Abstract
In some cases of infertility in women, human oocytes fail to mature when they reach the metaphase II (MII) stage. Mitochondria plays an important role in oocyte maturation. A large number of mitochondrial DNA (mtDNA), copied in oocytes, is essential for providing adenosine triphosphate (ATP) during oocyte maturation. The purpose of this study was to identify the relationship between transcript expression levels of the mitochondrial encoded gene (MT-CO1) and two nuclear encoded genes, nuclear respiratory factor 1 (NRF1) and mitochondrial transcription factor A (TFAM) in various stages of human oocyte maturation. Nine consenting patients, age 21–35 years old, with male factors were selected for ovarian stimulation and intracytoplasmic sperm injection (ICSI) procedures. mRNA levels of mitochondrial-related genes were performed by singlecell TaqMan® quantitative real-time polymerase chain reaction (qRT-PCR). There was no significant relationship between the relative expression levels in germinal vesicle (GV) stage oocytes (p = 0.62). On the contrary, a significant relationship was seen between the relative expression levels of TFAM and NRF1 and the MT-CO1 genes at the stages of metaphase I (MI) and MII (p = 0.03 and p = 0.002). A relationship exists between the transcript expression levels of TFAM and NRF1, and MT-CO1 genes in various stages of human oocyte maturation.
Keywords: Mitochondria, Oocyte maturation, Quantitative real-time polymerase chain reaction (qRT-PCR), single-cell
INTRODUCTION
Oocyte maturation and oocyte quality are important parameters in successful reproductive results of the assisted reproduction technologies (ART). Mitochondria are the most abundant organelles in the cytoplasmic oocyte [1]. They are necessary for adenosine triphosphate (ATP) generation through oxidativephosphorylation (OXPHOS), unlike any other process of the cell, and depend highly on the expression of proteins encoded by the mitochondrial and nuclear encoded gene [1,2]. Mitochondria are inherited exclusively from the mother [3]. Indeed, mitochondrial function is associated with mitochondrial DNA (mtDNA) [4,5]. The mitochondrial genome encodes essential proteins, which are crucial for the generation of ATP. These proteins are transcribed and translated in the mitochondrial matrix [6]. Human oocyte mitochondrion has only a single copy of the genome, which is representative of the mitochondrial number. However, it has been clearly documented that mtDNA number is expanded during oocyte growth [7].
Mitochondrial biogenesis is managed independently from the nuclear genome and depend on regulatory coordination between the nuclear and mitochondrial genomes [8]. However, little is known about the regulation of mitochondrial gene expression, compared with nuclear genes [8]. Regulatory coordination of the nuclear and mitochondrial genes are important in cell survival and energy homeostasis [9]. Two products of these genes, NRF1 and TFAM, are well-known essential ubiquitous factors for the mtDNA replication and expression [10]. The NRF1 gene contributes in regulating the expression of nuclear encoded components of the mitochondrial respiratory chain [11]. The human TFAM is a 25 kD protein with nuclear-encoded high-mobility group (HMG) box protein, which plays an important role through sequence binding to the heavy strand promoter (HSP) and light strand promoter (LSP) sites in the D-loop of human mtDNA, a control region that regulates mtDNA transcription and replication [12–14]. The TFAM gene has various functions, including packaging mtDNA into a nucleoid-like complex and maintaining and repairing mtDNA molecules [15,16]. NRF1 has also been shown to be linked to the promoters of the TFAM gene [17]. Their important roles have been demonstrated by transgenic experiments, showing that depletion of mtDNA in TFAM and NRF1 homozygous knockout mice resulted in the animals’ deaths [18,19]. Another factor, MT-CO1, is a terminal component and one of the three genes of mitochondrial respiratory chain, encoded by the mtDNA [19–21]. Transcription levels of the MT-CO1 gene may possibly be an indirect indicator of the mtDNA metabolic activity.
Active transcription of the mitochondrial genome has been demonstrated to initiate at various developmental stages, depending on the species [22]. Reports from many studies suggested that gene-specific transcription factors directly affect gene transcription in mitochondria [23]. However, relationship between the expression levels of nuclear and mitochondrial encoded genes during human oocyte maturation is not well understood. The aim of the current study was to quantify the relationship between relative expression levels of NRF1 and TFAM, and the MT-CO1 genes in single human oocytes at various stages of the human oocyte maturation from germinal vesicle (GV) stage to metaphase II (MII) stage.
