design of antiparallel coiled-coils.
Keywords: Coiled-coil, protein folding, protein structure
INTRODUCTION
The coiled-coil, consisting of two or more associating α-helices, is one of the most common protein structural elements.1–3 This motif plays critical roles in important biological processes such as transcription, motility, and membrane fusion. Designed coiled-coils have been applied to a variety of applications including development of therapeutic proteins, peptide materials, and self-assembling/self-propagating systems.4–7 Coiled-coil sequences are characterized by a regular pattern known as the heptad repeat, with positions denoted abcdefg where a and d are hydrophobic (generally aliphatic) residues.4,8,9 When sequences with a heptad repeat adopt an α-helical conformation, this pattern creates a stripe of hydrophobic residues along one side of the α-helix whose burial by packing of multiple α-helices against one another provides the driving force for folding and oligomerization. The typical core coiled-coil packing arrangement is termed “knobs-into-holes” packing whereby the side chain of a and d positions (“knobs”) insert into cavities formed by core (a or d) and peripheral (e and g) side chains of the opposing α-helix.9 The identity and attributes of the knobs and holes, as well as distortions in this optimized repeat pattern, can affect larger topological features such as superhelical diameter, pitch, and number of associating α-helices.10
Much effort has been devoted toward understanding subtle sequence variations that can affect coiled-coil topology. A prime example is the study of factors that influence parallel vs. antiparallel configuration. Although both parallel and antiparallel coiled-coils contain knobs-into-holes packing, the particular preferences and requirements among the “constellation” of side chain interactions differs.11–13 For example, in two-stranded parallel coiled-coils, a knobs insert into a’-d’-g’-d’ holes, but in antiparallel coiled-coils, the hole consists of residues at e’-a’-d’-a’ positions (prime indicates positions on the opposing α-helix). Several studies have illustrated how core steric matching, or combinations of appropriately matched core polar residues and electrostatic complementation among e and g positions of opposing α-helices can specify α-helical orientation preferences.15–17 In these examples, dramatic alterations to the basic core (either by inclusion of polar residues or much smaller or larger residues than normally found at core positions) are required to designate antiparallel topology. Other work has focused on using large datasets to predict sequences that are prone to adopt a parallel or antiparallel configuration.17–19
Here we describe an antiparallel coiled-coil X-ray structure of a peptide known as HA2-Del-L2seM. The sequence of this peptide is derived from the central trimeric, parallel coiled-coil of the low pH conformation of influenza HA2 (HA2-Del).20 This fact that HA2-Del-L2seM adopts an antiparallel configuration is somewhat surprising given that its sequence differs from a previously characterized parent peptide, a parallel trimer knowns as “HA2-Del”, by two conservative core Leu → selenomethionine (seMet) mutations.30 A mechanism for this switch in strand orientation is provided by comparison of the HA2-Del-L2seM and HA2-Del structures, and provides novel insight into aspects that can control α-helix orientation in coiled-coil proteins.
MATERIALS AND METHODS
Peptide Synthesis
HA2-Del-L2seM was synthesized by solid-phase peptide synthesis using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry on an ABI-433A peptide synthesizer with Rink Amide resin. Following synthesis, simultaneous side chain deprotection and cleavage of the peptide was achieved by treating the resin with a mixture of 95% trifluoroacetic acid (TFA), 2.5% 1,2-ethanedithiol, and 2.5% thioanisole for 3 hrs. The resin was removed by filtration, and the crude peptide precipitated in cold diethyl ether, pelleted by centrifugation, and then the resuspended in water/acetonitrile. The peptide was purified by reverse-phase HPLC on a Vydac C18 column (10 µm, 250 × 21.2 mm) with water/acetonitrile mobile phases containing 0.1% TFA. The purity was greater than 95% as judged by analytical HPLC, and identity was confirmed by MALDI-MS. The peptide was dissolved in either 10 mM sodium phosphate buffer (pH 7.0) or 10 mM sodium acetate buffer (pH 4.5) and used for subsequent studies. The peptide concentration was determined by absorbance at 280 nm.
X-Ray Crystallography
Diffraction quality crystals were grown by sitting drop vapor diffusion by mixing 1 µL of protein (concentration was 5.7 mg/mL in 10 mM NaH2PO4, pH 7.5) with 1 µL of reservoir solution. The reservoir solution contained 10% PEG 4000, 5% 2-propanol, 0.1 M HEPES:NaOH, pH 7.5. Crystals of HA2-DelEx-L2M with dimensions 0.1 × 0.1 × 0.2 mm3, were mounted in cryo-loops directly from the crystallization droplet and flash-cooled in liquid nitrogen. Prior to freezing, the droplet was supplemented with 20% glycerol as a cryoprotectant. Diffraction data were recorded on a Rayonix 225 HE CCD detector (Ryonyx, L.L.C., Evanston, IL, USA) with 0.979 Å wavelength radiation on the LRL-CAT beamline (Advanced Photon Source, Argone, IL). Intensities were integrated using the HKL2000 program and reduced to amplitudes using the SCALEPACK2MTZ program (see Table 1 for statistics).21–23 The structure was solved using Single Anomalous Dispersion (SAD) phasing and model building with PHENIX24. Final model building and refinement were performed with the programs COOT and REFMAC, respectively.25,26 The quality of the final structures was verified with composite omit maps, and stereochemistry was checked with MOLPROBITY.27 LSQKAB and SSM algorithms were used for structural superpositions.23,28 All other calculations were conducted using CCP4 program suite.23 The crystal structure of HA2-Del-L2seM was deposited with the Protein Data Bank under entry code 4S1X.
Table I.
