Abstract
Tumor immunoscoring is rapidly becoming a universal parameter of prognosis, and T-cells isolated from tumor masses are used for ex vivo amplification and readministration to patients to facilitate an antitumor immune response. We recently exploited the cancer genome atlas (TCGA) RNASeq data to assess T-cell receptor (TcR) expression and, in particular, discovered strong correlations between major histocompatibility class II (MHCII) and TcR-α constant region expression levels. In this article, we describe the results of searching TCGA exome files for TcR-α V-regions, followed by searching the V-region datasets for TcR-α-J regions. Both primary and metastatic breast cancer sample files contained recombined TcR-α V–J regions, ranging in read counts from 16–39, at the higher level. Among four such V–J rearrangements, three were productive rearrangements. Rearranged TcR-α V–J regions were also detected in TCGA–bladder cancer, –lung cancer, and –ovarian cancer datasets, as well as exome files representing bladder cancer, in Moffitt Cancer Center patients. These results suggest that a direct search of commonly available, conventional exome files for rearranged TcR segments could play a role in more sophisticated immunoscoring or in identifying particular T-cell clones and TcRs directed against tumor antigens.
Keywords: the cancer genome atlas, TCGA, T-cell receptor rearrangement, exome, breast cancer, bladder cancer
Introduction
The cancer microenvironment is usually considered to include infiltrating immune function cells, including innate and adaptive immune function cells.1 The impact of these cells is not comprehensively understood. In some cases, inflammation is considered to facilitate cancer metastasis, either by enhancing blood vessel formation, allowing the nurturing and spread of tumor cells,2 or by leading to cancer cell/immune cell fusions and metastasis via immune function or cell migration functions.3,4 In other cases, the immune function cell infiltrate is considered to represent an antitumor immune response and, in particular, may represent T-cells that are specific to tumor antigens, T-cells that could be amplified ex vivo for reintroduction into the patient, and augmentation of the immune response against the tumor.5–7
Considering the potential role of the T-cell in tumor development, efforts have been made to design exome capture processes specifically to identify recombined T-cell receptors (TcRs) in resected cancer specimens or in biopsy specimens.8 A routine identification of TcR recombinations could lead to identification of recombinations that generate proteins that interact with MHC-bound tumor antigens and may even lead to the generation of TcR tools that are independent of T-cells. However, exome capture processes that are designed specifically for TcRs are highly specialized and not commonly available, unlike total exome capture technologies, which include the germline versions of the unrearranged TcR gene segments. Thus, we developed a scripted algorithm for recovering TcR-α-variable (V) regions from standard whole exome files, followed by a search of the V-region files for recombined V–J regions.
Methods
A list of TcR-α-V and J regions was obtained from the National Center for Biotechnology Information and are present in the Supplementary File labeled, “Supporting online material Gill et al 2015”, under “Item 1: List of TcR-α V and J sequences”. Primary tumor whole exome sequence (WXS) samples for the cancer datasets indicated in the Results section were downloaded from the cancer genome atlas (TCGA). The program “FindV2” then found and extracted reads that matched portions of the V regions into individual tab separated values (TSV) files, ie, separate for each TCGA sample. The “FindV2” program is included in the Supplementary File labeled, “Supporting online material Gill et al 2015”, under “Item 2: Processing steps”. These TSV files were then searched using the program “SearchJ,” which extracted the sequences of the reads that matched any of the J regions, ranging from 10 base pairs to 30 base pairs. These extracted sequences were also placed in a separate TSV file and were then used to search the extracted V regions for the entire, original read representing a recombination. These reads were collected and organized by cancer and barcode, and their overall effects were analyzed through the International Immunogenetics Information System’s Variant Quest program to find occurrences of productive rearrangement within the reads.
In the Moffitt Cancer Center, for bladder cancer WXS preparations, patient samples were obtained via Moffitt Cancer Center approved protocol #18135 and University of South Florida IRB application approval #22538. Surgically resected, primary bladder tumors were micro-dissected, DNA was prepared, and sequencing was performed by the Functional Genomics Core Facility at the Moffitt Cancer Center and Research Institute (Tampa, FL). Briefly, 200 ng of DNA was used as input for the Agilent SureSelect XT Clinical Research Exome kit, which contains the exon targets of Agilent’s v5 “whole-exome kit, with increased coverage at 5000 disease-associated targets”. A genomic DNA library was constructed according to the manufacturer’s protocol, and the size and quality of the library were evaluated using the Agilent BioAnalzyer. An equimolar amount of library DNA was used for a whole-exome enrichment using the Agilent capture baits, and following quantitative polymerase chain reaction library quantification and quality control (QC) analysis on the BioAnalzyer, approximately 100,000,000 75-base paired-end sequences were generated using v2 chemistry on an Illumina NextSeq 500 sequencer.