MATERIALS AND METHODS
Sample Collection
Oocytes in various stages of human oocyte maturation were obtained from nine normal women (20–35 years old), undergoing intracytoplasmic sperm injection (ICSI) treatment because of male factor infertility. There was no evidence of ovarian pathology for any of the patients donating oocytes. Examination proceeded according to the following criteria: 1) medical history, clinical examination and common hormonal tests; 2) Rotterdam criteria to ensure polycystic ovary syndrome; 3) diseases related to the endocrine system, such as hyperprolactinemia, thyroid dysfunction; 4) possible surgical history of the reproductive system; 5) normal ovulatory period (25–35 days); 6) body mass index (BMI) between 18.3–22.2 kg/m2; 7) day 3 basal ultrasonography and hormone profile with follicle-stimulating hormone (FSH) <10.0 mIU/mL, estradiol <40.0 pg/mL and antral follicle count (AFC) >6; and 8) no history of smoking.
Treatment and Donation
Informed consent were obtained from the patients at the Infertility and Reproductive Health Research Center (IRHRC) at Taleghani Hospital of Shahid Beheshti University of Medical Sciences and Sarem Women’s Hospital, Tehran, Iran. The study protocol was approved by the Ethical Committee of Shahid Beheshti University of Medical Sciences, Tehran, Iran. All nine patients were treated according to the agonist protocol. The gonadotropin-releasing hormone (GnRH) agonist was begun in mid-luteal phase of the previous cycle (day 21), as determined by serum E2 and P concentrations. Stimulation of the follicular growth, performed by FSH recombination, was started after sufficient down-regulation.
This continued by daily injections according to individual endocrine and ovarian ultrasonic responses until at least one 18 mm diameter follicle showed. Ovulation was induced by human chorionic gonadotropin (HCG). Thirty-six hours later, vaginal puncture was performed under ultrasound-guided in vitro fertilization (IVF) and ICSI protocols using transvaginal probes for follow-up of the cumulus-oocyte complex (COC). First, mechanical procedures used to remove cumulus granulose cells by intermittent pipetting and then followed by enzymatic hyaluronidase treatment. Oocytes were subsequently washed in culture media (global media) before use. Collected oocytes were studied for nuclear maturity under a stereo microscope (Olympus, Tokyo, Japan). Morphological characteristics of the collected oocytes were classified into three categories based on the nuclear condition: 1) mature oocytes (MII) with first polar body (n = 8); 2) immature oocytes GV (n = 10); and 3) immature oocytes [metaphase I (MI)] without original polar body (n = 9).
Sample Processing for Gene Expression
Oocytes were processed using Ambion Single Cell-to-CT® Kit (Life Technologies, Foster City, CA, USA) according to the manufacturer’s protocol. Sibling oocytes were rapidly transferred to sterile RNase-free microtubes to minimize any changes in gene expression. Then, oocytes were transferred to lysis solution containing DNase I. The enzyme was inactivated using stop solution following 5 min. of incubation at room temperature and DNA digestion. Then, Single Cell VILO® RT Mix and Single Cell SuperScript® RT (Life Technologies, Carlsbad City, CA, USA) were added to each sample reaction. Reverse transcription-PCR (RT-PCR), was performed using 15.5 μL sample volume per reaction. After completing RT-PCR, PreAmp® reaction mixtures were prepared by adding 0.2 × pooled TaqMan® Gene expression assay to Single Cell PreAmp® Mix (Life Technologies) for each gene. Then, cDNA of each gene was specifically amplified through the following thermal cycling condition: 95 °C for 10 min. and then 17 cycles that included 15 seconds at 95 °C and 4 min. at 60 °C and a final 10 min. heat at 99 °C.
