Abstract
Background
The leading cause of end-stage renal disease in the US is diabetic kidney disease (DKD). Despite significant efforts to improve outcomes in DKD, the impact on disease progression has been disappointing. This has prompted clinicians and researchers to search for alternative approaches to identify persons at risk, and to search for more effective therapies to halt progression of DKD. The identification of novel therapies is critically dependent on a more comprehensive understanding of the pathophysiology of DKD, specifically at the molecular level. A more expansive and exploratory view of DKD is needed to complement more traditional research approaches that have focused on single molecules.
Summary
In recent years, sophisticated research methodologies have emerged within systems biology that should allow for a more comprehensive disease definition of DKD. Systems biology provides an interdisciplinary approach to describe complex interactions within biological systems, including how these interactions influence systems' functions and behaviors. Computational modeling of large, system-wide, quantitative data sets is used to generate molecular interaction pathways, such as metabolic and cell signaling networks.
Key Messages
Importantly, the interpretation of data generated by systems biology tools requires integration with enhanced clinical research data and validation using model systems. Such an integrative biological approach has already generated novel insights into pathways and molecules involved in DKD. In this review, we highlight recent examples of how combining systems biology with traditional clinical and model research efforts results in an integrative biology approach that significantly adds to the understanding of the complex pathophysiology of DKD.
Key Words: Diabetic kidney disease, Diabetic nephropathy, Deep phenotyping, Genome-phenome continuum, Systems biology
Introduction
Diabetic kidney disease (DKD) is a frequent microvascular complication of diabetes, and is associated with significant morbidity and cardiovascular mortality. In the US, DKD accounts for around 50% of incident cases of individuals with end-stage renal disease (ESRD) [1]. Globally, as the incidence of obesity increases, so too is the development of diabetes and associated DKD. Despite the growing population of persons at risk for this condition, our scientific efforts to understand the pathophysiology in an attempt to alter the disease course are failing. Unfortunately, neither the detection of early DKD nor the treatments currently available are significantly impacting the detection or progression of this devastating condition. Current practice relies on estimated glomerular filtration rate (eGFR) and urinary microalbumin excretion (albumin/creatinine ratio, ACR) to detect DKD, in lieu of routine histologic evaluation of renal tissue. However, by the time these biomarkers are detectable, significant renal structural damage has already occurred [2]. It is unclear whether this damage is reversible. Thus, there is a significant drive to understand a person's inherent risk factors, and to define the earliest alterations within the kidney that contribute to the development of DKD. This knowledge should help to identify persons at risk before they develop significant disease, and aid in the development of treatments that alter the disease course.
However, elucidation of this knowledge has proven to be a challenge, due in significant part to the complex physiology of the kidney. Each nephron within the kidney is composed of a glomerulus where blood is filtered, and a tubule where the filtrate is modified. Each component has a multitude of different cell types and functions: the glomerulus is composed of endothelial cells, mesangial cells, and podocytes, while tubules are composed of a variety of specialized epithelial cells with variable function dependent on their location along the tubular segment. In this intricate milieu, hypothesis-driven studies have been used to investigate genetic variants that influence DKD based on a compelling candidate-gene approach [3] including the angiotensin-converting enzyme insertion/deletion polymorphism [4], aldose reductase [5], and apolipoprotein E [6]. Unfortunately, the candidate gene approach suffers from inherent weaknesses such as inadequate sample sizes and susceptibility to false positives [7,8]. Disappointingly few candidate-gene studies have been independently replicated. Thus, the elaborate and complex nature of kidney function necessitates an integrative and multidisciplinary approach.
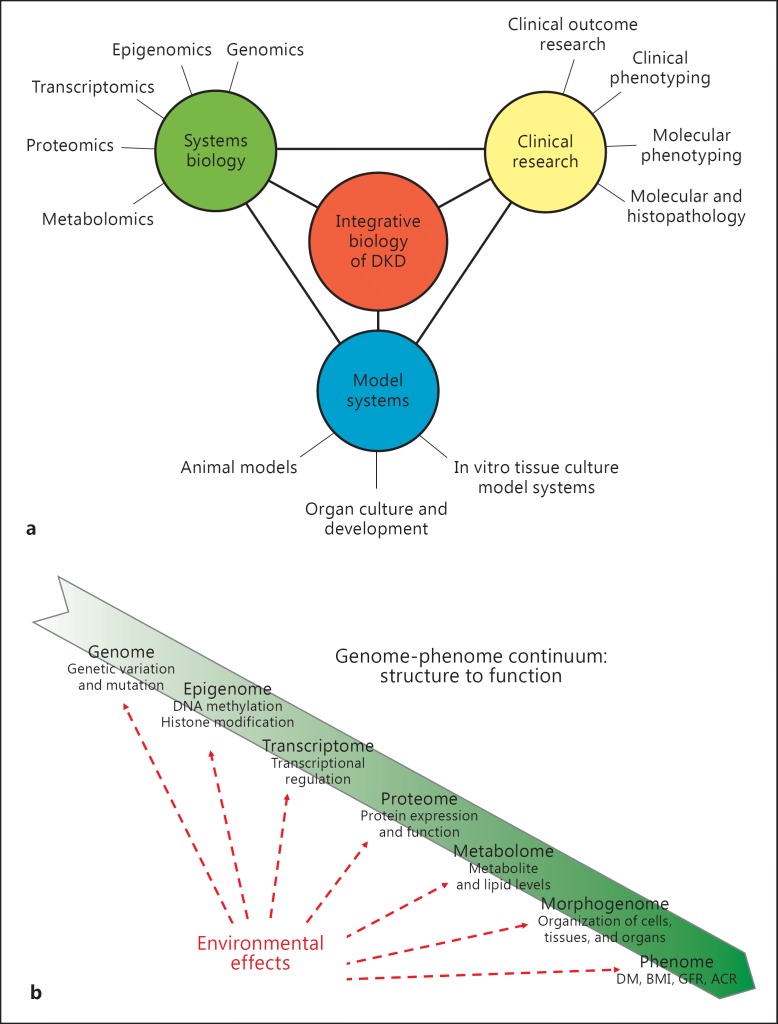
