Abstract
SELECTOR is a software package for studying the evolution of multiallelic genes under balancing or positive selection while simulating complex evolutionary scenarios that integrate demographic growth and migration in a spatially explicit population framework. Parameters can be varied both in space and time to account for geographical, environmental, and cultural heterogeneity. SELECTOR can be used within an approximate Bayesian computation estimation framework. We first describe the principles of SELECTOR and validate the algorithms by comparing its outputs for simple models with theoretical expectations. Then, we show how it can be used to investigate genetic differentiation of loci under balancing selection in interconnected demes with spatially heterogeneous gene flow. We identify situations in which balancing selection reduces genetic differentiation between population groups compared with neutrality and explain conflicting outcomes observed for human leukocyte antigen loci. These results and three previously published applications demonstrate that SELECTOR is efficient and robust for building insight into human settlement history and evolution.
Keywords: population genetics, spatially explicit simulation, MHC lineages, HLA, balancing selection
Introduction
Analysis of current genetic diversity enables inferences regarding the evolution of species. However, factors affecting genetic diversity (eg, demography, migration, genetic drift, and natural selection) interact to such an extent that they are challenging to disentangle, and distinct situations may lead to similar outcomes. Genetic data consequently require specific analytical tools. In this context, computer simulation is a powerful approach to investigate the joint effects of these various processes, since it allows simulation of complex scenarios. Although more complexity does not necessarily equal more realism,1 computer simulation has been shown to be an invaluable tool for understanding the evolutionary processes that might otherwise remain undetected.2–4
Until recently, spatially explicit simulations have been used mostly for the study of selectively neutral loci.5 A few studies have investigated the effects of positive or negative selection in a spatially explicit context,6,7 but none of these have considered multiallelic loci under balancing selection, the main characteristics of the major histocompatibility complex (MHC). Moreover, the interaction of balancing selection and population structure has been shown to be complex.8,9 Three types of balancing selection models have been proposed to explain the high diversity of MHC lineages as follows: (1) heterozygote advantage (HA),10 (2) rare allele advantage (RAA),11 and (3) fluctuating selection over space and time (FS).12 The relative role and contribution of these three nonmutually exclusive models in the evolution of MHC is intensely debated.12–15 Even though a spatial model with constant negative frequency-dependent selection (FDS) coefficients on specific human leukocyte antigen (HLA) alleles has previously been implemented,16 this framework is not flexible enough to test the alternative models aforementioned. HA and RAA selection have also been modeled,17–20 sometimes including sexual selection through disassortative mating,21 but without any spatial component. The island model22 has been used to integrate space and selection in order to assess the effects of migration on genetic differentiation when selection is at play,8,9,23,24 but this is not, strictly speaking, a spatially explicit model.
The study of the human MHC, namely, HLA, has great potential to unravel human evolution and settlement,25–30 as large worldwide databases are available for several genes.31 However, a spatially explicit computer simulation framework capable of taking HLA characteristics into account has not been previously available. In this context, we developed a new computer simulation program called SELECTOR. The originality of SELECTOR lies in the simulation of both population spatial demography and balancing selection at a multiallelic genetic locus. In particular, it merges three main evolutionary processes as follows: (1) population spatial structure and migration, (2) genetic drift, and (3) natural selection (balancing or positive). In SELECTOR, selection may either be uniform over the whole simulated geographic area or vary with latitude and/or longitude. Moreover, SELECTOR allows the user to perform model comparison and parameter estimation, as it can easily be integrated in an approximate Bayesian computation (ABC) estimation framework such as ABCtoolbox.32 In the work presented here, we describe the main principles of SELECTOR, validate the algorithms using simple models for which theoretical expectations can be computed, and finally illustrate its use through a specific application.
Program Implementation
SELECTOR can be used to simulate complex evolutionary models by taking into account demographic variation of population sizes over space and time, as well as the effect of selection at a given genetic locus. SELECTOR simulates the genes of diploid individuals, generation after generation (ie, forward in time) within a two-dimensional stepping-stone framework.33 The whole population is subdivided into numerous subunits called demes, where each deme has its own spatial coordinates and can exchange migrants with neighboring demes at each generation. Demographic increase in population within each deme follows a logistic growth model to account for competition for resources among the members of each deme.34 Demographic parameters such as migration, growth rates and carrying capacity can be modified independently in each deme at different periods of time during the simulation. A wide variety of demographic scenarios, including a succession of demographic events, can thus be tested. Even though the demographic and migration algorithms in SELECTOR are derived from those implemented in the program SPLATCHE (model no. 1: even number of migrant, SPLATCHE user manual, p. 21,35) SELECTOR simulates forward in time all the individuals and genes in the entire population and does not use a backward coalescent algorithm36 to reconstruct the genetic diversity of samples. This full forward approach has the major advantage of allowing the user to consider various types of natural selection on genetic lineages.
Demographic model
Each deme has its own demographic characteristics and is regulated independently using a logistic growth model as follows:
| (1) |
where Ni(t) and Ni(t − 1) are the number of individuals belonging to the current deme i at generation t and t − 1, respectively, ri is the growth rate by generation (the rapidity at which the population density increases or decreases), and Ki is the carrying capacity (the maximum number of individuals sustained by the resources of the deme). If ri = 0, the population does not grow, while if ri = 1, it doubles during the earliest phase of growth, then the increase slows down because of intrademe competition . If Ki = 0, the deme is not activated, which means that it cannot be occupied at any time (no growth, no immigration) even if some individuals are added into it at the beginning of the simulation.
Migration model
In every deme, the number of emigrants E from deme i at each generation t and in each direction j is computed as
| (2) |
where mij is the migration rate in that direction (from i to j), Ni(t) is the number of individuals in the current deme i at generation t, Bi is the number of neighbors (with K > 0) of the current deme i, and zij is the remaining fractional part of migration from preceding generations (refer the user’s manual for a numerical example). Eij is an integer (number of individuals) and equal among all neighboring demes, so that the remaining fractional part from Eij (i in direction j) is kept in memory for the next generation and computed as
| (3) |
Note that in SELECTOR, Nm defines the amount of emigration from one deme because it can be set independently for each deme using the parameters K and m. However, in a homogeneous area and at demographic equilibrium (ie, when N = K), the number of immigrants is equal to the number of emigrants. In SELECTOR, it is possible to modify the migration rate between any pair of demes. This refers to the routes in the SELECTOR user’s manual.37 Note that a deme with K = 0 is never considered as a target of migration and cannot receive immigrants.
Demographic processes
The order of demographic events is as follows:
Demographic regulation occurs in every deme i using Equation 1 leading to Ni(t).
The number of emigrants Eij(t) in all directions j is computed in every deme i using Equations 2 and 3 and the total number of emigrants is computed as .
