Abstract
MicroRNAs are identified as negative regulators in gene expression through silencing gene expression at the post-transcriptional and translational levels. Bone morphogenetic protein 9 (BMP9) is the most effective in inducing osteogenesis in the BMP family, the members of which were originally identified as osteoinductive cytokines. In the current study, the role of miR-23b in the progression of BMP9-induced C2C12 myoblasts was investigated. The results indicated that miR-23b was significantly downregulated in C2C12 myoblasts induced by BMP9. Overexpression of miR-23b significantly inhibited osteogenesis in the C2C12 myoblasts. In addition, it was observed that Runx2 was negatively regulated by miR-23b at the post-transcriptional level, via a specific target site within the 3′UTR of Runx2. Knockdown of Runx2 promoted miR-23b-induced inhibition of osteogenesis in C2C12 myoblasts. The expression of Runx2 was observed to be frequently upregulated in osteoblast cell lines and inversely correlated with miR-23b expression. Thus, the results of the present study suggest that miR-23b inhibits BMP9-induced C2C12 myoblast osteogenesis via targeting of the Runx2 gene, acting as a suppressor. The current study contributes to the understanding of the functions of BMP9 in ossification.
Keywords: bone morphogenetic protein 9, miR-23b, C2C12 myoblast cell, Runx2, osteogenesis
Introduction
Bone morphogenetic proteins (BMPs) belong to the transforming growth factor-β super family and were originally identified to induce ectopic bone growth and cartilage formation as osteoinductive cytokines (1,2). Subsequently, previous studies have demonstrated that the BMPs perform multiple functions in the regulation of cell growth, differentiation and apoptosis in various cell types (3–5). BMP9, a member of the BMP family, is a secreted protein that is expressed in the liver (6). BMP9 induces ectopic bone growth and hypertrophic chondrocyte formation, in addition to supporting the differentiation of mesenchymal cells (MSCs) into cartilage (7). Animal experiments have demonstrated that BMP9 has an increased capacity for the induction of osteogenesis in MSCs when compared with BMP2 and BMP7, which have been used in clinical therapy for bone defects and additional orthopedic diseases (8,9).
MicroRNAs (miRNAs) are a class of endogenous noncoding RNAs, of approximately 22 nucleotides in length, which act as post-transcriptional regulators in gene expression, via combining to the 3′-untranslated region (UTR) region of the targeted mRNA, causing it to degrade and inhibiting its translation (10). miRNAs are involved in diverse physiological and pathological processes and previous studies have identified that certain miRNAs may positively or negatively regulate osteogenesis and osteoclastogenesis (10–12). These results indicated that miRNAs may be therapeutic targets in the treatment of bone defects and additional orthopedic diseases.
In the present study, the expression of miR-23b was analyzed and the function in osteogenesis of BMP9-induced C2C12 myoblasts was investigated via the transfection of exogenous mimics and inhibitors of miR-23b.
Materials and methods
Cell culture
The C2C12 myoblast, HCT116 and HEK293T cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA), and were maintained in complete Dulbecco's modified Eagle's medium (DMEM; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were incubated at 37°C and a 5% CO2 in a humidified atmosphere.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from each sample using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Quality of the RNA was evaluated using the NanoDrop 1000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Inc.) and first-strand cDNA was synthesized using the PrimeScript™ RT reagent kit (Takara, Bio, Inc., Otsu, Japan). For miR-23b, the reverse transcription primer was used. SYBR Green II dye-based qPCR analysis was conducted using the Rotor-Gene 6000 Real-Time PCR machine (Corbett Research, Mortlake, Australia). The amplification program was set as follows: Initial denaturation at 94°C for 10 min; 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 15 sec and extension at 70°C for 15 sec; and a final extension step at 70°C for 5 min. The RT-qPCR was conducted using the 2−∆∆Ct method and each sample of 0.1 µg of cDNA was tested in triplicate (13). Glyceraldehyde 3-phosphate dehydrogenase and U6 were used as endogenous normalization controls. The primer sequences of the genes are presented in Table I.
Table I.