Data Collection and Refinement Statistics for the HA2-Del-L2M Crystal Structure
| PDB | 4S1X |
|---|---|
| Data Collection | |
| Wavelength (Å) | 20-1.9 |
| Space group | P21 |
| Unit cell dimensions (Å) | a = 34.45, b = 98.52 |
| c = 34.55 | |
| α γ= 90°, β = 89.96° | |
| Resolution range (Å) | 20-1.9 |
| Observed reflections | 90,591 |
| Unique reflections | 18,167 |
| Completeness (%)a | 99.6 (99.9)a |
| I/ I | 14.3 (3.0) |
| R-merge (I) b | 0.077 (0.741) |
| Structure Refinement | |
| Rcryst (%) c | 0.208 (0.214) a |
| Rfree (%) c | 0.245 (0.310) a |
| Protein nonhydrogen atoms | 1668 |
| Water molecules | 54 |
| Average B-factor (Å2) | 41.59 |
| RMS Deviations from Ideal Value | |
| Bonds (Å) | 0.008 |
| Angles (°) | 1.15 |
| Torsion angles (°) | 16.8 |
| Overall coordinate error based on R-factor | 0.035 |
| Ramachandran statistics (%) | |
| (for non-Gly/Pro residues) | |
| most favorable | 99.52 |
| additional allowed | 0.48 |
Values in parentheses indicate statistics for the high resolution bin.
Rmerge = ∑∑j|Ij(hkl) – <I(hkl)>|/ ∑∑j |<I(hkl)>|, where Ij is the intensity measurement for reflection j and <I> is the mean intensity over j reflections.
Rcryst/(Rfree) = ∑ ||Fo(hkl)| – |Fc(hkl)||/ ∑ |Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively. No s-cutoff was applied. 5% of the reflections were excluded from refinement and used to calculate Rfree.
Circular Dichroism
Measurements were performed on a Jasco J-815 spectrometer with a 1 mm quartz cuvette at 57 µM peptide concentration. CD wavelength scans were obtained with a 0.1 nm step size and a 2 sec averaging time. The raw signal was corrected for background and converted to molar ellipticity (θ).
Analytical Ultracentrifugation
Analysis was performed on a Beckman XL-1 analytical ultracentrifuge with a Ti60 rotor. The sample was loaded into double sector cells at 114 µM with reference buffer (10 mM sodium acetate, pH 4.5) in the second sector. Data were acquired at 58,000 rpm at 20 °C with 200 scans. Sedimentation boundaries were monitored by absorption at 280 nm. The sedimentation boundaries were directly fit as the derivative dc/dt using DCDT1 v2.4.029 to determine the sedimentation and diffusion coefficients, S and D, respectively from which was calculated the apparent molecular weight, MWapp. Sixty to 80 absorbance scans were globally analyzed for each experiment. The observed values were normalized to standard conditions of 20 °C and water (s20,w and D20,w) by correcting for buffer density and viscosity. Buffer density, viscosity, and v(bar) were calculated using Sednterp (http://sednterp.unh.edu/#).
RESULTS
Peptide Design
We previously described peptide HA2-Del, a variant of the central, trimeric coiled-coil region from the low pH structure of influenza hemagglutinin HA2 (Figure 1).20 HA2-Del was part of a designed series of peptides to probe the effects of a heptad repeat stutter that is found in HA2. The HA2-Del sequence contains elements of the WT HA2 coiled-coil but with four residues comprising the heptad repeat stutter deleted. We found that HA2-Del adopted a parallel, trimeric coiled-coil that is pH-dependent, forming structure at pH 4.5 but remaining unfolded at pH 7. This behavior was similar to variants that contain the WT stutter, but HA2-Del stability at pH 4.5 was enhanced despite its shorter length, and there was a stronger dependence on lower pH for HA2-Del coiled-coil formation. Both of these differences relative to WT HA2 (containing the stutter) were attributed to optimized knobs-into-holes packing in HA2-Del that resulted from removal of the stutter. An X-ray structure of HA2-Del provided insight into the molecular basis for the effects of removing the stutter.
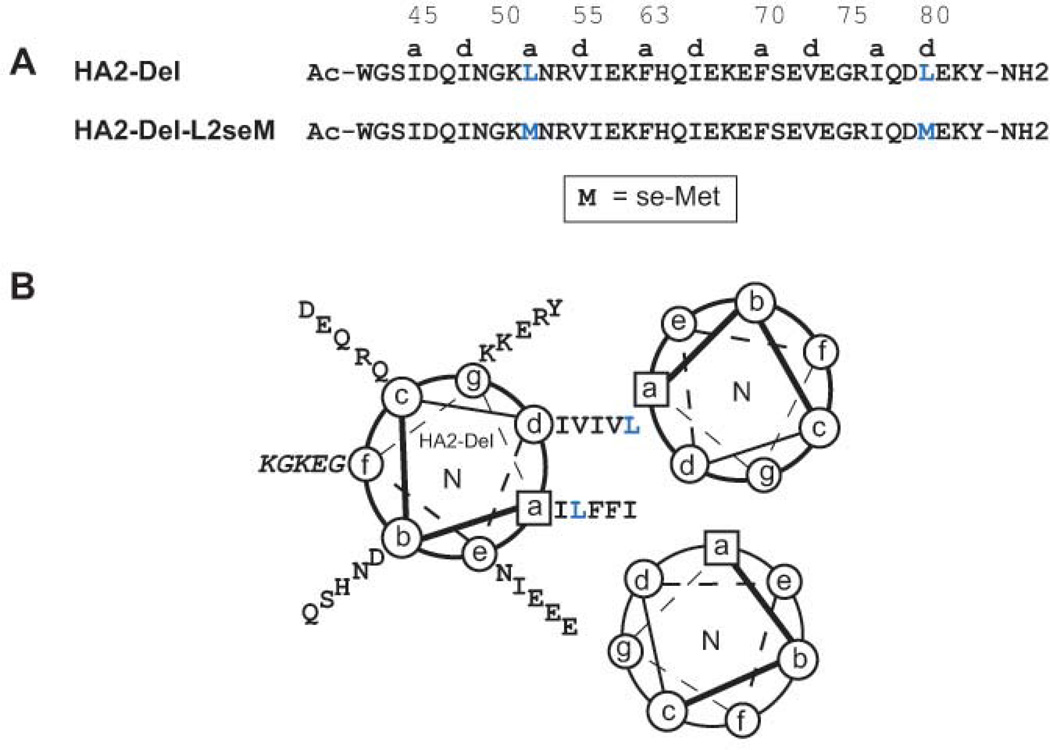
Figure 1. Peptide design.