An integrative biology approach to analyze biological systems as a unified whole has emerged to better characterize complex diseases such as DKD (fig. 1a). Substantial progress in experimental and computational tools is responsible for the evolution of these research strategies. The term ‘systems biology’ loosely defines an integrative approach towards knowledge discovery that combines large-scale data sets from diverse sources to gain a more holistic understanding of health and disease. This approach considers disease development to progress along the genome-phenome continuum (fig. 1b) whereby a sequence of regulatory cascades connects genetic predispositions with environmental influences to manifest a phenotype [9]. The technologies involved provide the means to measure changes in large numbers of components at different layers within this regulatory cascade and then to identify patterns of interactions of these components. Such data sets include genetic variations (genomics), epigenetic modifications (epigenomics), coding and non-coding RNAs (transcriptomics), proteins (proteomics), metabolites (metabolomics), detailed morphologic characterization (morphogenomics), and comprehensive clinical phenotyping (phenomics). This approach has garnered success towards a more comprehensive understanding of disease pathogenesis in several complex diseases, most prominently in oncology [10].
Fig. 1.
a An integrative biological approach to understanding the basis of DKD is necessary. This approach combines diverse sets of sophisticated expertise in an attempt to better characterize the pathophysiology of this complex disease. b Systems biology tools are used to describe the alterations seen in DKD at each point along the genome-phenome continuum. The goal is to integrate these data to visualize how alterations at specific points result in downstream events, and ultimately in the DKD phenotype.
In this review, we provide an overview of systems biology tools as applied to the study of DKD. Several previous reviews are excellent resources for the specifics of the techniques used [11,12,13,14,15,16,17]. We focus on the rationale of these ‘omics’ approaches and highlight pertinent examples of early progress integrating different layers within the genome-phenome continuum. Our goal is for clinicians and researchers to appreciate the strengths and necessity of an integrative biology approach to understand the complex pathophysiology of DKD.
Omics Approaches to DKD
Genomics
As well-described instances of genetic heritability of DKD were known, the development of sophisticated genomic sequencing methods held great promise for elucidating the genetic underpinnings of this disease. However, like other human diseases with complex traits such as hypertension, describing the genetic determinants underlying DKD has been challenging. Unlike monogenic disorders, there is no simple concordance between genetic variation and phenotype in DKD, and as such, gene mapping by linkage analysis is difficult. Thus, genome-wide association (GWA) studies were developed to correlate a dense set of single nucleotide polymorphisms (SNPs) across the genome to survey the most common genetic variation with a disease phenotype or trait across a population. An advantage is that no prior assumptions regarding the biology of the disease are required [7]; significant limitations are that they require a large sample size, a careful definition of the clinical trait, and significant costs. Indeed, single studies using GWA studies to investigate DKD including modest numbers of individuals have had limited success. Craig et al. [18] compared pooled DNA from ∼1,100 type 1 diabetes mellitus (T1DM) subjects, half with ESRD and half without DKD. McDonough et al. [19] performed an association analysis adjusting for admixture in ∼1,000 type 2 diabetes mellitus (T2DM) African-Americans with ESRD and in another ∼1,000 without T2DM or kidney disease. No association reached accepted significance thresholds in either study.
A promising approach, termed ‘systems genetics’, is designed to integrate genetic risk loci with other functional data sets and biological knowledge, and therefore overcome some of the limitations of straightforward GWA studies [13]. An example is illustrated by studies involving FRMD3. In an initial study, the GoKinD (Genetics of Kidneys in Diabetes) investigators examined 360,000 SNPs in 820 T1DM individuals with ESRD or proteinuria and compared them to 885 T1DM control individuals [20]. Implicated SNPs were then examined in 1,304 individuals from the DCCT (Diabetes Control and Complications Trial)/EDIC (Epidemiology of Diabetes Interventions and Complications) trial, identifying 2 novel loci (FRMD3 and CARS). Though neither reached genome-wide significance, Martini et al. [21] pursued the functional analysis of a candidate SNP (rs188747) located in the extended promoter region of the poorly annotated FRMD3 gene. In a cohort of individuals with early diabetic glomerulopathy, a gene coregulation network of several hundred transcripts correlating with FRMD3 expression contained several members of the BMP pathway. Promoter modeling demonstrated a shared regulatory transcriptional module suggestive of a common regulation of FRMD3 and BMP pathway genes, proposing a transcriptional link between the polymorphism and regulation of this signaling pathway. Thus, defining a putative functional context for the effects of a SNP is another analytical approach to investigate candidates of interest. By linking candidate genetic variants to transcripts within a defined coregulation network and pathway analysis, it is possible to identify a likely functional and regulatory context of disease-associated genetic variants [13].
