Biosynthesis of the labdane-related diterpenoids, a large superfamily of over 7,000 natural products, is characterized by the use of class II diterpene cyclases.[1] The vast majority of these biocatalysts are exclusively found in plants, stemming from repeated evolutionary diversification of the ent-copalyl diphosphate synthase (CPS) required for gibberellin phytohormone biosynthesis – i.e., via gene duplication and neofunctionalization.[2] As implied by this evolutionary scenario, these enzymes must exhibit catalytic plasticity that enables new products to readily arise. Consistent with this hypothesis, we have shown that substitution of smaller residues for the catalytic base group can lead to the incorporation of water, yielding hydroxylated products.[3] Here it is reported that larger aromatic replacement of the histidine from the catalytic base group leads to even more profound alteration of product outcome, rearrangement via a series of 1,2-hydride and methyl shifts. Coupled with additional mutational analysis and quantum chemical calculations, this clarifies the catalyzed reaction and underlying enzymatic structure-function relationships of the ancestral CPS.
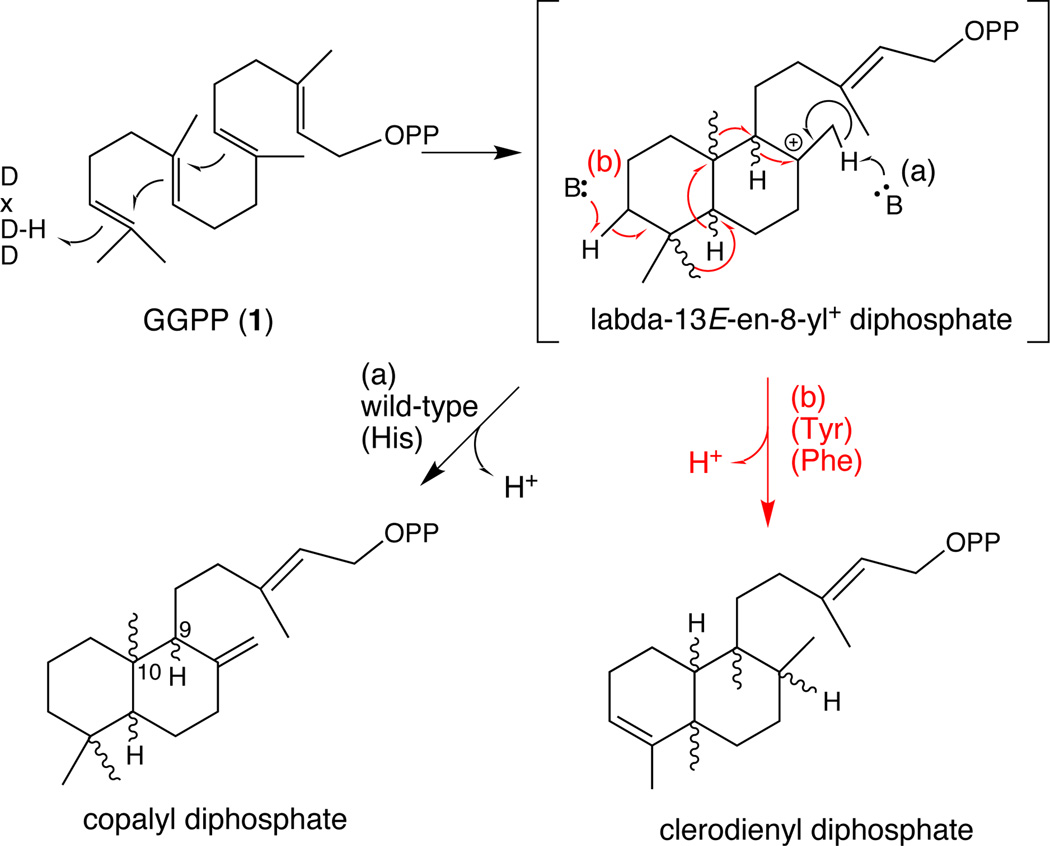
Class II diterpene cyclases catalyse cycloisomerization of the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP, 1), utilizing an acid/base mechanism wherein the middle aspartate of a highly conserved DxDD motif is employed as the general acid to protonate the terminal carbon-carbon double bond (C=C) of 1.[4] Sequential anti addition from the two internal C=C bonds leads to the bicyclic (trans-decalin) intermediate labda-13E-en-8-yl+ diphosphate. Typically this is deprotonated at C17 to yield copalyl diphosphate (CPP), which can differ in stereochemistry – e.g., the (9R,10R) ent-CPP (2) relevant to gibberellin biosynthesis.[1] However, other product outcomes are possible. Particularly relevant here, rather than deprotonation to produce CPP, a sequence of 1,2-hydride and methyl shifts can transpire, resulting in new skeleton backbones such as the fully rearranged clerodanes (Scheme 1).[1] Although class II diterpene cyclases have been characterized that produce rearranged labdane-related diterpenoids from bacteria (see Scheme S1 in the Supporting Information),[5] none are known from plants, and the enzymatic basis for such rearrangement is not known.