The number of immigrants in each deme i is computed as .
The density in every deme i is updated with the sum of emigrants and immigrants as
| (4) |
Note that the number of simulated generations must be either defined by the user, assuming that the generation time is known, or estimated for the organism under study, or a prior distribution may be defined in order to take into account uncertainty regarding the generation time.
Simulated genetic data
SELECTOR simulates the allele frequency trajectories in demes through the effects of genetic drift, migration, demographic variation, and selection. To achieve this, it records the two allelic variants of a given genetic locus for each simulated diploid individual. These variants are simply coded “ax” where x varies between 1 and nmax, the maximum number of allelic variants in the system (defined by the user as input parameters). At the beginning of a simulation, alleles taken from a list from 1 to nmax are randomly distributed in the first generation of individuals belonging to the source population(s). All alleles have the same probability 1/nmax of being drawn. This implies that initial allele frequencies vary among source populations, if there are more than one, and among independent simulations because different subsets of alleles are randomly drawn from the ancestral genetic pool of nmax alleles for each source and each simulation. Some alleles may not be represented in the source population(s), and this probability is inversely proportional to the initial population size. If the size of the source population is large, then the variation of initial genetic diversity among simulations is reduced. All source populations can either be constituted from the same or from different ancestral allelic pools (identical or different lists of alleles, respectively). The number of alleles can increase during the simulation if the mutation rate parameter is > 0, so that each new allele appearing by mutation increments ax by 1. Refer the SELECTOR user’s manual for more details.37 SELECTOR outputs either allele frequencies for the whole deme or for a sample of n individuals drawn randomly from the deme in a format directly usable by ARLEQUIN.38 The data output can then be used to compute various indices of genetic diversity and structure.
Transmission of gametes
Within each deme, the transmission of genes from one generation to the next is based on a Wright–Fisher’s model.22,39 For each individual at generation t, two alleles are drawn randomly from two different individuals at generation t − 1.
Effect of selection
Three different models of selection are implemented in SELECTOR. When selection applies (selection coefficient s > 0), the transmission of gametes from one generation to the next in each deme is modified as described here.
Symmetric overdominant selection (SOS), also called HA, simulates overdominant balancing selection, where all heterozygotes have the same selective advantage (fitness) over homozygotes.10,40 If the new genotype is heterozygote, it is accepted with the probability P = 1. If the new genotype is homozygote, it is either kept with the probability P = 1 − s, where s is the selection coefficient against homozygotes, or a new pair of alleles is drawn.
Frequency dependent selection (FDS), also called RAA, simulates a selection in favor of alleles that are less frequent in the population. A new allele ai is accepted with a probability P = 1 − f(ai) * s, where f(ai) is the current frequency of the allele ai in the deme and s is the selection coefficient against frequent alleles. If f(ai) tends to 0, then the probability of keeping ai is close to 1, while if f(ai) tends to 1, then this probability is close to 1 – s. If the allele is not retained, then a new one is drawn.
Dominant positive selection (DPS) simulates positive selection for one specific allele a1 among nmax alleles. Individuals who do not carry the selected allele have a fitness equal to 1 − s, whereas the carriers have a fitness of 1.41 If the new genotype has the selected allele, in a homozygote or heterozygote state, it is accepted with the probability P = 1. If the genotype does not have the selected allele, it is kept with the probability P = 1 − s, or a new genotype is drawn.
Program Validation
In order to validate the algorithm of allele transmission implemented in SELECTOR, we performed a series of simulations for which theoretical expectations can be analytically computed.
Evolution of allele frequencies within a single deme
In a single deme of size N individuals, we simulated the evolution of na alleles during t generations, in the absence of selection. Then, we computed the heterozygosity Ht within the deme and compared it with the expected heterozygosity given by the formula ,42 where H0 is the initial density at time 0. Here, the parameters K = N and r = 0 apply, since the population size is constant (ie, size is equal to carrying capacity).
We varied all three parameters (K, r, and na) in eight independent combinations and performed 1,000 simulations for each combination. Average and standard deviation over these 1,000 simulations have been computed and compared with the expected Ht (Table 1).
Table 1.
Comparison between simulated and expected (Ht) heterozygosity in a deme for various combinations of parameters na, H0, t, and N.
| SIMULATED WITH SELECTOR | EXPECTED Ht | DIFFERENCE | ||||
|---|---|---|---|---|---|---|
| na | H0 | t | N | |||
| 2 | 0.5 | 100 | 100 | 0.304 ± 0.18 | 0.303 | 0.001 |
| 2 | 0.5 | 1000 | 100 | 0.003 ± 0.03 | 0.003 | 0.000 |
| 2 | 0.5 | 100 | 1000 | 0.475 ± 0.03 | 0.476 | 0.003 |
| 2 | 0.5 | 1000 | 1000 | 0.300 ± 0.18 | 0.303 | 0.003 |
| 10 | 0.9 | 100 | 100 | 0.539 ± 0.16 | 0.545 | 0.006 |
| 10 | 0.9 | 1000 | 100 | 0.007 ± 0.05 | 0.006 | 0.001 |
| 10 | 0.9 | 100 | 1000 | 0.857 ± 0.02 | 0.856 | 0.001 |
| 10 | 0.9 | 1000 | 1000 | 0.550 ± 0.17 | 0.546 | 0.004 |
When comparing the simulated heterozygosity to the one expected theoretically under identical conditions (number of generations, deme size, and initial heterozygosity), we found that the average over 1,000 simulations was very close to the expected heterozygosity, differing by ≤0.6% in all cases. The small differences can be explained by the stochasticity of allele transmission in SELECTOR (see standard deviation), while the expected value is deterministic.
Evolution of allele frequencies within a series of interconnected demes
We assessed the validity of the migration algorithm by modeling four interconnected demes in a 2 × 2 stepping-stone area, in the absence of selection. At the beginning of the simulation, each deme had one specific allele with 100% frequency (a1 in deme 1, a2 in deme 2, etc.). We simulated 500 generations with a migration rate m = 0.01 between each pair of demes, and large deme size (10,000 diploid individuals) in order to minimize the effect of genetic drift. We then recorded the evolution of allele frequencies in the first deme and compared them with the frequencies theoretically expected with the formula where pt is the frequency expected at time t in a deme when the initial frequency of the allele in that deme is p0 and the average frequency of that allele over all demes is . This formula generates the expected allele frequencies for an island model with large deme size so that genetic drift can be neglected.43 Completion of 100 simulations of this model typically takes ~8 minutes 30 seconds in a single node from a Linux server.