Primer sequences of genes.
| Gene | Primer sequence (5′-3′) |
|---|---|
| Runx2 | F: GGTGAAACTCTTGCCTCGTC |
| R: AGTCCCAACTTCCTGTGCT | |
| ALP | F: TGCCTACTTGTGTGGCGTGAA |
| R: TCACCCGAGTGGTAGTCACAATG | |
| OCN | F: TCTGACAAAGCCTTCATGTCC |
| R: AAATAGTGATACCGTAGATGCG | |
| OPN | F: ACACTTTCACTCCAATCGTCC |
| R: TGCCCTTTCCGTTGTTGTCC | |
| miR-23b | RT: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAATCA |
| F: GAGGGTTCCTGGCATGC | |
| R: GTGCAGGGTCCGAGGT | |
| GAPDH | F: AATGTGTCCGTCGTGGATCTGA |
| R: AGTGTAGCCCAAGATGCCCTTC | |
| U6 | RT: AAAATATGGAACGCTTCACGAATTTG |
| F: CTCGCTTCGGCAGCACATATACT | |
| R: ACGCTTCACGAATTTGCGTGTC |
Runx2, runt-related transcription factor 2; ALP, alkaline phosphatase; OCN, osteocalcin; OPN, osteopontin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Plasmids construction
The 3′UTR constructs of runt-related transcription factor 2 (Runx2) were chemically synthesized, and were then cloned into the pMIR-REPORT miRNA Expression Reporter Vector System (Applied Biosystems; Thermo Fisher Scientific, Inc.) between the Hind III and Pme I (Promega Corporation, Madison, WI, USA) restriction enzyme sites subsequent to annealing by mixing with primers in boiling water for 10 min and allowed to cool. The 3′-UTR region fragments of the Runx2 containing the predicted binding sites of mmu-miR-23b and its mutant sequence were synthesized as presented in Table II, and the β-galactosidase (β-gal) reporter plasmid (Invitrogen; Thermo Fisher Scientific, Inc.) as a control.
Table II.
Synthesized sequences of Runx2-luc reporter.
| Gene | Sequence |
|---|---|
| Wild type | 5′-CTAGTCGGAAATATTGCTAAGCAATCTCAATTCCTTCAGGCATAATGTGATTTT-3′ |
| 3′-AGCTAAAATCACATTATGCCTGAAGGAATTGAGATTGCTTAGCAATATTTCCGA-5′ | |
| Mutant | 5′-CTAGTCGGAAATATTGCTAAGCAATCTCAATTCCTTCAGGCATAACTTGATTTT-3′ |
| 3′-AGCTAAAATCAAGTTATGCCTGAAGGAATTGAGATTGCTTAGCAATATTTCCGA-5′ |
Bold bases indicate the binding region of microRNA-23b.
Transfection of miRNA mimics, inhibitors and plasmids
The miRNA mimics (miR-23b mimic), inhibitors (miR-23b inhibitor) and negative controls of miR-23b were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China) and transfected into C2C12 at final concentrations of 50 nM per well in a 24-well plate with Entranster-R transfection reagent (Engreen, Beijing, China) following the manufacturer's instructions. The component was mixed in serum-free DMEM, and then the transfection was conducted in complete DMEM and refreshed 6 h subsequent to transfection. The plasmids were transfected into cells using Lipofectamine 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) as described previously (14).
Preparation of BMP9-conditioned medium
The recombinant adenovirus expressing BMP9 (Ad-BMP9) and short interfering (si)Runx2 (Ad-siRunx2) were generated previously using the AdEasy system, as developed by Dr T.C. He (The University of Chicago, Chicago, IL, USA) (15). BMP9-conditioned medium was prepared as previously described (16). Briefly, HCT116 cells were infected with an optimal titer of 0.2 µl Ad-BMP9. A total of 6 h later, the culture medium was refreshed with serum-free DMEM. The BMP9-conditioned medium was collected at 24 and 48 h subsequent to the exchange of the medium and it was used immediately with another stored at 4°C.