Scheme 1.
Relevant cyclization reactions.
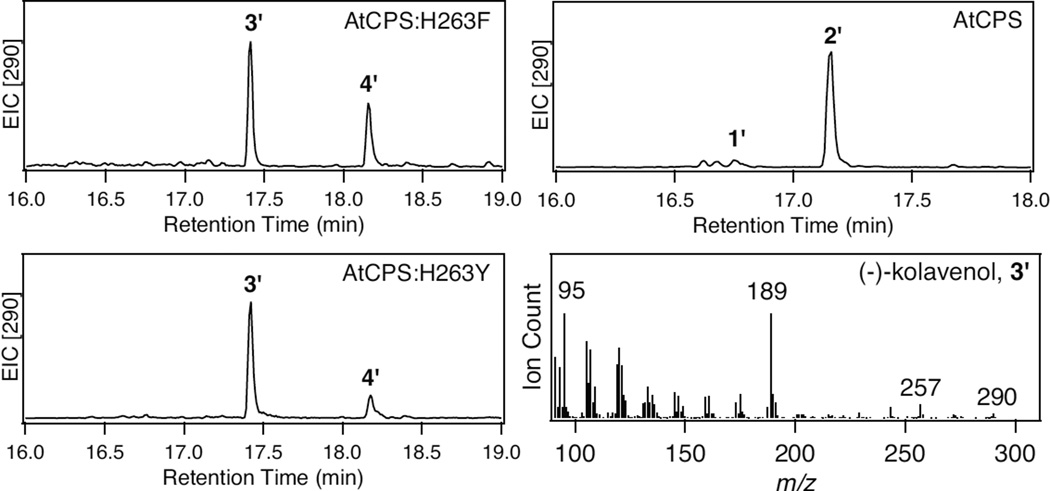
Recently, based on previously determined crystal structures,[6] the catalytic base group in the CPS from Arabidopsis thaliana (AtCPS) was identified.[3c] In this group the general base appears to be a water molecule, activated by ligation, in part by the side chains of a histidine and asparagine, which form a catalytic dyad conserved in all CPSs involved in gibberellin plant hormone biosynthesis (H263 and N322 in AtCPS). The role of the water was indicated by the fact that substitution of alanine for either residue, or both, led to addition of water to the ent-labda-13E-8-yl+ intermediate (A), with formation of ent-labda-13E-en-8-ol diphosphates, yielding both epimers (8α- and 8β- hydroxyls).[3c] Substitution of larger aliphatic residues for H263 in AtCPS led to increasingly inactive enzymes, which was interpreted as suggesting that a correctly positioned base (e.g., either the imidazole side chain of His or sufficient space for more water molecules) is required for catalysis. However, it is possible that such an electron-rich group is required to initiate the reaction, but not necessarily as a general base. This hypothesis was tested by substitution of larger aromatic residues for the catalytic His. Modeling indicated that a tryptophan would be too large, thus, phenylalanine and tyrosine were substituted for H263 in AtCPS (AtCPS:H263F and AtCPS:H263Y; see Supporting Information for the Experimental Section). Strikingly, novel enzymatic activity was observed, either in vitro or through expression in E. coli also engineered to produce 1 (Figure 1).[7] The new compound, following dephosphorylation and analysis by gas chromatography with mass spectral detection (GC-MS), exhibited a mass spectrum similar to the rearranged clerodane produced by a bacterial terpentedienyl diphosphate synthase.[5a] However, these compounds exhibited different retention times, demonstrating some (stereo)chemical difference between them (Figure S1). Therefore, approximately 1.5 mg of the dephosphorylated compound was purified and its structure investigated by NMR. This was found to correspond to a neoclerodane,[8] with comparison to previously reported spectral data and optical rotation demonstrating that this was (−)-kolavenol (see Table S1 and Figures S2–S4).[9] Therefore, these mutants, AtCPS:H263F and, particularly, AtCPS:H263Y, produce (−)-kolavenyl diphosphate (KPP, 3, Scheme 2). Thus, they not only catalyse bicyclization of 1, but also promote a subsequent series of 1,2- hydride and methyl shifts, leading to full rearrangement, and novel product outcome. Consistent with the good yields observed from the bacterial metabolic engineering system (Figure 1), kinetic analysis demonstrates that both mutants exhibit only relatively small decreases in catalytic efficiency relative to wild-type AtCPS (~10-fold), with 30-fold decreases in kcat offset by ~2-fold increases in substrate affinity, as well as ~5-fold decreases in substrate inhibition (see Table S2).
