Abstract
Background
We performed this retrospective study to identify the prevalence of KRAS mutation in Chinese populations and make a comprehensive investigation of the clinicopathological features of KRAS mutation in these patients.
Patients and methods
Patients from 2007 to 2013 diagnosed with primary lung adeno-carcinoma who received a radical resection were examined for KRAS, EGFR, HER2, BRAF mutations, and ALK, RET, and ROS1 fusions. Clinicopathological features, including sex, age, tumor–lymph node–metastasis stage, tumor differentiation, smoking status, histological subtypes, and survival information were analyzed.
Result
KRAS mutation was detected in 113 of 1,368 patients. Nine different subtypes of KRAS mutation were identified in codon 12, codon 13, and codon 61. KRAS mutation was more frequently found in male patients and former/current smoker patients. Tumors with KRAS mutation had poorer differentiation. Invasive mucinous adenocarcinoma predominant and solid predominant subtypes were more frequent in KRAS mutant patients. No statistical significance was found in relapse-free survival or overall survival between patients with KRAS mutation and patients with other mutations.
Conclusion
In Chinese populations, we identified KRAS mutation in 8.3% (113/1,368) of the patients with lung adenocarcinoma. KRAS mutation defines a molecular subset of lung adenocarcinoma with unique clinicopathological features.
Keywords: KRAS, NSCLC, surgery, prognosis
Introduction
Lung cancer remains the leading cause of cancer deaths worldwide.1 In the past few decades, lung cancer was binary classified into small-cell lung cancer and non-small-cell lung cancer (NSCLC). Nowadays, however, the discovery of the driver mutations and related target therapies has changed the way scientists and doctors are treating this disease. The 5-year survival rate of lung cancer remains low (only 16.8%) when taking all stages into account;2 some selected patients still benefit a lot from targeted therapy.3–6
KRAS protein functions as a GTPase that is essential for cell signaling. When extracellular stimuli, such as epidermal growth factor (EGF), activates the KRAS protein; the activated KRAS subsequently binds to a spectrum of downstream effector targets, and performs the signal transduction.7 Mutant KRAS protein, however, is stimulus independent. It can persistently activate the downstream effectors, such as the RAF-MEK-ERK cascade and PI3K-AKT-mTOR pathway, regardless of the absence of the upstream stimuli, and ultimately impact cell proliferation, survival, and metastasis.8
In NSCLC, the prevalence of KRAS mutation is 15%–32% according to the latest data.9 The mutation frequency varies among different ethnic populations. A relatively low frequency is identified in Asian and Latin America populations (15%–20%) compared with European population (20%–30%).10 This imbalance may partially attribute to the high frequency of EGFR mutation in Asian and Latin America populations, which are mutually exclusive with KRAS mutation, or other associated risk factors, such as tobacco use. Mutant KRAS gene is predominantly found in adenocarcinoma histology. In squamous histology tumors, it is a rare event. Many studies suggest that KRAS mutation is closely associated with cigarette smoking.11 Current or former smokers have a higher frequency of KRAS mutations than never smokers.11 Further analysis reveals that different smoking status leads to a different KRAS point mutation profile. Never smokers were significantly more likely than former or current smokers to have a transition mutation (G→A) rather than the transversion mutations known to be smoking-related (G→T or G→C; P<0.0001).12
In 1990, Slebos et al first reported that KRAS codon 12 point mutation was a strong and unfavorable prognostic factor based on an analysis of 69 lung adenocarcinoma patients.13 Since then, controversy remains on its prognostic significance for predicting the survival time in lung cancer. In 2005, a systematic review and meta-analysis revealed that KRAS mutation was of poor prognostic significance for survival in patients with NSCLC. Recently, a meta-analysis of 12 randomized trials confirmed that KRAS mutation was associated with poor prognosis for NSCLC, particularly for those with advanced stage disease or who received second or later line therapy or treated with EGFR TKIs.14
Although many studies analyzed the prevalence, clinicopathological features, and prognosis of KRAS mutation, contradictory conclusions have appeared and controversy remains.9–11 Besides, data from Asian population were insufficient. Therefore, here, we performed the largest retrospective study of KRAS mutation in patients with lung adenocarcinoma in an Asian ethnic group – to identify the prevalence of KRAS mutation in Chinese population, describe the clinicopathological features of these patients, and analyze their survival with other types of gene mutations.
Methods
Patients and samples
Patients were selected retrospectively at the Department of Thoracic Surgery, Fudan University Shanghai Cancer Center from 2007 to 2013. Patients who received neoadjuvant chemotherapy were excluded. All the qualified patients had primary lung cancer and received a radical resection. (Patients with stage I to stage III underwent complete surgical excision. As for stage IV patients, all of them had solitary or surgically resectable metastasis. Both the primary lesion and the metastasis were surgically removed through one or two surgeries). Two independent pathologists (Yuan Li and Xuxia Shen) pathologically confirmed their tumor samples as lung adenocarcinoma according to the new WHO classification of lung tumors. Tumor tissues and normal para-carcinoma tissues were sampled just after the surgical resection and immediately stored in liquid nitrogen.
This study was conducted in line with the Declaration of Helsinki and approved by the Institutional Review Board of the Fudan University Shanghai Cancer Center. Written informed consent was obtained from all patients to allow their biological samples to be genetically analyzed. The experimental protocol of this study was performed strictly in accordance to the guidelines.
