Abstract
Inflammatory agonists differentially activate gene expression of the chemokine family of proteins in endothelial cells (EC). TNF is a weak inducer of the chemokine CXCL11, while TNF and IFN-γ costimulation results in potent CXCL11 induction. The molecular mechanisms underlying TNF plus IFN-γ-mediated CXCL11 induction are not fully understood. We have previously reported that the protein arginine methyltransferase PRMT5 catalyzes symmetrical dimethylation of the NF-κB subunit p65 in EC at multiple arginine residues. Methylation of Arg30 and Arg35 on p65 is critical for TNF induction of CXCL10 in EC. Here we show that PRMT5-mediated methylation of p65 at Arg174 is required for induction of CXCL11 when EC are costimulated with TNF and IFN-γ. Knockdown of PRMT5 by RNAi reduced CXCL11 mRNA and protein levels in costimulated cells. Reconstitution of p65 Arg174Ala or Arg174Lys mutants into EC that were depleted of endogenous p65 blunted TNF plus IFN-γ-mediated CXCL11 induction. Mass spectrometric analyses showed that p65 Arg174 arginine methylation is enhanced by TNF plus IFN-γ costimulation, and is catalyzed by PRMT5. Chromatin immunoprecipitation assays (ChIP) demonstrated that PRMT5 is necessary for p65 association with the CXCL11 promoter in response to TNF plus IFN-γ. Further, reconstitution of p65 Arg174Lys mutant in EC abrogated this p65 association with the CXCL11 promoter. Finally, ChIP and Re-ChIP assays revealed that symmetrical dimethylarginine-containing proteins complexed with the CXCL11 promoter were diminished in p65 Arg174Lys-reconstituted EC stimulated with TNF and IFN-γ. In total, these results indicate that PRMT5-mediated p65 methylation at Arg174 is essential for TNF plus IFN-γ-mediated CXCL11 gene induction. We therefore suggest that the use of recently developed small molecule inhibitors of PRMT5 may present a therapeutic approach to moderating chronic inflammatory pathologies.
Introduction
Activation of endothelial cells (EC) by inflammatory substances results in stimuli-specific induction of proteins that participate in EC-leukocyte interactions and leukocyte recruitment to inflammatory foci [1]. We have previously reported that arginine methylation of transcription factors catalyzed by PRMT5 potentiates expression of the leukocyte adhesion molecules VCAM1 and E-selectin, and the chemokines CXCL10 and CX3CL1 in response to TNF [2, 3]. PRMT5 is a member of the protein arginine methyltransferase (PRMT) family and catalyzes the covalent addition of methyl groups to the two terminal nitrogen atoms of protein-incorporated arginine. Type I and II arginine methyltransferases can add a single methyl group to arginine, producing monomethylarginine (MMA). Type I PRMT add a second methyl group to the same nitrogen atom to produce di-ω-N,N-dimethylarginine (asymmetric dimethylarginine, ADMA). In contrast, type II PRMT are capable of adding a second methyl group to the opposite terminal nitrogen atom of arginine, producing a product with methyl adducts on both terminal nitrogen residues (di-ω-N,N’-dimethylarginine; symmetrical dimethylarginine, SDMA) [4–6]. MMA, SDMA, or ADMA at a particular site may result in opposing transcriptional outcomes depending on cellular context [7, 8]. The presence of ADMA or SDMA is therefore not definitively predictive of transcriptional activity.
PRMT5 is at type II enzyme is the primary enzyme responsible for formation of SDMA in mammals [9, 10]. Addition of a methyl group to the arginine side chain increases steric bulk and hydrophobicity, and eliminates potential hydrogen bond donors, but does not alter arginine’s positive charge [9, 11, 12]. These chemical changes imparted by PRMT5 function to enhance or impede protein-substrate interactions by modulating interaction surfaces [6, 8, 13–20].
Combinations of posttranslational modifications (PTMs) such as arginine methylation, phosphorylation, and acetylation enable a “PTM code” that transduces context-specific information to the nucleus, facilitating nuanced control over transcriptional responses [21–25]. Symmetrical dimethylation of the transcription factors p53 [26–28] and NF-κB p65 [3, 29] by PRMT5 have been shown by multiple groups to enhance expression of specific genes. We reported previously that expression of CXCL10 in response to TNF in EC requires PRMT5-catalyzed arginine methylation of the transcription factor p65 at Arg30 and Arg35 [3]. These residues are located in the proximal region of the p65 rel homology domain and are part of the p65 DNA-binding core. Arg35 in particular directly interacts with the 3´ subsite of the κB promoter element [30]. Methylation of these [21] residues likely enhances p65-DNA binding by facilitating hydrophobic interactions between the methyl groups and DNA base pairs [3, 29, 30].
Our previous study also identified other residues methylated by PRMT5 on p65, such as Arg174, a residue accessible to the cytosol that is located in a region of the rel homology domain important for mediating protein-protein interactions [31]. In the current study, we posited that p65 Arg174 methylation is important for mediating stimuli-specific chemokine gene expression. To test this hypothesis, we first suppressed PRMT5 levels in EC using RNAi, and stimulated the cells with TNF, IFN-γ, and TNF plus IFN-γ to identify chemokines requiring PRMT5 for induction. We discovered that PRMT5 is critical for induction of the chemokine CXCL11 when costimulated with TNF plus IFN-γ. CXCL11 is transcribed upon stimulation by EC, astrocytes, monocytes, neutrophils, and keratinocytes [32]. Ligation of CXCL11 to the classical CXCR3 receptor enriched on activated Th1-type (type-1 helper) CD4+ and CD8+ cytotoxic T-lymphocytes (CTL), CD4+ and CD8+ memory cells, natural killer (NK), natural killer T cells (NKT), dendritic cells, and some B cells results in recruitment of leukocyte populations to inflamed sites [33–35]. CXCL11 further increases the polarity of lymphocytes at inflammatory lesions by antagonizing the CCR3 receptor enriched on Th2-type lymphocytes [36]. CXCL11 participates in numerous pathologies including atherosclerosis, organ transplant, inflammatory arthritis, inflammatory bowel disease, psoriasis, asthma, hematopoietic malignancies, and responses to infection [33, 34, 37, 38].
Additional characterization of the role of PRMT5 in the induction of CXCL11 revealed a requirement for methylation of p65 at Arg174. Together with our previous report [3], our results show that arginine methylation of p65 residues by PRMT5 comprises a critical aspect of the PTM code governing the specificity of inflammatory chemokine gene expression.
Materials and Methods
Ethics Statement
HUVEC are isolated from umbilical collected by MetroHealth Hospital and Hillcrest Hospital. The cords are not linked to any patient identification, and isolated EC are pooled. The Cleveland Clinic Foundation Institutional Review Board has confirmed that our use of discarded, de-identified human tissue is exempt from review under National Institutes of Health guidelines.
Cell Culture and Reagents
Primary human EC were isolated from human umbilical cords [39]. Cells from multiple individuals were pooled and used at early passages (P2-4). Cells were cultured in MCDB 107 medium (Sigma Life Sciences) supplemented with 15% FBS (Atlas Biologicals), 150 μg/ml endothelial cell growth supplement (ECGS), and 90 μg/ml heparin (Sigma-Aldrich). Recombinant TNF-α (R&D Systems) was used at 2 ng/ml. Recombinant IFN-γ (R&D Systems) was used at 450 U/ml. Cells were serum starved in MCDB media for 3 hours prior to the addition of agonists. Antibodies used in immunoblotting were anti-PRMT5 (Millipore 07–405), anti-p65 (Millipore 06–418), anti-dimethyl-arginine symmetric (SYM10, Millipore, 07–412), anti-CXCL11 (R&D Systems MAB672), anti-MCP1 (Cell Signaling Technology #2027), and anti-GAPDH (Cell Signaling Technology #2118). ChIP antibodies included anti-FLAG M2 (Sigma F1804) and anti-SDMA (SYM10). Additional details of the antibodies used are provided in S1 Table.