Figure 1.
Effect of H263Y and H263F mutations on AtCPS product outcome. Wild-type also shown for comparative purposes. Extracted ion chromatographs (m/z = 290) from GC-MS analyses of the dephosphorylated products (numbering as in text, with prime′notation used to indicate that these are dephosphorylated derivatives). Also presented is the mass spectrum for the novel peak (3′, kolavenol).
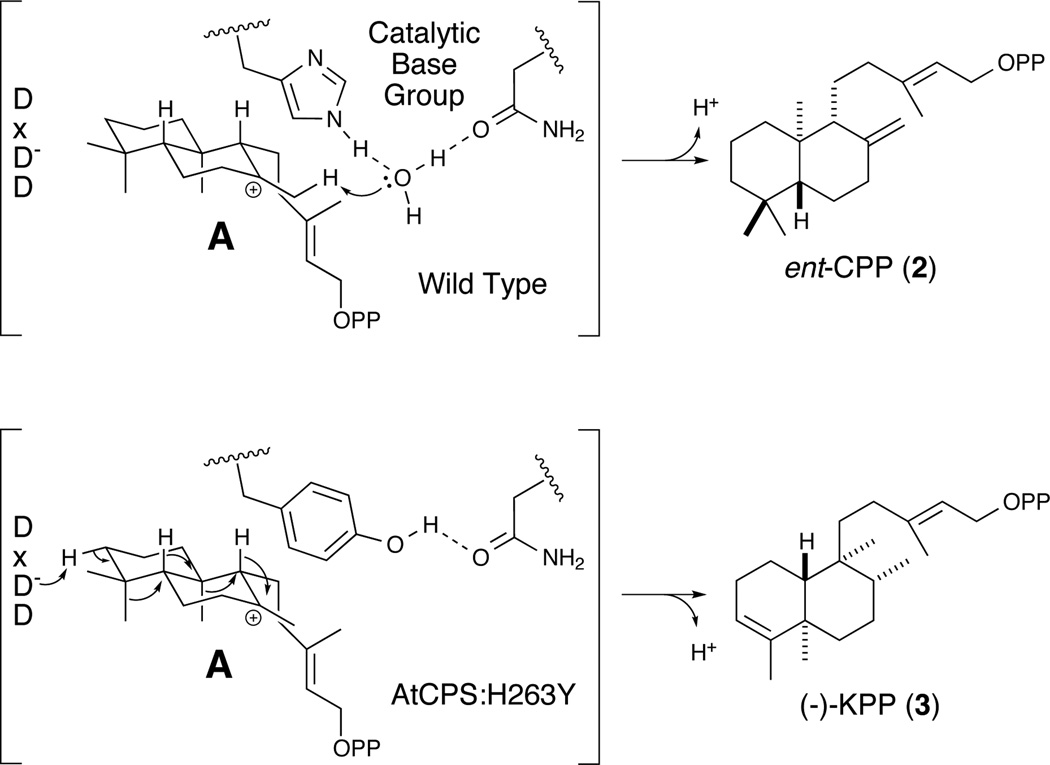
Scheme 2.
Reactions catalysed by wild-type and the H263Y mutant of tCPS.
Formation of (−)-KPP (3) requires removal of a proton from the same carbon that is protonated to initiate the bicyclization reaction. This suggests that the same proton used to initiate bicyclization might be removed following the subsequent rearrangement. To test this mechanistic hypothesis, labelling studies were performed with in vitro enzymatic assays in D2O with wild-type and the H263Y mutant enzymes. Upon overnight incubation, dephosphorylation of the products, and analysis by GC-MS, it was found that (−)-kolavenol derived from 3 produced by the mutant enzyme in D2O retained a molecular weight of 290 Da – i.e., deuterium is not incorporated, while ent-copalol derived from 2 formed by the wild-type enzyme in D2O exhibited a molecular weight of 291 Da – i.e., as expected, deuterium is incorporated (Figure S5). This result demonstrates that formation of 3 proceeds via removal of the same proton originally added to initiate (bi)cyclization. Although this result does not rule out other possible bases (e.g., another water molecule), it seems likely that the middle aspartate of the DxDD motif (D379 in AtCPS) that acts as the catalytic acid also acts as the catalytic base to remove the proton in formation of 3.