Though systems genetics is an ideal mechanism to incorporate genetic and functional data, much work still needs to be done to identify potential genetic loci of DKD. One main limitation of a GWA study approach is that although it is able to identify SNPs associated with a clinical trait, linking SNPs with molecular function is not trivial. Thus, to further characterize the pathophysiological context of a particular SNP, its effect on gene, protein, and metabolite expression is sought. Expression quantitative trait locus (eQTL) analysis is a mechanism to determine the statistical association of a genotype with transcript expression. A SNP associated with the expression of a nearby gene is called a cis-eQTL, which likely tags a causal SNP that changes transcript abundance. A SNP associated with transcript expression of distant genes is defined as a trans-eQTL, which may identify remote enhancers or repressors or gene-gene interaction. In addition, clinical measures may be used concomitantly with eQTL analysis to further clarify genetic variants.
GENIE (Genetics of Nephropathy: an International Effort) has extended initial GWA studies' observations by performing a meta-analysis utilizing eQTL mapping to assess the functional context of SNPs profiled in almost 7,000 subjects with T1DM [22]. The analysis revealed loci and their association with neighboring transcripts (1 in AFF3, 1 near SP3, and 1 between RGMA and MCTP2) that were associated with ESRD, and a fourth locus (within the intronic region of ERBB4) when the definition of DKD also included macroalbuminuria. The FIND (Family Investigation of Nephropathy in Diabetes) network has followed a parallel approach in a study of 6,197 patients with T2DM and advanced kidney disease across 4 ethnicities in the US [23].
An additional collaborative approach that is currently being pursued by investigators of the SUMMIT (Surrogate Markers for Micro- and Macro-Vascular Hard Endpoints for Innovative Diabetes Tools) and JDRF-DNCRI (Juvenile Diabetes Research Foundation-Diabetic Nephropathy Collaborative Research Initiative) studies is to combine GWA studies involving more than 25,000 T1DM participants characterized for DKD status and extend the variants to be interrogated.
Though whole-genome sequencing of populations has become feasible [24], it requires substantial resources for data generation and analysis; moreover, the interpretation of noncoding variants linked to clinical traits in complex diseases is challenging. A more feasible approach than whole-genome sequencing is whole-exome sequencing, which focuses investigation on the protein-coding regions of the genome [25]. Because the human exome comprises only a small fraction of the entire genome, human exome sequencing allows much greater sequence coverage at reduced cost and in less time. Functional predictions of the coding variants are more feasible and can allow effective filtering of variants to a single putative causal variant. This approach is gaining impetus in investigations of DKD. Cooke Bailey et al. [26] used publicly available whole-exome sequencing data from the NHLBI GO ESP (National Heart Lung Blood Institute Grand Opportunity Exome Sequencing Project) database. Thirty-one coding SNPs in 19 genes previously implicated in kidney disease were used to interrogate the genotypes of individuals with ESRD in T2DM. Though their associations were not dramatic, several SNPs were replicated in both the African-American and Euro-American data sets.
Thus far, identified causative genetic loci and variants in DKD explain only a small fraction of the known heritability. However, this is consistent with most other complex traits, where less than half of the total trait heritability can be ascribed to the sum of genetic effects [27]. With falling costs of sequencing, and collaborative efforts to pool data, GWA studies are expected to reveal additional genetic risk loci. A systems genetics approach integrating genetic and functional data is of great use in validating candidate loci and greatly enriches our physiologic understanding. However, since other nongenetic elements are thought to contribute to the risk of DKD, including untested rare variants as well as gene-gene and gene-environment interactions, other omics approaches are essential to complement the genomics approach.
Epigenomics
In both T1 and T2DM complication research, recent studies in humans, animal models, and cells in culture point to a ‘metabolic memory’ of a hyperglycemic environment [28]. For example, even when blood glucose levels are tightly controlled, diabetic individuals may still suffer from chronic kidney disease and progress to kidney failure driven by initial hyperglycemia [29]. To explain this effect, investigators have turned to studies of epigenetics. Epigenetics describes heritable and self-perpetuating but reversible changes that affect gene expression during cell division, without alterations in the DNA sequence [30]. This includes covalent modifications such as histone acetylation or DNA methylation, which alter chromatin density and accessibility of the transcriptional machinery to DNA. These modifications are conserved during somatic cell division and are important in the preservation of cell identity. Noncoding RNAs, such as microRNAs (miRNAs), are also considered to be part of the epigenome. Such epigenetic factors likely function as mediators between genes and the environment, by creating a memory of the environmental signal as an adaptation or response [31]. Indeed, this is of particular interest in studies of diabetic complications as changes in metabolite levels can affect gene expression by inducing covalent modifications of nucleic acids and histone complexes [30].