Figure 1A shows that the evolution of allele frequencies through time within a deme converges toward an equilibrium frequency of 25% for all four alleles, as expected theoretically. The differences between the theoretical and the simulated curves are due to the differences between the two models: the theoretical curves have been obtained for a deterministic island model with an infinite size, while the simulated curves have been generated by a stochastic stepping-stone model with a finite (but large) size. Figure 1A shows that the frequency of allele a1 (blue curve), which was fixed in deme 1 at the beginning of the simulation, decreases, while the frequency of allele a4 (yellow curve), initially fixed in deme 4 without any direct connection with deme 1, takes longer to reach equilibrium compared with the two alleles fixed in the neighboring demes, 2 and 3, which directly exchange migrants. The orange and green curves represent a2 and a3, respectively.
Figure 1.
Comparison between simulated frequencies (sim, solid lines) and frequencies expected theoretically (exp, dotted lines). (A) Evolution of allele frequencies in the absence of selection within four interconnected demes. (B) Evolution of two alleles within a single deme under the effect of DPS on allele a1. (C) Evolution of two alleles within a single deme under the effect of SOS and FDS. (D) Schematic representation of the grid of demes where simulations are performed for the theoretical application. Each color represents one group of demes (blue, red, green, and yellow). Dark demes represent demes where genetic sampling is done. Migration between adjacent demes from different groups occurs at rate minter and is symbolized by black arrows, while gray arrows symbolize migration between adjacent demes from the same group, which occurs at rate mintra. For the sake of clarity, only a few arrows are represented but migration occurs between all pairs of demes.
Evolution of allele frequencies under DPS
In a single deme of size N = 10,000 (again to avoid too rapid genetic drift), we simulated the evolution of the frequencies of two alleles, a1 and a2, where a1 is under positive selection (DPS). The fitness of the various genotypes is computed as wa1a1 = 1, wa1a2 = 1, and wa2a2 = 1 − s, where s is the selection coefficient. The initial frequency of both alleles at time t0 is 0.5, but a1 is positively selected (s = 0.1) for 100 generations. We compared the simulated frequencies with those computed using the classical equation of allele frequencies in diploids under selection for survivorship.43
Figure 1B shows that the frequency of the allele a1 under positive selection increases gradually through time, in accordance with theoretical expectations. The small variation is explained by the stochasticity of the simulated algorithm. The result of a single simulation is displayed. Completing 100 simulations of this model typically takes ~30 seconds in a single node from a Linux server.
Evolution of allele frequencies under overdominant selection
We further simulated the evolution of allele frequencies in a biallelic system (na = 2) where heterozygotes are favored compared with homozygotes (SOS: wa1a1 = 1 − s, wa1a2 = 1, and wa2a2 = 1 − s) in a single deme of size N = 10,000. The initial frequencies of alleles a1 and a2 are 0.1 and 0.9, respectively, and we simulated selection for 200 generations when s = 0.1. We also simulated the evolution of these frequencies with the same parameters but using FDS. Then, we compared the simulated frequencies with those computed using the classical equation of allele frequencies in diploids under selection for survivorship.43 Completing 100 simulations of this model typically takes ~60 seconds in a single node from a Linux server.
Figure 1C shows that the frequencies of the two alleles converge to an equilibrium frequency of 0.5 if the size of the population is large enough to neglect drift, for both balancing selection models (SOS or FDS), as expected from theory.
Application
The interaction of balancing selection and population structure is challenging to study.9 It has, for example, been shown that genes under balancing selection are less sensitive to population subdivision than neutral ones and are thus expected to show limited differentiation among demes compared with neutral genes under the same demographic conditions.8,23,44 However, this conflicts with most observations,15,45 with some exceptions.46 In human populations, discordant results have been obtained; some studies have not found any reduction of interpopulation differentiation,44,47 while others have found some low interpopulation or intercontinental differentiation for almost all HLA loci, except DPB1.48–50 Because Schierup et al.23 theoretically showed that the reduction of genetic differentiation under balancing selection, compared to neutrality, is more pronounced when gene flow between demes is low, with almost no difference when gene flow is large, we believe that spatial gene flow heterogeneity between demes may explain these contrasting HLA results.
We tested this hypothesis by assessing the effects of heterogeneous gene flow between populations on measures of genetic differentiation under balancing selection. We used SELECTOR to simulate the evolution of allele frequencies in interconnected demes, when SOS is at play. Thus, we designed a virtual square area made up of 256 demes (16 × 16 demes, Fig. 1D). This area was divided into four identical groups of demes (square area of 64 demes each, Fig. 1D) connected by a migration rate minter, while the rate between demes of the same group is mintra. We performed several tens of thousands of simulations, varying input parameters such as K (carrying capacity), r (growth rate), mintra (migration rate between each pair of neighboring demes belonging to the same group), minter (migration rate between each pair of neighboring demes belonging to different groups), na (number of alleles), and s (coefficient of selection against homozygotes). Either all the demes were fully occupied at carrying capacity during the whole simulation (constant population size) or they were populated after a population expansion from one single deme, where this deme was located in group A and the initial population size starting the population expansion was 50 individuals. Population density within demes increased logistically at rate r (0.5 for all simulations) until it reached K (two choices, either 50 or 500). Migration occurred at rate m = 0.2 (20% of individuals belonging to each deme spread over the neighboring demes at each generation), even when demes had reached demographic equilibrium (carrying capacity). Genetic isolation between groups was not complete, and gene transfers could occur at a reduced migration rate (either by 40, 20, or 10 folds). We simulated a locus with 20 different lineages associated with symmetrical HA, with a selection coefficient between 0 (no selection) and 0.2 (very strong selection), and we randomly drew eight samples of 100 genes in each group of demes, at regular spatial intervals (dark colored demes in Fig. 1D).