Finally, a test file was prepared for verification of the code developed for this study. The code indicated in the Supplementary File, together with the test file, can be used to obtain a TcR-α V–J recombination. The test file was prepared from the cancer cell line encyclopedia WXS file, C836.ZR-75-1.2.bam.tsv, which ordinarily has no rearranged TcR-α V–J regions. “C836.ZR-75-1.2.bam.tsv” was downloaded from the CGHub site. The test file is present in the Supplementary File labeled, “Gill SOM test set five reads”. The files can be searched with the J string CCGGTAACCAGTTCTATTTTGGGACAGGGA to verify recovery of five copies of the V–J sequence.
Results
To obtain an initial indication of the usefulness of TCGA DNA sequence files (http://cancergenome.nih.gov/) for detecting rearranged TcR-α V–J regions, we conducted a low-stringency recovery of all TcR-α-V regions in skin, cutaneous melanoma (SKCM) and breast cancer (BRCA) whole genome sequence (WGS) and WXS files downloaded from CGHub (scripted algorithms and an example V recovery file in the Supplementary File; see also Methods). The BRCA WGS files included three blood-normal files. The BRCA and SKCM WGS files and the SKCM WXS files represented both metastatic samples and primary tumor samples. The BRCA WXS files represented only primary tumor files.
The V region files outputted from the initial processing step were searched for a match with TcR-α J regions. The J regions that matched were deposited into files (example J file in Supplementary File), and these J regions were then used to re-search the V region files to create summary files representing all TcR-α V–J recombinations detected for each barcode studied, ie, the specific reads representing the recombinations. The summary files for the entire TCGA portion of this study are provided in the Supplementary File.
This first approach with the SKCM and BRCA TCGA files indicated that a large variety of TcR-α rearrangements could be detected in both WXS and WGS sequence files, with the majority of V–J rearrangements present in the BRCA WXS files (Table 1). For all but one barcode, the V–J rearrangements in each SKCM and BRCA file were completely unique, ie, only one BRCA barcode file contained a rearrangement that coupled the same J with two distinct V regions.
Table 1.
Summary of results of search for TcR-α V–J rearrangements in TCGA BRCA and SKCM files. Process steps are provided in the Supplementary File.
| TYPES OF TCGA FILES | NO. OF WGS FILES | NO. OF WXS FILES | NO. OF SAMPLES WITH J’S 14 NUCLEOTIDES AND ABOVE | TOTAL NO. OF V–J COMBINATIONS | NO. OF SAMPLES WITH MORE THAN ONE V–J COMBINATION | NO. OF SAMPLES WITH MORE THAN A TOTAL OF 20 READS FOR ALL V–J COMBINATIONS | NO. OF SAMPLES WITH MORE THAN ONE V PER J |
|---|---|---|---|---|---|---|---|
| Patient samples | |||||||
| BRCA-WGS | 14 | 7 | 13 | 4 | 1 | 0 | |
| 1BRCA-WXS | 17 | 13 | 45 | 9 | 4 | 1 | |
| BRCA Totals | 14 | 17 | 20/31 | 58/31 | 13/31 | 5/31 | 1/31 |
| SKCM-WGS | 9 | 6 | 10 | 2 | 1 | 0 | |
| SKCM-WXS | 4 | 0 | 0 | 0 | 0 | 0 | |
| SKCM Totals | 9 | 4 | 6/13 | 10/13 | 2/13 | 1/13 | 0/13 |
| Cell lines (CCLE) | |||||||
| 1Breast cancer | 13 | 3 | 4 | 0 | 0 | 0 | |
| Melanoma | 16 | 2 | 2 | 0 | 0 | 0 |
Notes:
Bold indicates comparisons of detection rates for BRCA-WXS versus breast cancer cell lines. P < 0.002, for No. of J’s with 14 nucleotides and above; P < 0.001, for total number of V–J combinations; and P <0.0004 for number of samples with more than one V–J combination. (Student’s t-test, one-tailed distribution, unequal variances).