Western blotting
Western blotting was performed according to a standard protocol as previously described (17). C2C12 cells in dishes were lysed with radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China), and the total protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electro-blotted onto a polyvinylidene difluoride membrane (EMD Millipore, Billerica, CA, USA). The membranes were then incubated with an optimal concentration of the following primary antibodies: Anti-Runx2 (1:1,000) and anti-β-actin (1:5,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Protein bands were visualized using the Quantity One software 4.5.2 (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Alkaline phosphatase (ALP), Alizarin Red S (ARS) staining and the quantitative ALP assay
ALP activity was assessed using a modified Great EscAPe SEAP Chemiluminescence kit (Clontech Laboratories, Inc., Mountain View, CA, USA) and/or histochemical staining assay [using a mixture of 0.1 mg/ml naphthol AS-MX phosphate (Sigma-Aldrich, St. Louis, MO, USA) and 0.6 mg/ml Fast Blue BB salt (Sigma-Aldrich)] as described (18). For quantitative ALP measurement, the ALP activity was determined at a wavelength of 405 nm (E6080; Promega Corporation) in triplicate and the results were repeated in at least three independent experiments.
ARS staining was conducted to evaluate mineralized matrix nodules as described previously (19). Briefly, cells were cultured in the presence of ascorbic acid (50 µg/ml; Beijing Solarbio Science and Technology Co., Ltd., Beijing, China) and β-glycerophosphate (10 mmol/l; Beijing Solarbio Science and Technology Co., Ltd.). Cells were fixed with 0.05% (v/v) glutaraldehyde (Chongqing Chuandong Chemical Group Co., Ltd., Chongqing, China) at room temperature for 10 min. Subsequent to washing with distilled water, the fixed cells were incubated with 0.4% ARS (Sigma-Aldrich) for 5 min, followed by extensive washing with distilled water. The staining of calcium mineral deposits was recorded under bright field microscopy (T-DH; Nikon Corporation, Tokyo, Japan).
Dual luciferase reporter assay
A total of 0.4 µg β-gal reporter plasmid or pMir-Runx2 plasmid and 0.3 µg mimics were co-transfected into 293T cells using Lipofectamine 2000 reagent. Cells were harvested at 48 h subsequent to transfection and assayed for firefly luciferase activity using the Dual-Glo™ Luciferase Assay system (Promega Corporation). Firefly luciferase activity was normalized to β-gal.
Bio-informatics prediction
To predict the target genes of miR-23b during the C2C12 myoblast differentiation induced by BMP9, the following three miRNA target prediction databases were used: TargetScan (http://www.targetscan.org), PicTar (http://www.pictar.org) and miRbase (http://www.mirbase.org).
Statistical analysis
The results represent the average of three independent experiments and the data are presented as the mean ± standard deviation. Statistical significance was determined using Student's paired t-test, and P<0.05 was considered to indicate a statistically significant difference.
Results
miR-23b expression reduces in the early stage of BMP9-induced osteoblast differentiation of C2C12 myoblasts
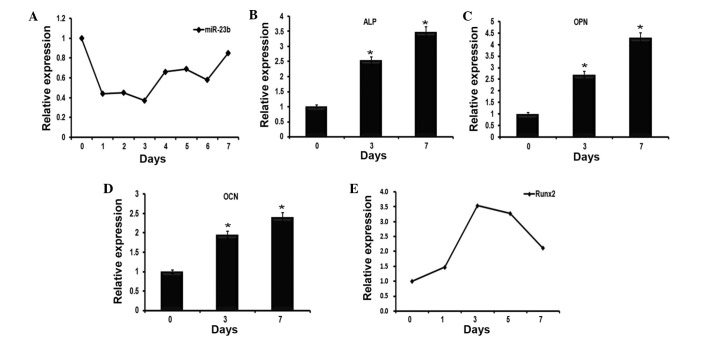
Several miRNAs including miR-21, miR-23b and miR-155 have been previously identified to be expressed during the early stages of MSC osteogenesis by microarray data analysis (data not shown). Given that the role of miR-23b in the regulation of osteogenesis remains to be defined, the current study investigated whether miR-23b serves a role in regulating BMP9-induced osteogenic differentiation. By RT-qPCR, it was identified that the expression levels of miR-23b were downregulated during the early stages of BMP9-induced C2C12 osteogenic differentiation, which reached its minimum on day 3 (Fig. 1A). To further confirm the effect of BMP9, RT-qPCR was used to detect the mRNA expression levels of osteoblast-specific genes over 7 days, including ALP (Fig. 1B), OCN (Fig. 1C), OPN (Fig. 1D) and Runx2 (Fig. 1E). Similar to the results of a previous study, BMP9 was observed to promote the expression of these genes in C2C12 myoblasts (20).