We measured genetic differentiation at the end of each simulation by computing the indices FCT, FSC, and FST based on allele frequencies through an Analysis of molecular variance (AMOVA),51 the average pairwise genetic distances Dintra between samples within the four groups of demes, and the average pairwise genetic distances Dinter between samples belonging to different groups. We used the coancestry coefficient as a measure of genetic distance.52 We also used multidimensional scaling analyses (MDS) to visualize pairwise genetic distances between the subsets of data.53,54
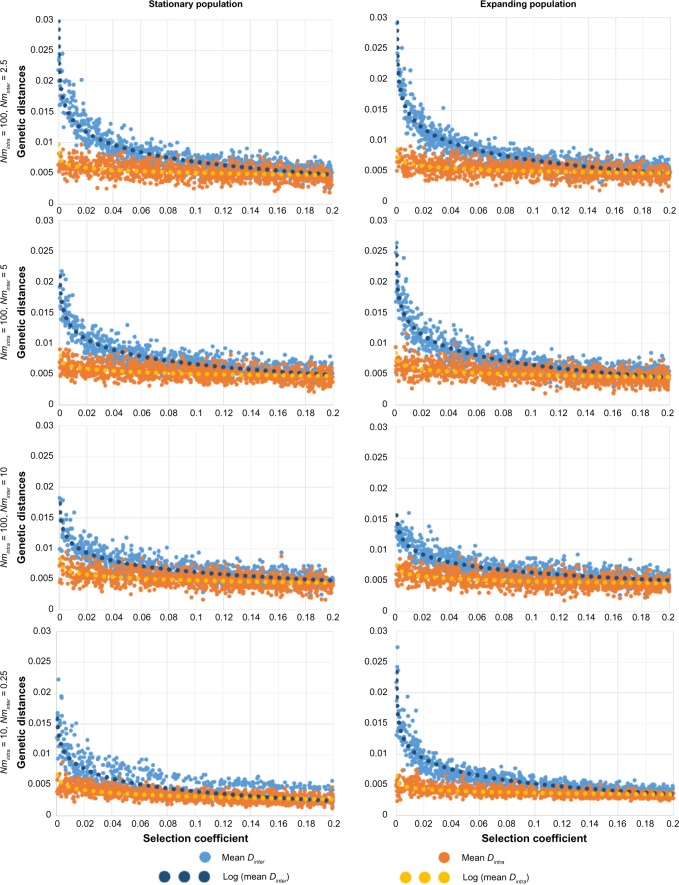
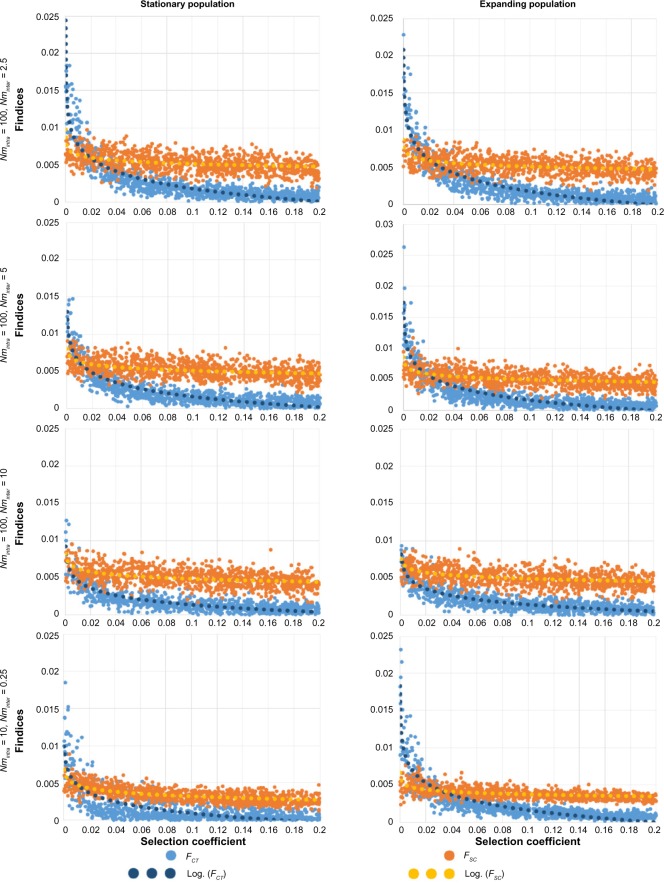
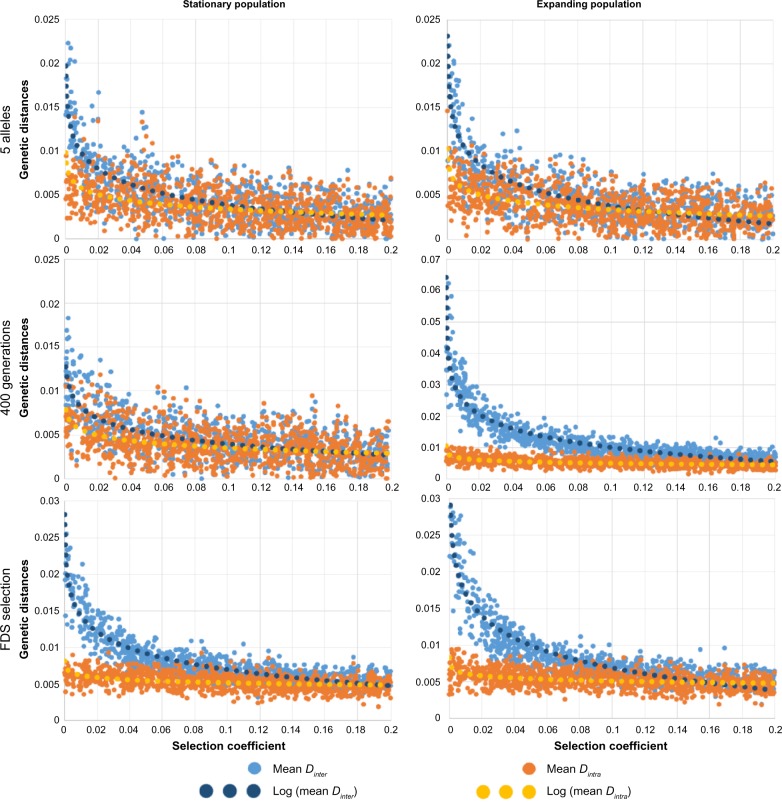
Our results show that, as theoretically expected,8,23,44 genetic differentiation between demes is globally reduced under balancing selection compared with neutrality (Figs. 2 and 3). They also support previous results, suggesting that this reduction is generally less pronounced when gene flow is high (increasing Nminter from line 1 to line 3 in Figs. 2 and 3). In addition, our results reveal new patterns of population differentiation that emerge when mixing various gene flow intensities in a spatially structured population. When gene flow between groups of populations is reduced compared with within groups (Nminter <Nmintra), we observe that the average genetic distance Dinter between groups tend to be much more affected by selection than the intragroup average genetic distance Dintra (Fig. 2). When balancing selection increases, then Dinter gradually decreases until reaching Dintra (sometimes even becoming smaller, Fig. 4), thereby diluting the signal of genetic isolation. This can also be seen when looking at fixation indices, the FCT being much more affected by balancing selection than the FSC. FCT may even become smaller than FSC if balancing selection is relatively strong (Fig. 3). This effect is more pronounced in an expanding population compared with a stationary one and less pronounced when the number of alleles is lower or the time frame is shorter (Fig. 4, line 1 and line 2). This effect is consistently observed through parameter values and the choice of balancing selection models (FDS instead of SOS, Fig. 4, line 3).
Figure 2.