Transfection, Constructs, and RNAi
Cells were transfected 12–18 h following passage at 70–80% confluency. At this time complete cell media containing 15% FBS was removed, and replaced with a transfection mixture containing Targefect F-2 (5 μl/ml) and peptide enhancer (5 μl/ml) (Targeting Systems) in serum-free Dulbecco’s modified eagle’s medium (DMEM; Sigma-Aldrich). siRNA was transfected at 50 nM, whilst cDNA was transfected at 250 ng/ml. The transfection mixture was incubated with the cells at 37°C for 4 h, at which point it was removed and replaced with complete growth media containing serum. Cells were maintained in this media for 24–48 h to allow for protein expression or knockdown. Details of our wild type p65, p65 Arg174Ala and Arg174Lys cDNA constructs can be found in our previous publication [3]. In brief, human p65 cDNA (Addgene construct 21966) was inserted into pCDNA3 with an N-terminal FLAG epitope tag (AA: DYKDDDDK) in frame with the start codon and expressed under the control of the CMV promoter. Arg174 was mutated using the PCR-based GeneArt Mutagenesis System (Life Technologies). We repeated all experiments using two different siRNA sequences for each target to minimize the potential of off-target effects. siRNAs complementary to the PRMT5 coding sequence were sense 5´-GAGGGAGUUCAUUCAGGAAUU-3´, and PRMT5 3´ UTR, sense 5´-GCUCAAGCCACCAAUCUAUUU-3´. siRNAs targeting the NF-κB p65 3´ UTR were sense 5´-GGAUUCAUUACAGCUUAAUUU-3´ and sense 5´-GCUCUUUCUACUCUGAACUUU-3´. All siRNAs were designed using the Whitehead Institute siRNA design tool (http://sirna.wi.mit.edu/) and synthesized by Ambion containing the Silencer Select modifications [3]. Nontargeting Silencer Select siRNA was used as a control (Ambion).
Immunoblotting
Lysates for immunoblots were prepared by addition of 2X Laemmli buffer. After boiling for 5 min, proteins were resolved on 10% bis-tris acrylamide gels by SDS-PAGE, and transferred to PVDF membranes using a semi-dry transfer apparatus (Bio-Rad). Membranes were blocked with either 5% nonfat milk in 1X TBST or protein-free TBS blocking buffer (Pierce). Primary antibody incubation occurred overnight at 4°C with gentle agitation. Multiple washes were performed with 1X TBST. Secondary antibodies were HRP-conjugated. Chemiluminescence was activated using Western Lightning Plus ECL Substrate (PerkinElmer). All immunoblots were quantified via the Image Studio Lite program (LI-COR Biosciences) and normalized to a loading control.
RNA Purification and Quantitative Real Time PCR
Cells were lysed in 350 μl of Buffer RLT containing 1% 2-mercaptoethanol (Sigma-Aldrich) per each well of a 6-well plate following agonist exposure. Total RNA was isolated with the RNeasy kit (Qiagen). SuperScript First-Strand reagents (Life Technologies) were used to produce cDNA from 1 μg of RNA. The resulting cDNA was diluted 3-fold with water. qRT-PCR was performed using Fast SYBR Green Reagent (Life Technologies) using an ABI StepOnePlus machine (Life Technologies) with 40 cycles of amplification. Primers were separated by an intron on the corresponding gDNA and amplified 100–150 base pairs. Results were normalized to GAPDH according to the –ΔΔCt method. Primer sequences are provided in S2 Table. Amplification products were validated with sequencing, and analysis of amplification and melt curves.
Quantitative Mass Spectrometry
p65 and PRMT5 siRNAs were transfected into EC. After 40 hours, cells were serum starved and treated with TNF and IFN-γ then lysed in 1X RIPA. FLAG-p65 was immunoprecipitated using anti-FLAG antibodies. Proteins were separated by PAGE and the gel was silver-stained. p65 bands were excised and digested in-gel with trypsin overnight at room temperature. Fragments were assessed using capillary column LC-tandem mass spectrometry using a self-packed 9 cm x 75 μm inner diameter Phenomenex Jupiter C18 reversed-phase capillary chromatography column and Finnigan LTQ (linear trap quadrapole) ion trap mass spectrometer (ThermoFinnigan). Collision-induced dissociation (CID) spectra were analyzed using the NCBI human non-redundant protein database. Targeted experiments were also performed involving selected reaction monitoring (SRM) of specific p65 peptides, including unmodified and arginine methylated forms. Chromatograms derived from these data were assessed in relation to known fragmentation patterns. Peak areas were used to quantitate the extent of methylation.
Chromatin Immunoprecipitation and Sequential ChIP (Re-ChIP)
Cells for ChIP were cultured in 10 cm dishes. The ChIP protocol was followed according to the instructions included in the ChIP kit (Millipore 17–295). Crosslinked protein-DNA complexes were formed by incubating the cells with 1% formaldehyde (Sigma-Aldrich) in MCDB media for 10 min at 37°C. Following sonication, immunoprecipitation, and crosslink reversal the immunoprecipitated DNA was purified with the PrepEase DNA Cleanup kit (Affymetrix). In the Re-ChIP experiments the complexes from the initial IP (anti-FLAG) were eluted at 37°C for 30 minutes with 10 mM DTT. Following centrifugation the supernatant was removed to a fresh tube and diluted 20 times in Re-ChIP buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl (pH 8.1). The second IP antibody (anti-SDMA) or antiserum was then added, followed by crosslink reversal and DNA purification as outlined above. DNA products from the immunoprecipitation were quantified by qRT-PCR relative to input. Primers were designed to amplify chemokine proximal promoters containing κB site(s) and provided in S3 Table.
Statistical Analysis
All experiments were repeated with an n = 3–4 and are presented as mean ± standard error of the mean (SEM). One- or two-way analysis of variance (ANOVA) were performed. Pairwise comparisons were assessed with Student’s t-test, and corrected with Bonferroni post hoc tests. A p value of ≤ 0.05 was considered significant.
Results and Discussion
In our previous publication, we identified that symmetrical dimethylarginine (SDMA) formation catalyzed by PRMT5 on Arg30 and Arg35 of p65 is necessary for induction of CXCL10 by TNF in EC [3]. Methylation of these residues is part of a signaling mechanism enabling transcription of the chemokines CXCL10 and CX3CL1, and not other pro-inflammatory chemokines, such as CXCL8/IL-8 or CCL2/MCP1. We also discovered methylation at other p65 arginine residues, such as Arg174, and postulated that arginine methylation of this residue may be necessary to induce other pro-inflammatory chemokines in a stimulus-dependent manner. In particular, we explored the effects of EC costimulation by TNF plus IFN-γ as this combination of agonists synergistically induces CXCL10 [40–42] and CXCL11 [43–45] expression. Coincident stimulation of EC with TNF and IFN-γ is physiologically relevant as both factors are simultaneously present in localized regions of inflammation under pathological conditions, such as atherosclerosis [46].
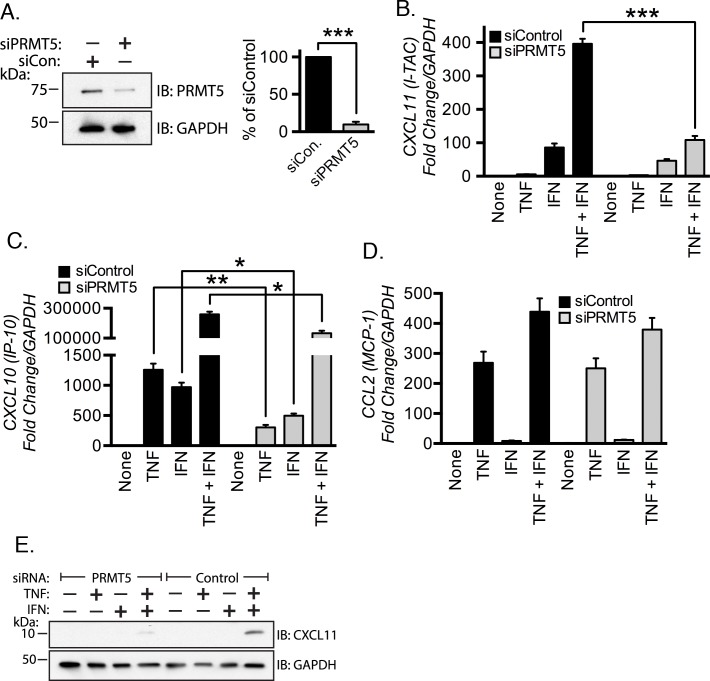
We tested our hypothesis by measuring chemokine mRNA expression levels in control versus PRMT5-depleted EC. Cells were serum-starved for 48 h post siRNA transfection and then stimulated with TNF, IFN-γ, and TNF plus IFN-γ. Total RNA was isolated and real-time PCR was performed to measure chemokine mRNA levels. Knockdown of PRMT5 efficiently suppressed PRMT5 protein levels by greater than 90% (Fig 1A, p < 0.001). We found that transcription of CXCL11 mRNA was significantly blunted in PRMT5-depleted EC in response to TNF plus IFN-γ costimulation (~30% of the mRNA elicited from the control cells, p < 0.05, Fig 1B). PRMT5-depletion did not significantly alter CXCL11 mRNA abundance in response to IFN-γ stimulation alone (p > 0.05). We observed a significant decrease in CXCL10 mRNA induction when both agonists were presented individually, and in combination (Fig 1C, all p < 0.05). TNF plus IFN-γ-mediated induction of CCL2 (Fig 1D) was not significantly affected by PRMT5 depletion. We verified a positive role for PRMT5 in CXCL11 induction at the protein level by immunoblot (Fig 1E). These results indicate that PRMT5 is necessary for CXCL11 induction in response to TNF and IFN-γ costimulation.