These results provide insights into the overall reaction mechanism of class II diterpene cyclases. Substitution of larger aromatics for His263 still promotes protonation and bicyclization, which presumably results from not only enforcement of the folding of 1 into the correct conformation, but also interactions of the aromatic π electrons with the resulting ent-labda-13E-en-8-yl+ intermediate (A).[10] As previously described,[3c] substitution of larger aliphatic residues (i.e., valine and isoleucine) for H263 simply resulted in decreasing yields of the same ent-labda-13E-en-8α-ol diphosphate (4) primary product as formed by AtCPS:H263A (i.e., with AtCPS:H263V) or essentially complete loss of any product (i.e., with AtCPS:H263I). Although this may be due in part to improper folding (of the protein and/or substrate), no rearranged compounds were detected from reactions with any of these mutants. The ability of larger aromatic side chains at this position (residue 263 in AtCPS) to promote the production of 3 presumably then arises from steric blockage of access to the carbocation centre of A (e.g., by a water molecule for production of 4). Notably, the AtCPS:H263Y mutant leads to more specific production of 3 (77%), apparently at the expense of 4, than the AtCPS:H263F mutant (55% of 3). This may be due to the potential ability of the tyrosine hydroxyl group to form a hydrogen bond with N322, generating a more stable enzyme and/or, possibly even more importantly, increased effectiveness in blocking the binding of water at this position in the active site (Scheme 2 and Figure S6).
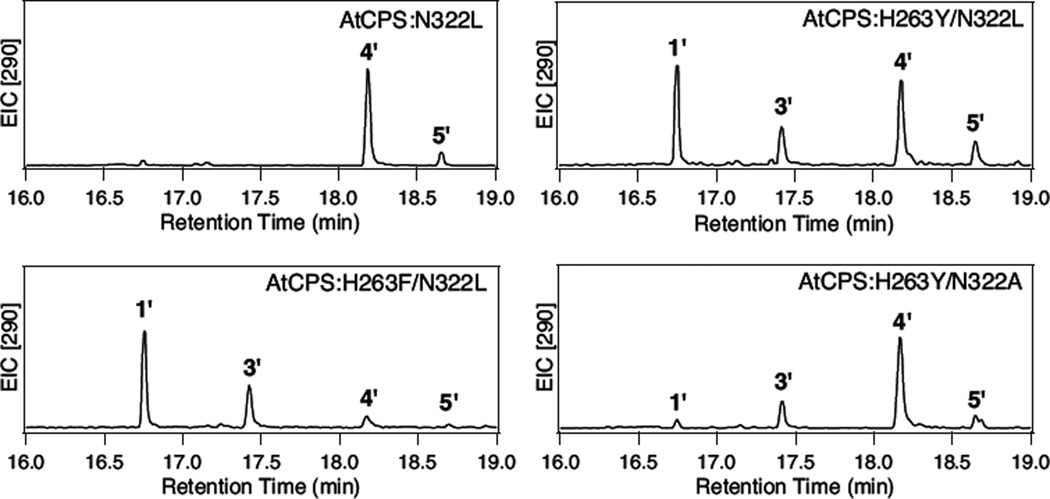
The role of the Asn from the catalytic dyad (N322 in AtCPS) in the observed production of 3 was probed by additional mutagenesis. Substitution of leucine alone (i.e., AtCPS:N322L) led to only production of unrearranged products, predominantly the hydroxylated 4, along with small amounts of the epimeric ent-labda-13E-en-8β-ol diphosphate (5). However, in the context of aromatic substitutions for H263, significant amounts of 3 were observed as well, particularly with AtCPS:H263F/N322L (75%, versus the 33% of 3 observed with AtCPS:H263Y/N322L). Significant loss of yield in the bacterial metabolic engineering system was observed with these two double mutants (Figure 2). While this could be restored to some extent by alanine substitution instead (e.g., AtCPS:H263Y/N322A), this also reduced the production of 3 relative to 4. Intriguingly, these results differ from those observed for substitution of H263, which seems to support the hypothesis that either an aromatic side chain or one sufficiently small enough to allow water to bind in its place is required at this position to initiate bicyclization, with only a minimal (if any) role for the functionality of the asparagine residue from the catalytic base group, apart from serving as an activating ligand for the water that normally serves as the general base.
Figure 2.
Effect of N322L mutation, in the absence or presence of H263Y or F mutations, on AtCPS product outcome. H263Y/N322A double-mutant also is shown for comparative purposes. Extracted ion chromatographs (m/z = 290) from GC-MS analyses of the dephosphorylated products (numbering as in text, with prime′notation used to indicate that these are dephosphorylated derivatives).