Gene Expression Analysis
Quantitative RT-PCR was used to quantify mRNA transcript levels of TFAM, MT-CO1 and NRF1 genes using hypoxanthine phosphoribosyltransferase 1 (HPRT1) mRNA transcripts as endogenous reference. Relative mRNA expression levels of the studied genes were quantified using TaqMan® qRT-PCR analysis (Applied Biosystems, Foster City, CA, USA) in 48-well plates. Relative gene expression was calculated using the 2-ΔΔCT formula. The Ct value of each transcript was calculated via TaqMan® (Applied Biosystems) gene expression assay for transcripts.
Designing Primers and Probes
The specifications of the forward and reverse primers and Taq-Man® (Applied Biosystems) probes are listed in Table 1.
Table 1.
Primers and TaqMan® (Applied Biosystems) probe sequences used in quantitative real-time polymerase chain reaction.
| Gene | Forward Primer Sequences (5′>3′) | Reverse Primer Sequences (5′>3′) |
|---|---|---|
| NRF1 | GGC ACT GTC TCA CTT ATC CAG GT T | CAG CCA CGG CAG AAT AAT TCA |
| TFAM | AAG ATT CCA AGA AGC TAA GGG TGA | CAG AGT CAG ACA GAT TTT TTC CAG TTT |
| MT-CO1 | GAG CTG CTG TTC GGT GTC C | TGC CAG TGG TAG AGA TGG TTG |
| HPRT1 | TGG ACT AAT TAT GGA CAG GAC TGA AC | GCA CAC AGA GGG CTA CAA TGTG |
| Gene | Probe Sequences (5′>3′) | |
| NRF1 | FAM-ACC ACG GTC ACC GTT GCC CAA-BHQ1 | |
| TFAM | FAM-CAC CGC AGG AAA AGC TGA AGA CTG TAA AG-BHQ1 | |
| MT-CO1 | FAM-CAA TAC CGC AAC CGC ATT GCC AT-BHQ1 | |
| HPRT1 | FAM-CTC CCA TCT CCT TCA TCA CAT CTC GAG C-BHQ1 | |
Statistical Analysis
Analysis of variance (ANOVA) with the post-hoc Tukey test and the Statistical Package for the Social Sciences (SPSS Inc., Chicago, IL, USA) software, version 16.0, were used to determine differences between the mean values.