Fig 1. PRMT5 promotes expression of CXCL11 in EC costimulated with TNF and IFN-γ.
(A) PRMT5-specific siRNA depleted protein levels in endothelial cells (EC) by ~90% as measured by immunoblotting (left panel) and densitometry (right panel). Expression of the chemokines CXCL11 (B), CXCL10 (C) and CCL2 (D) were measured by real-time PCR of RNA isolated from EC transfected with the indicated siRNAs and subsequently stimulated with TNF, IFN-γ, or TNF plus IFN-γ for 3 hours. An immunoblot showing CXCL11 protein expression in PRMT5 or control siRNA-transfected cells after 3 h of stimulation is shown in (E). *, p < 0.05; ***, p < 0.005; error bars represent S.E. (n = 3–4).
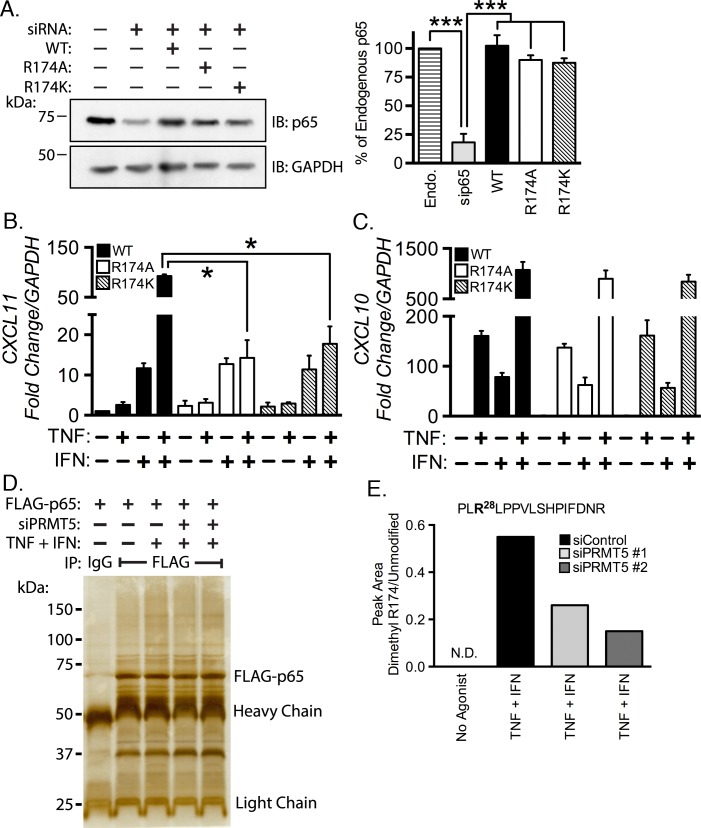
We then asked whether Arg174 must be present in order to transactivate CXCL11. We cotransfected siRNA targeting the 3´ UTR of p65 along with cDNA encoding wild type p65, p65 Arg174Ala, or p65 Arg174Lys. PRMT5 is incapable of methylating alanine and lysine residues [5, 47–49]. Transfection of p65 siRNA depleted ~80% of p65, and reconstitution of wild type or either Arg174 point mutant recapitulated endogenous levels of p65 (Fig 2A, quantified in right panel). EC reconstituted with p65 mutants were then stimulated with TNF, IFN-γ, or TNF plus IFN-γ, and CXCL11 mRNA levels were measured with quantitative real-time PCR. Wild type p65 reconstituted-EC costimulated with TNF and IFN-γ showed synergistic CXCL11 induction. The synergistic induction of CXCL11 by TNF or IFN-γ in wild type-reconstituted EC was ~92 fold increased over unstimulated cells. Importantly, CXCL11 induction was dramatically reduced in cells reconstituted with either Arg174Ala (~14 fold increase over unstimulated, p < 0.05) or p65 Arg174Lys (~18 fold increase over unstimulated, p < 0.05; Fig 2B). CXCL11 mRNA levels were not significantly different in wild type or Arg174 mutant reconstituted EC upon stimulation with TNF or IFN-γ in isolation. These results suggest that p65 Arg174 is critical for the synergistic induction of CXCL11 in response to TNF and IFN-γ costimulation. In contrast, expression of CXCL10 in cells expressing p65 WT or either Arg174 mutant stimulated with either agonist alone or in combination was comparable to EC expressing wild type p65 (Fig 2C). This finding was unsurprising given our previous findings that methylation of Arg30 and Arg35, but not Arg174, is necessary to transactivate CXCL10 [3].
Fig 2. TNF and IFN-γ mediated CXCL11 induction requires PRMT5 catalyzed p65 dimethylation at Arg174.
siRNA targeting the 3´ UTR of p65 was cotransfected in EC with cDNA encoding wild type, Arg174Ala, or Arg174Lys p65. Knockdown of endogenous p65 and reconstitution with p65 constructs is shown by immunoblot and densitometric analysis (A). EC reconstituted with wild type and p65 mutants were treated with the indicated pro-inflammatory agonists for 3 hours followed by RNA collection and real-time PCR analysis to determine the mRNA levels of CXCL11 (B) and CXCL10 (C). In (D), FLAG-p65 was immunoprecipitated from EC transfected with two different PRMT5 siRNAs (sequence 1, lane 4; sequence 2, lane 5) following stimulation with TNF and IFN-γ for 30 minutes. Immunoprecipitates were separated by PAGE and the gel was silver-stained. FLAG-p65 bands were excised for mass spectrometric analysis. (E) Mass spectrometric quantitation of the peak area of dimethylated Arg174 present in the tryptic 172PLRLPPVLSHPIFDNR185 peptide relative to the unmodified peptide. The 28 in the peptide sequence refers to the +28 Da mass shift consistent with dimethylation (2 x 14 Da) of Arg174.*, p < 0.05; ***, p < 0.005; error bars represent S.E (n = 3).
We next sought to determine whether Arg174 methylation levels are modulated in response to costimulation with TNF and IFN-γ. We introduced wild type FLAG-p65 cDNA along with control or PRMT5 siRNA. After 40 hours, we stimulated the cells, prepared lysates, and immunoprecipitated p65 using the FLAG tag. IP products were separated using PAGE and silver-stained (Fig 2D). The 65 kDa bands were excised, digested with trypsin, and fragmented by collision induced dissociation. A mass spectra of the 172PLRLPPVLSHPIFDNR185 peptide containing dimethylarginine at Arg174 is provided in S1 Fig. Peptides were also subjected to mass spectrometric selected reaction monitoring experiments to quantitate relative levels of arginine methylation. We found that p65 Arg174 dimethylation was undetected under unstimulated conditions, but was present in ~0.6 percent of the 172PLRLPPVLSHPIFDNR185 peptide (Fig 2E). We did not detect monomethylation with or without treatment with TNF and IFN-γ. Additionally, we determined that Arg174 dimethylation was catalyzed by PRMT5, as depletion of PRMT5 using two different sequences in TNF plus IFN-γ treated cells diminished Arg174 methylation to ~50% and ~33% of the control transfected, costimulated levels (Fig 2E), respectively. Together, these findings suggest a requirement for PRMT-mediated p65 Arg174 methylation in the expression of CXCL11, but not CXCL10, when EC are costimulated with TNF and IFN-γ. We do not contend that Arg174 methylation is the only p65 PTM necessary for CXCL11 activation, as other modifications such as phosphorylation, may also contribute to CXCL11 induction.