We next recovered the TcR-α V–J recombinations present in seven BRCA metastasis files (Table 2), which had a higher density of rearrangements: 54 rearrangements for seven files (Table 2) versus 45 rearrangements present in seventeen primary BRCA tumor files (Table 1). The basis for the difference cannot be known at this time, because of the many differences in the preparation of the TCGA samples and the WXS files. However, this difference does raise the question of whether the BRCA metastatic files had a higher number of TcR-α V–J rearrangements, because the metastatic samples were taken from lymph nodes, leading to the inclusion of more T-cells in sample preparation?
Table 2.
Summary of results of search for TcR-α V–J rearrangements in TCGA BRCA WXS metastasis files.
| TOTAL NUMBER OF FILES | NO. OF SAMPLES WITH J’S 14 NUCLEOTIDES AND ABOVE | TOTAL NO. OF V–J COMBINATIONS | NO. OF SAMPLES WITH MORE THAN ONE V–J COMBINATION | NO. OF SAMPLES WITH MORE THAN A TOTAL OF 20 READS FOR ALL V–J COMBINATIONS | NO. OF SAMPLES WITH MORE THAN ONE V PER J |
|---|---|---|---|---|---|
| 7 | 6 | 54 | 5 | 4 | 2 |
We hypothesize that the above-indicated TcR-α V–J recombinations represent T-cells that have infiltrated the biopsies or surgical resections of the above-indicated cancers, such that even a close dissection of the tumor for WXS did not remove all of the T-cells. To test this hypothesis, we processed the WXS files of 16 BRCA and 15 melanoma cell lines. Comparison of the BRCA cell lines and the BRCA samples, for three related parameters (number of Js with 14 nucleotides or above; total number of V–J combinations and for number of distinct V–J combinations), indicated that the BRCA samples have significantly more detectable V–J combinations than those found in the cell lines, consistent with the idea that the detection of the V–J recombinations in the BRCA tissue samples represents infiltrating lymphocytes. There were not enough WXS SKCM tissue samples to make the same comparison, but the small detection rates of recombined V–J segments in the melanoma and breast samples were similar.
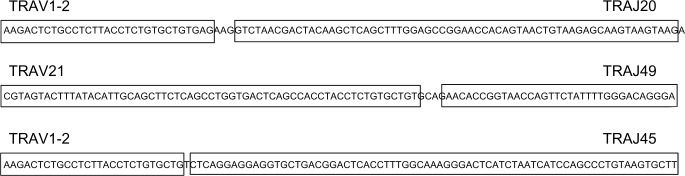
We next addressed the question of whether the TcR-α V–J rearrangements could encode proteins. We used the processing steps available at http://www.imgt.org/to analyze several V–J recombinations for productive, in-frame rearrangements that did not include stop codons.9 We analyzed the V–J recombinations from the primary tumor and metastatic BRCA WXS files with the most reads, representing the following read counts: 28, 28, 16, and 39, respectively (Table 3). Three out of these four V–J rearrangements were determined to be productive (Fig. 1).
Table 3.
Structures of BRCA TcR-α V–J rearrangements represented by comparatively large numbers of reads.
| TCGA BARCODE | V–J REARRANGEMENT READ SEQUENCE | NUMBER OF READS | IMGT ASSESSMENT* |
|---|---|---|---|
| TCGA-BH-A0DV-01A | CTGCCCTTGTGAGCGACTCCGCTTTGTACTTCTG TGCTGTGAGAGAGGGGATAGCAGCTATAAATTGATC TTCGGGAGTGGGACCAGACTGCTGGTCAGG |
28 | Unproductive, stop codons, out-of-frame junction; TRAV3, J12 |
| TCGA-E9-A1NH-01A-11D | AAGACTCTGCCTCTTACCTCTGTGCTGTGAGA AGGTCTAACGACTACAAGCTCAGCTTTGGAGCC GGAACCACAGTAACTGTAAGAGCAAGTAAGTAAGA |
28 | Productive, no stop codons, in-frame junction; TRAV1-2, J20 |
| TCGA-E2-A15A-06A-11D | CGTAGTACTTTATACATTGCAGCTTCTCAGCCTGG TGACTCAGCCACCTACCTCTGTGCTGTGCAGAACA CCGGTAACCAGTTCTATTTTGGGACAGGGA |
39 | Productive, no stop codons, in-frame junction; TRAV21, J49 |
| TCGA-E2-A15A-06A-11D | AAGACTCTGCCTCTTACCTCTGTGCTGTCTCAGGA GGAGGTGCTGACGGACTCACCTTTGGCAAAGGGAC TCATCTAATCATCCAGCCCTGTAAGTGCTT |
16 | Productive, no stop codons, in-frame junction; TRAV1-2, J45 |
Notes: Top two V–J rearrangements represent primary BRCA samples; the bottom two V–J rearrangement represents a metastatic BRCA sample.