Pairwise FST within population groups (in orange) and between population groups (in blue) for 1,000 simulations and varying selection coefficients (symmetric HA). Dotted lines represent logarithmic regression lines. The left column shows the results for a stationary population, while the right column shows the results for an expanding population. Nminter increases from line 1 to line 3 and K is changed from 500 to 50 in line 4, compared to line 1 while all the other parameters remain identical (na = 20 alleles, r = 0.5, and m = 0.2).
Figure 3.
FCT (in orange) and FSC (in blue) values for 1,000 simulations and varying selection coefficients (symmetric HA). Dotted lines represent logarithmic regression lines. The left column shows the results for a stationary population, while the right column shows the results for an expanding population. Nminter increases from line 1 to line 3 and K is changed from 500 to 50 in line 4, compared to line 1 while all the other parameters remain identical (na = 20 alleles, r = 0.5, and m = 0.2).
Figure 4.
Pairwise FST within population groups (in red) and between population groups (in blue) for 1,000 simulations and varying selection coefficients (symmetric HA). Dotted lines represent logarithmic regression lines. The left column shows the results for a stationary population, while the right column shows the results for an expanding population. All simulations have been obtained with the parameters Nmintra = 100, Nminter = 2.5 as in the first line of Figure 2. Only five alleles have been set in the first line (instead of 20 in Figure 2), 400 generations in the second line (instead of 2,000), and the FDS model for balancing selection in the third line (instead of SOS). Note the differences in the scale of the Y-axis in various graphs.
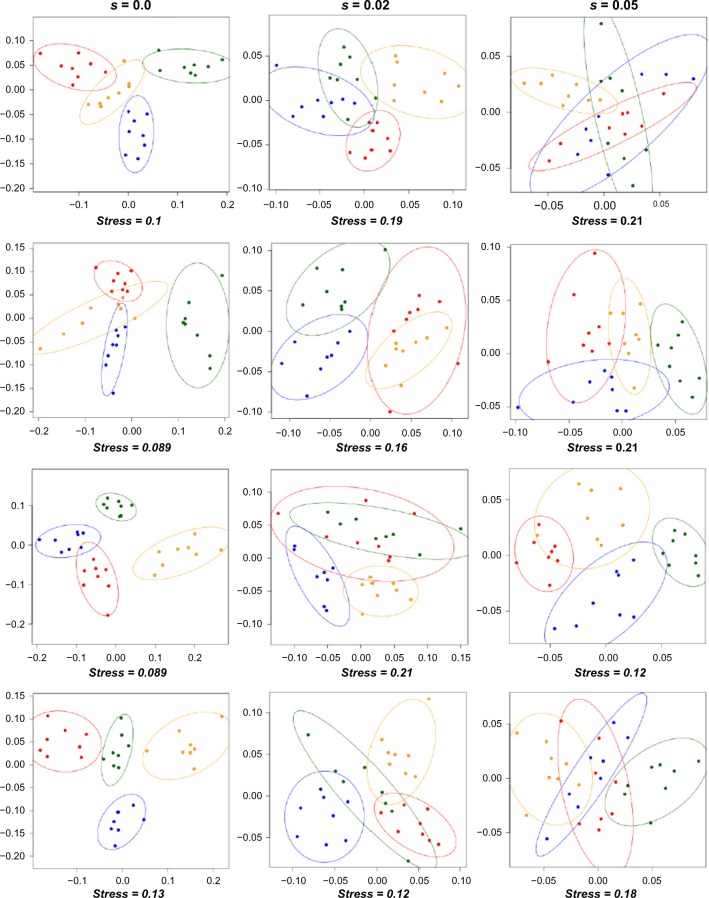
When balancing selection occurs, genetic distances between populations that are not spatially isolated are only slightly reduced compared with neutral expectations, whereas genetic distances between spatially isolated populations are strongly reduced (ie, they are inversely proportional to the gene flow across the geographical barrier). This effect translates into a distortion of the graphical representation of the genetic relationships between populations. For instance, Figure 5 shows 12 examples (randomly taken) of MDS performed on the simulated population samples, with s = 0.0, 0.02, or 0.05. In the absence of selection (s = 0.0), the genetic differentiation between geographic groups of populations is clearly visible in most of the cases, reflecting the strong barrier to gene flow between them (Nminter = Nmintra/40). In contrast, when selection is at play (s > 0.0), genetic differentiation between geographic groups vanishes and the groups tend to overlap on the MDS. This effect increases with selection, and there is almost no genetic differentiation between the four groups of demes when balancing selection is strong (s = 0.05), despite a strong reduction of gene flow between them. Inversely, the genetic differentiation between populations within the geographic groups is almost independent of the amount of balancing selection and does not change much with or without the action of selection. This is due to the fact that the effect of migration is stronger than that of selection within groups, while it is the opposite between groups.
Figure 5.
Examples of MDS obtained in three series of independent simulations. The four MDS on the left pane have been generated from simulations without selection (s = 0%), while the four on the middle have been generated with a coefficient of balancing selection s = 2% and the four on the right pane with a coefficient of balancing selection s = 5%. Each dot represents one sample and each color (red, blue, yellow, and green) a group of eight samples located in the same area (color code is identical to Figure 1D). In all, 90% confidence limits for ellipses around the centroid of each group are displayed using the ordiellipse function of the VEGAN package in R.
Here, we used SELECTOR to investigate patterns of genetic differentiation between spatially distributed populations, accounting for isolation by distance,55 where gene flow is not uniform (accounting for partial barriers to migration, such as mountains, rivers, or seas). These two condition properties constitute one of the novelties of SELECTOR compared with previous approaches. In a virtual square area subdivided into four groups of demes separated by a partial barrier to migration, we simulated the evolution of allele frequencies in populations (demes) during 2,000 generations and we recorded indices of genetic differentiation such as genetic distances D and fixation indices FSC and FCT. Our results confirm that genetic differentiation between demes is globally reduced under balancing selection compared with neutrality (Figs. 2–4). This could be explained by two factors: (1) under overdominance, the demes tend to share the same set of alleles, as rare alleles entering a deme have a higher fitness than alleles already present and (2) overdominant selection tends to increase within-deme diversity relative to the total diversity because alleles are kept at more equal frequency than for neutral alleles. Our results also confirm that the difference between overdominant selection and neutrality is more pronounced when gene flow is reduced, because a strong gene flow aids the mixing of alleles between demes and thus tends to erase the effect of balancing selection. More strikingly, new patterns of population differentiation emerge when mixing various gene flow intensities in a spatially structured population (Figs. 2 and 3). When a partial barrier to gene flow exists between geographic groups (Nminter < Nmintra, eg, separation between two different continents), the average genetic distance Dinter between groups tends to be more affected by selection than the intragroup average genetic distance Dintra. Consequently, when significant genetic differentiation is detected through the analysis of neutral loci, this differentiation may not leave any signal on loci under balancing selection. But when looking at populations not separated by a barrier (belonging to the same continent), no differences are found between the two types of loci. Our results are robust to the parameters of the model as long as Nminter remains smaller than Nmintra (Fig. 4).