Figure 1.
Expression profiles of miR-23b and osteogenesis-associated genes during the early stages of BMP9-induced osteogenic differentiation of C2C12 myoblasts. (A) C2C12 myoblast cell lines were stimulated to undergo osteoblast differentiation using BMP9. RT-qPCR was used to examine the expression levels of miR-23b, which were normalized to U6 as a control. The data demonstrated that miR-23b was reduced over seven days. (B–D) Cells underwent the same treatment as in (A), then were collected at 0, 3 and 7 days. The expression levels of (B) ALP, (C) OCN and (D) OPN were evaluated by RT-qPCR. n=3, *P<0.05, vs. 0 day. (E) RT-qPCR detected the expression of Runx2 in BMP9-induced C2C12 myoblasts over 7 days. miR-23b, microRNA-23b; BMP9, bone morphogenetic protein 9; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; ALP, alkaline phosphatase; OCN, osteocalcin; OPN, osteopontin; Runx2, runt-related transcription factor 2.
Downregulation of miR-23b could promote osteoblast differentiation of C2C12 myoblasts induced by BMP9
RT-qPCR analyzed the intracellular miR-23b content subsequent to transfection of exogenous synthetic molecules expressing or inhibiting miR-23b, and the control, termed miR-23b mimics, miR-23b inhibitor and miR-C, respectively. Subsequent to induction of C2C12 by BMP9 for 7 days, the intracellular miR-23b level was identified to be significantly elevated by transfection with the miR-23b mimics, whereas the miR-23b inhibitor led to a reduction in miR-23b content, miR-NC had a non-significant effect (Fig. 2A). ALP (Fig. 2B) and ARS (Fig. 2D) staining demonstrated that the ALP level and calcium deposition in miR-23b-transfected C2C12 cells induced by BMP9 were significantly lower than those in the negative control group. On the contrary, these levels were much higher in the miR-23b inhibitor-transfected group. The quantitative ALP assay, which was performed using the same method as for ALP staining at the same time points subsequent to transfection with miR-23b, the miR-23b inhibitor and miR-NC, implied that ALP activity was inhibited by 27% compared with the control group on day 7 following miR-23b transfection. The miR-23b inhibitor enhanced ALP levels by 2.4-fold when compared with the miR-NC group (Fig. 2C). In conclusion, the above results indicated that the overexpression of miR-23b was able to repress BMP9-induction of C2C12 osteogenesis, however downregulation of miR-23b promoted the osteoblast differentiation of C2C12 myoblasts induced by BMP9.
Figure 2.
Downregulated expression of miR-23b promotes BMP9-induced osteogenic differentiation of C2C12 myoblasts. (A) Downregulated efficiency of miR-23b inhibitor following transfection into C2C12 cells 6 h subsequent to Ad-BMP9 infection for 6 h; n=3, **P<0.01 vs. the BMP9 group. (B) ALP staining of BMP9-induced C2C12 myoblasts at day 7 of osteogenic differentiation with treatment with exogenous miR-23b mimics and inhibitors (magnification, ×200). (C) The quantitative ALP assay was a parallel experiment to ALP staining, n=3, *P<0.05 vs. the BMP9. (D) Alizarin Red S staining was used to evaluate the late stage osteogenic marker and BMP9-induced C2C12 myoblasts at day 14 of osteogenic differentiation in cells treated with exogenous miR-23b (magnification, ×200). miR-23b, microRNA-23b; BMP9, bone morphogenetic protein 9; ALP, alkaline phosphatase.