RESULTS
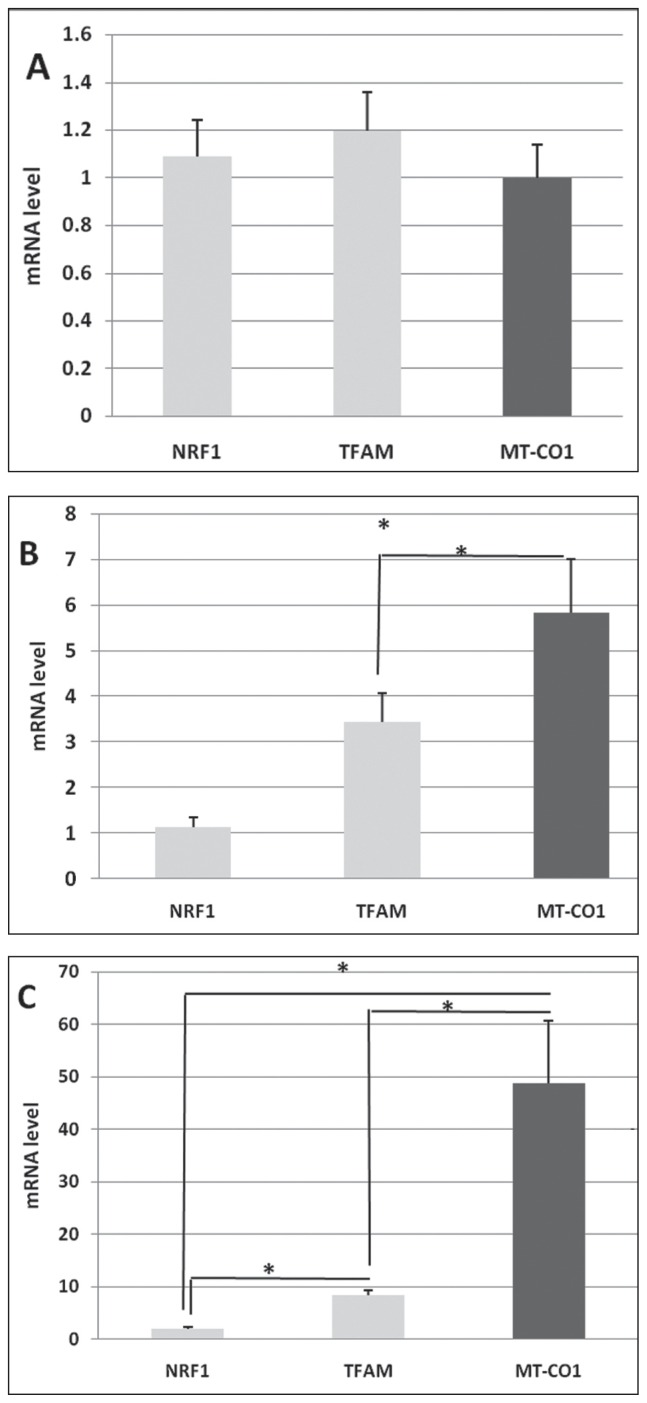
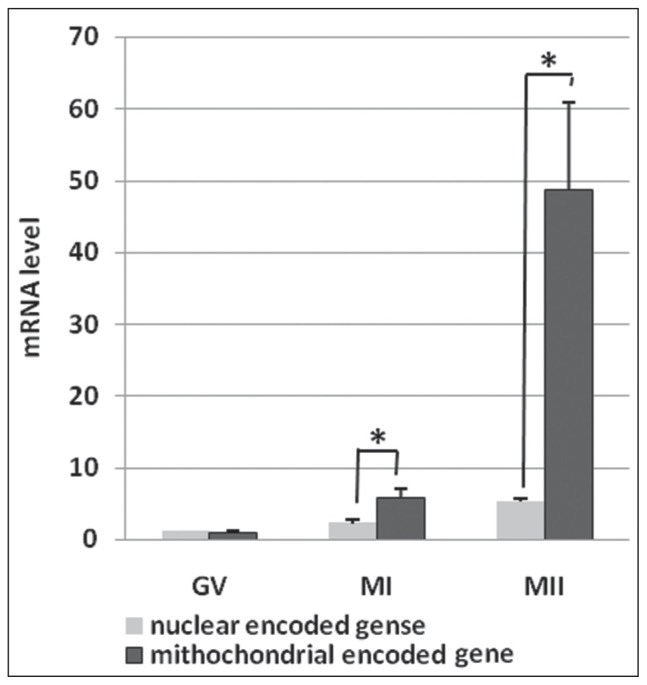
For the first time, the present study showed detailed temporal expression of mRNAs associated to mitochondria during human oocyte maturation. Results showed that the mean expression levels of TFAM and NRF1 were relatively low as well as the MT-CO1 genes in GV-stage oocytes. No significant difference was seen between the expression levels of all studied genes (p = 0.62) (Figure 1A). Significant differences were detected in the mean expression levels of TFAM and NRF1, and the MT-CO1 genes when the oocyte maturation began and transited to the MI stage (p = 0.03) (Figure 1B). At the MII stage, significant differences were reported between the expression levels of TFAM and NRF1, and the MT-CO1 (p = 0.002) genes (Figure 1C). Furthermore, significant differences were seen between the expression levels of NRF1 and TFAM, and the MT-CO1 genes at the MI and MII stages (Figure 2).
Figure 1.
Relationship between mitochondrial related mRNA expression levels of TFAM, NRF1 and MTCO1 genes at GV, MI, MII stages of human oocyte maturation (A, B, C, respectively). A: p = 0.62; B: p = 0.03; C: p = 0.002. Superscript asterisk (*) shows statically significant differences (<0.05). Data are presented as means ± standard deviation (SD).
Figure 2.
Relationship between the mean relative expression levels of nuclear encoded (TFAM, NRF1) and the mitochondrial encoded (MT-CO1) genes at various stages of the human oocyte maturation. GV: p = 0.58; MI: p = 0.23; MII: p = 0.012.
Superscript asterisk (*) shows statically significant differences (<0.05). Data are presented as means ± standard deviation (SD).