(A) Sequence of HA2-Del and HA2-Del-L2seM. HA2 numbering shown on top; since HA2-Del contains a four-residue deletion relative to the WT HA2, the numbering skips from 58 to 63 in order to maintain consistency with HA2. The two Leu→seMet mutations are indicated in blue. (B) Helical wheel diagram of HA2-Del and packing arrangement in a parallel trimer.
During the course of our studies on HA2-Del, we were prompted to produce several selenomethione- (seMet-) containing mutants of this peptide in order to aid with phasing of X-ray diffraction data. One such analog, HA2-Del-L2seM, contains core a and d Leu→seMet mutations at positions 52 and 80 (HA2 numbering). HA2-Del-L2seM was produced by standard solid phase peptide synthesis with N- and C-termini blocked by acetyl and amide groups, respectively. MALDI-MS of the peptide product yielded a [MH+]obs of 4750.9 Da, consistent with the expected [MH+]exp of 4747.9 Da.
X-ray Structure of HA2-Del-L2seM
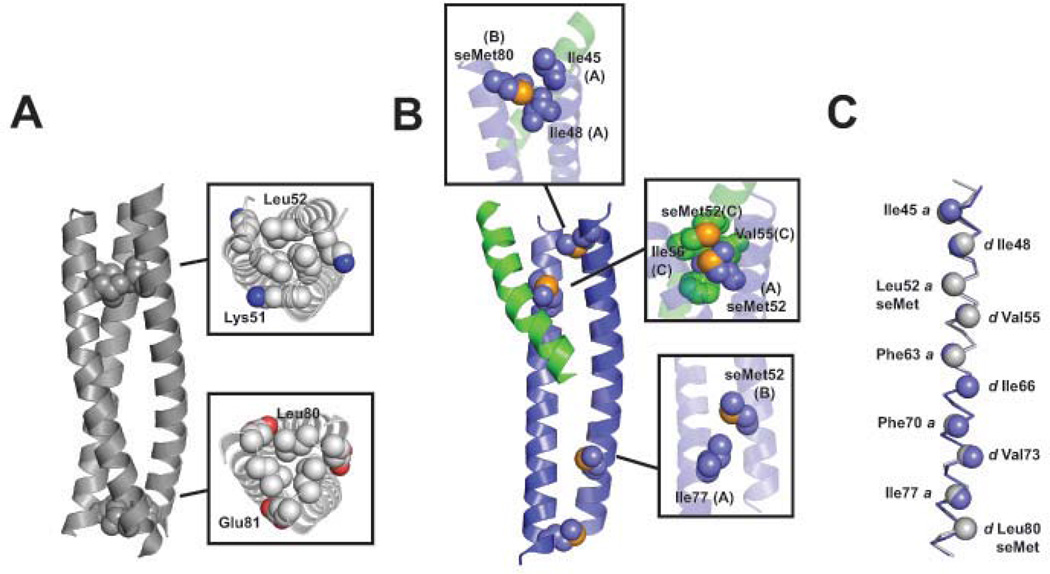
Crystals of HA2-Del-L2seM diffracted readily and the anomalous signal from selenium atoms was used for direct phase determination. Data were processed initially in space group P41 but, upon refinement, it became clear that P41 is an apparent space group due to pseudo-symmetry, and the actual space group is P21 (Table 1). The final three-dimensional structure of HA2-Del-L2seM was refined at 1.9 Å resolution. Surprisingly, the asymmetric unit contained two well-ordered core antiparallel two-stranded coiled-coils instead of the expected parallel three-stranded coiled-coil as in the HA2-Del parent structure.20 Additional α-helical density was apparent near each of the two antiparallel dimers in the asymmetric unit, indicating that a third chain binds to each dimer in irregular fashion (Figure 2). The electron density for the third α-helix is much weaker than the corresponding densities for the first two chains, suggesting only partial occupancy and significant disorder. Electron density corresponding to the N-terminal segment of this third chain, residues Ile45-Phe70, could be assigned but other portions were not visible. The C-terminal end of this partially bound third α-helix is presumably disordered, pointing away from the midsection of the core antiparallel dimer (Figure 1D).