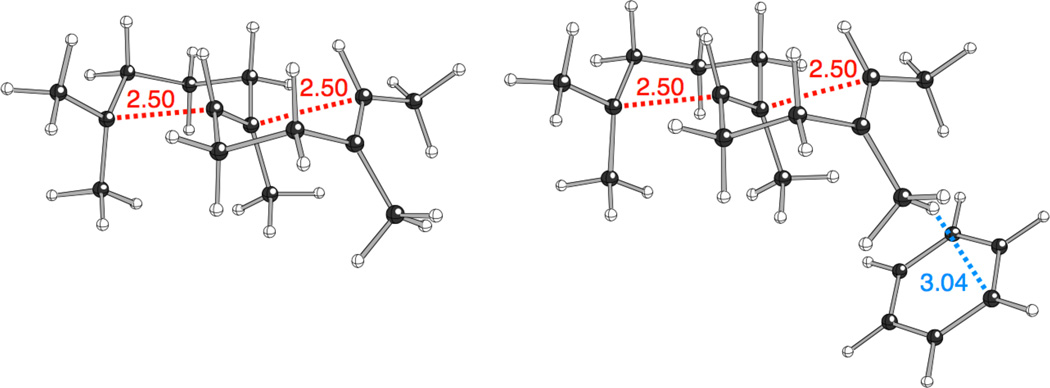
Structural studies and quantum chemical analysis of the interactions of the π-faces of aromatic groups with carbocations indicates that C-H•••π interactions may play important roles in terpene synthase catalyzed reactions.[10–11] However, the ability of an aromatic group in the catalytic base group to stabilize the bicyclic carbocationic intermediate A does not immediately suggest how this group might also promote initiation of the reaction. Accordingly, this effect was further probed by quantum chemical calculations (see Supporting Information for details), the application of which to terpene cyclization reactions has been validated by experimental tests of mechanistic predictions.[12] Here three model systems were examined: the geranylgeran-15-yl+ diphosphate formed by initial protonation (the fully optimized conformer most suited for cyclization was used; the tail leading to the OPP group was removed to simplify this series of calculations), a carbocation in which the two C–C bonds that form during bicyclization were constrained to 2.50 Å (to approximate partially formed bonds, although these likely do not form synchronously in the enzymatic reaction; Figure 3, left), and A. A theozyme-based approach[10, 13] was then used to examine the effect of complexation by an aromatic group, modelled as a benzene. The position of the benzene molecule was fully optimized in all three systems (Figure S7). This optimization was initiated with the benzene near to the C17 methyl group of A, with one aromatic carbon held in line with the C8–C17 bond of A at a distance of 4.05 Å in the case of the protonated GGPP (to prevent the benzene from wandering towards the distal carbocation center, which would not be possible in the enzyme). These calculations (carried out with several different theoretical methods) indicate that A can be selectively stabilized (versus protonated GGPP) by 3–5 kcal/mol, the carbocation with partial C–C bonds can be selectively stabilized by 1–3 kcal/mol and the basicity of the π-bond in GGPP that is protonated increases by ~1 kcal/mol (Figure S7). While these results are preliminary, in the sense that alternative orientations of the components of the substrate–theozyme complex have not yet been examined and the remainder of the enzyme active site is not included, they nevertheless show that the presence of an aromatic group in the catalytic base group can promote cyclization in both a thermodynamic and kinetic sense.
Figure 3.
Constrained carbocation, free (left) and in the presence of benzene (right). Constrained distances (Å) are shown in red. The closest H•••Cbenzene distance is shown in blue.
Formation of the fully rearranged 3 almost certainly reflects the intrinsic reactivity of A. Consistent with previous studies demonstrating that addition of a C=C to a carbocation to form a cyclohexane is quite exothermic,[14] the quantum chemical calculations reported here indicate that bicyclization is highly exothermic (by >30 kcal/mol). Accordingly, the formation of A is expected to be irreversible, but its trans-decalin geometry also permits a series of antiparallel 1,2-hydride and methyl migrations via tertiary carbocations (Scheme 1).