DISCUSSION
Oocytes require a large supply of ATP for maturation to support critical processes such as spindle formation, chromatid separation and cell division after fertilization [24–26]. Mitochondrial number increases during oocyte maturation at a direct ratio to increase copy number of mtDNA [27–29]. For the first time, this study identified the relationship between the relative expression levels of nuclear and mitochondrial encoded genes at various stages of human oocyte maturation using single-cell qRT-PCR. This method evaluated the relative gene expression levels in single oocytes without isolating RNA. In the present study, relationship between the relative expression levels of TFAM and NRF1, and the MT-CO1 genes was significant in MI and MII stage oocytes. Therefore, it can be concluded that nuclear-mitochondrial relationships occur by active transcriptions during human oocyte maturation.
No relationship was observed between mRNA expression levels of nuclear and mitochondrial encoded genes in immature oocytes (GV stage). However, contrary to the findings of the current study in GV-stage oocytes, a high expression level of TFAM was observed by increasing the relative expression level of the MT-CO1 gene. In MII-stage oocytes, a significant difference was detected between the relative mRNA levels of TFAM and NRF1, and the MT-CO1 genes. This can be explained by the response of regulation mechanism to changes in energy demand associated with a large number of mtDNA copies at this stage. Previous studies have shown that a relative expression level of TFAM results in further direct mtDNA replication [18,30, 31]. Furthermore, studies have reported that NRF1 transactivates the promoters of a number of mitochondrial-linked genes such as TFAM, including genes that encode respiratory chain subunits [32].
Results showed a high expression level of the nuclear encoded TFAM gene with important mitochondrial encoded MT-CO1 gene upregulation in MI- and MII-stage oocytes, which could confirm the activation of mtDNA heavy strand promoter by TFAM, causing the relative expression of mRNA encoding mitochondrial proteins such as cytochrome oxidase subunit 1 (COX1). This causes further differentiation of mitochondria in MI and MII stages [5,33–35]. It is commonly accepted that NRF1, TFAM and MT-CO1 genes serve as markers of mitochondrial differentiation [36–38]. Moreover, it seems that the increased expression of NRF1 positively affects TFAM binding to regulatory regions of the MT-CO1 promoter; therefore, TFAM is well known as a main mitochondrial transcription factor, regulating mtDNA copy number and mtDNA transcription activity [39].
However, a relationship has been observed between the upregulated gene expression of NRF1 and TFAM, and the MT-CO1 in MII-stage oocytes, compared to GV-stage human oocytes. Similarly, previous studies have reported that TFAM reduction resulted in decreased mitochondrial COX1 expression to lower than 30.0% of the normal expression level and caused embryonic lethality [25]. Another study has revealed that delayed expression of mtDNA replication factors result in delayed mtDNA replication and reduced development of oocyte competence [27].Other studies have also demonstrated that the homozygous knockout embryos of TFAM (TFAM [−/−]) die due to deficiencies in impaired respiratory chain [17,23]. Therefore, it may be concluded that increase in transcription levels in oocyte mitochondria results in increase in mtDNA due to demanded ATP levels during human oocyte maturation, which are important steps in oocyte maturation [40–42]. In conclusion, the present study has supported the fact that the relationship between the relative expression level of nuclear encoded genes (TFAM, NRF1) and the mitochondrial-encoded gene (MT-CO1) is associated with human oocyte maturation and subsequently, oocyte quality. However, further investigations with appropriate methodology are required to better understand unknown aspects of this relationship.