RNA extraction
Total RNA was extracted from the tumor and normal paracarcinoma tissue as per standard protocols (RNeasy Mini Kit, Qiagen, Hilden, Germany), respectively. Total RNA samples were reverse transcribed into complementary DNA using a Revert Aid First Strand cDNA Synthesis Kit (Fermentas, St Leon-Rot, Germany).
Mutation analysis
KRAS (exons 2–3), EGFR (exons 18–22), HER2 (exons 18–21), and BRAF (exons 11–15) were amplified by polymerase chain reaction (PCR) using KOD-plus DNA polymerase and cDNAs.15 (The primers are listed in Table S1). Direct dideoxynucleotide method sequencing was performed to analyze the gene mutations. A combined strategy of reverse transcriptase PCR and quantitative real-time PCR was performed to detect ALK, RET, and ROS1 fusions. Fluorescent in situ hybridization was used as a validation for these fusion genes.16–18
Clinicopathological features
Clinical information including sex, age at diagnosis, pathologic tumor–lymph node–metastasis stage (according to the seventh edition of the lung cancer staging classification system) tumor differentiation, and smoking status was collected. Histologic subtypes of adenocarcinoma were confirmed based on the newest International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society multidisciplinary classification of lung adenocarcinoma. Survival and tumor relapse information was collected every 3 or 6 months after surgery in the clinic or by telephone.
Statistical analysis
Pearson’s chi-squared test or Fisher’s exact test (when the count in any cell of a contingency table was less than required) was used on categorical variables. The Kaplan–Meier method was performed to estimate the survival curve. Log-rank test was performed to compare the survival data. Multivariate analysis (Cox regression analysis) was used to assess the effect of covariates on relapse-free survival (RFS) and overall survival. We used SPSS for Windows (version 19.0) (IBM Corporation, Armonk, NY, USA) to analyze data. All tests were two-tailed and a P-value <0.05 indicated statistical significance.
Results
Clinicopathological features
A total of 1,368 patients from 2007 to 2013 were qualified in this retrospective study cohort. The average age was 59.7 years, ranging from 22.4 to 84.1 years. There were 623 (45.5%) males and 745 (54.5%) females. Of them, 369 were current smokers (27.0%), 79 were former smokers (5.8%), and 920 were never smokers (67.2%). The numbers of patients in pathological tumor–lymph node–metastasis stages I to IV were 749 (54.8%), 167 (12.2%), 425 (31.0%), and 27 (2.0%), respectively. In adenocarcinoma subtypes, acinar predominant (43.1%) was the most frequent, followed by solid predominant (18.6%), and papillary predominant (15.6%). More detailed clinicopathological features are sum-marized in Table 1.
Table 1.
Clinicopathological features of 1,368 lung adenocarcinoma patients
| Variables | KRAS | EGFR | HER2 | BRAF | ALK/RET/ROS1 | WT and others |
|---|---|---|---|---|---|---|
| Mutation type, n (%) | 113 (8.26) | 837 (61.18) | 32 (2.34) | 20 (1.46) | 102 (7.46) | 264 (19.30) |
| Age, years | ||||||
| <60, n (%) | 63 (56) | 414 (49) | 23 (72) | 11 (55) | 64 (73) | 116 (44) |
| ≥60, n (%) | 50 (44) | 423 (51) | 9 (28) | 9 (45) | 38 (27) | 148 (56) |
| Mean | 59.46 | 59.95 | 54.43 | 57.76 | 56.66 | 60.87 |
| Standard deviation | 9.04 | 9.96 | 9.85 | 9.47 | 9.81 | 10.72 |
| Sex | ||||||
| Male, n (%) | 90 (80) | 303 (36) | 2 (6) | 12 (60) | 39 (6) | 177 (67) |
| Female, n (%) | 23 (20) | 534 (64) | 30 (94) | 8 (40) | 63 (94) | 87 (33) |
| Smoker | ||||||
| Never, n (%) | 34 (30) | 657 (78) | 32 (100) | 8 (40) | 75 (100) | 114 (43) |
| Current/former, n (%) | 79 (70) | 180 (22) | 0 (−) | 12 (60) | 27 (−) | 150 (57) |
| Differentiation | ||||||
| Well, n (%) | 11 (10) | 131 (16) | 9 (28) | 4 (20) | 7 (27) | 30 (11) |
| Moderate, n (%) | 52 (46) | 494 (59) | 14 (44) | 8 (40) | 55 (46) | 102 (39) |
| Poor, n (%) | 50 (44) | 212 (25) | 9 (28) | 8 (40) | 40 (27) | 132 (50) |
| Pathological stage | ||||||
| I, n (%) | 56 (50) | 496 (59) | 20 (63) | 11 (55) | 39 (61) | 127 (48) |
| II, n (%) | 22 (19) | 90 (11) | 1 (3) | 2 (10) | 16 (3) | 36 (14) |
| III, n (%) | 33 (29) | 235 (28) | 11 (34) | 7 (35) | 42 (33) | 97 (37) |
| IV, n (%) | 2 (2) | 16 (2) | 0 (−) | 0 (−) | 5 (3) | 4 (2) |
| Pathological subtype | ||||||
| AIS, n (%) | 0 (−) | 15 (1.8) | 3 (9.4) | 0 (−) | 1 (1.0) | 9 (3.4) |
| MIA, n (%) | 2 (1.8) | 25 (3.0) | 2 (6.3) | 0 (−) | 0 (−) | 4 (1.5) |
| Lepidic, n (%) | 5 (4.4) | 92 (11.0) | 1 (3.1) | 3 (15.0) | 3 (2.9) | 17 (6.4) |
| Acinar, n (%) | 34 (30.1) | 430 (51.4) | 13 (40.6) | 5 (25.0) | 28 (27.5) | 79 (30.0) |
| Papillary, n (%) | 14 (12.4) | 139 (16.6) | 4 (12.5) | 3 (15.0) | 18 (17.6) | 35 (13.3) |
| Micropapillary, n (%) | 0 (−) | 18 (2.2) | 0 (−) | 0 (−) | 1 (1.0) | 2 (0.8) |
| Solid, n (%) | 34 (30.1) | 90 (10.8) | 4 (12.5) | 5 (25.0) | 30 (29.4) | 91 (34.5) |
| IMA, n (%) | 23 (20.4) | 18 (2.2) | 5 (15.6) | 2 (10.0) | 19 (18.6) | 15 (5.7) |
| Other, n (%) | 1 (0.9) | 10 (1.2) | 0 (−) | 2 (10.0) | 2 (2.0) | 12 (4.5) |
Note: Other includes enteric subtype and unknown.