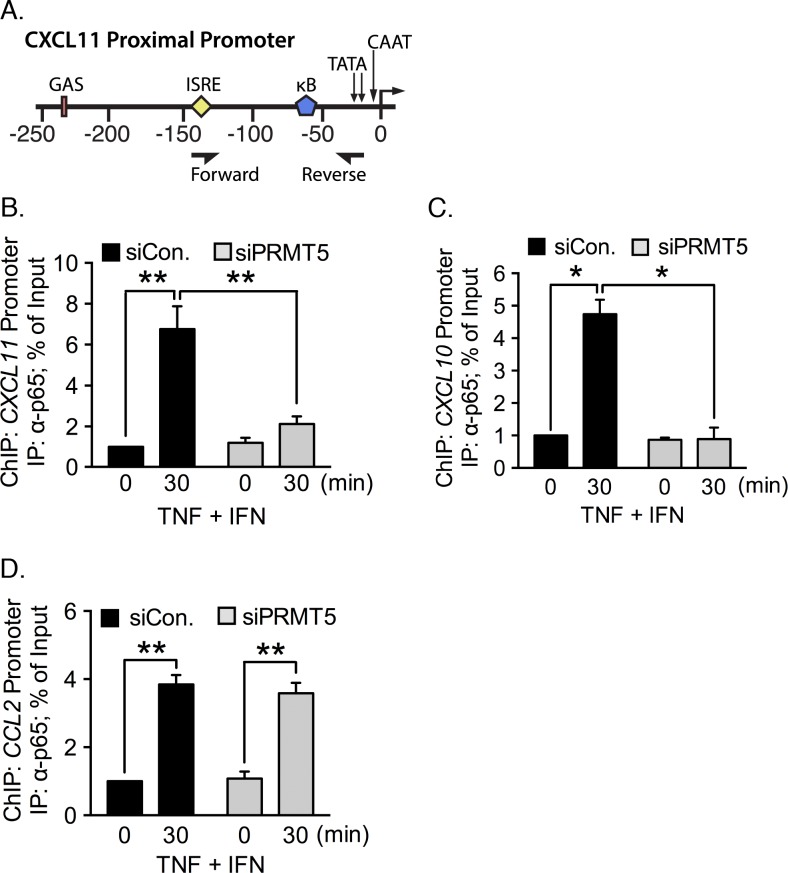
We next performed ChIP to determine whether PRMT5 is obligatory for p65 association with the CXCL11 promoter following TNF plus IFN-γ costimulation. We immunoprecipitated chromatin from PRMT5-intact or -depleted EC using an anti-p65 antibody. We then amplified CXCL11 promoter fragments using primers encompassing the κB site (Fig 3A). p65 was found to be associated with the CXCL11 promoter upon TNF plus IFN-γ exposure (~7 fold increase) but PRMT5-depletion decreased p65 binding to the CXCL11 promoter significantly (~2 fold increase, p < 0.01) as compared with the unstimulated condition (Fig 3B). We were unable to detect p65 on the CXCL10 promoter in PRMT5-depleted EC (Fig 3C). In contrast, p65 association with the CCL2 (MCP1) promoter following costimulation was unaffected by PRMT5 knockdown (Fig 3D), in line with our observations that PRMT5 is not involved in CCL2 expression in response to TNF, IFN-γ, or TNF plus IFN-γ stimulation (Fig 1D). These results reveal that PRMT5 activity is critical for p65 recruitment to the CXCL10 and CXCL11 promoters when EC are costimulated with TNF and IFN-γ.
Fig 3. PRMT5 knockdown reduces association of p65 with the CXCL11 promoter.
The location of major regulatory elements and primer binding sites used for the chromatin immunoprecipitation (ChIP) assay of the CXCL11 promoter are illustrated in (A). IFN-γ response elements include the gamma interferon activation site (GAS) and interferon-sensitive response element (ISRE). The major TNF responsive element is the NF-κB binding site. ChIP assays were performed to assess p65 binding with the proximal promoter encompassing the κB binding site(s) of CXCL11 (B), CXCL10 (C), and CCL2 (D) in PRMT5-intact or -depleted EC stimulated with TNF plus IFN-γ for 30 minutes. *, p < 0.05; **, p < 0.01; error bars represent S.E. (n = 3).
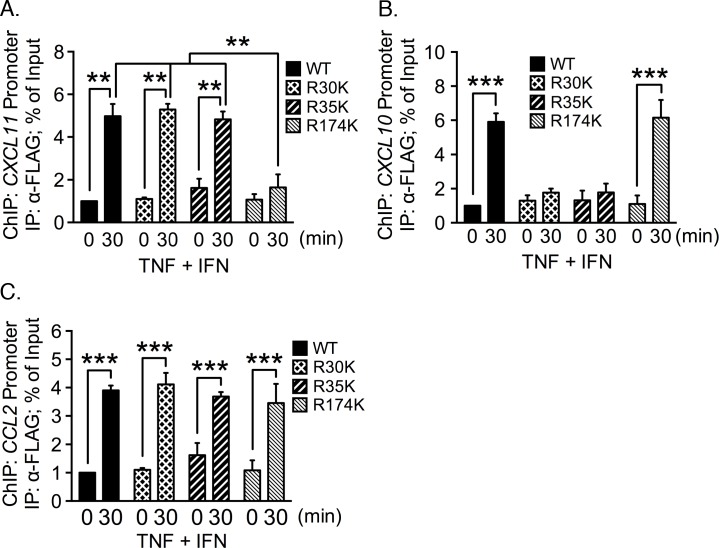
Additional ChIP assays were performed to ascertain whether p65 Arg174Lys is recruited to the CXCL11 promoter. Mutation of arginine-to-lysine is a conservative mutation that preserves the positive charge at amino acid 174 but precludes the possibility of methylation by PRMT5. Cells were cotransfected with siRNA targeting the 3´ UTR of p65 along with cDNA encoding either FLAG-tagged wild type p65 or Arg174Lys. DNA-protein complexes were immunoprecipitated with anti-FLAG antibody and probed with primers that amplify the CXCL11 promoter. We detected a significant increase in the association of wild type p65 with the CXCL11 promoter following costimulation (Fig 4A). However, the level of CXCL11 promoter immunoprecipitated from cells reconstituted with p65 Arg174Lys was not statistically different from unstimulated cells. In contrast, CXCL10 (Fig 4B) and CCL2 (Fig 4C) promoter fragments were enriched equally well in costimulated cells expressing either wild type p65 or Arg174Lys p65. We conclude that there is a requirement for p65 Arg174 for the expression of CXCL11, but not CXCL10 or CCL2.
Fig 4. Mutation of Arg174 to lysine abrogates p65 association with the CXCL11 promoter.
Endogenous p65 was depleted with siRNA targeting the 3´ UTR of p65 in EC. p65 was reconstituted in these cells by transfecting cDNA encoding FLAG-tagged wild type p65 or the FLAG-tagged Arg174Lys mutant. Following cell stimulation, the protein-DNA complexes were immunoprecipitated with an anti-FLAG antibody. Enrichment of the CXCL11 (A), CXCL10 (B), and CCL2 (C) promoters by the IP was quantified by real-time PCR. *, p < 0.05; ***, p < 0.005; error bars represent S.E. (n = 3).