Figure 1.
Structures of the productive TcR-α V–J rearrangements representing the three BRCA samples in Table 3.
We recently characterized RNASeq data for TcR-α constant region expression among eight cancer datasets represented by TCGA.10 We ranked the datasets based on an immunoscore that included the correlation of MHCII and TCR-α expression, among other related parameters. This ranking indicated that the bladder cancer (BLCA) dataset, along with the BRCA dataset, represented one of the highest immunoscores. Thus, we repeated the above analyses on 12 BLCA TCGA WXS files representing primary samples. Recombined TcR-α V–J regions were detected, but the results were of a relatively poor quality, with low read numbers and smaller length reads (Tables 4 and 5).
Table 4.
Summary of results of the search for TcR-α V–J rearrangements in TCGA BLCA, LUAD, and OVCA primary tumor files.
| TOTAL NUMBER OF FILES | NO. OF SAMPLES WITH J’S 14 NUCLEOTIDES AND ABOVE | TOTAL NO. OF V–J COMBINATIONS | NO. OF SAMPLES WITH MORE THAN ONE V–J COMBINATION | NO. OF SAMPLES WITH MORE THAN A TOTAL OF 20 READS FOR ALL V–J COMBINATIONS | NO. OF SAMPLES WITH MORE THAN ONE V PER J |
|---|---|---|---|---|---|
| BLCA | |||||
| 12 | 8 | 16 | 5 | 1 | 0 |
| LUAD | |||||
| 10 | 3 | 7 | 2 | 1 | 0 |
| OVCA | |||||
| 9 | 6 | 12 | 4 | 0 | 0 |
Table 5.
Structures of example, productive TCGA–BLCA, –LUAD, –OVCA and Moffitt Cancer Center BLCA TcR-α V–J recombinations. For a summary of the Moffitt Cancer Center WXS analysis, see Supplementary File, “Gil SOM Moffitt Cancer Center bladder cancer WXS summary with read depth”.
| TCGA BARCODE | V–J REARRANGEMENT READ SEQUENCE | NUMBER OF READS | IMGT ASSESSMENT* |
|---|---|---|---|
| TCGA-BLCA | |||
| TCGA-2F-A9KO-01A-11D | GTGATACAGGCCTCTACCTCTGTGCA GGGGTTGATAACACCAATGCAGGCAA ATCAACCTTTGGGGATGGGACTAC |
4 | Productive, no stop codons, in-frame junction; TRAV27, J27 (low V-region score) |
| TCGA-4Z-AA7N-01A-11D | CCAGTGATTCAGCCACCTACCTCTGTG CCGTCCCCAATACTGGAGGCTTCAAAA CTATCTTTGGAGCAGGAACAAG |
4 | Productive, no stop codons, in-frame junction; TRAV12, J9 (Low V-region score). |
| TCGA-LUAD | |||
| TCGA-67-3771-01A-01D | GTCGTGGACTCAGCAGTATACTTC TGTGCTCTGAGTGGTAGGAACAGAGA TGACAAGATCATCTTTGGAAAAGGG ACACGACTTCATATTCTCCCCAGTAA |
6 | Productive, no stop codons, in-frame junction; TRAV19, J30 |
| TCGA-44-2659-01A-01D | GCCCATATGAGCGACGCGGCTGAGTAC TTCTGTGCTGTGAGTCTTTATGGGAACA ACAGACTCGCTTTTGNGAAGGGGAACC AAGTGGTGGTCATACCAAG |
14 | Productive, no stop codon, in-frame junction; TRAV8, J7 |
| TCGA-OVCA | |||
| TCGA-04-1542-01A-01D | AGATCTCAGACTCACAGCTGGGGG ACACTGCGATGTATTTCTGTGCTTTC ATGAAGGGTTATGGAGGAAGCCAAG GAAATCTCATCTTTGGAAAAGGCACT |
2 | Productive, no stop codon, in-frame junction; TRAV38, J42 |
| TCGA-09-1666-01A-01D | AGATCTCAGACTCACAGCTGGGGG ACACTGCGATGTATTTCTGTGCTTTC ATGAAGGGTTATGGAGGAAGCCAAG GAAATCTCATCTTTGGAAAAGGCACT |