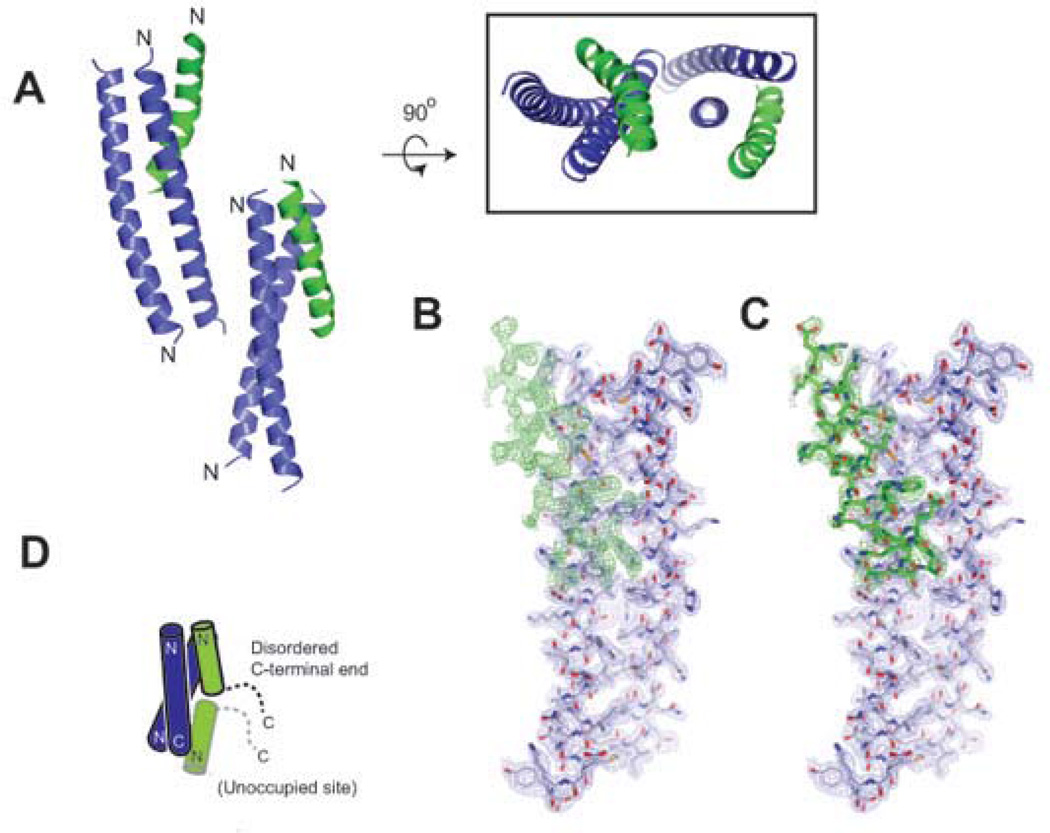
Figure 2. X-ray structure of HA2-Del-L2seM.
(A) Side and top view of the overall arrangement of asymmetric unit containing two antiparallel core dimers (blue), each associated with a third partially ordered helix (green) in irregular fashion. The N-terminus of each chain is indicated. (B) Electron density for the third α-helix is weaker than for the core antiparallel dimer. After exhausting refinement of the antiparallel dimer, weaker unaccounted density (green) became apparent. (C) Residues Ile45-Phe70 fit into the density with only partial occupancy. (D) Potential C2 symmetric tetramer, with partial binding of two α-helices on the antiparallel core dimer. The C-terminal end of the observed third chain (green with black outline) was disordered. Positioning of a potential (but unoccupied) binding site (green with grey outline) is shown. Although this additional binding site is shown here in head-to-head arrangement with the other green α-helix, it is possible that this could bind in the other orientation.
The root mean square deviation (RMSD) values between four well-ordered chains are 1.2 Å for Cα atoms, and 1.6 Å for all atoms. A potential reason for partial disorder of the third α-helix in this irregular trimer is because the well-ordered antiparallel dimer has two-fold symmetry, and the third α-helix can bind to it in two mutually exclusive orientations. The crystal lattice selects only one of them, and since the sequence is not optimized for this binding mode, the site is only partially occupied. In solution, the stable two-stranded antiparallel coiled-coil binding groove could potentially accommodate two of the HA2-Del-L2seM fragments (Ile45-Phe70) at either end without overlaps, therefore forming a staggered four-stranded coiled-coil with C2 symmetry (Figure 2d).
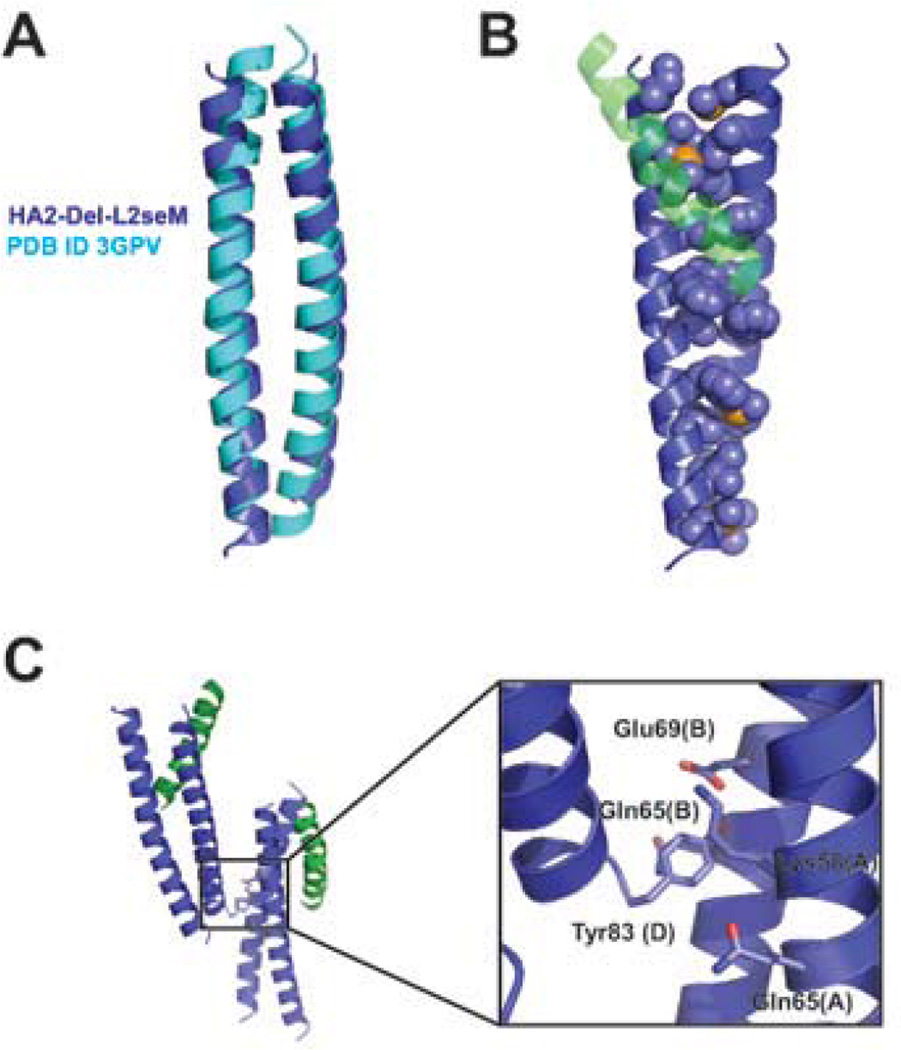
Within a single α-helical pairing that forms the well-defined antiparallel dimer, there is a distorted interhelical packing arrangement yielding decreased superhelical pitch relative to a model antiparallel coiled-coil (e.g., PDB ID 3GPV) (Figure 3a). This distorted packing arrangement leads to non-canonical core interactions within the dimer, generally the side chains participate in interdigitating interactions but the canonical a knob into e’-a’-d’-a’ hole and d knob into g’-d’-a’-d’ hole constellations are disrupted.11–14 Instead, the translation of the hydrophobic a/d stripe along the α-helical axis, caused by the imperfect 7.2 residues/2 turns parameter of an α-helix vs. 7 residue heptad repeat, results in exposure of a and d residues on either end of the α-helices (Figure 3b). These exposed residues form the hydrophobic groove into which the partially resolved α-helix packs at one end of the well-defined antiparallel dimer. This interface involves hydrophobic a and d residues on all three α-helices, critical a/d contacts include: Ile45, Ile48, seMet52, Val55, Phe63 on Chain A of one symmetry partner (of the well-defined dimer); Phe63, Ile66, Phe70, Val73, Ile77, and seMet80 on Chain B (well-defined dimer); and Ile48, seMet52, Val55, Phe63, Ile66, and Phe70 on Chain C (partially ordered third α-helix) (Figure 3b). In addition, a peripheral residue, Ile56 (e position), of both Chain A and Chain B also contributes to this hydrophobic interface, as do a number of other peripheral interactions (a full list of contacts between Chain C and Chains A and B is provided in the Supporting Information). The interface between the two symmetry-related dimers appears to consist mainly of burial of the C-terminal Tyr83 (g position) on one of the well-defined dimers (Chain D) into a pocket comprised of aliphatic portions of Lys58, Gln65 on Chain A, and Gln65 and Glu69 on Chain B (Figure 3c) on the antiparallel dimer symmetry mate.