Abbreviations: AIS, adenocarcinoma in situ; IMA, invasive mucinous adenocarcinoma; MIA, minimally invasive adenocarcinoma; WT, wild type.
Gene mutation spectrum
There were 837 patients who harbored EGFR mutation, accounting for approximately 60% of the population. KRAS mutation was detected in 113 (8.3%) of all the patients. Thirty-two (2.3%) patients had HER2 mutation, 20 (1.5%) patients had BRAF mutation, and 102 (7.5%) patients had ALK, RET, or ROS1 fusion genes (Figure 1). No patients with KRAS mutations had a concomitant mutation in EGFR, HER2, BRAF, or ALK/RET/ROS1.
Figure 1.
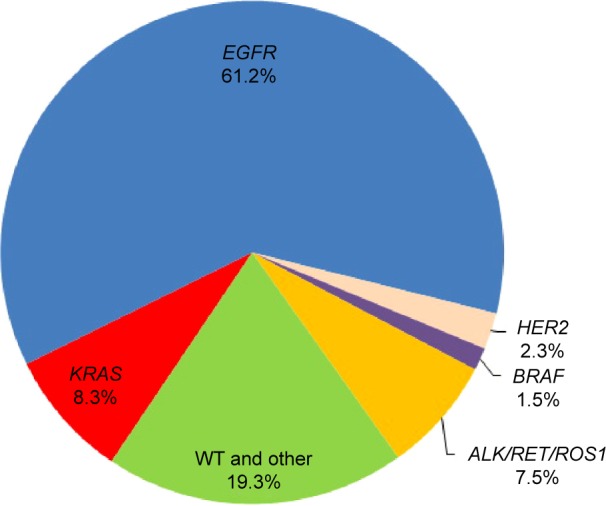
Frequency of gene mutations in 1,368 patients with lung adenocarcinoma.
Abbreviation: WT, wild type.
KRAS mutation subtypes
Nine different subtypes of KRAS mutation were identified, including five types of G12* mutations, two types of G13* mutations, and two types of Q61* mutations. G12C (GGT→TGT) was the most frequent amino acid substitution seen in this cohort, accounting for 33.6% of the patients followed by G12D (GGT→GAT) mutation (23.9%), G12V (GGT→GTT) mutation (22.1%), and G12A (GGT→GCT) mutation (7.1%). Seventy percent of patients (79/113) harbored transversion mutations (G12A, G12C, G12V, G13C, Q61H) (Figure 2).
Figure 2.
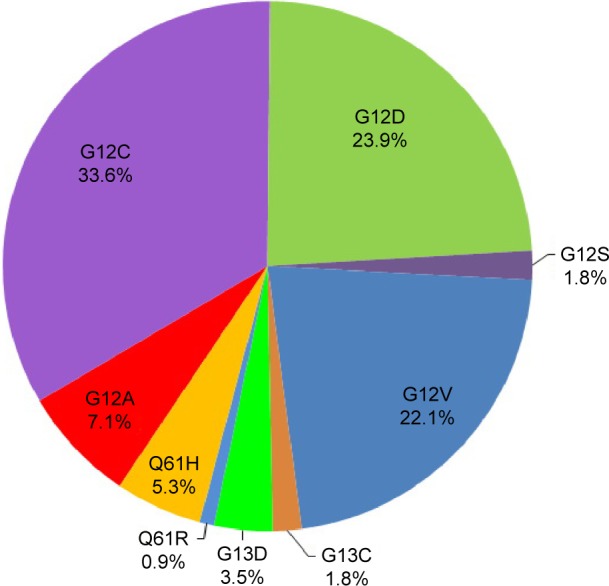
Frequency of KRAS mutation subtypes in 113 KRAS mutation patients.
KRAS mutation and clinicopathological variables
In this study, KRAS mutation was more frequently found in male patients (odds ratio 5.30; 95% confidence interval (CI) 3.31–8.49; P<0.001) and former/current smoke patients (odds ratio 5.58; 95% CI 3.67–8.49; P<0.001). Tumors with KRAS mutation were poorly differentiated (odds ratio 1.69; 95% CI 1.15–2.50; P=0.008). Among all the adenocarcinoma subtypes, invasive mucinous adenocarcinoma predominant (20.4% vs 4.7%, P<0.001) and solid predominant (30.1% vs 17.5%, P=0.004) were more frequent in KRAS mutant patients compared with KRAS wide-type patients (Table 2).