2 | Productive, no stop codon and in-frame junction; TRAV38, J42 (overlaps with OVCA barcode 1542 above) |
| Moffitt cancer center BLCA | |||
| Patient-1 | ATCTCAGACTCACAGCTGGGGGA TGCCGCGATGTATTTCTGTGCTTATA GGAGCGCGTACTCTTATGGAGGAAGC CAAGGAAATCTCATCTTTGGAAAAGG |
2 | Productive, no stop codon and in-frame junction; TRAV24, J49 |
| Patient-2 | AGTGATTCAGCCACCTACCTCTGTGCAA TGAGGAACACAGGCTTTCAGAAACTTG TATTTGGAACTGGCACCCGAC |
3 | Productive, no stop codon and in-frame junction; TRAV12; J8 |
Thus, we processed ovarian cancer (OVCA) and lung adenocarcinoma (LUAD) WXS files for recombined TcR-α V–J regions, in an attempt to obtain a wider range of indications of available read numbers. However, the read numbers of the detected and recombined TcR-α V–J regions, in the OVCA and LUAD TCGA samples, were also lower than those detected in the BRCA samples. We obtained WXS of bladder cancer patients from the Moffitt Cancer Center, ie, independently of TCGA, where the average read depth was about 140, in comparison to approximately an average of 20 reads for the TCGA samples. However, even in the case of the Moffitt Cancer Center patients, the productive, TcR-α V–J recombinations were detected with read numbers lower than the highest numbers noted in the BRCA cases.
Finally, the above approaches have indicated that there are significantly more detections of recombined TcR-α V–J segments in the BRCA samples, per sample, than in any of the other TCGA cancer datasets (Fig. 2).
Figure 2.
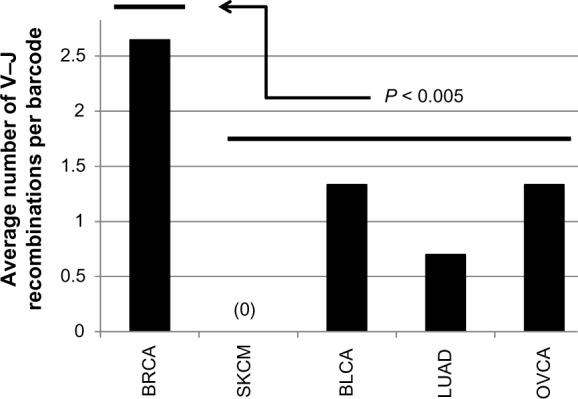
Summary bar graph of data from Tables 1 and 4, indicating the differences in detection of TcR-α V–J rearrangements in the indicated TCGA datasets, per sample.
Discussion
The above data indicate that it is possible to detect productively rearranged TcR-α V–J segments in primary and metastatic TCGA WXS and WGS files. Based on the comparison with cell line WXS files, and many other data in the scientific literature, it is likely that most of the recombined TcR-α V–J segments represent tumor infiltrating lymphocytes (TILs).6 However, a small number of recombined TcR-α V–J segments were detected in the BRCA and melanoma cell lines, consistent with proposals that some cancer cells may represent cancer cell-immune function cell fusions.3,4,11,12 Such fusions may explain some of the metastatic cancer migration patterns, for example, migration to the lymph nodes.
In the above analysis, the TCGA BRCA samples had the most robust collections of recombined TcR-α V–J segments. While many technical issues make it impossible to compare the TCGA samples in a way that leads to any conclusions regarding the immunological make-up or development of the different cancers, the above-described data provide an indication that, with a prospective study that can control for the many variables among the TCGA samples, there is an opportunity to take advantage of standard WXS approaches to evaluate recombined TcR-α V–J segments in cancer.