Discussion
The rapid development of computational techniques has led to studies of new aspects of genetic evolution. In particular, simulation approaches permit exploration of realistic evolutionary scenarios that are too complex to be studied analytically, and these approaches have the power to integrate models affected by various processes (including genetic, environmental, cultural, and demographic processes). In this context, we present SELECTOR, a program that simulates genetic lineages under selection in a spatially explicit population framework. SELECTOR was primarily adapted to HLA loci because of the large worldwide datasets available for these genes and the need of a simulation program able to take into consideration their specific characteristics (multiple alleles and overdominant balancing selection) in a population genetic framework. We have validated the algorithms of SELECTOR by showing that its outputs are those expected theoretically in simple situations, but the power of SELECTOR resides in the simulation of more complex scenarios for which expectations cannot be computed analytically.30,56,57 While SELECTOR has primarily been designed to investigate the joint effects of selection and demography on HLA markers, as exemplified in the study by Di et al.30, it has also been applied to assess the effects of restricted gene flow on selected and neutral loci.57 SELECTOR can also be used to test comparable evolutionary hypotheses on other markers, such as lactase persistence.56
Here, we used SELECTOR to investigate patterns of genetic differentiation that could explain contrasting outcomes obtained on HLA data, for which some studies show a reduced interpopulation differentiation compared with neutral loci,48–50 while others show no difference.44,47 Our results suggest that, if gene flow is high between populations, the effect of balancing selection is negligible compared with that of demography.30,47 In contrast, when gene flow is reduced among populations, such as where there is a partial or strong geographic barrier, the effect of selection is visible, as is the case for the Strait of Gibraltar.57 The same logic may also explain why in the study by Sanchez-Mazas,50 the variance of components among continents is significantly reduced (t-test: P-value = 0.036, Table 2) between HLA under putative balancing selection (HLA-A, -B, -C, -DRB1, -DQA1, and -DQB1) compared with nearly neutral loci (HLA-DPB1, DNA markers, and STR), while the variance components between populations within continents is not (t-test: P-value = 0.337, Table 2). Indeed, smaller Nm between continents would allow for balancing selection to be detected, while larger Nm within continents would not. This question deserves to be investigated further through additional quantitative analyses.
Table 2.
Hierarchical analysis of genetic variance showing the variance components (%) taken from the study by Sanchez-Mazas.50
| LOCUS | NUMBER OF POPULATIONS | WITHIN POPULATIONS | AMONG POPULATIONS WITHIN CONTINENTS | AMONG CONTINENTS |
|---|---|---|---|---|
| HLA-A | 81 | 88.5 | 6.2 | 5.3 |
| HLA-C | 59 | 92 | 4.1 | 3.9 |
| HLA-B | 69 | 92.5 | 4.3 | 3.2 |
| HLA-DRB1 | 91 | 91.3 | 5.1 | 3.6 |
| HLA-DQA1 | 46 | 88.3 | 4.6 | 7.1 |
| HLA-DQB1 | 69 | 89.3 | 5.4 | 5.3 |
| HLA-DPB1a | 49 | 84 | 6.3 | 9.7 |
| DNA markersa | 14 | 84.4 | 4.7 | 10.8 |
| STRa | 52 | 87.6 | 3.1 | 9.2 |
Note:
Loci considered as evolving nearly neutrally.
Here, we mainly used a model of overdominant selection as a first approximation to the mode of balancing selection acting on HLA loci, which explains the maintenance of numerous alleles.11 Interestingly, these results do not differ significantly when either a model of HA (SOS) or a model of RAA (FDS, Fig. 4) is applied. However, the patterns of selective pressure may have been more complex, resulting from a combination of the following: (1) overdominant and positive selection at some alleles in response to the presence of specific pathogens58; and/or (2) variability through space due to various pathogen environments59; and/or (3) variable through time due to climatic variation. New selection models, such as divergent allele advantage20 or selection varying in time and space,15 and more detailed relationships between selection, demography, and environment could be the next improvements of SELECTOR. Indeed, one of the interests of the simulation approach is that models can be improved and carefully investigated to assess specific combinations of parameter values and processes that may bring deeper understanding of observed patterns of genetic diversity. Another improvement of our approach would be the incorporation of molecular or multilocus data to analyze genomic information.
SELECTOR has been inspired by the program SPLATCHE,35 and both programs have similar basic demographic processes. The major difference between the two processes is that the forward-in-time process implemented in SELECTOR, while being computationally more demanding than SPLATCHE’s backward-in-time coalescent approach, allows incorporating natural selection processes, while SPLATCHE simulates neutral loci only. However, the similarity between the two programs renders their respective outputs easily comparable, and they can consequently be used to associate genetic patterns of selected genes, such as HLA, to neutral molecular markers.57 The evolution of allele frequencies in all demes can be obtained in a simple tabulated text format that is easily readable. In addition, final allele frequencies in a series of samples specified by the user are output in ARLEQUIN format (currently 3.5).38 and can thus be easily analyzed to compute various intra- or interpopulation genetic statistics. SELECTOR has been developed in C++, which makes it computationally efficient and particularly suitable for research purposes. Thanks to its Linux version, SELECTOR may be incorporated through a bash pipeline into the ABC approach,60 such as with ABCtoolbox,32 in order to estimate parameters and formally compare models. This powerful method permits assessment of the relative probabilities of contrasting evolutionary scenarios, as well as estimation of the best parameter values.30,56,57 Input parameters of SELECTOR can be directly drawn from prior distributions defined by the user (see SELECTOR user manual)37 and summary statistics computed from the output using ARLEQUIN. Since it is an approximation technique, ABC avoids the calculation of a likelihood function, and thus allows evaluation of complex evolutionary scenarios that would otherwise be intractable by full likelihood approaches. Even though ABC requires a large number of simulations, which depend on the number of parameters to be estimated and on the prior ranges used,61–63 it has been shown to outperform approximate maximum-likelihood approaches.64 We believe that SELECTOR provides a versatile and computationally efficient framework to investigate such scenarios.
When compared with other forward-in-time simulation approaches, SELECTOR allows simulating (1) various natural selection mechanisms (not available in SPLATCHE), (2) spatially explicit scenarios, in comparison with “simuPOP,”65 and (3) more than three populations with sophisticated patterns of gene flow between populations and subpopulations, in comparison with “dadi”.66 SELECTOR is also more research oriented, being faster than dadi or simuPOP since it is written in C++, which is compiled into a binary executable file, instead of interpreted Python.