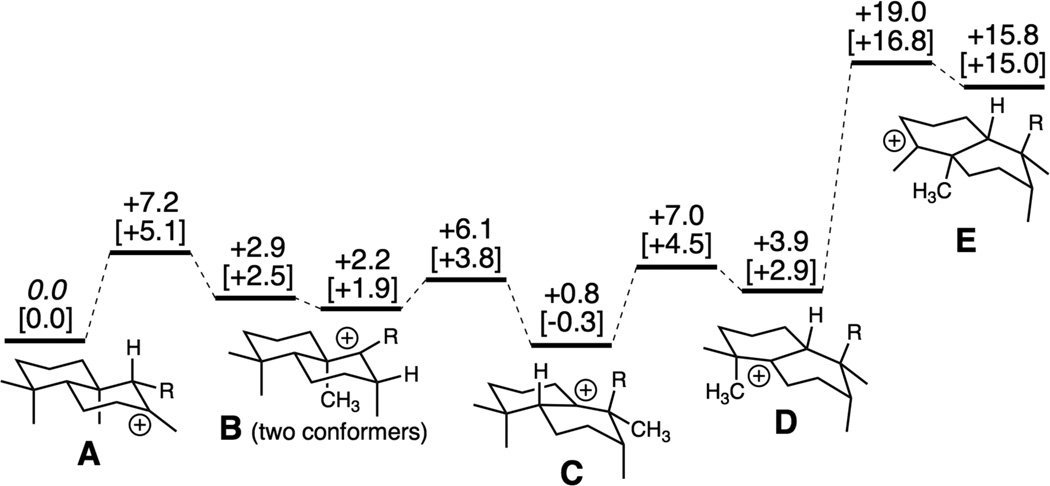
These rearrangements were probed by quantum chemical calculations. The relevant intermediates begin with A, which undergoes 1,2-hydride migration to form the corresponding 9-yl+ (B), with subsequent 1,2-methyl migration forming an ent-halima-13E-en-10-yl+ (C), another 1,2-hydride migration forms the corresponding 5-yl+ (D), and subsequent 1,2-methyl migration forms the fully rearranged ent-clerodan-13E-en-4-yl+ (E). Notably, quantum chemical calculations predict that the barriers for the first H/Me/H 1,2-shifts are 2–7 kcal/mol, with only small differences in energy for the corresponding intermediates (i.e., A–D), while there is a substantial energetic barrier of nearly 15 kcal/mol for the final Me shift, and formation of E is highly endothermic, approximately +12 kcal/mol relative to D (Figures 4 and S8). Intriguingly, these results suggest that intermediates A–D exist in equilibrium, with formation of the fully rearranged 3 critically dependent on the absence of any other suitably located general base in the AtCPS active site other than D379, the residue that acted as the general base initiating the overall reaction by protonation, and which presumably quenches E by removal of that same proton.
Figure 4.
Energies of carbocation minima and transition state structures involved in hydride and methyl shifts en route to 3. Relative energies (B3LYP/6-31+G(d,p) and mPW1PW91/6-31+G(d,p)//B3LYP/6-31+G(d,p) [in brackets]) are shown in kcal/mol; in this model, R = (CH2)2C(Me)=CHCH2OPO3H2.
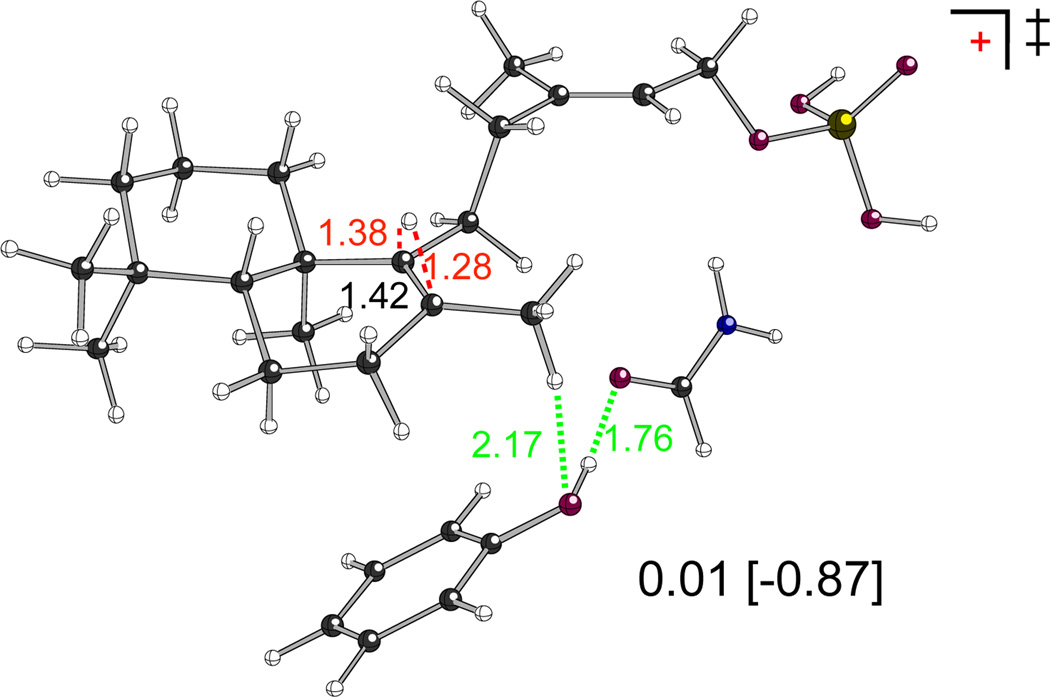
The ability of tyrosine substitution for the catalytic histidine to promote rearrangement was further probed by quantum chemical calculations, again using a theozyme-based approach. Here, the tyrosine was modelled as phenol, with hydrogen-bonding to a formamide model of the asparagine, and the hydroxyl group positioned near the C8 methyl group that would be deprotonated in formation of the usual product 2. In this arrangement, these functional groups remove the barrier to rearrangement (Figure 5), and provide selective stabilization for B, such that the initial 1,2-hydride shift becomes exothermic (by 4–5 kcal/mol). Again, these results are preliminary in the sense that alternative orientations of the components of the substrate–theozyme complex have not yet been examined, but nevertheless clearly show that the hydride shift can be promoted by the functional groups present in the mutant.