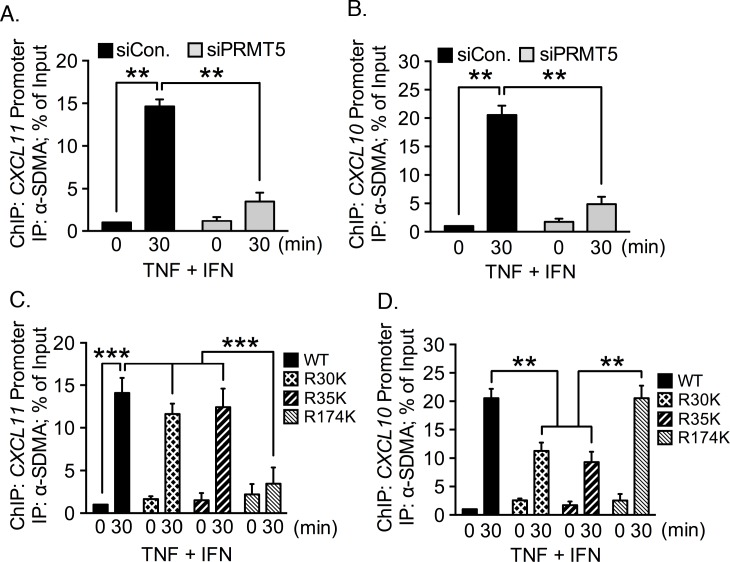
In our previous publication we showed that p65 contains SDMA catalyzed by PRMT5 [3]. With that information in mind, our next question in the current project asked whether we could detect SDMA-containing proteins associated with the CXCL11 promoter in TNF plus IFN-γ costimulated cells. We immunoprecipitated chromatin from PRMT5 siRNA-transfected, costimulated cells using an anti-SDMA antibody. We were able to detect a robust enrichment of the CXCL11 promoter following stimulation, but this enrichment was lost in the absence of PRMT5 (Fig 5A). Results were similar when we assayed the immunoprecipitated DNA for the CXCL10 promoter: we were able to detect SDMA-associated CXCL10 promoter after stimulation in the endogenous condition, but not when PRMT5 was depleted (Fig 5B). We therefore concluded that SDMA-catalyzed by PRMT5 is associated with both the CXCL10 and CXCL11 promoters after simultaneous exposure to TNF and IFN-γ.
Fig 5. PRMT5 depletion diminishes detection of SDMA-containing proteins associated with the CXCL11 promoter.
EC were transfected with PRMT5-specific or control siRNA and stimulated with TNF plus IFN-γ. ChIP was performed using an antibody specific for SDMA. Enrichment of the CXCL11 (A) and CXCL10 (B) promoters was quantified by qRT-PCR. (C-D) ChIP was performed on EC reconstituted with FLAG-tagged wild type p65 or the indicated p65 mutants from TNF plus IFN-γ stimulated EC. Protein-DNA complexes were immunoprecipitated with anti-FLAG antibody. Enrichment of the CXCL11 (C), and CXCL10 (D) promoters was quantified by qRT-PCR. **, p < 0.01; error bars represent S.E. (n = 3).
We next sought to determine whether arginine methylation of p65 amino acid 174 is necessary for CXCL11 induction. We reconstituted EC with wild type, Arg30Lys, Arg35Lys, and Arg174Lys p65, and performed ChIP with an anti-SDMA antibody. Results indicated that the CXCL11 promoter was pulled down when wild type p65 and the p65 Arg30Lys and Arg35Lys mutants were expressed in costimulated EC (Fig 5C). When p65 Arg174Lys was reconstituted, the CXCL11 promoter levels were not statistically different from the unstimulated condition. This suggests that p65 with lysine at position 174 does not associate with the CXCL11 promoter.
The ChIP immunoprecipitates were also probed for the presence of the CXCL10 promoter. We found that the CXCL10 promoter was enriched from both wild type and Arg174Lys reconstituted cells upon costimulation (Fig 5D, p < 0.005). When the Arg30Lys and Arg35Lys mutants were expressed, levels of CXCL10 promoter dropped to ~50% of the levels observed with the wild type and Arg175Lys mutant (Fig 5D, p < 0.005). These results are consistent with our previous finding that methylation of p65 Arg30 and Arg35 are necessary for CXCL10 induction in EC. Additionally, these results indicate that Arg30 and Arg35 may be methylated independently, as mutation of either Arg30 or Arg35 to lysine does not reduce methylation of the other residue.
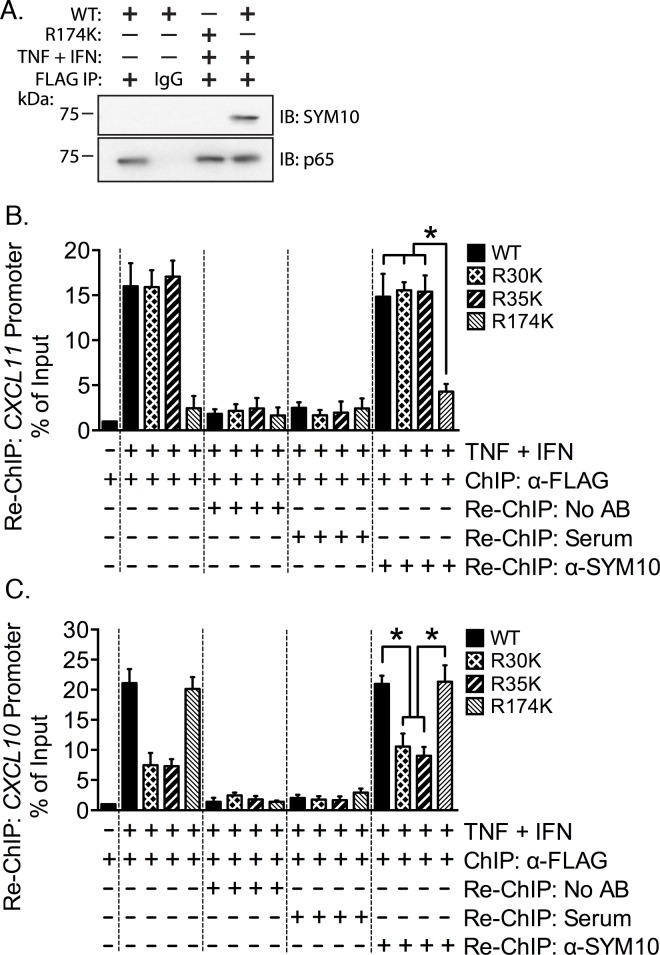
To confirm that the SDMA antibody is detecting methylation at p65 Arg174 we reconstituted wild type and Arg174Lys mutants in EC, and used the FLAG epitope to immunoprecipitate p65 proteins, which where then probed with anti-SDMA antibody (SYM10; Fig 6A). We are able to detect arginine methylation in response to TNF plus IFN-γ only when wild type p65 is present (lane 4), but not when the Arg174Lys was transfected (lane 3). These results indicate that the symmetrical dimethylation of p65 detected by the anti-SDMA antibody is likely modification of Arg174.
Fig 6. p65 Arg174 methylation is present on the CXCL11 promoter following costimulation.
(A) EC were depleted of endogenous p65 and reconstituted with wild type or Arg174Lys mutant p65. Cells were stimulated with TNF plus IFN-γ for 30 minutes and lysed in RIPA buffer. Wild type and mutant p65 was immunoprecipitated using the FLAG epitope or IgG. Immunoprecipitates were immunoblotted for p65 and for the SDMA modification. (B-C) Cells were reconstituted with N-FLAG wild type, Arg30Lys, Arg35Lys, and Arg174Lys p65 constructs, and simultaneously exposed to TNF and IFN-γ. ChIP was performed using anti-FLAG antibody. Sequential ChIP (Re-ChIP) was carried out with addition of normal rabbit serum or anti-SYM10 antibody. Following crosslink reversal, DNA was purified and promoter fragment content was quantitated using qRT-PCR relative to input.
We next sought to assess whether p65 and SDMA are detected on the same fragments of the CXCL11 promoter. We utilized a sequential ChIP (Re-ChIP) approach where we reconstituted wild type and point mutant p65, then immunoprecipitated p65 using the FLAG-tag. After removal of the anti-FLAG antibody, we immunoprecipitated the protein-chromatin complexes a second time using the anti-SDMA antibody. The CXCL11 promoter was present at background levels in the absence of cell costimulation with TNF and IFN-γ following immunoprecipitation of wild type p65 (Fig 6B, bar 1). We found CXCL11 promoter enrichment when wild type, Arg30Lys, and Arg35Lys were expressed in costimulated cells where p65 was immunoprecipitated with the anti-FLAG antibody (Fig 6B, bars 2–4). In contrast, we detected the CXCL11 promoter when Arg174Lys p65 was expressed only at background levels (bar 5).