Conclusion
Here, we have presented the simulation program SELECTOR and demonstrated that it is a powerful and robust tool for investigating the combined effects of selection and demography on the genetic variability of MHC loci.30,57 Moreover, its versatility makes it invaluable for tackling other evolutionary questions and gaining insights into the genetic evolution of human beings and other organisms. SELECTOR is freely available for research and teaching purposes at http://ua.unige.ch/en/agp/tools/selector/.
Availability and Requirements
SELECTOR is written in C++, runs on MS windows or Linux, and is freely available for academic purposes at http://ua.unige.ch/en/agp/tools/selector/.
Acknowledgments
The authors thank four anonymous reviewers for helping to improve the manuscript.
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1909 words, excluding any confidential comments to the academic editor.
FUNDING: This work has been financially supported by the Swiss National Science Foundation (SNSF, Switzerland) grants 31003A-144180 (to ASM) and 31003A_156853 (to MC) and the Marie Curie initial training network BEAN. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: MC, AS-M. Analyzed the data: MC. Wrote the first draft of the manuscript: MC. Contributed to the writing of the manuscript: MC, PG, DD, JMN, AS-M. Agree with manuscript results and conclusions: MC, PG, DD, JMN, AS-M. Jointly developed the structure and arguments for the paper: MC, PG, DD, JMN, AS-M. Made critical revisions and approved final version: MC, PG, DD, JMN, AS-M. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Wakeley J. Recent trends in population genetics: more data! More math! Simple models? J Hered. 2004;95(5):397–405. doi: 10.1093/jhered/esh062. [DOI] [PubMed] [Google Scholar]
- 2.Ray N, Currat M, Excoffier L. Intra-deme molecular diversity in spatially expanding populations. Mol Biol Evol. 2003;20(1):76–86. doi: 10.1093/molbev/msg009. [DOI] [PubMed] [Google Scholar]
- 3.Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: massive introgression by local genes. Evolution. 2008;62(8):1908–20. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 4.Edmonds CA, Lillie AS, Cavalli-Sforza LL. Mutations arising in the wave front of an expanding population. Proc Natl Acad Sci U S A. 2004;101(4):975–9. doi: 10.1073/pnas.0308064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arenas M. Simulation of molecular data under diverse evolutionary scenarios. PLoS Comput Biol. 2012;8(5):e1002495. doi: 10.1371/journal.pcbi.1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itan Y, Powell A, Beaumont MA, Burger J, Thomas MG. The origins of lactase persistence in Europe. PLoS Comput Biol. 2009;5(8):e1000491. doi: 10.1371/journal.pcbi.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peischl S, Dupanloup I, Kirkpatrick M, Excoffier L. On the accumulation of deleterious mutations during range expansions. Mol Ecol. 2013;22(24):5972–82. doi: 10.1111/mec.12524. [DOI] [PubMed] [Google Scholar]
- 8.Muirhead CA. Consequences of population structure on genes under balancing selection. Evolution. 2001;55(8):1532–41. doi: 10.1111/j.0014-3820.2001.tb00673.x. [DOI] [PubMed] [Google Scholar]
- 9.Schierup MH. The number of self-incompatibility alleles in a finite, subdivided population. Genetics. 1998;149(2):1153–62. doi: 10.1093/genetics/149.2.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doherty PC, Zinkernagel RM. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature. 1975;256(5512):50–2. doi: 10.1038/256050a0. [DOI] [PubMed] [Google Scholar]
- 11.Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124(4):967–78. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedrick PW. Pathogen resistance and genetic variation at MHC loci. Evolution. 2002;56(10):1902–8. doi: 10.1111/j.0014-3820.2002.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 13.Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci. 2010;277(1684):979–88. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer D, Thomson G. How selection shapes variation of the human major his-tocompatibility complex: a review. Ann Hum Genet. 2001;65(pt 1):1–26. doi: 10.1046/j.1469-1809.2001.6510001.x. [DOI] [PubMed] [Google Scholar]
- 15.Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol. 2003;16(3):363–77. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- 16.Fix AG. Gene frequency clines in Europe: demic diffusion or natural selection? J R Anthropol Inst. 1996;2:625–43. [Google Scholar]
- 17.Borghans JA, Beltman JB, De Boer RJ. MHC polymorphism under host-pathogen coevolution. Immunogenetics. 2004;55(11):732–9. doi: 10.1007/s00251-003-0630-5. [DOI] [PubMed] [Google Scholar]
- 18.Ejsmond MJ, Babik W, Radwan J. MHC allele frequency distributions under parasite-driven selection: a simulation model. BMC Evol Biol. 2010;10:332. doi: 10.1186/1471-2148-10-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ejsmond MJ, Radwan J. MHC diversity in bottlenecked populations: a simulation model. Conserv Genet. 2011;12(1):129–37. [Google Scholar]
- 20.Satta Y. Effects of intra-locus recombination on HLA polymorphism. Hereditas. 1997;127:105–12. doi: 10.1111/j.1601-5223.1997.00105.x. [DOI] [PubMed] [Google Scholar]
- 21.Jan Ejsmond M, Radwan J, Wilson AB. Sexual selection and the evolutionary dynamics of the major histocompatibility complex. Proc Biol Sci. 2014;281(1796):20141662. doi: 10.1098/rspb.2014.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright S. Evolution in Mendelian populations. Genetics. 1931;16(2):0097–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schierup MH, Vekemans X, Charlesworth D. The effect of subdivision on variation at multi-allelic loci under balancing selection. Genet Res. 2000;76(1):51–62. doi: 10.1017/s0016672300004535. [DOI] [PubMed] [Google Scholar]
- 24.Nagylaki T, Lou Y. Patterns of multiallelic polymorphism maintained by migration and selection. Theor Popul Biol. 2001;59(4):297–313. doi: 10.1006/tpbi.2001.1526. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Mazas A, Meyer D. The relevance of HLA sequencing in population genetics studies. J Immunol Res. 2014;2014:971818. doi: 10.1155/2014/971818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Mazas A, Fernandez-Vina M, Middleton D, et al. Immunogenetics as a tool in anthropological studies. Immunology. 2011;133(2):143–64. doi: 10.1111/j.1365-2567.2011.03438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solberg OD, Mack SJ, Lancaster AK, et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol. 2008;69(7):443–64. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buhler S, Sanchez-Mazas A. HLA DNA sequence variation among human populations: molecular signatures of demographic and selective events. PLoS One. 2011;6(2):e14643. doi: 10.1371/journal.pone.0014643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez-Mazas A, Buhler S, Nunes JM. A new HLA map of Europe: regional genetic variation and its implication for peopling history, disease-association studies and tissue transplantation. Hum Hered. 2013;76(3–4):162–77. doi: 10.1159/000360855. [DOI] [PubMed] [Google Scholar]