Figure 3. Interhelical interfaces.
(A) There is a larger superhelical pitch for the HA2-Del-L2seM core antiparallel dimer in comparison to a model antiparallel coiled-coil from MerR (PDB ID 3GPV). (B) Side chains in a and d positions of the core antiparallel dimer are exposed at either end, creating a hydrophobic groove against which the partially-ordered third α-helix binds in the crystal lattice (shown as opaque green) at the top end, and an unoccupied hydrophobic surface on the bottom end. (C) Interface between symmetry-related antiparallel dimers involves insertion of Tyr83 into a cluster of residues.
Effects of the Two Leu→seMet Mutations
Comparison of the parent parallel trimer (HA2-Del) with the HA2-Del-L2seM structure provides some insight into why the two conservative Leu→seMet mutations have such a significant impact on relative α-helix topology (Figure 4). Both Leu52 (a position) and Leu80 (d position) on HA2-Del participate in core packing interactions with the corresponding residues on opposing α-helices, forming a- and d-layer arrangement as has been previously described for parallel trimeric coiled-coils. At position Leu52 in particular, the a-layer is tightly packed with an outer layer comprised of methylenes from Lys51 residues (g position). Although the packing is not as tight at Leu80, which is near the end of the α-helix rather than the center, a similar outerlayer interaction is formed with the aliphatic portion of neighboring Glu81 (e position). In both cases, mutation from Leu→seMet in HA2-Del-L2seM increases the length of the core side chain and likely precludes this tight packing arrangement. Instead, the side chains of seMet participate in offset interactions within the antiparallel dimer, and one of the seMet residues (seMet52 of Chain A) extends into a pocket on the third partially-bound α-helix consisting of seMet52, Val55, Ile56, and Phe63. However, the partial occupancy of this third α-helix highlights the fact that these interactions are not optimized.
Figure 4. Comparison of interactions in HA2-Del and HA2-Del-L2seM.
Overall structures of HA2-Del (A, PDB ID 4P67) and HA2-Del-L2seM (B) with close-up views of interactions discussed in the text. (C) The relative positioning of core a and d residues (identity indicated) is similar within a single α-helix.
These packing disruptions also likely cause the switch from parallel to antiparallel α-helix orientation. Specifically, the terminal Leu80 of the parent HA2-Del peptide is tightly nestled with the corresponding residues from adjacent α-helices, but this packing would be sterically disfavored with two or more methinonine residues. Instead, the C-terminally located seMet80 of HA2-Del-L2seM participates in an extended interaction with Ile45 and Ile48 on the N-terminal end of the adjacent α-helix and thus an antiparallel arrangement, at least among two of the α-helices, is able to satisfy requirements to bury the bulkier seMet residue at this end. The packing of seMet52 is mostly peripheral to the adjacent α-helix that forms the well-defined dimer, but interactions with Ile77 appear to bury at least part of the seMet methylenes.