Given the convenience and “big-data” opportunities of using conventional WXS files for detection and evaluation of recombined TcR-α V–J segments, several goals would be important, eg, i) determination of any consistency, cancer specificity, or cancer subset specificity in segment selection for TILs, ii) a biochemical assay for TIL segment efficiencies for binding to MHC-bound epitopes, and iii) determination of whether identification of recombined TcR-α V–J segments provided for more sophisticated immunoscoring, either for prognoses or ex vivo expansion of TILs for administration to the patient.
Supplementary Material
Gill et al., 2015, supporting online material.
Item 1, p. 2: TcR-α V and J sequences.
Item 2, p. 17 Processing steps.
Item 3, attached Excel files.
Acknowledgments
Authors gratefully acknowledge the support of the taxpayers of the State of Florida, the Anna Valentine Grant Program, the USF research computing facility, especially Dr. Tony Green, the Moffitt Cancer Center Functional Genomics core facility, especially Mr. Sean Yoder, and Margaret Penichet, for extensive regulatory committee assistance.
Footnotes
ACADEMIC EDITOR: J. T. Efird, Editor in Chief
PEER REVIEW: Eight peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1367 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by the Anna Valentine Grant Program. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: GB, WS, JM, SB. Analyzed the data: TG, MS. Wrote the first draft of the manuscript: TG, GB. Contributed to the writing of the manuscript: MS, WS, JM, SB. Agree with manuscript results and conclusions: TG, MS, SB, JM, WS, GB. Jointly developed the structure and arguments for the paper: GB, SB, WS. Made critical revisions and approved final version: MS, GB. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Skornick Y, Topalian S, Rosenberg SA. Comparative studies of the long-term growth of lymphocytes from tumor infiltrates, tumor-draining lymph nodes, and peripheral blood by repeated in vitro stimulation with autologous tumor. J Biol Response Mod. 1990;9(4):431–8. [PubMed] [Google Scholar]
- 2.Coussens LM, Tinkle CL, Hanahan D, Werb Z. MMP-9 supplied by bone marrow-derived cells contributes to skin carcinogenesis. Cell. 2000;103(3):481–90. doi: 10.1016/s0092-8674(00)00139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawelek JM. Viewing malignant melanoma cells as macrophage-tumor hybrids. Cell Adh Migr. 2007;1(1):2–6. [PMC free article] [PubMed] [Google Scholar]
- 4.Lloyd MC, Szekeres K, Brown JS, Blanck G. Class II transactivator expression in melanoma cells facilitates T-cell engulfment. Anticancer Res. 2015;35(1):25–9. [PubMed] [Google Scholar]
- 5.Rosenberg SA. IL-2: the first effective immunotherapy for human cancer. J Immunol. 2014;192(12):5451–8. doi: 10.4049/jimmunol.1490019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandolfino MC, Labarriere N, Tessier MH, et al. High-scale expansion of melanoma-reactive TIL by a polyclonal stimulus: predictability and relation with disease advancement. Cancer Immunol Immunother. 2001;50(3):134–40. doi: 10.1007/PL00006683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chacon JA, Sarnaik AA, Pilon-Thomas S, Radvanyi L. Triggering co-stimulation directly in melanoma tumor fragments drives CD8 tumor-infiltrating lymphocyte expansion with improved effector-memory properties. Oncoimmunology. 2015;4(12):e1040219. doi: 10.1080/2162402X.2015.1040219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linnemann C, Heemskerk B, Kvistborg P, et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat Med. 2013;19(11):1534–41. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- 9.Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- 10.Butler SN, Blanck G. Immunoscoring by correlating MHC class II and TCR expression: high level immune functions represented by the KIRP dataset of TCGA. Cell and Tissue Research. 2016;363(2):491–6. doi: 10.1007/s00441-015-2261-1. [DOI] [PubMed] [Google Scholar]
- 11.Lazova R, Chakraborty A, Pawelek JM. Leukocyte-cancer cell fusion: initiator of the warburg effect in malignancy? Adv Exp Med Biol. 2011;714:151–72. doi: 10.1007/978-94-007-0782-5_8. [DOI] [PubMed] [Google Scholar]
- 12.De Baetselier P, Roos E, Brys L, et al. Nonmetastatic tumor cells acquire metastatic properties following somatic hybridization with normal cells. Cancer Metastasis Rev. 1984;3(1):5–24. doi: 10.1007/BF00047690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gill et al., 2015, supporting online material.
Item 1, p. 2: TcR-α V and J sequences.
Item 2, p. 17 Processing steps.
Item 3, attached Excel files.