Footnotes
Declaration of Interest. The authors greatly appreciate the Sarem Women’s Hospital and Shahid Beheshti University of Medical Sciences, Tehran, Iran, for their support and for funding this thesis. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
REFERENCES
- 1.Reynier P, May-Panloup P, Chretien M, Morgan C, Jean M, Savagner F, et al. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Reprod. 2001;7(5):425–429. doi: 10.1093/molehr/7.5.425. [DOI] [PubMed] [Google Scholar]
- 2.Montoya J, López-Pérez MJ, Ruiz-Pesini E. Mitochondrial DNA transcription and diseases: Past, present and future. Biochim Biophys Acta. 2006;1757(9–10):1179–1189. doi: 10.1016/j.bbabio.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Ratchford AM, Chi MM, Schoeller E, Frolova A, Schedl T, et al. Maternal diabetes causes mitochondrial dysfunction and meiotic defects in murine oocytes. Mol Endocrinol. 2009;23(10):1603–1612. doi: 10.1210/me.2009-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Blerkom J. Mitochondria in early mammalian development. Semin Cell Dev Biol. 2009;20(3):354–364. doi: 10.1016/j.semcdb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Van Blerkom J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion. 2011;11(5):797–813. doi: 10.1016/j.mito.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Zhao J, Li Y. Adenosine triphosphate content in human unfertilized oocytes, undivided zygotes and embryos unsuitable for transfer or cryopreservation. J Int Med Res. 2012;40:734–739. doi: 10.1177/147323001204000238. [DOI] [PubMed] [Google Scholar]
- 7.Chiaratti MR, Meirelles FV. Mitochondrial DNA copy number, a marker of viability for oocytes. Biol Reprod. 2010;83(1):1–2. doi: 10.1095/biolreprod.110.084269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leigh-Brown S, Enriquez JA, Odom DT. Nuclear transcription factors in mammalian mitochondria. Genome Biol. 2010;11(7):1–9. doi: 10.1186/gb-2010-11-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotterill M, Harris SE, Collado Fernandez E, Lu J, Huntriss JD, Campbell BK, et al. The activity and copy number of mitochondrial DNA in ovine oocytes throughout oogenesis in vivo and during oocyte maturation in vitro. Mol Hum Reprod. 2013;19(7):444–450. doi: 10.1093/molehr/gat013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Curr Top Dev Biol. 2007;77:21–49. doi: 10.1016/S0070-2153(06)77002-8. [DOI] [PubMed] [Google Scholar]
- 11.Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J Cell Biochem. 2006;97(43):673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- 12.Lenka N, Vijayasarathy C, Mullick J, Avadhani NG. Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex. Prog Nucleic Acid Res Mol Biol. 1998;61:309–344. doi: 10.1016/s0079-6603(08)60830-2. [DOI] [PubMed] [Google Scholar]
- 13.Scarpulla RC. Transcriptional activators and co-activators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002;286(1):81–89. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- 14.Clayton DA. Nuclear-mitochondrial intergenomic communication. Biofactors. 1998;7(3):203–205. doi: 10.1002/biof.5520070307. [DOI] [PubMed] [Google Scholar]
- 15.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13(9):935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 16.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, et al. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24(22):9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang D, Hamasaki N. Mitochondrial transcription factor A in the maintenance of mitochondrial DNA. Ann N Y Acad Sci. 2005;1042:101–108. doi: 10.1196/annals.1338.010. [DOI] [PubMed] [Google Scholar]
- 18.Larsson N, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18(3):231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 19.Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21(2):644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002;1576(1–2):1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 21.Evans MJ, Scarpulla RC. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J Biol Chem. 1998;264(24):14361–14368. [PubMed] [Google Scholar]
- 22.Evans MJ, Scarpulla RC. NRF-1: A trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990;4(6):1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 23.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, et al. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31(6):1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stojkovic M, Machado SA, Stojkovic P, Zakhartchenko V, Hutzler P, Gonzalves PB. Mitochondrial distribution and adenosine triphosphate content of bovine oocytes before and after in vitro maturation: Correlation with morphological criteria and developmental capacity after in vitro fertilization and culture. Biol Reprod. 2001;64(3):904–909. doi: 10.1095/biolreprod64.3.904. [DOI] [PubMed] [Google Scholar]