Although incorporation of the two seMet side chains alters the optimal packing arrangement required for the parallel trimer, it is interesting to note that many of the side chains within a single α-helix are in similar locations as the parent HA2Del peptide with the exception of the C-terminal end containing the seMet80. At the C-terminal end, the overall decreased superhelical pitch of the HA2-Del trimer relative to the HA2-Del-L2seM dimer causes an overall “straightening” of this end of the α-helix. However, relative position of most a/d positions within a single α-helix are unchanged and this is likely why binding of the antiparallel dimer with the partially disordered α-helix is accommodated in the crystal lattice. Many aspects of the hydrophobic groove into which the third α-helix would bind in the HA2-Del trimer are maintained for one partner of the antiparallel dimer in HA2-Del-seMet.
Solution Characterization
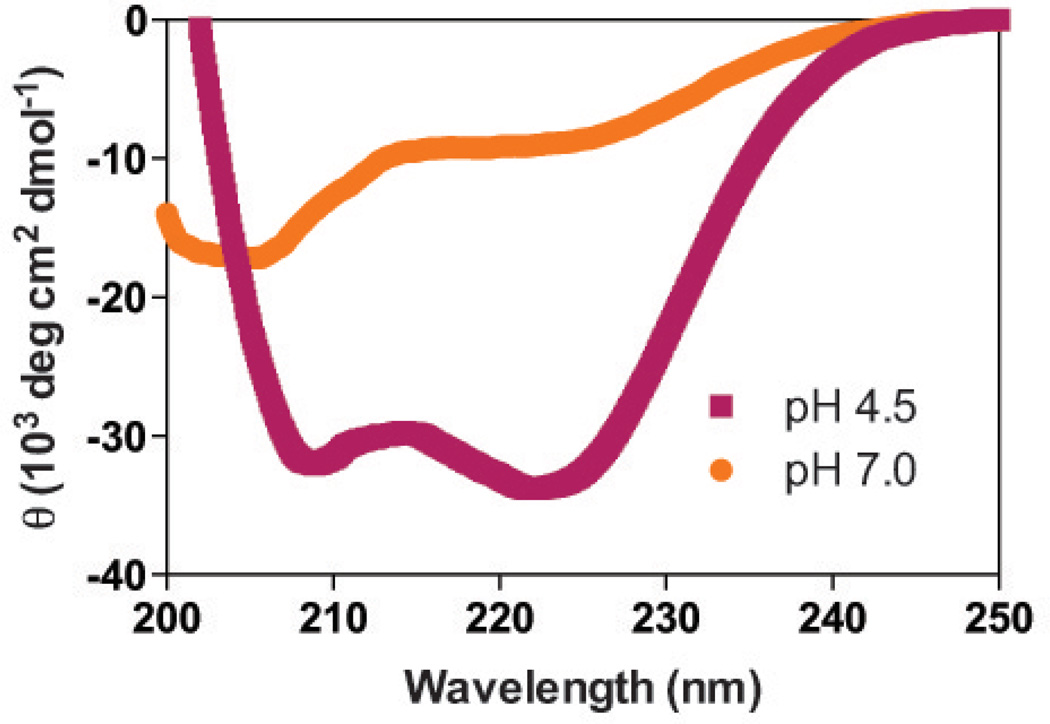
Circular dichroism (CD) indicates that HA2-Del-L2seM forms a pH-dependent α-helical structure, with α-helicity much more pronounced at lower pH (4.5) than at neutral pH (7.0). This pH-dependent behavior was also reported for HA2-Del and is consistent with the low-pH-activated fusion behavior of influenza HA2, which requires formation of this coiled-coil for membrane fusion in low pH environments of the endosome.20,30,31 In other systems, we have attributed such pH-dependent behavior to potentially repulsive side chain-side chain interactions among two or more Glu or Asp residues (termed Ani-Ani interactions) or a histidine and a cationic residue (His-Cat).32–36 Alterations in pH affect the protonation states of Glu/Asp or His side chains, changing the charge state and thus proximity of such residues to other charged residues in either pre- or post-fusion conformations can dictate which state is preferred under various pH conditions. For low pH-dependent stability of the coiled-coil from Ebola virus, Marburg virus, and CAS virus glycoprotein GP2, Ani-Ani interactions at core or surface positions destabilize the coiled-coil at neutral pH.33–36 Such repulsive interactions are attenuated or eliminated at lower pH, due to protonation of Glu/Asp residues and elimination of their negative charge. However, analysis of the HA2-Del-L2seM structure reveals no Ani-Ani interactions. Instead, there appears to be a single salt bridge between Glu69 and Lys82 on opposing α-helices of the antiparallel dimer and potentially repulsive interactions between Lys51 and Arg76 again on opposing α-helices. Glu69 was implicated as a potential residue involved in network of interactions involving Glu74 and Glu67 in the WT HA2 peptide and here potentially could mediate pH-dependence via salt bridge with Lys68.20
Velocity analytical ultracentrifugation (AUC) at pH 4.5 indicate that HA2-Del-L2seM sediments as an ideal single species with observed MW of 19.2 kDa (see Supporting Information), corresponding most closely to a tetramer (19 kDa). Based on the X-ray structure, it is possible that two antiparallel α-helical dimers associate in solution in the same way as do the symmetry-related twins in the crystal lattice to form a define tetramer. Alternatively, a third α-helix could bind the antiparallel dimer in the same mode as the third partially-bound α-helix of the crystal lattice, and a forth α-helix at another site on the dimer. However, this is conjecture only as CD and AUC do not provide any information about relative α-helix topology in solution.