Although the structurally and mechanistically analogous triterpene cyclases also often catalyse rearrangement reactions similar to those observed here, the underlying mechanism remains unknown for these enzymes as well. It has been suggested that triterpene rearrangement may depend on organization of a π-electron gradient and/or positioning of the catalytic base.[15] Our results suggest that such rearrangement largely relies on the latter – i.e., positioning of the catalytic base, as simply replacing the histidine from the catalytic base group in AtCPS with a larger aromatic residue, particularly tyrosine, leads to predominant formation of the fully rearranged 3. Significantly, the results reported here provide even further insight into such catalysis. Specifically, bicyclization is enabled by the ability of such aromatic substitution to enforce correct folding of 1 within the active site, yet block deprotonation, while also providing some selective stabilization of the initial bicyclic intermediate A and other structures along the reaction coordinate for protonation/bicyclization via carbocation–π interactions. The exothermic nature of bicyclization of 1 to A then leads, in the absence of a suitable base, to a series of 1,2-hydride and methyl shifts to the fully rearranged intermediate E, wherein the carbocation is proximal to the middle Asp of the DxDD motif that acted as the catalytic acid, and which seems likely to be the only remaining general base.
Finally, it should be noted that the AtCPS:H263Y mutant appears to represent the first identified (−)-kolavenyl diphosphate synthase, offering potential access to labdane-related diterpenoids requiring such rearrangement. For example, a number of diterpenoids derived from 3 are of pharmaceutical interest, such as salvinorin A, a known hallucinogen that, along with salvinicins A and B, are potential treatments for Alzheimer’s disease, with the first two operating as κ-opioid agonists, while the latter is a μ-opioid antagonist; as well as barbatins A–C, which showed significant cytotoxic effects against several cancer lines.[16],[17] Indeed, even (−)-kolavenol itself has been shown to exhibit antitumor activity.[18] Finally, the ability of this single residue change (requiring change of only a single nucleotide) to lead to such a dramatic change in product outcome highlights the plasticity of class II diterpene cyclases, which presumably underlies the observed diversification of LRD natural products.
Supplementary Material
Figure 5.
The barrier for the 1,2-hydride shift that initiates rearrangement of ent-labda-13E-8-yl+ (A) is removed in the presence of a Tyr/Asn model (computed energies of the transition state – corresponding to TS (A–B1) in Figure 4 – relative to A in kcal/mol with B3LYP/6-31+G(d,p) and mPW1PW91/6-31+G(d,p)//B3LYP/6-31+G(d,p) [in brackets] are shown, as are distances in Å); see SI for details).
Footnotes
We would like to thank the W.M. Keck Metabolomics Research Laboratory for the generous use of their analytical equipment and facility. This work was supported by a grant from the NIH (GM076324 to R.J.P.) and NSF (CHE-0957416 and CHE-030089 [supercomputing resources via XSEDE] to D.J.T.).
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Kevin C. Potter, Roy J. Carver Department of Biochemistry, Biophysics & Molecular Biology, Iowa State University, Ames, IA 50011 USA
Dr. Jiachen Zi, Roy J. Carver Department of Biochemistry, Biophysics & Molecular Biology, Iowa State University, Ames, IA 50011 USA
Dr. Young J. Hong, Department of Chemistry University of California, Davis Davis, CA 95616 USA
Samuel Schulte, Roy J. Carver Department of Biochemistry, Biophysics & Molecular Biology Iowa State University Ames, IA 50011 USA.