To explore this angle, Ko et al. [32] looked at cytosine methylation in microdissected renal tubular epithelial cells from individuals with DKD. They found differentially methylated regions enriched in consensus binding sites for renal transcription factors in putative enhancer regions of a set of genes related to kidney fibrosis; moreover, these changes correlated with gene transcript levels. These findings raise the possibility that dysregulated cytosine methylation plays a role in the predisposition and development of DKD.
miRNAs are also being actively pursued as modulators of DKD expression. They are short, single-stranded RNAs that post-transcriptionally regulate gene expression by binding to 3′ untranslated regions of target messenger RNAs (mRNAs), leading to inhibition of translation or to decay. One particularly attractive feature of miRNAs is that they are present in urinary exosomes, and thus offer the possibility of being biomarkers for DKD. Recent studies in animals and tissue culture models showed that several miRNAs are involved in the pathogenesis of DKD, including miR-451, miR-195, miR-192, the miR-29 family, and miR-21 [33]. Additionally, Argyropoulos et al. [34] showed that in individuals with T1DM with different stages of DKD, profiles of 27 urinary miRNAs differed according to the stage of disease. These miRNAs mapped to pathways involved in growth factor signaling and renal fibrosis. More recently, Lai et al. [35] showed that miR-21 seems to have a protective role in glomerular injury[;] miR-21-deficient TGF-β1 transgenic mice had increased proteinuria, decreased renal function, and decreased podocyte numbers compared with miR-21 wild-type TGF-β1 transgenic littermates. The authors went on to show that in American Indians with DKD, ACR was positively associated with glomerular miR-21 expression. These findings suggest that miR-21 inhibits TGF-β1 signaling and functions.
Epigenetics is a relatively new area of study; yet initial studies in DKD suggest promising results in this arena. Efforts to advance this field of research are actively being pursued by the Human Epigenome Project and ENCODE (Encyclopedia of DNA Elements) with the hope that other chromatin factors and epigenetic mechanisms will be revealed. As epigenetic changes are highly cell type specific, cell-based studies are critical. Though the initial ENCODE project does not include kidney cell-specific lineages, epithelial and endothelial cell lines are included. Ongoing studies using a systems genetics approach that integrates genetic risk with epigenetic profiles may yield insights into the puzzle of ‘missing heritability’ in our current GWA analyses [30,36].
Transcriptomics
Transcriptomics is the analysis of mRNA expression on a genome-wide scale. Compared to the genome, i.e. the entity of genetic information, the transcriptome is far more dynamic and may vary extensively between cells, tissues, and physiologic or pathologic conditions. In practice, expression profiling involves isolating mRNA from tissue and quantifying individual transcript expression signals by microarray or RNA sequencing. Comparison of gene expression mRNA levels between diseased and control tissues reveals relative expression changes of the set of genes. Correlative analysis can be performed to determine gene expression relative to the severity of disease. Expression data can also be linked with prior knowledge (literature search tools or protein interaction databases) and aggregated into functional categories, leading to the identification of molecular disease processes represented by the transcriptional changes. Importantly, mRNA levels do not necessarily show a direct correlation with protein level, with translational regulation, post-translational modifications, and protein degradation being additional components involved in protein regulation.
Not only does a transcriptomic approach provide a mechanism to discover new pathways and genes involved in disease, but it also can be used to define the molecular pattern at any point along the continuum of disease. This is of particular interest in DKD, as it offers an opportunity to identify individuals early in the disease course before traditional markers of disease including GFR and ACR are affected. Moreover, it may help to identify those individuals at increased risk for disease progression, as well as subpopulations that may respond to a particular therapy.
Comprehensive profiling of gene expression in the human kidney has provided novel insights into both physiologic and pathogenetic mechanisms in DKD. Analytical strategies have been developed using published gene associations to organize differential gene expression patterns into defined pathways. Berthier et al. [37] performed glomerular and tubulointerstitial gene expression profiling on type 2 diabetics, with early and late DKD, and revealed a highly regulated Janus kinase-signal transducer and activator transcription (JAK-STAT) pathway in individuals with DKD compared to controls. Schmid et al. [38] screened tubulointerstitial gene expression from human renal biopsies and employed bioinformatic analyses to identify a specific nuclear factor-κB promoter module (NFKB_IRFF_01) in the inflammatory stress response of progressive DKD, uncovering potential therapeutic targets for further investigation.
Interestingly, key members of the JAK-STAT pathway were not regulated in a mouse model of DKD, though it should be noted that humans had progressive disease, while the murine model was nonprogressive. Hodgin et al. [39] identified cross-species, shared transcriptional networks in glomeruli of early type 2 human DKD and 3 murine models of DKD (streptozotocin DBA/2, C57BLKS db/db, and eNOS-deficient C57BLKS db/db strains). Shared transcriptional mechanism defined by network matching algorithms identified nodes reflecting established pathogenic mechanisms of diabetes complications including JAK-STAT and VEGFR signaling pathways. In addition, nodes and pathways unique to each human-murine model pairing were defined. This highlights the importance of selecting murine models most relevant to the human disease process.
Transcriptomics is currently one of the most mature and comprehensive omics tools for studying DKD, due to the extensive experience with available sequencing and analytical methods. Progress integrating data sets with those emerging from other omics platforms is already occurring, as in the case of FRMD3 described above. However, one of the crucial limitations of transcriptional profiling in DKD is the access to renal tissue, especially at earlier time points within the disease process. This time point is of specific interest, since these data are likely to reveal DKD-specific pathogenic pathways. Protocol biopsies are helping to fill this void, as will be discussed below.