Table 2.
Features of patients with lung adenocarcinoma harboring KRAS mutation
|
KRAS mutation (n=113)
|
KRAS WT (n=1,255)
|
P-value | |||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Sex | P<0.001* | ||||
| Male | 90 | 80 | 533 | 42 | |
| Female | 23 | 20 | 722 | 58 | |
| Age | P=0.245 | ||||
| <60 years | 63 | 56 | 628 | 50 | |
| ≥60 years | 50 | 44 | 627 | 50 | |
| Smoker | P<0.001* | ||||
| Never | 34 | 30 | 886 | 71 | |
| Former/current | 79 | 70 | 369 | 29 | |
| Differentiation | P=0.008* | ||||
| Well/moderate | 63 | 56 | 854 | 68 | |
| Poor | 50 | 44 | 401 | 32 | |
| Pathological stage | P=0.626 | ||||
| I/II | 78 | 69 | 838 | 67 | |
| III/IV | 35 | 31 | 417 | 33 | |
| IMA predominant | P<0.001* | ||||
| Yes | 23 | 20 | 59 | 5 | |
| No | 90 | 80 | 1,196 | 95 | |
| Solid predominant | P=0.004* | ||||
| Yes | 34 | 30 | 220 | 18 | |
| No | 79 | 70 | 1,035 | 82 | |
Note:
Indicates statistical significance.
Abbreviations: IMA, invasive mucinous adenocarcinoma; WT, wild type.
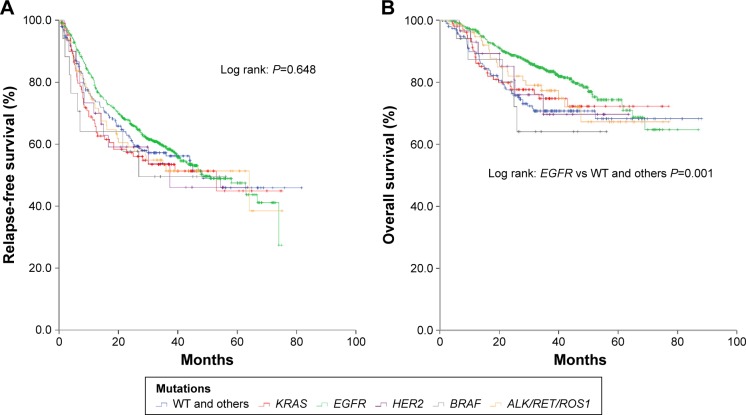
Survival analysis
In all, 1,131 patients were involved in survival analysis, including 108 patients harboring KRAS mutation. Follow-up started from October 2007 to June 2015.
The follow-up time ranged from 1 to 88 months. The median RFS for all patients and KRAS mutant patients was 49 months (95% CI 40–57 months) and 53 months (95% CI 16–90 months), respectively. No statistical significance was found in RFS between patients with KRAS mutation and patients with other mutations (Figure 3A). Patients with EGFR mutation lived longer than mutation wide type patients (P=0.001). No statistical significance was found between patients with KRAS mutation and patients with other mutations in overall survival (Figure 3B). In multivariate analysis, former/current smoke patients (hazard ratio [HR] 1.32; 95% CI 1.09–1.58; P=0.004), advanced stage (HR 3.36; 95% CI 2.76–4.08; P<0.001), and poor differentiation in tumor (HR 3.07; 95% CI 2.09–4.50; P<0.001) were risk factors in recurrence. The occurrence of KRAS mutation in patients did not mean an unfavorable prognosis; nevertheless, male patients, patients with advanced stage, or patients with poor differentiation in tumor lived shorter than others (Table 3).
Figure 3.
Relapse-free survival (A) and overall survival (B) in all patients.
Note: Kaplan–Meier curve for relapse-free survival and overall survival in 1,131 patients with lung adenocarcinoma.
Abbreviation: WT, wild type.
Table 3.
Univariate and multivariate survival analysis of 1,131 patients with lung adenocarcinoma
| Variable | Category | RFS analysis
|
OS analysis
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate
|
Multivariate
|
Univariate
|
Multivariate
|
||||||||||
| HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | ||
| Age, years | ≥60/<60 | 1.03 | (0.86–1.23) | 0.77 | NA | NA | NA | 1.18 | (0.91–1.53) | 0.219 | 1.30 | (0.99–1.69) | 0.052 |
| Sex | Male/female | 1.60 | (1.33–1.91) | <0.001* | NA | NA | NA | 1.77 | (1.36–2.30) | <0.001* | 1.34 | (1.02–1.75) | <0.033* |
| Smoker | Yes/no | 1.76 | (1.47–2.11) | <0.001* | 1.32 | (1.09–1.58) | 0.004* | 1.75 | (1.34–2.27) | <0.001* | NA | NA | NA |
| Pathological stage | III, IV/I, II | 4.31 | (3.59–5.17) | <0.001* | 3.36 | (2.76–4.08) | <0.001* | 4.06 | (3.11–5.31) | <0.001* | 2.94 | (2.21–3.92) | <0.001* |
| Differentiation | Moderate/well | 2.86 | (2.00–4.09) | <0.001* | 2.24 | (1.56–3.22) | <0.001* | 3.73 | (1.95–7.15) | <0.001* | 2.78 | (1.44–5.37) | 0.002* |
| Poor/well | 5.91 | (4.10–8.51) | <0.001* | 3.07 | (2.09–4.50) | <0.001* | 9.94 | (5.21–18.99) | <0.001* | 5.17 | (2.64–10.12) | <0.001* | |
| KRAS mutation | Mutation/WT | 1.18 | (0.88–1.60) | 0.266 | NA | NA | NA | 1.26 | (0.83–1.92) | 0.271 | NA | NA | NA |
Note:
Indicates statistical significance.