DISCUSSION
Previous efforts to explore residue-specific interactions that designate antiparallel coiled-coil formation have focused on polar or electronic matching at core or flanking sites, or hydrophobic steric matching among large and small side chains at core positions.14–16 However, it is clear from large-scale sequence and host-guest studies in model antiparallel coiled-coils that subtle core effects can also contribute to relative α-helix orientation.11–13, 17–19 The work described herein provides an example of how seemingly conservative differences between Leu and seMet residues at two positions within a coiled-coil sequence can have a dramatic effect on relative strand topology. Comprehensive host-guest analysis for antiparallel coiled-coil mutations by Gellman and coworkers focused on those residues found most commonly at the core, Leu, Ile, and Val; surprisingly little is known about preferences for Met or seMet at these positions despite the abundance of Met at many intermolecular interfaces.11–13, 37 In one study, a core Met residue was identified from a phage library to an appropriate pairing to a core Ala on an apposing α-helix.38 In the case of HA2-Del-L2seMet, whether or not there is a dependence on the large selenium as opposed to sulfur of methionine was not explored. However, the X-ray structure of HA2-Del-L2seM demonstrates that the longer, linear aliphatic side chain of seMet is likely a significant cause of the effects observed here.
The partial occupancy of a third α-helix associated in a hydrophobic groove with each antiparallel dimer suggests that the two Leu→seMet alterations precluded parallel trimer formation but nonetheless, other features of the coiled-coil that promote trimer formation (such as a hybrophobic groove) are maintained. It is interesting that this constraint can be satisfied within the crystal lattice by partial binding of a third α-helix. In addition, the interaction between the two antiparallel dimer symmetry mates, whose contacts are a result of the change in both topology and α-helix orientation, is quite surprising. Based on the crystal packing arrangement, it seems there are two plausible possibilities for formation of a well-defined tetramer in solution. The first is that the two antiparallel dimers associate in the same way as the symmetry mates in the X-ray structure. In this model, it is anticipated that the location of binding for the third α-helix remains free in solution. Although a solution KD was not measured for binding of this third α-helix, the weak electron density for this segment indicates only partial occupancy and is consistent with weak binding. The peptide concentration of the crystallization (2.9 mg/mL ~ 600 µM) was at least 5-fold higher than the CD and AUC experiments (57 µM and 114 µM; the peptide concentration likely increased during the crystallization process), and thus it is possible that the third α-helix does not bind at the lower concentrations, which would explain why higher order oligomers are not observed. However, if this model were correct, it would be somewhat surprising because the interface between the two symmetry mates is not extensive (insertion of a single Tyr83 into a pocket). Typical interhelical interactions in larger coiled-coils span multiple turns of the α-helix and involve interdigitating or knobs-into-holes interactions.8–10
The second possibility is that two hydrophobic grooves on the antiparallel dimer exist but the ordered crystal lattice permits observation of partial binding in only one site. This would create a tetramer consisting of the core antiparallel dimer and two α-helices offset, including one that binds in the site that is partially occupied in the crystal structure. A second unoccupied hydrophobic groove on the surface of the antiparallel core dimer is apparent, a result of the inherent symmetry of the dimer, and consisting of similar residues that comprise the bound site. Since the partially-bound third α-helix N-terminus resides at the central portion of the groove in which it occupies, it is conceivable that another α-helix could bind at the adjacent groove with either head-to-head or head-to-tail topology relative to the third α-helix. However, no evidence for this fourth binding mode was observed in the X-ray structure, and the fact that the third α-helix has weak electron density is not consistent with a well-defined oligomer in solution. The crystals were grown at pH 7.5, and CD data show that α-helical structure is attenuated at neutral pH relative to pH 4.5, at which condition the AUC data were obtained. It is therefore possible that stronger associations of the core antiparallel dimer with additional α-helices occurs at lower pH, giving rise to a well-defined tetramer that was not visualized in the crystal lattice at pH 7.5. The solution experiments performed here do not provide unambiguous assignment of relative α-helical topology and thus it is still possible that the solution tetramer forms an alternative arrangement.
Although coiled-coils have been studied extensively, most of the analysis has focused on parallel coiled-coil systems, despite the fact that antiparallel coiled-coils are highly abundant.1–4 The HA2-Del-L2seM structure reported here provides a model system to study factors influence stability of such systems, and principles obtained from this analysis can be integrated to further improve prediction and design algorithms.
Supplementary Material
Figure 5. Circular dichroism of HA2-Del-L2seM.
Acknowledgments
We thank Michael Brenowitz for assistance with analytical ultracentrifugation experiments and Rafael Toro for assistance with crystallization. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Experimental support from the LRL-CAT team is gratefully acknowledged.