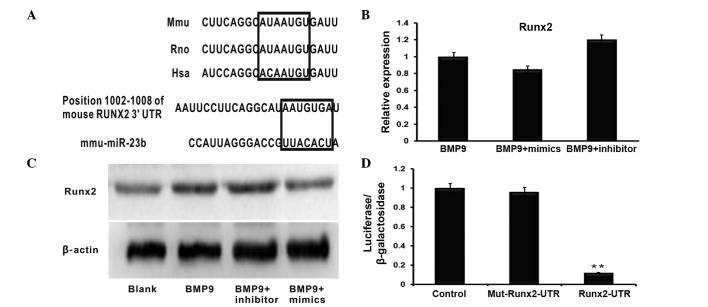
Runx2 is a direct target of miR-23b
To investigate the mechanism of miR-23b regulating BMP9-induced osteogenesis, biological information analysis was used to search the potential targets of miR-23b. According to the bioinformatics databases used, Runx2 was identified as a potential target of miR-23b in the C2C12 osteogenesis induced by BMP9 (Fig. 3A). The predicted binding site of mmu-miR-23b is located at position 1002–1008 of the 3′UTR of the Runx2 mRNA. The miR-23b mimics and inhibitors were individually transfected into BMP9-induced C2C12 cells. The RT-qPCR analysis indicated that Runx2 mRNA levels were not significantly altered in different transfection groups (Fig. 3B). However, the Runx2 protein level was repressed by miR-23b mimics, and promoted by the miR-23b inhibitor when compared with the control group (Fig. 2C). To verify the predicted binding sites, the firefly lucif-erase reporter system containing the wild type 3′-UTR binding site (Runx2-3′-UTR-WT) and the mutant 3′-UTR binding site (Runx2-3′-UTR-mt) was constructed and co-transfected with the miR-23b mimics. As presented in Fig. 2D, the luciferase activity assay detected that co-transfection of miR-23b mimics and the Runx2-3′-UTR-WT plasmid reduced the luciferase expression level compared with the control group. Furthermore, the co-transfection of miR-23b and Runx2-3′-UTR-mt plasmid had no clear effect on luciferase levels. In summary, the results suggested that Runx2 mRNA is a direct target of miR-23b.
Figure 3.
miR-23b directly binds to the 3′UTR of Runx2. (A) Schematic diagram illustrating the the binding sites of mmu-miR-23b and Runx2 as predicted by the target prediction databases. (B) Reverse transcription-quantitative polymerase chain reaction demonstrated that neither the miR-23b mimic nor the miR-23b inhibitor had significant effects on Runx2 mRNA levels. n=3, P>0.05 (C) Western blot analysis indicated that the Runx2 expression level was promoted by the downregulation of miR-23b, however the overexpression of miR-23b via the transfection of the miR-23b mimics inhibited its expression. (D) All data demonstrated co-transfection of miR-23b and its WT 3′-UTR binding site, termed Runx2-3′UTR-WT, significantly reduced the luciferase activity, whereas miR-23b had no effect on the Mut 3′-UTR binding region. n=3, **P<0.01. miR-23b, microRNA-23b; UTR, untranslated region; Runx2, runt-related transcription factor 2; BMP9, bone morphogenetic protein 9; WT, wild type; Mut, mutant.