- 25.May-Panloup P, Vignon X, Chrétien MF, Heyman Y, Tamassia M, Malthiéry Y, et al. Increase of mitochondrial DNA content and transcripts in early bovine embryogenesis associated with upregulation of mtTFA and NRF1 transcription factors. Reprod Biol Endocrinol. 2005;3:65. doi: 10.1186/1477-7827-3-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asin-Cayuela J, Gustafsson CM. Mitochondrial transcription and its regulation in mammalian cells. Trends Pharmacol Sci. 2007;32(3):111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Elshourbagy SH, Spikings EC, Freitas M, St John JC. Mitochondria directly influence fertilisation outcome in the pig. Reproduction. 2006;131(2):233–245. doi: 10.1530/rep.1.00551. [DOI] [PubMed] [Google Scholar]
- 28.Watson AJ. Oocyte cytoplasmic maturation: A key mediator of oocyte and embryo developmental competence. J Anim Sci. 2007;85(13):E1–E3. doi: 10.2527/jas.2006-432. [DOI] [PubMed] [Google Scholar]
- 29.Wilding M, Dale B, Marino M, Di Matteo L, Alviggi C, Pisaturo ML, et al. Mitochondrial aggregation patterns and activity in human oocytes and preimplantation embryos. Hum Reprod. 2001;16(5):909–917. doi: 10.1093/humrep/16.5.909. [DOI] [PubMed] [Google Scholar]
- 30.Pejznochova M, Tesarova M, Hansikova H, Magner M, Honzik T, Vinsova K, et al. Mitochondrial DNA content and expression of genes involved in mtDNA transcription, regulation and maintenance during human fetal development. Mitochondrion. 2010;10(4):321–329. doi: 10.1016/j.mito.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18(9):3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Q, Wu G, Lai L, Park K, Cabot R, Cheong H, et al. Translocation of active mitochondria during pig oocyte maturation, fertilization and early embryo development in vitro. Reproduction. 2001;122(1):155–163. [PubMed] [Google Scholar]
- 33.Forbes D, Jackman M, Bishop A, Thomas M, Kambadur R, Sharma M. Myostatin auto regulates its expression by feedback loop through Smad7 dependent mechanism. J Cell Physiol. 2006;206(1):264–272. doi: 10.1002/jcp.20477. [DOI] [PubMed] [Google Scholar]
- 34.Grumbach IM, Chen W, Mertens SA, Harrison DG. A negative feedback mechanism involving nitric oxide and nuclear factor kappa-B modulates endothelial nitric oxide synthase transcription. J Mol Cell Cardiol. 2005;39(4):595–603. doi: 10.1016/j.yjmcc.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Machatkova M, Jeseta M, Hulinska P, Knitlova D, Nemcova L, Kanka J. Characteristics of bovine oocytes with different meiotic competence in terms of their mitochondrial status and expression of nuclear encoded factors. Reprod Domest Anim. 2012;47(5):806–814. doi: 10.1111/j.1439-0531.2011.01972.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang LY, Wang DH, Zou XY, Xu CM. Mitochondrial functions on oocytes and preimplantation embryos. J Zhejiang Univ Sci B. 2009;10(7):483–492. doi: 10.1631/jzus.B0820379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catalá MG, Izquierdo D, Uzbekova S, Moraty R, Roura M, Romaguera R, Papillier P, et al. Brilliant cresyl blue stain selects largest oocytes with highest mitochondrial activity, maturation-promoting factor activity and embryo developmental competence in prepubertal sheep. Reproduction. 2011;142(4):517–527. doi: 10.1530/REP-10-0528. [DOI] [PubMed] [Google Scholar]
- 38.Bilodeau-Goeseels S, Schultz G. Changes in the relative abundance of various housekeeping gene transcripts in in vitro-produced early bovine embryos. Mol Reprod Dev. 1997;47(4):413–420. doi: 10.1002/(SICI)1098-2795(199708)47:4<413::AID-MRD7>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 39.Scarpulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann N Y Acad Sci. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dairaghi DJ, Shadel GS, Clayton DA. Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim Biophys Acta. 1995;1271(1):127–134. doi: 10.1016/0925-4439(95)00019-z. [DOI] [PubMed] [Google Scholar]
- 41.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 42.Duran HE, Simsek-Duran F, Oehninger SC, Jones HW, JR, Castora FJ. The association of reproductive senescence with mitochondrial quantity, function, and DNA integrity in human oocytes at different stages of maturation. Fertil Steril. 2011;96(2):384–388. doi: 10.1016/j.fertnstert.2011.05.056. [DOI] [PubMed] [Google Scholar]