We included a no antibody control to assess whether the anti-FLAG antibody was successfully dissociated from the chromatin during the Re-ChIP procedure. We did not detect the CXCL11 promoter with this control (Fig 6B, bars 6–9), indicated that we were able to remove the antibody with buffer exchange. Following buffer exchange, we proceeded with the Re-ChIP portion of the experiment, using either the anti-SDMA antibody (SYM10) or normal rabbit serum, as SYM10 is presented in antiserum format (bars 10–13). We did not enrich the CXCL11 promoter in the Re-ChIP stage of the experiment using the serum regardless of whether wild type or any of the mutants were present.
Enrichment of the CXCL11 promoter was detected equal to roughly 15% of the input material when wild type, Arg30Lys, and Arg35Lys p65 were present (bars 14–16). Crucially, we found that the level of CXCL11 was significantly diminished when Arg174Lys was expressed to ~5% of input (p < 0.05; bar 17). In full, these results indicate that the methylation associated with the CXCL11 promoter is predominately symmetrical dimethylation of p65 at Arg174. Arginine methylation of other p65 arginine residues, such as Arg30 or Arg35, is only a minor component of p65 methylation associated with this promoter under TNF plus IFN-γ costimulated conditions.
We also quantitated the presence of the CXCL10 promoter in this experiment (Fig 6C). p65 is not associated with the promoter in the absence of TNF and IFN-γ (bar 1). A robust increase in promoter association was observed when wild type and Arg174Lys were expressed in stimulated cells and immunoprecipitated with the anti-FLAG antibody (~20% of input). When Arg30Lys and Arg35Lys were present, CXCL10 promoter was enriched to ~7% of input (p < 0.05; bars 2–5). Only background levels of CXCL10 promoter were found following buffer exchange to remove the anti-FLAG antibody (bars 6–9). Levels of CXCL10 promoter were also at background when rabbit serum was applied in the Re-ChIP. Quantitation of the promoter showed following Re-ChIP with the SYM10 antibody showed that wild type and Arg174Lys p65 pulled down CXCL10 promoter equal to ~20% of input, but that Arg30Lys and Arg35Lys mutant expression was associated with lower quantities of the promoter, ~8–10% of input (p < 0.05; bars 14–17). These results are consistent with our previous findings that methylation of Arg30 and Arg35 is necessary for CXCL10 induction, but that Arg174 methylation is not required [3]. It is interesting to note that mutation of either Arg30 or Arg35 to lysine reduces, but does not eliminate, detection of methylated p65 associated with this promoter, supporting a model where methylation of Arg30 and Arg35 are independent.
Given the sum of our experiments reported here we conclude that PRMT5-catalyzed p65 methylation at Arg174, Arg30 and Arg35, is critical for p65 recruitment to the CXCL11 promoter and the subsequent synergistic induction of CXCL11 in response to TNF plus IFN-γ costimulation. This requirement does not exist for the synergistic induction of CXCL10. Our findings are consistent with a mechanism whereby concurrent stimulation of EC by both TNF and IFN-γ activate PRMT5. PRMT5 is then recruited to p65, where it catalyzes dimethylation of Arg174. Arginine methylation of p65 at Arg174 is necessary for p65 recruitment to the CXCL11 promoter. Given the location of Arg174 on the p65 molecule is probable that modification of this residue facilitates a protein-protein interaction that enhances CXCL11 transcription.
We do not currently know the mechanism linking activation of cytokine receptors with p65 methylation by PRMT5. One possible mechanism could involve activation of the JAK kinases by ligation of the IFN-γ receptor by IFN-γ. PRMT5 was first discovered in human cells as a JAK2 binding protein [50], and it may also interact with JAK1 and JAK3. JAK proteins have been shown to phosphorylate PRMT5 [51]. Phosphorylation of PRMT5 by a JAK may upregulate PRMT5 methyltransferase activity, or enhance it’s affinity for p65, leading to catalysis of SDMA formation at Arg174. TNF signaling would then mobilize the p65 methylated at Arg174 to the nucleus, where it associates with transcription cofactors and the CXCL11 promoter. It is not clear whether IFN-γ alone is sufficient to trigger methylation at p65 at Arg174, or whether Arg174 methylation is a product of synergism between the TNF and IFN-γ pathways. We also can not rule out contribution of other PRMT enzymes in the methylation of Arg174, including PRMT1 and PRMT4, both of which are found in EC. PRMT1 and PRMT4 catalyze formation of ADMA, which may differ in transcriptional outcome to the SDMA modification catalyzed by PRMT5 that we report here.
Our findings suggest that the PRMT5-mediated methylation imparts an indexing signal to p65 that enables stimulus-specific chemokine expression. Differential combinations of such modifications enable p65 to interact with DNA or other components of specific transcriptional complexes in a context-specific manner, resulting in unique patterns of gene expression [22, 52]. Such a PTM code coupling extracellular stimuli to transcriptional outcomes is essential to the regulation of complex physiological and pathological processes such as inflammation. CXCL10 and CXCL11 are both found at high levels in human atheroma, as is TNF, and IFN-γ. CXCL10 and CXCL11 have redundant functions in that both ligate CXCR3 and promote chemotaxis of leukocytic populations and smooth muscle cells, and are angiostatic for EC [46, 53, 54]. Th1-type T-cells recruited by CXCL10 and CXCL11 exacerbate atherosclerosis by producing copious amounts of IFN-γ and proteinases that reduce collagen maturation [55]. However, CXCL10 and CXCL11 differ in their expression in diseases states such as atherosclerosis. Atheroma-associated EC, smooth muscle cells, and macrophages robustly produce CXCL10. CXCL11 is secreted by EC, macrophages, and lesional EC microvessels [56]. These cell types are found in discrete regions of the plaque, resulting in the accumulation of T-cells in the shoulder regions and fibrous cap [56, 57]. Varying levels of expression of both CXCL10 and CXCL11 by these cell types may be influenced by distinct regulatory and biochemical properties of these ligands. CXCL10 expression is elicited by the widest variety of agonists, including IFN-α/β/γ, and is most sensitive to TNF. In contrast, CXCL11 expression is weakly transcribed by TNF, but is induced by IFN-β/γ [33]. CXCL11 has greater potency than CXCL10 [34, 58, 59] as it is able to trigger receptor internalization, calcium mobilization, and chemotaxis at lower doses. These differences in CXCR3 ligand expression patterns according to cell type, agonist potency, and receptor affinity appear likely to exert fine control of T-cell recruitment within the lesion. Given our findings, the newly discovered specific PRMT5 inhibitors [60–62] should be investigated in animal models of chronic inflammatory disease as potential therapeutic agents to attenuate pathological progression.
Supporting Information
MS/MS spectra of the 633 Da triply charged ion identified in the tryptic digestion of NF-κB p65. The mass of this peptide is consistent with the addition of two methyl groups to the 172PLRLPPVLSHPIFDNR185 peptide. This spectra contains several unmodified C-terminal y ions, all of which are consistent with dimethylation at Arg174.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Chad Braley for isolation and culture of HUVEC used in this study.