Runx2 knockdown reduces the effect of miR-23b
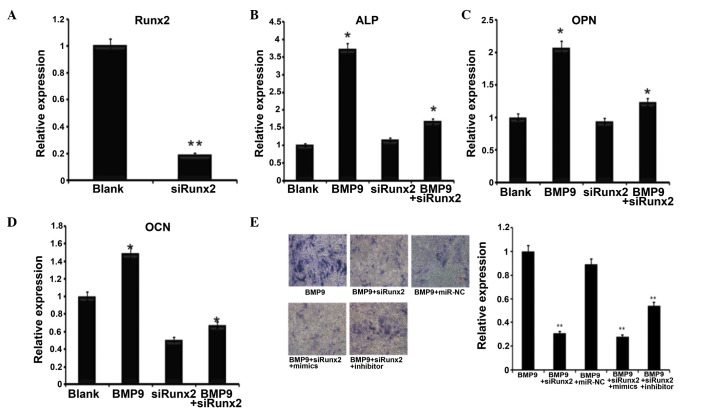
To verify the association between miR-23b and Runx2 further, Ad-sim-Runx2 was transfected into the C2C12 cells induced by BMP9 (Fig. 4A), the role of Runx2 in osteogenic differentiation induced by BMP9 was identified by the mRNA expression of ALP (Fig. 4B), OCN (Fig. 4C) and OPN (Fig. 4D). The expression of these osteoblast-specific genes was significantly downregulated following the knockdown of Runx2. Subsequently, the miR-23b mimics and inhibitors were transfected into the BMP9-induced C2C12 cells with Runx2 knockdown. ALP staining and the quantitative ALP assay were then subsequently conducted, in order to detect the effects of miR-23b in Runx2-knockdown C2C12 cells. As indicated, transfection of the miR-23b inhibitor in Runx2-knockdown C2C12 cells had no significant impact on the promotion of osteogenesis when compared with the group only transfected with the miR-23b inhibitor. In the Runx2-knockdown C2C12 cells, the miR-23b inhibitor promoted C2C12 osteogenesis by greater than 1.5-fold when compared with the control. ALP staining and the quantification results indicated that the osteogenesis of Runx2-knockdown C2C12 cells was likely to be enhanced by the miR-23b inhibitor, as presented in Fig. 4E. Taken together, these results indicate that the knockdown of Runx2 inhibits the effects of miR-23b on the osteogenesis of BMP9-induced C2C12 cells.
Figure 4.
Runx2 affected BMP9-induced differentiation of C2C12 cells. (A) Ad-siRunx2 was used to knockdown the expression of Runx2. n=3, **P<0.01. (B-D) Co-infected Ad-siRunx2 and Ad-BMP9 cells were collected at 3 days. (B) ALP, (C) OCN and (D) OPN expression levels were detected by reverse transcription-quantitative polymerase chain reaction, n=3, *P<0.05 vs. the blank group. (E) ALP staining of BMP9-induced C2C12 myoblasts at day 7 of osteogenic differentiation treated with Ad-siRunx2 followed by transfection with mimics and inhibitors (magnification, ×200). **P<0.01 vs. BMP9 group. Runx2, runt-related transcription factor 2; BMP9, bone morphogenetic protein 9; Ad, adenovirus; si, short interfering; ALP, alkaline phosphatase; OCN, osteocalcin; OPN, osteopontin.
Discussion
While BMP9 has been demonstrated to exert potent osteogenic activity (21,22), the detailed molecular mechanisms underlying BMP9 action remain to be fully elucidated. The current study aimed to investigate the possible effects of miRNA inhibition on BMP9-induced osteogenic differentiation due to the fact that epigenetic regulation serves an important role in regulating osteogenesis (23).
The present study investigating the molecular mechanisms of BMP9 predominantly focused on transcriptional regulation, while the post-transcriptional mechanism has not been thoroughly studied (24). miRNAs are endogenous, non-coding RNAs that are negative regulators of their target genes at the post-transcriptional level (12). miR-23b belongs to the miR-23a/24/27a cluster which is located on chromosome 19p13.12 and the miR-23b/27b/24-1 cluster which is located on chromosome 9q22.32 (25,26). The clusters have been previously reported to be enhanced in acute lymphoblastic leukemia, acute myeloid leukemia, glioblastoma, hepatocellular carcinoma, gastric cancer, pancreatic cancer and uterine leiomyoma (27–29). The majority of these studies have focused on the function of miR-23b as an oncogene in non-small cell lung cancer while ignoring its role in bone formation (26).
In the current study, it was demonstrated that miR-23b reduced activity of the early osteogenic marker ALP in BMP9-induced C2C12 cells using miR-23b mimics, in addition to reducing late osteogenic marker matrix mineralization. The dual-luciferase reporter assays demonstrated that miR-23b reduced BMP9-induced bone formation possibly via suppression of Runx2 translation. Runx2 is considered as the most important early osteogenic transcriptional factor (30). Ad-siRunx2 was employed to evaluate its important role in the course miR-23b restrained BMP9-inducing osteogenesis.
In conclusion, the observations suggest that miR-23b serves an important role in BMP9-mediated osteogenic signaling through negative regulation. Thus, miR-23b inhibitors may be used as a novel therapeutic strategy in bone fracture healing.