Proteomics
The proteome is the entire set of proteins expressed within an organism, system, cell, or subcellular compartment. As proteins are the backbone of the physiologic pathways of cells, it follows that protein profiles vary according to the particular needs of the tissues in both health and disease. Proteomic analysis depends on protein extraction and fractionation, high-throughput mass spectrometry, and protein identification. Protein identification is pursued using web-based databases that detail amino acid sequences of expressed proteins, based on genes identified in human and murine genomes. Large data sets have been generated from healthy and diseased tissue, but interpretation remains an issue. Identification is inherently biased for more abundant proteins, and for certain characteristics that make some peptides technically more easily identifiable. Comparison of studies is complicated by the use of different methods for protein capture and separation and different types of mass spectrometer equipment, as well as by a lack of standardization of sample processing and analysis [11]. Despite the technical difficulties, several studies have yielded useful insights.
To date, most proteomic studies in DKD have focused on analysis of urine, searching for biomarkers of disease. Some have resulted in the identification of candidate predictors for the development of DKD in T2DM individuals. For example, as part of the VADT (Veterans Affairs Diabetes Trial), Bhensdadia et al. [40] performed liquid chromatography-mass spectrometry on urine from T2DM individuals with varying degrees of GFR and albuminuria to identify best predictors of early renal function loss. After identifying several candidates in individuals with normoalbuminuria, and testing those candidates in urine from individuals with early DKD (CKD stage 2 or better, ACR <300 mg/g), urine haptoglobin was defined as the best molecular outcome predictor. When urine haptoglobin-to-creatinine ratio was included in a model to predict early renal functional decline (that included ACR), predictive performance improved. Another urinary proteomic study by Zürbig et al. [41] also searched for markers of early DKD and progression in a longitudinal cohort of 35 diabetic (T1 and T2DM) individuals who were initially normoalbuminuric. They found that urinary collagen α-1(I and III) fragments decreased 3–5 years before the onset of macroalbuminuria.
More recently, a few proteomics studies have surfaced that used kidney tissue from individuals with DKD. In one example, Nakatani et al. [42] performed proteomic analyses on laser-captured, micro-dissected glomeruli from formalin-fixed paraffin-embedded autopsy kidney tissue from 10 individuals with DKD (average eGFR 59.9 ml/min/1.73 m2, SD ±7.5; ACR not included) and 10 nondiabetic controls without kidney disease. A total of 170 proteins were identified, 100 of which were differentially expressed in DKD samples as compared to controls (55 overexpressed and 45 repressed). The investigators went on to validate one of the proteins identified by the analysis, nephronectin, by performing immunohistochemistry on a second, larger group of autopsy specimens, demonstrating a positive correlation of histologic glomerular sclerosis with nephronectin immunostaining of the extracellular matrix.
Though proteomic analysis of urine has yielded several candidate biomarkers of DKD [43], studies involving proteomic analysis of DKD kidney tissue are just emerging. So far, the focus has been on individual or small groups of proteins, limiting the ability to look at the interactions of the proteins from a systems standpoint. However, combining tissue-derived proteomic profiling with transcriptional profiling of the same sample will allow simultaneous assessment of a gene set's transcriptional and post-translational profile. Though the samples represent a single snapshot in time, this information is likely to provide a unique vantage for gene control mechanisms involved in DKD.
Metabolomics
The metabolome is the collection of endogenous, chemically diverse small molecule metabolites (<1,500 Da) within a biological system. This includes lipids, steroids, carbohydrates, nucleotides, amino acids, and organic acids. Thus, metabolomics is a method to measure the functional output of a cell, or organ, and usually incorporates mass spectrometry-based techniques or NMR spectroscopy. Because metabolites vary widely in concentration, and can be affected by exogenous substances such as food or medications and thus vary between individuals, analytical assessment of the metabolome is challenging. No single methodology provides an accurate measurement of the entire metabolome. To date, few metabolomic studies on DKD have been published [12].
Niewczas et al. [44] performed mass spectrometry-based global metabolic profiling on T2DM individuals, searching for plasma metabolites associated with the risk for progression to ESRD. The cases consisted of a nested cohort of 40 individuals within the Joslin Study of Genetics of Type 2 Diabetes and Kidney Complications. They all had essentially normal renal function at baseline, but developed ESRD during the subsequent 8–12 years. Matched control cases were those individuals who did not progress to ESRD. Interestingly, the metabolomic platform identified 16 metabolites previously classified as uremic solutes that were found to be elevated at baseline in the cases that progressed to ESRD. Additionally, cases showed a significant depletion of essential amino acids and derivatives, raising the possibility of impaired tubular reabsorption. Amino acid-derived acylcarnitines related to the urea cycle were increased, while precursors were depleted, suggesting enhanced mitochondrial amino acid β-oxidation.
As illustrated by Niewczas et al. [44], these data can highlight previously unappreciated metabolic pathways of importance in DKD pathogenesis and help focus further investigations, especially in combination with other omic platforms. For example, one could reapproach the proteomic and transcriptomic profiles of DKD and specifically interrogate the data sets related to the urea cycle and to amino acid transporters in the renal tubules. Thus, metabolomic profiling in DKD provides a functional view of the kidney that adds context to the single-time point snapshot of proteomic and transcriptomic profiling.