Contract grant sponsor: National Institutes of Health
Contract grant numbers: R01-AI090249; U54-GM094662; P30-CA13330
ABBREVIATIONS
- Fmoc
N-(9-fluorenyl)methoxycarbonyl
- TFA
trifluoroacetic acid
- seMet
selenomethionine
- CD
circular dichroism
- AUC
analytical ultracentrifugation
- RMSD
root mean square deviation
REFERENCES
- 1.Woolfson DN. The design of coiled-coil structures and assemblies. Adv. Prot. Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 2.Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Curr. Opin. Struct. Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lupas AN, Gruber M. The structure of α-helical coiled coils. Adv. Prot. Chem. 2005;70:73–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 4.Regan L, DeGrado WF. Characterization of a helical protein designed from first principles. Science. 1998;241:976–978. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- 5.Root MJ, Kay MS, Kim PS. Protein design of an HIV-1 entry inhibitor. Science. 2001;291:884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher JM, Harniman RL, Barnes FR, Boyle AL, Collins A, Mantell J, Sharp TH, Antognozzi M, Booth PJ, Linden N, Miles MJ, Sessions RB, Verkade P, Woolfson DN. Self-assembling cages from coiled-coil peptide modules. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee DH, Granja JR, Martinez JA, Severin K, Ghadiri MR. A self-replicating peptide. Nature. 1997;389:706–709. doi: 10.1038/382525a0. [DOI] [PubMed] [Google Scholar]
- 8.O’Shea EK, Lumb KJ, Kim PS. Peptide ‘Velcro’: Design of a heterodimeric coiled coil. Curr. Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 9.Crick FHC. The packing of α-helices: Simple coiled-coils. Acta Crystallographica. 1953;6:689–697. [Google Scholar]
- 10.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 11.Hadley EB, Testa OD, Woolfson DN, Gellman SH. Preferred side-chain constellations at antiparallel coiled-coil interfaces. Proc. Natl. Acad. Sci. USA. 2008;105:530–535. doi: 10.1073/pnas.0709068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steinkruger JD, Bartlett GJ, Hadley EB, Fay L, Woolfson DN, Gellman SH. The d'--d--d' vertical triad is less discriminating than the a'--a--a' vertical triad in the antiparallel coiled-coil dimer motif. J. Am. Chem. Soc. 2012;134:2626–2633. doi: 10.1021/ja208855x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinkruger JD, Bartlett GJ, Woolfson DN, Gellman SH. Strong contributions from vertical triads to helix-partner preferences in parallel coiled coils. J. Am. Chem. Soc. 2012;134:15652–15655. doi: 10.1021/ja3063088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnarr NA, Kennan AJ. Strand orientation by steric matching: a designed antiparallel coiled-coil trimer. J. Am. Chem. Soc. 2004;126:14447–14451. doi: 10.1021/ja047496v. [DOI] [PubMed] [Google Scholar]
- 15.Gurnon DG, Whitaker JA, Oakley MG. Design and characterization of a homodimeric antiparallel coiled-coil. J. Am. Chem. Soc. 2003;125:7518–7519. doi: 10.1021/ja0357590. [DOI] [PubMed] [Google Scholar]
- 16.McClain DL, Woods HL, Oakley MG. Design and characterization of a heterodimeric coiled coil that forms exclusively with an antiparallel relative helix orientation. J. Am. Chem. Soc. 2001;123:3151–3152. doi: 10.1021/ja004099l. [DOI] [PubMed] [Google Scholar]
- 17.Apgar JR, Gutwin KN, Keating AE. Predicting helix orientation for coiled-coil dimers. Proteins. 2008;72:1048–1065. doi: 10.1002/prot.22118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szczepaniak K, Lach G, Bujnicki JM, Dunin-Horkawicz S. Designability landscape reveals sequence features that define axial helix rotation in four-helical homo-oligomeric antiparallel coiled-coil structures. J. Struct. Biol. 2014;188:123–133. doi: 10.1016/j.jsb.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Ramos J, Lazaridis T. Computational analysis of residue contributions to coiled-coil topology. Protein Sci. 2011;20:1845–1855. doi: 10.1002/pro.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins CD, Malashkevich VN, Almo SC, Lai JR. Influence of a heptad repeat stutter on the pH-dependent conformational behavior of the central coiled-coil from influenza hemagglutinin HA2. Proteins. 2014;82:2220–2228. doi: 10.1002/prot.24585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.French S, Wilson K. On the treatment of negative intensity observations. Acta Crystallogr. A. 1978;34:517–525. [Google Scholar]
- 23.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams PD, Afonine PV, Bunkóczi G, Chen VB, IW Davis IW, Echolsn N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 29.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 30.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 31.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 32.Harrison JS, Higgins CD, O'Meara MJ, Koellhoffer JF, Kuhlman BA, Lai JR. Role of electrostatic repulsion in controlling pH-dependent conformational changes of viral fusion proteins. Structure. 2013;21:1085–1096. doi: 10.1016/j.str.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison JS, Higgins CD, Chandran K, Lai JR. Designed protein mimics of the Ebola virus glycoprotein GP2 α-helical bundle: stability and pH effects. Protein Sci. 2011;20:1587–1596. doi: 10.1002/pro.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrison JS, Koellhoffer JF, Chandran K, Lai JR. Marburg virus glycoprotein GP2: pH-dependent stability of the ectodomain α-helical bundle. Biochemistry. 2012;51:2515–2525. doi: 10.1021/bi3000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koellhoffer JF, Malashkevich VN, Harrison JS, Toro R, Bhosle RC, Chandran K, Almo SC, Lai JR. Crystal structure of the Marburg virus GP2 core domain in its postfusion conformation. Biochemistry. 2012;51:7665–7675. doi: 10.1021/bi300976m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koellhoffer JF, Dai Z, Malashkevich VN, Stenglein MD, Liu Y, Toro R, Harrison JS, Chandran K, Derisi JL, Almo SC, Lai JR. Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species. J. Mol. Biol. 2014;426:1452–1468. doi: 10.1016/j.jmb.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gellman SH. On the role of methionine residues in the sequence-independent recognition of nonpolar protein surfaces. Biochemistry. 1991;30:6633–6636. doi: 10.1021/bi00241a001. [DOI] [PubMed] [Google Scholar]
- 38.Lai JR, Fisk JD, Weisblum B, Gellman SH. Hydrophobic core repacking in a coiled-coil dimer via phage display: insights into plasticity and specificity at a protein-protein interface. J. Am. Chem. Soc. 2004;126:10514–10515. doi: 10.1021/ja047710f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.