Acknowledgments
The current study was supported by the National Natural Science Foundation of China (grant no. 31200971) and the Program of the Ministry of Science and Technology of Yu-Zhong District (grant no. 20130136). The authors would like to thank Dr Tong-Chuan He (University of Chicago, Chicago, IL, USA) for donating the Ad-BMP9 and Ad-siRunx2.
References
- 1.Urist MR. Bone: Formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- 2.Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. Novel regulators of bone formation: Molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 3.Okla M, Ha JH, Temel RE, Chung S. BMP7 drives human adipogenic stem cells into metabolically active beige adipocytes. Lipids. 2015;50:111–120. doi: 10.1007/s11745-014-3981-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei Y, Wu Y, Zeng B, Zhang H. Effects of sodium fluoride treatment in vitro on cell proliferation, BMP-2 and BMP-3 expression in human osteosarcoma MG-63 cells. Biol Trace Elem Res. 2014;162:18–25. doi: 10.1007/s12011-014-0148-8. [DOI] [PubMed] [Google Scholar]
- 5.Jiqing C, Yaqin L, Yingyin L, Fei C, Huili Z, Yuling Z, Juan Y, Shanwei F, Cheng Z. BMP4 inhibits myogenic differentiation of bone marrow-derived mesenchymal stromal cells in mdx mice. Cytotherapy. 2015;17:1213–1219. doi: 10.1016/j.jcyt.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Brown MA, Zhao Q, Baker KA, Naik C, Chen C, Pukac L, Singh M, Tsareva T, Parice Y, Mahoney A, et al. Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J Biol Chem. 2005;280:25111–25118. doi: 10.1074/jbc.M503328200. [DOI] [PubMed] [Google Scholar]
- 7.Kang Q, Sun MH, Cheng H, Peng Y, Montag AG, Deyrup AT, Jiang W, Luu HH, Luo J, Szatkowski JP, et al. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther. 2004;11:1312–1320. doi: 10.1038/sj.gt.3302298. [DOI] [PubMed] [Google Scholar]
- 8.Tsuda H, Wada T, Yamashita T, Hamada H. Enhanced osteoinduction by mesenchymal stem cells transfected with a fiber-mutant adenoviral BMP2 gene. J Gene Med. 2005;7:1322–1334. doi: 10.1002/jgm.777. [DOI] [PubMed] [Google Scholar]
- 9.Chen P, Vukicevic S, Sampath TK, Luyten FP. Osteogenic protein-1 promotes growth and maturation of chick sternal chondrocytes in serum-free cultures. J Cell Sci. 1995;108:105–114. doi: 10.1242/jcs.108.1.105. [DOI] [PubMed] [Google Scholar]
- 10.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 11.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 12.Papaioannou G, Mirzamohammadi F, Kobayashi T. MicroRNAs involved in bone formation. Cell Mol Life Sci. 2014;71:4747–4761. doi: 10.1007/s00018-014-1700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Mo RH, Zaro JL, Ou JH, Shen WC. Effects of Lipofectamine 2000/siRNA complexes on autophagy in hepatoma cells. Mol Biotechnol. 2012;51:1–8. doi: 10.1007/s12033-011-9422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He TC, Zhou S, Da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu DJ, Zhao Y-Z, Wang J, He JW, Weng YG, Luo JY. Smads, p38 and ERK1/2 are involved in BMP9-induced osteogenic differentiation of C3H10T1/2 mesenchymal stem cells. BMB Rep. 2012;45:247–252. doi: 10.5483/BMBRep.2012.45.4.247. [DOI] [PubMed] [Google Scholar]