Phenomics
The phenome is a description of the physical and biochemical traits of an organism. According to Houle et al. [45], phenomics is ‘the acquisition of high-dimensional phenotypic data on an organism-wide scale’. This is especially pertinent to the study of DKD. Because DKD is a complex trait, GFR and ACR arguably are not adequate parameters to describe the phenotypic complexity of an individual's kidney disease, especially when attempting to categorize disease for further study. Indeed, there has been a push for ‘deep phenotyping’, that is, both a precise and a comprehensive analysis of the phenotype across the spectrum of human diseases [46]. Such efforts to classify individuals into subpopulations based on their disease susceptibility, or molecular subclass of disease, may not only assist in earlier diagnoses, but also predict responses to a particular treatment.
Deep phenotyping of individuals requires significant resources and trained personnel. Individuals at risk need to be identified and recruited. Information is obtained on multiple levels in a standardized fashion, including comprehensive assessment of an individual's ethnogenetic background, detailed and comprehensive clinical and laboratory information, and sophisticated morphologic renal tissue measurements. Ideally, these data are obtained in a prospective manner with regular assessments over decades. Additionally, blood, urine, and renal tissue are collected at routine time points during the observation, to be available for omic platform evaluations.
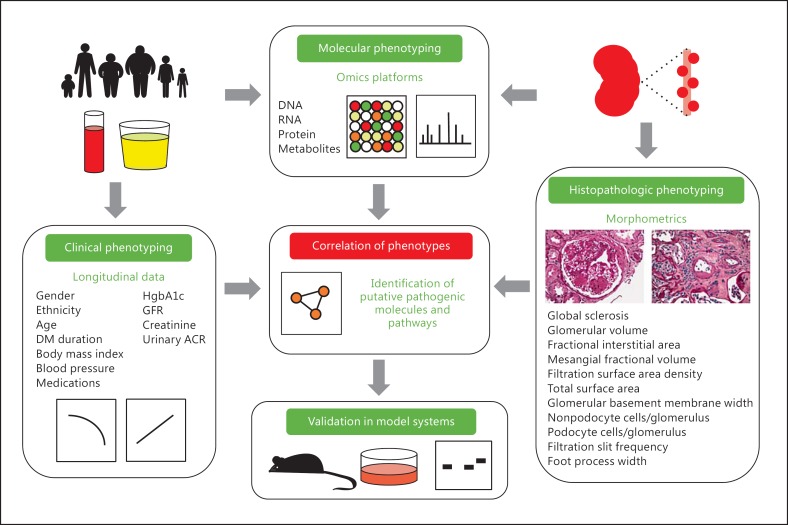
Benefits of such an approach are already apparent from multiple ongoing longitudinal studies in T1 and T2DM, including the Joslin cohorts and the Pima Indian cohorts of Arizona; this Native American population has high rates of obesity and T2DM and unusually high rates of early development of DKD. They have been followed for more than 4 decades in the fashion described above (fig. 2). One interesting example of how such deep phenotyping has already benefited DKD investigations comes from Muller et al. [47] regarding a particular genetic variant in PFKFB2 (rs17258746) associated with body mass index. They found that lower gene expression of PFKFB2 correlated with higher percent body fat and body mass index, while lower gene expression in kidney tissue correlated with DKD. Thus, not only were the authors able to correlate longitudinal clinical parameters to a genetic risk locus, but they were then able to use adipose and renal tissue to examine gene expression. This cohort represents a relatively small, genetically homogenous group of individuals. In some aspects, this is ideal and more likely to reveal genetic associations. However, the generalizability to persons of other ethnic backgrounds will need to be carefully assessed, and underlines the importance of expanding the deep phenotyping used in studies of DKD to persons of all ethnicities.
Fig. 2.
Deep phenotyping is combined with molecular analysis in an attempt to more specifically associate clinical and histopathologic features seen in DKD with resulting molecular alterations. The molecular alterations (such as the transcription level of a particular gene) can then be correlated with a clinical outcome (such as GFR). This approach can be used to identify genes, molecules, and pathways that are associated with disease, which can then be validated in model systems.
Concluding Remarks
For a complex disease such as DKD, an integrative biological approach is an absolute necessity to significantly advance knowledge of the pathophysiology of this disease. Omics technologies offer an organizational and methodological platform to identify key genes and pathways involved. The maturation of various integrative biology approaches is already contributing to research capabilities, and this is expected to expand. However, crucial components of the future success of these endeavors are deep phenotyping and access to renal tissue.
Disclosure Statement
The authors declare no conflicts in relationship to the data presented in this review.
Acknowledgements
The authors are supported by grants K08-DK089119, R24-DK082841, and P30-DK081943 from the National Institutes of Health. They thank the many colleagues and collaborators who have contributed to the advancement of this field, and apologize for limited reference inclusion due to space restrictions. We especially thank Alice Bram for her assistance with illustrations.