Abbreviations: CI, confidence interval; HR, hazard ratio; NA, not applicable; OS, overall survival; RFS, relapse-free survival; WT, wild type.
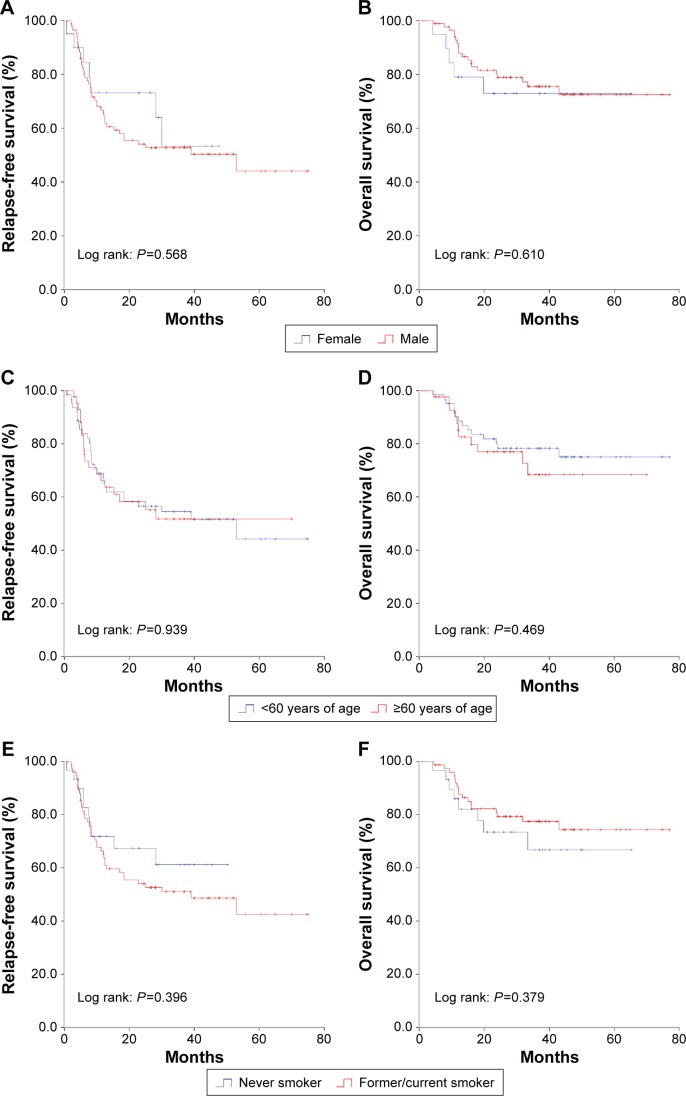
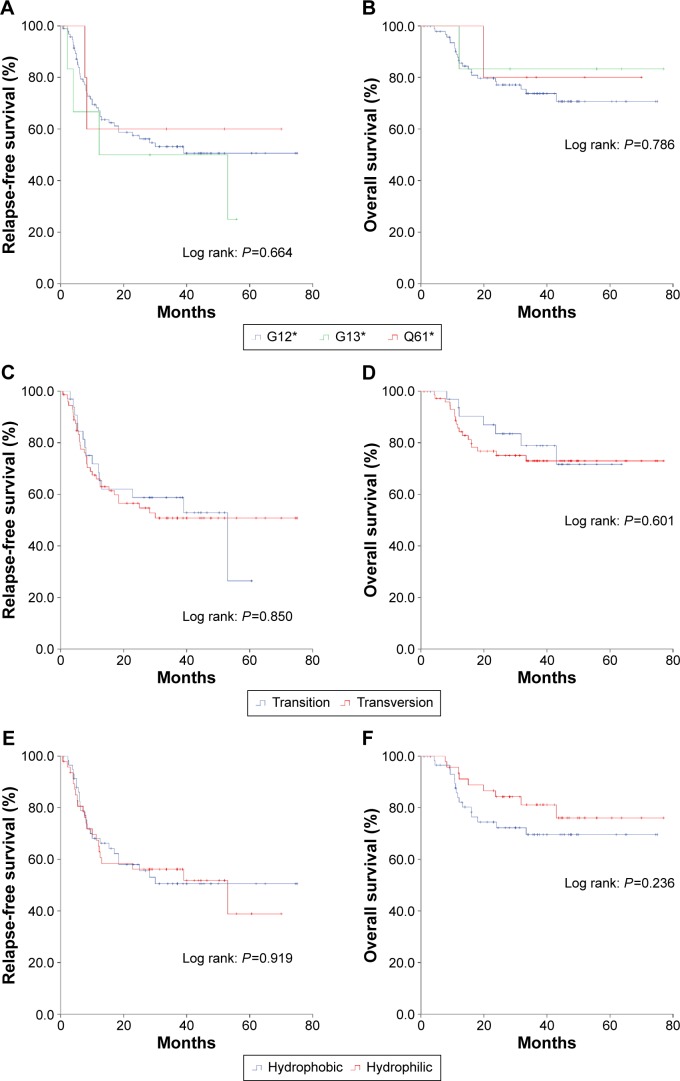
Survival analysis of KRAS subtypes
In 108 qualified patients with KRAS mutation, the median RFS was 53 months, while the median overall survival was not reached. No significant difference was found in survival among patients with different KRAS point mutations. Sex (Figure 4A and B), age (Figure 4C and D), and smoking status (Figure 4E and F) did not affect survival time either. Those patients with advanced stage or whose tumor was poorly differentiated got an earlier recurrence and shorter survival (Table 3). Mutation sites in KRAS did not affect the prognosis (Figure 5A and B). There was no difference in survival between patients with KRAS transition mutations and patients with KRAS transversion mutations (Figure 5C and D). When the KRAS mutation was divided into two groups (hydrophobic alterations [G12C and G12V] and hydrophilic alterations [such as G12D]), no significant difference was found in either RFS or overall survival (Figure 5E and F). Multivariate analysis revealed that advanced stage was the only risk factor that indicated shorter recurrence (HR 2.66; 95% CI 1.51–4.70; P=0.001), and smoking status, sex, age, or different KRAS subtypes did not affect the outcome (Table 3).