Abbreviations
- PRMT5
protein arginine methyltransferase 5
- p65
NF-κB p65 (RelA)
- EC
endothelial cell
- SDMA
symmetrical dimethylarginine
- PCR
quantitative real-time
- TNF
tumor necrosis factor-α
- IFN
interferon-gamma (IFN-γ)
- CXCL10
C-X-C motif chemokine 10 (IP-10)
- CXCL11
C-X-C motif chemokine 11 (I-TAC)
- CCL2
chemokine C-C motif ligand 2 (MCP-1)
- CX3CL1
C-X3-C motif chemokine ligand 1 (fractalkine)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Heart, Lung, and Blood Institute (NHLBI, http://www.nhlbi.nih.gov/) grant P01-HL29582 (to P.E.D.). Support for the collection of HUVEC was made possible by the Case Western Reserve University/Cleveland Clinic CTSA Grant UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS, https://ncats.nih.gov/). The Orbitrap Elite instrument was purchased with a National Center for Research Resources (NCRR, http://www.nih.gov/about/almanac/organization/NCRR.htm) shared instrument grant (1S10RR031537-01). NHLBI, NCATS, and NCRR are components of the United States National Institutes of Health (NIH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9(10):692–703. Epub 2009/10/28. 10.1038/nri2634 . [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay S, Harris DP, Adams GN, Lause GE, McHugh A, Tillmaand EG, et al. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol Cell Biol. 2012;32(7):1202–13. Epub 2012/01/25. 10.1128/mcb.05977-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris DP, Bandyopadhyay S, Maxwell TJ, Willard B, DiCorleto PE. Tumor necrosis factor (TNF)-alpha induction of CXCL10 in endothelial cells requires protein arginine methyltransferase 5 (PRMT5)-mediated nuclear factor (NF)-kappaB p65 methylation. The Journal of biological chemistry. 2014;289(22):15328–39. Epub 2014/04/23. 10.1074/jbc.M114.547349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, et al. Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci U S A. 2012;109(44):17960–5. Epub 2012/10/17. 10.1073/pnas.1209814109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M, Xu RM, Thompson PR. Substrate Specificity, Processivity, and Kinetic Mechanism of Protein Arginine Methyltransferase 5. Biochemistry. 2013. Epub 2013/07/20. 10.1021/bi4005123 . [DOI] [PubMed] [Google Scholar]
- 6.Wilczek C, Chitta R, Woo E, Shabanowitz J, Chait BT, Hunt DF, et al. Protein arginine methyltransferase Prmt5-Mep50 methylates histones H2A and H4 and the histone chaperone nucleoplasmin in Xenopus laevis eggs. The Journal of biological chemistry. 2011;286(49):42221–31. Epub 2011/10/20. 10.1074/jbc.M111.303677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Green RD, Kouzarides T. Distinct transcriptional outputs associated with mono- and dimethylated histone H3 arginine 2. Nat Struct Mol Biol. 2009;16(4):449–51. Epub 2009/03/10. 10.1038/nsmb.1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng S, Moehlenbrink J, Lu YC, Zalmas LP, Sagum CA, Carr S, et al. Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol Cell. 2013;52(1):37–51. Epub 2013/10/01. 10.1016/j.molcel.2013.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S. Protein arginine methyltransferases: Evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113(1):50–87. . [DOI] [PubMed] [Google Scholar]
- 10.Wang M, Fuhrmann J, Thompson PR. Protein arginine methyltransferase 5 catalyzes substrate dimethylation in a distributive fashion. Biochemistry. 2014;53(50):7884–92. Epub 2014/12/09. 10.1021/bi501279g . [DOI] [PubMed] [Google Scholar]
- 11.Boisvert FM, Chenard CA, Richard S. Protein interfaces in signaling regulated by arginine methylation. Science's STKE: signal transduction knowledge environment. 2005;2005(271):re2 Epub 2005/02/17. 10.1126/stke.2712005re2 . [DOI] [PubMed] [Google Scholar]
- 12.Nussinov R, Tsai CJ, Xin F, Radivojac P. Allosteric post-translational modification codes. Trends Biochem Sci. 2012;37(10):447–55. Epub 2012/08/14. 10.1016/j.tibs.2012.07.001 . [DOI] [PubMed] [Google Scholar]
- 13.Cho EC, Zheng S, Munro S, Liu G, Carr SM, Moehlenbrink J, et al. Arginine methylation controls growth regulation by E2F-1. The EMBO journal. 2012;31(7):1785–97. Epub 2012/02/14. 10.1038/emboj.2012.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azzouz TN, Pillai RS, Dapp C, Chari A, Meister G, Kambach C, et al. Toward an assembly line for U7 snRNPs: interactions of U7-specific Lsm proteins with PRMT5 and SMN complexes. The Journal of biological chemistry. 2005;280(41):34435–40. . [DOI] [PubMed] [Google Scholar]
- 15.Chuang TW, Peng PJ, Tarn WY. The exon junction complex component Y14 modulates the activity of the methylosome in biogenesis of spliceosomal small nuclear ribonucleoproteins. The Journal of biological chemistry. 2011;286(11):8722–8. Epub 2011/01/07. 10.1074/jbc.M110.190587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001;7(5):1111–7. . [DOI] [PubMed] [Google Scholar]
- 17.Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21(24):8289–300. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miranda TB, Khusial P, Cook JR, Lee JH, Gunderson SI, Pestka S, et al. Spliceosome Sm proteins D1, D3, and B/B ' are asymmetrically dimethylated at arginine residues in the nucleus. Biochem Biophys Res Commun. 2004;323(2):382–7. . [DOI] [PubMed] [Google Scholar]
- 19.Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, et al. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol. 2011;18(12):1414–20. Epub 2011/11/22. 10.1038/nsmb.2185 . [DOI] [PubMed] [Google Scholar]
- 20.Hsu JM, Chen CT, Chou CK, Kuo HP, Li LY, Lin CY, et al. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat Cell Biol. 2011;13(2):174–81. Epub 2011/01/25. 10.1038/ncb2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prabakaran S, Lippens G, Steen H, Gunawardena J. Post-translational modification: nature's escape from genetic imprisonment and the basis for dynamic information encoding. Wiley interdisciplinary reviews Systems biology and medicine. 2012;4(6):565–83. Epub 2012/08/18. 10.1002/wsbm.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venne AS, Kollipara L, Zahedi RP. The next level of complexity: crosstalk of posttranslational modifications. Proteomics. 2014;14(4–5):513–24. Epub 2013/12/18. 10.1002/pmic.201300344 . [DOI] [PubMed] [Google Scholar]
- 23.Lothrop AP, Torres MP, Fuchs SM. Deciphering post-translational modification codes. FEBS Lett. 2013;587(8):1247–57. Epub 2013/02/14. 10.1016/j.febslet.2013.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, et al. A transcriptional switch mediated by cofactor methylation. Science. 2001;294(5551):2507–11. . [DOI] [PubMed] [Google Scholar]
- 25.Sims RJ 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9(10):815–20. Epub 2008/09/12. 10.1038/nrm2502 . [DOI] [PubMed] [Google Scholar]
- 26.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10(12):1431–U122. 10.1038/ncb1802 . [DOI] [PubMed] [Google Scholar]
- 27.Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009;37(15):4965–76. 10.1093/nar/gkp516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Chitnis N, Nakagawa H, Kita Y, Natsugoe S, Yang Y, et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer discovery. 2015;5(3):288–303. Epub 2015/01/15. 10.1158/2159-8290.cd-14-0625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei H, Wang B, Miyagi M, She Y, Gopalan B, Huang DB, et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-kappaB. Proc Natl Acad Sci U S A. 2013;110(33):13516–21. Epub 2013/08/02. 10.1073/pnas.1311784110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391(6665):410–3. Epub 1998/02/05. 10.1038/34956 . [DOI] [PubMed] [Google Scholar]
- 31.Huang B, Yang XD, Lamb A, Chen LF. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cellular signalling. 2010;22(9):1282–90. Epub 2010/04/07. 10.1016/j.cellsig.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohan K, Ding Z, Hanly J, Issekutz TB. IFN-gamma-inducible T cell alpha chemoattractant is a potent stimulator of normal human blood T lymphocyte transendothelial migration: differential regulation by IFN-gamma and TNF-alpha. J Immunol. 2002;168(12):6420–8. Epub 2002/06/11. . [DOI] [PubMed] [Google Scholar]