References
- 1.United States Renal Data System . USRDS 2014 Annual Data Report: Atlas of End-Stage Renal Disease in the United States. Bethesda: National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; 2014. [Google Scholar]
- 2.Caramori ML, Fioretto P, Mauer M. The need for early predictors of diabetic nephropathy risk: is albumin excretion rate sufficient? Diabetes. 2000;49:1399–1408. doi: 10.2337/diabetes.49.9.1399. [DOI] [PubMed] [Google Scholar]
- 3.O'Seaghdha CM, Fox CS. Genome-wide association studies of chronic kidney disease: what have we learned? Nat Rev Nephrol. 2012;8:89–99. doi: 10.1038/nrneph.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruggenenti P, Bettinaglio P, Pinares F, Remuzzi G. Angiotensin converting enzyme insertion/deletion polymorphism and renoprotection in diabetic and nondiabetic nephropathies. Clin J Am Soc Nephrol. 2008;3:1511–1525. doi: 10.2215/CJN.04140907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heesom AE, Hibberd ML, Millward A, Demaine AG. Polymorphism in the 5′-end of the aldose reductase gene is strongly associated with the development of diabetic nephropathy in type I diabetes. Diabetes. 1997;46:287–291. doi: 10.2337/diab.46.2.287. [DOI] [PubMed] [Google Scholar]
- 6.Chowdhury TA, Dyer PH, Kumar S, Gibson SP, Rowe BR, Davies SJ, Marshall SM, Morris PJ, Gill GV, Feeney S, et al. Association of apolipoprotein ε2 allele with diabetic nephropathy in Caucasian subjects with IDDM. Diabetes. 1998;47:278–280. doi: 10.2337/diab.47.2.278. [DOI] [PubMed] [Google Scholar]
- 7.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drawz PE, Sedor JR. The genetics of common kidney disease: a pathway toward clinical relevance. Nat Rev Nephrol. 2011;7:458–468. doi: 10.1038/nrneph.2011.85. [DOI] [PubMed] [Google Scholar]
- 9.Kretzler M, Cohen CD. Integrative biology of renal disease: toward a holistic understanding of the kidney's function and failure. Semin Nephrol. 2010;30:439–442. doi: 10.1016/j.semnephrol.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 11.Starkey JM, Tilton RG. Proteomics and systems biology for understanding diabetic nephropathy. J Cardiovasc Transl Res. 2012;5:479–490. doi: 10.1007/s12265-012-9372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sas KM, Karnovsky A, Michailidis G, Pennathur S. Metabolomics and diabetes: analytical and computational approaches. Diabetes. 2015;64:718–732. doi: 10.2337/db14-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller BJ, Martini S, Sedor JR, Kretzler M. A systems view of genetics in chronic kidney disease. Kidney Int. 2012;81:14–21. doi: 10.1038/ki.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasuda Y, Cohen CD, Henger A, Kretzler M. Gene expression profiling analysis in nephrology: towards molecular definition of renal disease. Clin Exp Nephrol. 2006;10:91–98. doi: 10.1007/s10157-006-0421-z. [DOI] [PubMed] [Google Scholar]
- 15.He JC, Chuang PY, Ma'ayan A, Iyengar R. Systems biology of kidney diseases. Kidney Int. 2012;81:22–39. doi: 10.1038/ki.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodgin JB, Cohen CD. Experimental approaches to the human renal transcriptome. Semin Nephrol. 2010;30:455–467. doi: 10.1016/j.semnephrol.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Tsui IF, Chari R, Buys TP, Lam WL. Public databases and software for the pathway analysis of cancer genomes. Cancer Inform. 2007;3:379–397. [PMC free article] [PubMed] [Google Scholar]
- 18.Craig DW, Millis MP, DiStefano JK. Genome-wide SNP genotyping study using pooled DNA to identify candidate markers mediating susceptibility to end-stage renal disease attributed to type 1 diabetes. Diabet Med. 2009;26:1090–1098. doi: 10.1111/j.1464-5491.2009.02846.x. [DOI] [PubMed] [Google Scholar]
- 19.McDonough CW, Palmer ND, Hicks PJ, Roh BH, An SS, Cooke JN, Hester JM, Wing MR, Bostrom MA, Rudock ME, et al. A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int. 2011;79:563–572. doi: 10.1038/ki.2010.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, Ng DP, Placha G, Canani LH, Bochenski J, et al. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009;58:1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martini S, Nair V, Patel SR, Eichinger F, Nelson RG, Weil EJ, Pezzolesi MG, Krolewski AS, Randolph A, Keller BJ, et al. From single nucleotide polymorphism to transcriptional mechanism: a model for FRMD3 in diabetic nephropathy. Diabetes. 2013;62:2605–2612. doi: 10.2337/db12-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandholm N, Salem RM, McKnight AJ, Brennan EP, Forsblom C, Isakova T, McKay GJ, Williams WW, Sadlier DM, Mäkinen VP, et al. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 2012;8:e1002921. doi: 10.1371/journal.pgen.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyengar SK, Sedor JR, Freedman BI, Kao WH, Kretzler M, Keller BJ, Abboud HE, Adler SG, Best LG, Bowden DW, et al. Family Investigation of Nephropathy and Diabetes (FIND) Genome-wide association and trans-ethnic meta-analysis for advanced diabetic kidney disease: Family Investigation of Nephropathy and Diabetes (FIND) PLoS Genet. 2015;11:e1005352. doi: 10.1371/journal.pgen.1005352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Hum Mol Genet. 2010;19(R2):R145–R151. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooke Bailey JN, Palmer ND, Ng MC, Bonomo JA, Hicks PJ, Hester JM, Langefeld CD, Freedman BI, Bowden DW. Analysis of coding variants identified from exome sequencing resources for association with diabetic and non-diabetic nephropathy in African Americans. Hum Genet. 2014;133:769–779. doi: 10.1007/s00439-013-1415-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirschhorn JN. Genetic approaches to studying common diseases and complex traits. Pediatr Res. 2005;57:74R–77R. doi: 10.1203/01.PDR.0000159574.98964.87. [DOI] [PubMed] [Google Scholar]