- 30.Di D, Sanchez-Mazas A, Currat M. Computer simulation of human leukocyte antigen genes supports two main routes of colonization by human populations in East Asia. BMC Evol Biol. 2015;15:240. doi: 10.1186/s12862-015-0512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nunes JM, Buhler S, Roessli D, Sanchez-Mazas A, Collaboration HN. The HLA-net GENE[RATE] pipeline for effective HLA data analysis and its appli-cation to 145 population samples from Europe and neighbouring areas. Tissue Antigens. 2014;83(5):307–23. doi: 10.1111/tan.12356. [DOI] [PubMed] [Google Scholar]
- 32.Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinformatics. 2010;11:116. doi: 10.1186/1471-2105-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura M. “Stepping-stone” model of population. Annu Rep Natl Inst Genet. 1953;3:62–3. [Google Scholar]
- 34.Verhulst PF. Notice sur la loi que la population suit dans son accroissement. Curr Math Phys. 1838;10:113. [Google Scholar]
- 35.Ray N, Currat M, Foll M, Excoffier L. SPLATCHE2: a spatially-explicit simulation framework for complex demography, genetic admixture and recombination. Bioinformatics. 2010;26(23):2993–4. doi: 10.1093/bioinformatics/btq579. [DOI] [PubMed] [Google Scholar]
- 36.Hudson RR. Gene Genealogies and the Coalescent Process. Vol. 7. Oxford: Oxford University Press; 1990. [Google Scholar]
- 37.SELECTOR (version 1.0) User Manual [computer program]. Version 12015
- 38.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10(3):564–7. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 39.Fisher RA. The Genetical Theory of Natural Selection. The Clarendon Press; Oxford: 1930. [Google Scholar]
- 40.Lewontin R, Ginzburg LR, Tuljapurkar SD. Heterozis as an explanation for large amounts of genic polymorphism. Genetics. 1978;88(1):149–70. doi: 10.1093/genetics/88.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedrick PW. Genetics of Populations. Second ed. Sudbury, Massachussets: Jones and Bartlett; 2000. [Google Scholar]
- 42.Gillespie JH. Population Genetics A Concise Guide. Baltimore, Maryland: The Johns Hopkins University Press; 2004. [Google Scholar]
- 43.Hartl DL, Clark AG. Principles of Population Genetics. Fourth ed. Sunderland, Massachusetts: Sinauer Associates, Inc; 2007. [Google Scholar]
- 44.Takahata N. Allelic genealogy and human evolution. Mol Biol Evol. 1993;10(1):2–22. doi: 10.1093/oxfordjournals.molbev.a039995. [DOI] [PubMed] [Google Scholar]
- 45.Piertney SB, Oliver MK. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96(1):7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- 46.Glemin S, Gaude T, Guillemin ML, Lourmas M, Olivieri I, Mignot A. Balancing selection in the wild: testing population genetics theory of self-incompatibility in the rare species Brassica insularis. Genetics. 2005;171(1):279–89. doi: 10.1534/genetics.104.035915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer D, Single RM, Mack SJ, Erlich HA, Thomson G. Signatures of demographic history and natural selection in the human major histocompatibility complex Loci. Genetics. 2006;173(4):2121–42. doi: 10.1534/genetics.105.052837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Begovich AB, Moonsamy PV, Mack SJ, et al. Genetic variability and linkage disequilibrium within the HLA-DP region: analysis of 15 different populations. Tissue Antigens. 2001;57(5):424–39. doi: 10.1034/j.1399-0039.2001.057005424.x. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez-Mazas A. African diversity from the HLA point of view: influence of genetic drift, geography, linguistics, and natural selection. Hum Immunol. 2001;62(9):937–48. doi: 10.1016/s0198-8859(01)00293-2. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez-Mazas A. An apportionment of human HLA diversity. Tissue Antigens. 2007;69(suppl 1):198–202. doi: 10.1111/j.1399-0039.2006.00802.x. [DOI] [PubMed] [Google Scholar]
- 51.Excoffier L, Smouse P, Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–91. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reynolds J, Weir BS, Cockerham CC. Estimation for the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–79. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kruskal J. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrics. 1964;29:1–27. [Google Scholar]
- 54.Kruskal J. Non metric multidimensional scaling: a numerical method. Psychometrics. 1964;29:115–29. [Google Scholar]
- 55.Malécot G. The decrease of relationship with distance. Cold Spring Harbor Symp Quant Biol. 1955;20:52–3. [Google Scholar]
- 56.Gerbault P, Moret C, Currat M, Sanchez-Mazas A. Impact of selection and demography on the diffusion of lactase persistence. PLoS One. 2009;4(7):e6369. doi: 10.1371/journal.pone.0006369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Currat M, Poloni ES, Sanchez-Mazas A. Human genetic differentiation across the Strait of Gibraltar. BMC Evol Biol. 2010;10:237. doi: 10.1186/1471-2148-10-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez-Mazas A, Lemaitre JF, Currat M. Distinct evolutionary strategies of human leucocyte antigen loci in pathogen-rich environments. Philos Trans R Soc Lond B Biol Sci. 2012;367(1590):830–9. doi: 10.1098/rstb.2011.0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prugnolle F, Manica A, Charpentier M, Guegan JF, Guernier V, Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol. 2005;15(11):1022–7. doi: 10.1016/j.cub.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 60.Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162(4):2025–35. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Csillery K, Blum MG, Gaggiotti OE, Francois O. Approximate Bayesian computation (ABC) in practice. Trends Ecol Evol. 2010;25(7):410–8. doi: 10.1016/j.tree.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 62.Bertorelle G, Benazzo A, Mona S. ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol Ecol. 2010;19(13):2609–25. doi: 10.1111/j.1365-294X.2010.04690.x. [DOI] [PubMed] [Google Scholar]
- 63.Marjoram P, Tavare S. Modern computational approaches for analysing molecular genetic variation data. Nat Rev Genet. 2006;7(10):759–70. doi: 10.1038/nrg1961. [DOI] [PubMed] [Google Scholar]
- 64.Arenas M. Advances in computer simulation of genome evolution: toward more realistic evolutionary genomics analysis by approximate Bayesian computation. J Mol Evol. 2015;80(3–4):189–92. doi: 10.1007/s00239-015-9673-0. [DOI] [PubMed] [Google Scholar]
- 65.Peng B, Amos CI. Forward-time simulations of non-random mating populations using simuPOP. Bioinformatics. 2008;24(11):1408–9. doi: 10.1093/bioinformatics/btn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5(10):e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]