- 33.Van Raemdonck K, Van den Steen PE, Liekens S, Van Damme J, Struyf S. CXCR3 ligands in disease and therapy. Cytokine Growth Factor Rev. 2014. Epub 2014/12/17. 10.1016/j.cytogfr.2014.11.009 . [DOI] [PubMed] [Google Scholar]
- 34.Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunology and cell biology. 2011;89(2):207–15. Epub 2011/01/12. 10.1038/icb.2010.158 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zernecke A, Shagdarsuren E, Weber C. Chemokines in atherosclerosis: an update. Arteriosclerosis, thrombosis, and vascular biology. 2008;28(11):1897–908. Epub 2008/06/21. 10.1161/atvbaha.107.161174 . [DOI] [PubMed] [Google Scholar]
- 36.Loetscher P, Pellegrino A, Gong JH, Mattioli I, Loetscher M, Bardi G, et al. The ligands of CXC chemokine receptor 3, I-TAC, Mig, and IP10, are natural antagonists for CCR3. The Journal of biological chemistry. 2001;276(5):2986–91. Epub 2000/12/09. 10.1074/jbc.M005652200 . [DOI] [PubMed] [Google Scholar]
- 37.Lazzeri E, Romagnani P. CXCR3-binding chemokines: novel multifunctional therapeutic targets. Current drug targets Immune, endocrine and metabolic disorders. 2005;5(1):109–18. Epub 2005/03/22. . [DOI] [PubMed] [Google Scholar]
- 38.Lacotte S, Brun S, Muller S, Dumortier H. CXCR3, inflammation, and autoimmune diseases. Annals of the New York Academy of Sciences. 2009;1173:310–7. Epub 2009/09/18. 10.1111/j.1749-6632.2009.04813.x . [DOI] [PubMed] [Google Scholar]
- 39.Scarpati EM, DiCorleto PE. Identification of a thrombin response element in the human platelet-derived growth factor B-chain (c-sis) promoter. The Journal of biological chemistry. 1996;271(6):3025–32. . [DOI] [PubMed] [Google Scholar]
- 40.Clarke DL, Clifford RL, Jindarat S, Proud D, Pang LH, Belvisi M, et al. TNF alpha and IFN gamma Synergistically Enhance Transcriptional Activation of CXCL10 in Human Airway Smooth Muscle Cells via STAT-1, NF-kappa B, and the Transcriptional Coactivator CREB-binding Protein. J Biol Chem. 2010;285(38):29101–10. 10.1074/jbc.M109.0999952 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majumder S, Zhou LZ, Chaturvedi P, Babcock G, Aras S, Ransohoff RM. p48/STAT-1alpha-containing complexes play a predominant role in induction of IFN-gamma-inducible protein, 10 kDa (IP-10) by IFN-gamma alone or in synergy with TNF-alpha. J Immunol. 1998;161(9):4736–44. . [PubMed] [Google Scholar]
- 42.Lombardi A, Cantini G, Mello T, Francalanci M, Gelmini S, Cosmi L, et al. Molecular mechanisms underlying the pro-inflammatory synergistic effect of tumor necrosis factor alpha and interferon gamma in human microvascular endothelium. European journal of cell biology. 2009;88(12):731–42. Epub 2009/09/29. 10.1016/j.ejcb.2009.07.004 . [DOI] [PubMed] [Google Scholar]
- 43.Helbig KJ, Ruszkiewicz A, Semendric L, Harley HA, McColl SR, Beard MR. Expression of the CXCR3 ligand I-TAC by hepatocytes in chronic hepatitis C and its correlation with hepatic inflammation. Hepatology. 2004;39(5):1220–9. Epub 2004/05/04. 10.1002/hep.20167 . [DOI] [PubMed] [Google Scholar]
- 44.Kraft M, Riedel S, Maaser C, Kucharzik T, Steinbuechel A, Domschke W, et al. IFN-gamma synergizes with TNF-alpha but not with viable H. pylori in up-regulating CXC chemokine secretion in gastric epithelial cells. Clinical and experimental immunology. 2001;126(3):474–81. Epub 2001/12/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazanet MM, Neote K, Hughes CC. Expression of IFN-inducible T cell alpha chemoattractant by human endothelial cells is cyclosporin A-resistant and promotes T cell adhesion: implications for cyclosporin A-resistant immune inflammation. J Immunol. 2000;164(10):5383–8. Epub 2000/05/09. . [DOI] [PubMed] [Google Scholar]
- 46.Tedgui A, Mallat Z. Cytokines in atherosclerosis: pathogenic and regulatory pathways. Physiological reviews. 2006;86(2):515–81. Epub 2006/04/08. 10.1152/physrev.00024.2005 . [DOI] [PubMed] [Google Scholar]
- 47.Baldwin GS, Carnegie PR. Isolation and partial characterization of methylated arginines from the encephalitogenic basic protein of myelin. The Biochemical journal. 1971;123(1):69–74. Epub 1971/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brostoff S, Eylar EH. Localization of methylated arginine in the A1 protein from myelin. Proc Natl Acad Sci U S A. 1971;68(4):765–9. Epub 1971/04/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kakimoto Y, Akazawa S. Isolation and identification of N-G,N-G- and N-G,N'-G-dimethyl-arginine, N-epsilon-mono-, di-, and trimethyllysine, and glucosylgalactosyl- and galactosyl-delta-hydroxylysine from human urine. The Journal of biological chemistry. 1970;245(21):5751–8. Epub 1970/11/10. . [PubMed] [Google Scholar]
- 50.Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. The Journal of biological chemistry. 1999;274(44):31531–42. Epub 1999/10/26. . [DOI] [PubMed] [Google Scholar]
- 51.Liu F, Zhao X, Perna F, Wang L, Koppikar P, Abdel-Wahab O, et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell. 2011;19(2):283–94. Epub 2011/02/15. 10.1016/j.ccr.2010.12.020 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12(8):695–708. Epub 2011/07/21. 10.1038/ni.2065 . [DOI] [PubMed] [Google Scholar]
- 53.Gerszten RE, Mach F, Sauty A, Rosenzweig A, Luster AD. Chemokines, leukocytes, and atherosclerosis. The Journal of laboratory and clinical medicine. 2000;136(2):87–92. Epub 2000/08/17. 10.1067/mlc.2000.108154 . [DOI] [PubMed] [Google Scholar]
- 54.Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nat Rev Immunol. 2006;6(7):508–19. . [DOI] [PubMed] [Google Scholar]
- 55.Ovchinnikova O, Robertson AK, Wagsater D, Folco EJ, Hyry M, Myllyharju J, et al. T-cell activation leads to reduced collagen maturation in atherosclerotic plaques of Apoe(-/-) mice. Am J Pathol. 2009;174(2):693–700. 10.2353/ajpath.2009.080561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mach F, Sauty A, Iarossi AS, Sukhova GK, Neote K, Libby P, et al. Differential expression of three T lymphocyte-activating CXC chemokines by human atheroma-associated cells. The Journal of clinical investigation. 1999;104(8):1041–50. Epub 1999/10/19. 10.1172/jci6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonasson L, Holm J, Skalli O, Bondjers G, Hansson GK. Regional accumulations of T cells, macrophages, and smooth muscle cells in the human atherosclerotic plaque. Arteriosclerosis. 1986;6(2):131–8. . [DOI] [PubMed] [Google Scholar]
- 58.Cole KE, Strick CA, Paradis TJ, Ogborne KT, Loetscher M, Gladue RP, et al. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. The Journal of experimental medicine. 1998;187(12):2009–21. Epub 1998/06/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer M, Hensbergen PJ, van der Raaij-Helmer EM, Brandacher G, Margreiter R, Heufler C, et al. Cross reactivity of three T cell attracting murine chemokines stimulating the CXC chemokine receptor CXCR3 and their induction in cultured cells and during allograft rejection. Eur J Immunol. 2001;31(8):2521–7. Epub 2001/08/14. . [DOI] [PubMed] [Google Scholar]
- 60.Alinari L, Mahasenan KV, Yan F, Karkhanis V, Chung JH, Smith EM, et al. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood. 2015;125(16):2530–43. Epub 2015/03/07. 10.1182/blood-2014-12-619783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11(6):432–7. Epub 2015/04/29. 10.1038/nchembio.1810 . [DOI] [PubMed] [Google Scholar]
- 62.Smil D, Eram MS, Li F, Kennedy S, Szewczyk MM, Brown PJ, et al. Discovery of a Dual PRMT5-PRMT7 Inhibitor. ACS medicinal chemistry letters. 2015;6(4):408–12. Epub 2015/04/22. 10.1021/ml500467h [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MS/MS spectra of the 633 Da triply charged ion identified in the tryptic digestion of NF-κB p65. The mass of this peptide is consistent with the addition of two methyl groups to the 172PLRLPPVLSHPIFDNR185 peptide. This spectra contains several unmodified C-terminal y ions, all of which are consistent with dimethylation at Arg174.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.