- 28.Reddy MA, Tak Park J, Natarajan R. Epigenetic modifications in the pathogenesis of diabetic nephropathy. Semin Nephrol. 2013;33:341–353. doi: 10.1016/j.semnephrol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villeneuve LM, Natarajan R. Epigenetic mechanisms. Contrib Nephrol. 2011;170:57–65. doi: 10.1159/000324944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Susztak K. Understanding the epigenetic syntax for the genetic alphabet in the kidney. J Am Soc Nephrol. 2014;25:10–17. doi: 10.1681/ASN.2013050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirbahai L, Chipman JK. Epigenetic memory of environmental organisms: a reflection of lifetime stressor exposures. Mutat Res Genet Toxicol Environ Mutagen. 2014;764-765:10–17. doi: 10.1016/j.mrgentox.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Ko YA, Mohtat D, Suzuki M, Park AS, Izquierdo MC, Han SY, Kang HM, Si H, Hostetter T, Pullman JM, et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14:R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DiStefano JK, Taila M, Alvarez ML. Emerging roles for miRNAs in the development, diagnosis, and treatment of diabetic nephropathy. Curr Diabetes Rep. 2013;13:582–591. doi: 10.1007/s11892-013-0386-8. [DOI] [PubMed] [Google Scholar]
- 34.Argyropoulos C, Wang K, McClarty S, Huang D, Bernardo J, Ellis D, Orchard T, Galas D, Johnson J. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS One. 2013;8:e54662. doi: 10.1371/journal.pone.0054662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai JY, Luo J, O'Connor C, Jing X, Nair V, Ju W, Randolph A, Ben-Dov IZ, Matar RN, Briskin D, et al. MicroRNA-21 in glomerular injury. J Am Soc Nephrol. 2015;26:805–816. doi: 10.1681/ASN.2013121274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddy MA, Natarajan R. Recent developments in epigenetics of acute and chronic kidney diseases. Kidney Int. 2015;88:250–261. doi: 10.1038/ki.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berthier CC, Zhang H, Schin M, Henger A, Nelson RG, Yee B, Boucherot A, Neusser MA, Cohen CD, Carter-Su C, et al. Enhanced expression of Janus kinase-signal transducer and activator of transcription pathway members in human diabetic nephropathy. Diabetes. 2009;58:469–477. doi: 10.2337/db08-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Gröne HJ, et al. Modular activation of nuclear factor-κB transcriptional programs in human diabetic nephropathy. Diabetes. 2006;55:2993–3003. doi: 10.2337/db06-0477. [DOI] [PubMed] [Google Scholar]
- 39.Hodgin JB, Nair V, Zhang H, Randolph A, Harris RC, Nelson RG, Weil EJ, Cavalcoli JD, Patel JM, Brosius FC, 3rd, et al. Identification of cross-species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes. 2013;62:299–308. doi: 10.2337/db11-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhensdadia NM, Hunt KJ, Lopes-Virella MF, Michael Tucker J, Mataria MR, Alge JL, Neely BA, Janech MG, Arthur JM. Urine haptoglobin levels predict early renal functional decline in patients with type 2 diabetes. Kidney Int. 2013;83:1136–1143. doi: 10.1038/ki.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zürbig P, Jerums G, Hovind P, Macisaac RJ, Mischak H, Nielsen SE, Panagiotopoulos S, Persson F, Rossing P. Urinary proteomics for early diagnosis in diabetic nephropathy. Diabetes. 2012;61:3304–3313. doi: 10.2337/db12-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakatani S, Wei M, Ishimura E, Kakehashi A, Mori K, Nishizawa Y, Inaba M, Wanibuchi H. Proteome analysis of laser microdissected glomeruli from formalin-fixed paraffin-embedded kidneys of autopsies of diabetic patients: nephronectin is associated with the development of diabetic glomerulosclerosis. Nephrol Dial Transplant. 2012;27:1889–1897. doi: 10.1093/ndt/gfr682. [DOI] [PubMed] [Google Scholar]
- 43.Mayer P, Mayer B, Mayer G. Systems biology: building a useful model from multiple markers and profiles. Nephrol Dial Transplant. 2012;27:3995–4002. doi: 10.1093/ndt/gfs489. [DOI] [PubMed] [Google Scholar]
- 44.Niewczas MA, Sirich TL, Mathew AV, Skupien J, Mohney RP, Warram JH, Smiles A, Huang X, Walker W, Byun J, et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int. 2014;85:1214–1224. doi: 10.1038/ki.2013.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11:855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 46.Robinson PN. Deep phenotyping for precision medicine. Hum Mutat. 2012;33:777–780. doi: 10.1002/humu.22080. [DOI] [PubMed] [Google Scholar]
- 47.Muller YL, Piaggi P, Hanson RL, Kobes S, Bhutta S, Abdussamad M, Leak-Johnson T, Kretzler M, Huang K, Weil EJ, et al. A cis-eQTL in PFKFB2 is associated with diabetic nephropathy, adiposity and insulin secretion in American Indians. Hum Mol Genet. 2015;24:2985–2996. doi: 10.1093/hmg/ddv040. [DOI] [PMC free article] [PubMed] [Google Scholar]