Brandi Malchow, Roy J. Carver Department of Biochemistry, Biophysics & Molecular Biology Iowa State University Ames, IA 50011 USA.
Prof. Dr. Dean J. Tantillo, Department of Chemistry University of California, Davis Davis, CA 95616 USA
Prof. Dr. Reuben J. Peters, Email: rjpeters@iastate.edu, Roy J. Carver Department of Biochemistry, Biophysics & Molecular Biology Iowa State University Ames, IA 50011 USA.
References
- 1.Peters RJ. Nat. Prod. Rep. 2010;27:1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zi J, Mafu S, Peters RJ. Annu Rev Plant Biol. 2014;65:259–286. doi: 10.1146/annurev-arplant-050213-035705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Criswell J, Potter K, Shephard F, Beale MB, Peters RJ. Org. Lett. 2012;14:5828–5831. doi: 10.1021/ol3026022. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mafu S, Potter KC, Hillwig ML, Schulte S, Criswell J, Peters RJ. Chem Commun (Camb) 2015;51:13485–13487. doi: 10.1039/c5cc05754j. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Potter K, Criswell J, Peters RJ. Angew. Chem. Int. Ed. 2014;53:7198–7202. doi: 10.1002/anie.201402911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prisic S, Xu J, Coates RM, Peters RJ. ChemBioChem. 2007;8:869–874. doi: 10.1002/cbic.200700045. [DOI] [PubMed] [Google Scholar]
- 5.a) Hamano Y, Kuzuyama Y, Itoh N, Furihata K, Seto H, Dairi T. J. Biol. Chem. 2002;277:37098–37104. doi: 10.1074/jbc.M206382200. [DOI] [PubMed] [Google Scholar]; b) Nakano C, Okamura T, Sato T, Dairi T, Hoshino T. Chem Commun (Camb) 2005;2005:1016–1018. doi: 10.1039/b415346d. [DOI] [PubMed] [Google Scholar]
- 6.a) Köksal M, Potter K, Peters RJ, Christianson DW. Biochim Biophys Acta. 2014;1840:184–190. doi: 10.1016/j.bbagen.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Köksal M, Hu H, Coates RM, Peters RJ, Christianson DW. Nat Chem Biol. 2011;7:431–433. doi: 10.1038/nchembio.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cyr A, Wilderman PR, Determan M, Peters RJ. J. Am. Chem. Soc. 2007;129:6684–6685. doi: 10.1021/ja071158n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozama A, Prisinzano TE. Bioorg Med Chem Lett. 2009;19:5490–5495. doi: 10.1016/j.bmcl.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Misra R, Pandey RC, Dev S. Tetrahedron. 1979;35:985–987. [Google Scholar]; b) Hubert T, Wiemer D. Phytochemistry. 1985;24:1197–1198. [Google Scholar]; c) Monti H, Tiliacos N, Faure R. Phytochemistry. 1999;51:1013–1015. [Google Scholar]
- 10.Hong YJ, Tantillo DJ. Chem. Sci. 2013;4:2512–2518. [Google Scholar]
- 11.Christianson DW. Chem. Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 12.a) Zu L, Xu M, Lodewyk MW, Cane DE, Peters RJ, Tantillo DJ. J. Am. Chem. Soc. 2012;134:11369–11371. doi: 10.1021/ja3043245. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tantillo DJ. Nat Prod Rep. 2011;28:1035–1053. doi: 10.1039/c1np00006c. [DOI] [PubMed] [Google Scholar]
- 13.Hong YJ, Tantillo DJ. J Org Chem. 2007;72:8877–8881. doi: 10.1021/jo701526x. [DOI] [PubMed] [Google Scholar]
- 14.Jenson C, Jorgensen WL. J Am Chem Soc. 1997;119:10846–10854. [Google Scholar]
- 15.Wendt KU. Angew. Chem. Int. Ed. 2005;44:3966–3971. doi: 10.1002/anie.200500804. [DOI] [PubMed] [Google Scholar]
- 16.Shirota O, Nagamatsu K, Sekita S. J Nat Prod. 2006;69:1782–1786. doi: 10.1021/np060456f. [DOI] [PubMed] [Google Scholar]
- 17.Dai SJ, Tao JY, Liu K, Jiang YT, Shen L. Phytochemistry. 2006;67:1326–1330. doi: 10.1016/j.phytochem.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 18.Ohsaki A, Lu Y, Ito S, Edatsugi H, Iwata D, Komoda Y. Bioorg Med Chem Lett. 1994;4:2889–2892. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.