Figure 4.
Relapse-free survival and overall survival of sex (A and B), age (C and D), and smoking status (E and F) in KRAS mutation patients.
Figure 5.
Relapse-free survival and overall survival of codon (A and B), transition/transversion (C and D), and hydrophobic/hydrophilic (E and F) subtypes in KRAS mutation patients.
Discussion
In the present study, KRAS mutation was detected in 8.3% (113/1,368) of lung adenocarcinoma patients in Chinese population. This number was relatively lower compared with the data from Asian population. European population, however, had a much higher mutation rate of KRAS gene.9,10 Besides, KRAS mutation was predominantly found in male patients and smokers. KRAS mutation and other driver mutations, such as EGFR, HER2, and BRAF, were mutually exclusive. Poor differentiation appeared in almost half of the tumors with KRAS mutation, compared with less than a third of the non-KRAS mutant tumors. This suggested that KRAS mutation probably led to an unfavorable prognosis. Meanwhile, the proportion of invasive mucinous adenocarcinoma predominant subtype and solid predominant subtype was significantly higher in KRAS mutation. Considering these two subtypes, especially the solid predominant subtype as indicators of poor prognosis, KRAS mutation might be a late event in lung cancer. Keohavong et al19 reported that specific KRAS point mutations (such as G12V) were associated with poorer outcome. Recently, a meta-analysis which synthesized 12 randomized trials also confirmed that KRAS mutations were related to poor survival benefit for NSCLC.14 Our data, however, did not show this pattern in survival analysis.
KRAS point mutation in human cancers takes place primarily at residues G12, G13, or Q61, with single amino acid substitution. In lung cancer, KRAS mutation occurs predominantly at residues G12 or G13.7,9,12 Mutations can be further divided into transversion and transition subtypes according to its biological features and prognostic impact. Riely’s discovery of different smoking status leading to a different KRAS point mutation profile suggested that there might be an explicit mechanism of tobacco carcinogens on KRAS mutation.12 Conflicting data also arouse the suspicion on cigarette smoke.20 In animal experiments, scientists revealed quite a similar pattern in lung tumors from mice treated with BaP, 5-methylchrysene, and benzo[b] fluoranthene, which are carcinogens commonly found in tobacco smoke.21 However, detailed analysis showed that the methylguanine pathway of nicotine-derived nitrosamine ketone, also known as 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) metabolic results in a high percentage of GGT→GAT mutations (transition) in codon 12 of KRAS,22 while pyridyloxobutylation leads to more G→T mutation (transversion) in codon 12.23 The relative significance of these pathways in human lung cells remains unknown, and further investigations are needed.
Remarkably, Ihle et al24 classified KRAS mutation into hydrophobic (G12C and G12V) and hydrophilic alteration (G12D) subgroups. In vitro experiments demonstrated different patterns of downstream signal transduction and response to targeted therapies between these two subgroups. Specifically, mutant G12C or G12V KRAS protein would activate Ral signaling and decrease growth factor-dependent Akt activation, while mutant G12D KRAS protein would predominantly activate the PI3K and MEK/ERK pathways. Such results could be partially explained by the changes in spatial conformation after mutation. Finally, tumor behavior and drug sensitivity varied in different subgroups of KRAS mutation.25 Although in general, KRAS codon 12 point mutation was proven to indicate a poor prognosis, a more specific classification was required to assess its biological malignancy and therapeutic efficacy. In clinical practice, individualized treatment was recommended concerning patients harboring KRAS mutation.
Nowadays, scientists also focus on KRAS mutation co-occurring with other gene mutations. Lung adenocarcinomas harboring STK11/LKB1, TP53, or CDKN2A/B mutation as well as KRAS mutation identified distinct clinicopathological features and different therapeutic responses.26 This insight would direct the treatment strategy not only pointing at a single KRAS gene, but a set of related genes.
There are no known direct KRAS targeted agents. Survival of patients harboring KRAS mutation is almost disappointing with or without chemotherapy, targeted therapy, or other treatment. Some researchers came to a conclusion that KRAS status might indicate poor response to EGFR TKIs.27–31 This evidence should be considered with caution due to the small numbers involved in the analyses. Meanwhile, meta-analysis showed that KRAS mutations are highly specific negative predictors of response to single-agent EGFR TKIs in advanced NSCLC.32 Limited success was developed on the treatment of KRAS mutation in the past decade; however, delightful consequences emerged.