- 17.Lv Z, Yang D, Li J, Hu M, Luo M, Zhan X, Song P, Liu C, Bai H, Li B, et al. Bone morphogenetic protein 9 overexpression reduces osteosarcoma cell migration and invasion. Mol Cells. 2013;36:119–126. doi: 10.1007/s10059-013-0043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song T, Wang W, Xu J, Zhao D, Dong Q, Li L, Yang X, Duan X, Liang Y, Xiao Y, et al. Fibroblast growth factor 2 inhibits bone morphogenetic protein 9-induced osteogenic differentiation of mesenchymal stem cells by repressing Smads signaling and subsequently reducing Smads dependent up-regulation of ALK1 and ALK2. Int J Biochem Cell Biol. 2013;45:1639–1646. doi: 10.1016/j.biocel.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Luo J, Tang M, Huang J, He BC, Gao JL, Chen L, Zuo GW, Zhang W, Luo Q, Shi Q, et al. TGFbeta/BMP type I receptors ALK1 and ALK2 are essential for BMP9-induced osteogenic signaling in mesenchymal stem cells. J Biol Chem. 2010;285:29588–29598. doi: 10.1074/jbc.M110.130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Li L, Dong Q, Wang Y, Feng Q, Ou X, Zhou P, He T, Luo J. Activation of PKA/CREB signaling is involved in BMP9-induced osteogenic differentiation of mesenchymal stem cells. Cell Physiol Biochem. 2015;37:548–562. doi: 10.1159/000430376. [DOI] [PubMed] [Google Scholar]
- 21.Luu HH, Song WX, Luo X, Manning D, Luo J, Deng ZL, Sharff KA, Montag AG, Haydon RC, He TC. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J Orthop Res. 2007;25:665–677. doi: 10.1002/jor.20359. [DOI] [PubMed] [Google Scholar]
- 22.Ye G, Li C, Xiang X, Chen C, Zhang R, Yang X, Yu X, Wang J, Wang L, Shi Q, Weng Y. Bone morphogenetic protein-9 induces PDLSCs osteogenic differentiation through the ERK and p38 signal pathways. Int J Med Sci. 2014;11:1065–1072. doi: 10.7150/ijms.8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng L, Zhong X. Epigenetic regulation of drug metabolism and transport. Acta Pharm Sin B. 2015;5:106–112. doi: 10.1016/j.apsb.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C, Weng Y, Yuan T, Zhang H, Bai H, Li B, Yang D, Zhang R, He F, Yan S, et al. CXCL12/CXCR4 signal axis plays an important role in mediating bone morphogenetic protein 9-induced osteogenic differentiation of mesenchymal stem cells. Int J Med Sci. 2013;10:1181–1192. doi: 10.7150/ijms.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jahid S, Sun J, Edwards RA, Dizon D, Panarelli NC, Milsom JW, Sikandar SS, Gümüs ZH, Lipkin SM. miR-23a promotes the transition from indolent to invasive colorectal cancer. Cancer Discov. 2012;2:540–553. doi: 10.1158/2159-8290.CD-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donadelli M, Dando I, Fiorini C, Palmieri M. Regulation of miR-23b expression and its dual role on ROS production and tumour development. Cancer Lett. 2014;349:107–113. doi: 10.1016/j.canlet.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Ma G, Dai W, Sang A, Yang X, Gao C. Upregulation of microRNA-23a/b promotes tumor progression and confers poor prognosis in patients with gastric cancer. Int J Clin Exp Pathol. 2014;7:8833–8840. [PMC free article] [PubMed] [Google Scholar]
- 28.Aghaee-Bakhtiari SH, Arefian E, Naderi M, Noorbakhsh F, Nodouzi V, Asgari M, Fard-Esfahani P, Mahdian R, Soleimani M. MAPK and JAK/STAT pathways targeted by miR-23a and miR-23b in prostate cancer: Computational and in vitro approaches. Tumour Biol. 2015;36:4203–4212. doi: 10.1007/s13277-015-3057-3. [DOI] [PubMed] [Google Scholar]
- 29.Chiyomaru T, Seki N, Inoguchi S, Ishihara T, Mataki H, Matsushita R, Goto Y, Nishikawa R, Tatarano S, Itesako T, et al. Dual regulation of receptor tyrosine kinase genes EGFR and c-Met by the tumor-suppressive microRNA-23b/27b cluster in bladder cancer. Int J Oncol. 2015;46:487–496. doi: 10.3892/ijo.2014.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu TM, Lee EH. Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Eng Part B Rev. 2013;19:254–263. doi: 10.1089/ten.teb.2012.0527. [DOI] [PMC free article] [PubMed] [Google Scholar]