A Phase II randomized trial showed that selumetinib plus docetaxel had promising efficacy, albeit with a higher number of adverse events than with docetaxel alone, in previously treated advanced KRAS-mutant NSCLC.33 Patients in the experiment group had a longer progression-free survival and 37% of them had an objective response.
By blocking one of the most important downstream pathways of KRAS, an MEK1/2 inhibitor (AZD6244) combined with cisplatin showed an antitumor effect in KRAS-dependent lung cancer cells and animal models.34 Clinical studies of applications of MEK1/2 inhibitors in cisplatin-based chemotherapy for lung cancer patients harboring KRAS mutation are urgently required.
Ostrem et al35 developed a small molecule that irreversibly bound to a common oncogenic mutant, K-Ras (G12C), while it had no effect on the wild-type protein. Once bound, the inhibitor–KRAS compound subverted the native nucleotide preference to favor GDP over GTP and therefore impaired binding to the downstream Raf protein. If it is possible to make this molecule available in clinics in the future, it will make KRAS mutation targetable and may bring benefit to patients.
Furthermore, oncogenic drivers, such as mutated KRAS can be targeted with synthetic lethality approaches.36 Although synthetic lethality till now is a concept and has not yet been applied in clinical practice, it is a promising rationale and will ultimately become a therapeutic approach.
Conclusion
In Chinese population, we identified KRAS mutation in 8.3% (113/1,368) of the patients with lung adenocarcinoma. KRAS mutation defines a molecular subset of lung adenocarcinoma with unique clinicopathological features. KRAS mutations were more frequent in male patients, former/current smokers, and patients harboring KRAS mutations who showed poor differentiation in tumor tissues.
Supplementary material
Table S1.
Primers used in this study
| Primers for detecting KRAS mutation | ||
| Target | Forward primer (5′>>3′) | Reverse primer (5′>>3′) |
| KRAS exon 2–3 | AGGCCTGCTGAAAATGACTG | TGGTGAATATCTTCAAATGATTTAGT |
| Primers for detecting EGFR mutation | ||
| Target | Forward primer (5′>>3′) | Reverse primer (5′>>3′) |
| EGFR exon 18–22 | TGAAGGCTGTCCAACGAATG | AGGCGTTCTCCTTTCTCCAG |
| Primers for detecting HER2 mutation | ||
| Target | Forward primer (5′>>3′) | Reverse primer (5′>>3′) |
| HER2 exon 18–21 | CCCTCTGACGTCCATCATCT | GCAGGGTCTGGACTGAAGAA |
| Primers for detecting BRAF mutation | ||
| Target | Forward primer (5′>>3′) | Reverse primer (5′>>3′) |
| BRAF exon 11–15 | TCAGAAGACAGGAATCGAATGA | GATGACTTCTGGTGCCATCC |
Acknowledgments
This work was funded by the National Natural Science Foundation of China (81330056, 81401886, 81401891, 81422029, 81472173, and 81372525), the Key Project of Science and Technology Commission of Shanghai Municipality (JGGG1302), and Shen-Kang Center Project (SKMB1201).
Footnotes
Author contributions
Haiquan Chen is the guarantor of the manuscript and contributed to conception and study design, analysis of data, and review and revision of the manuscript. Difan Zheng, Rui Wang, and Yang Zhang contributed to conception and study design, acquisition and analysis of data, and writing and revision of the manuscript. Yunjian Pan and Xinghua Cheng contributed to acquisition of data and writing and revision of the manuscript. Chao Cheng and Xuxia Shen contributed to analysis of data and revision of the manuscript. Shanbo Zheng, Hang Li, Ranxia Gong, and Yuan Li: contributed to acquisition of data and revision of the manuscript. Yihua Sun contributed to conception and study design, and review and revision of the manuscript. All authors gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ettinger DS, Wood DE, Akerley W, et al. Non-small cell lung cancer, version 6.2015. J Natl Compr Canc Netw. 2015;13(5):515–524. doi: 10.6004/jnccn.2015.0071. [DOI] [PubMed] [Google Scholar]
- 3.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer – molecular and clinical predictors of outcome. N Engl J Med. 2005;353(2):133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 4.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 5.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368(25):2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 6.Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 7.Roberts PJ, Stinchcombe TE. KRAS mutation: should we test for it, and does it matter? J Clin Oncol. 2013;31(8):1112–1121. doi: 10.1200/JCO.2012.43.0454. [DOI] [PubMed] [Google Scholar]
- 8.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 9.Yu HA, Sima CS, Shen R, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol. 2015;10(3):431–437. doi: 10.1097/JTO.0000000000000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arrieta O1, Cardona AF, Martín C, et al. Updated frequency of EGFR and KRAS mutations in non-small-cell lung cancer in Latin America: The Latin-American Consortium for the Investigation of Lung Cancer (CLICaP) J Thorac Oncol. 2015;10(5):838–843. doi: 10.1097/JTO.0000000000000481. [DOI] [PubMed] [Google Scholar]
- 11.Mao C, Qiu LX, Liao RY, et al. KRAS mutations and resistance to EGFR-TKIs treatment in patients KRAS mutations in NSCLC with non-small cell lung cancer: a meta-analysis of 22 studies. Lung Cancer. 2010;69:272–278. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 12.Riely GJ, Kris MG, Marks JL, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14(18):5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–565. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- 14.Ying M, Zhu XX, Zhao Y, Li DH, Chen LH. KRAS mutation as a biomarker for survival in patients with non-small cell lung cancer, a meta-analysis of 12 randomized trials. Asian Pac J Cancer Prev. 2015;16(10):4439–4445. doi: 10.7314/apjcp.2015.16.10.4439. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Ren Y, Fang Z, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol. 2010;28:4616–4620. doi: 10.1200/JCO.2010.29.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R, Pan Y, Li C, et al. The use of quantitative real-time reverse transcriptase PCR for 5′ and 3′ portions of ALK transcripts to detect ALK rearrangements in lung cancers. Clin Cancer Res. 2012;18:4725–4732. doi: 10.1158/1078-0432.CCR-12-0677. [DOI] [PubMed] [Google Scholar]
- 17.Pan Y, Zhang Y, Li Y, et al. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: a comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer. 2014;84(2):121–126. doi: 10.1016/j.lungcan.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Rui W, Lei W, Yuan L, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res. 2014;20(15):4107–4114. doi: 10.1158/1078-0432.CCR-14-0284. [DOI] [PubMed] [Google Scholar]
- 19.Keohavong P, DeMichele MA, Melacrinos AC, et al. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clin Cancer Res. 1996;2:411–418. [PubMed] [Google Scholar]
- 20.Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18(22):6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nesnow S, Ross JA, Mass MJ, Stoner GD. Mechanistic relationships between DNA adducts, oncogene mutations, and lung tumorigenesis in strain A mice. Exp Lung Res. 1998;24:395–405. doi: 10.3109/01902149809087376. [DOI] [PubMed] [Google Scholar]
- 22.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 23.Ronai ZA, Gradia S, Peterson LA, Hecht SS. G to A transitions and G to T transversions in codon 12 of the Ki-ras oncogene isolated from mouse lung tumors induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and related DNA methylating and pyridyloxobutylating agents. Carcinogenesis. 1993;14:2419–2422. doi: 10.1093/carcin/14.11.2419. [DOI] [PubMed] [Google Scholar]
- 24.Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104(3):228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garassino MC, Marabese M, Rusconi P, et al. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non-small-cell lung cancer. Ann Oncol. 2011;22(1):235–237. doi: 10.1093/annonc/mdq680. [DOI] [PubMed] [Google Scholar]
- 26.Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS – mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han SW, Kim TY, Jeon YK, et al. Optimization of patient selection for gefitinib in non-small cell lung cancer by combined analysis of epidermal growth factor receptor mutation, K-ras mutation, and Akt phosphorylation. Clin Cancer Res. 2006;12:2538–2544. doi: 10.1158/1078-0432.CCR-05-2845. [DOI] [PubMed] [Google Scholar]
- 29.Felip E, Rojo F, Reck M, et al. A phase II pharmacodynamic study of erlotinib in patients with advanced non-small cell lung cancer previously treated with platinum-based chemotherapy. Clin Cancer Res. 2008;14:3867–3874. doi: 10.1158/1078-0432.CCR-07-5186. [DOI] [PubMed] [Google Scholar]
- 30.Miller VA, Riely GJ, Zakowski MF, et al. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol. 2008;26:1472–1478. doi: 10.1200/JCO.2007.13.0062. [DOI] [PubMed] [Google Scholar]
- 31.Roberts PJ, Stinchcombe TE, Der CJ, Socinski MA. Personalized medicine in non-small-cell lung cancer: Is KRAS a useful marker in selecting patients for epidermal growth factor receptor-targeted therapy? J Clin Oncol. 2010;28:4769–4777. doi: 10.1200/JCO.2009.27.4365. [DOI] [PubMed] [Google Scholar]
- 32.Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: A systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 33.Janne PA, Shaw AT, Pereira JR, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14(1):38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- 34.Kim EY, Kim A, Kim SK, Chang YS. AZD6244 inhibits cisplatin-induced ERK1/2 activation and potentiates cisplatin-associated cytotoxicity in K-ras G12D reclinical models. Cancer Lett. 2015;358(1):85–91. doi: 10.1016/j.canlet.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 35.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras (G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503(7477):548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLornan DP, List A, Mufti GJ. Applying synthetic lethality for the selective targeting of cancer. N Engl J Med. 2014;371(18):1725–1735. doi: 10.1056/NEJMra1407390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Primers used in this study
| Primers for detecting KRAS mutation | ||
| Target | Forward primer (5′>>3′) | Reverse primer (5′>>3′) |
| KRAS exon 2–3 | AGGCCTGCTGAAAATGACTG | TGGTGAATATCTTCAAATGATTTAGT |
| Primers for detecting EGFR mutation | ||
| Target | Forward primer (5′>>3′) | Reverse primer (5′>>3′) |
| EGFR exon 18–22 | TGAAGGCTGTCCAACGAATG | AGGCGTTCTCCTTTCTCCAG |
| Primers for detecting HER2 mutation | ||
| Target | Forward primer (5′>>3′) | Reverse primer (5′>>3′) |
| HER2 exon 18–21 | CCCTCTGACGTCCATCATCT | GCAGGGTCTGGACTGAAGAA |
| Primers for detecting BRAF mutation | ||
| Target | Forward primer (5′>>3′) | Reverse primer (5′>>3′) |
| BRAF exon 11–15 | TCAGAAGACAGGAATCGAATGA | GATGACTTCTGGTGCCATCC |