Abstract
Previous studies have demonstrated that thioredoxin 1 (Trx1) exerts neuroprotective effects against cerebral ischemia/reperfusion injury caused by oxidative stress. While Trx1 is known to maintain the anti-oxidant activity of 2-Cys peroxiredoxins (Prdxs), the underlying mechanisms of its protective effects have remained to be elucidated, which was the aim of the present study. For this, an in vitro ischemic model of hypoxemia lasting for 4 h, followed by 24 h of reperfusion was used. Primary astrocytes from neonatal rats were pre-treated with small interfering RNA targeting Trx1 prior to oxygen glucose deprivation/reperfusion (OGD/R). MTS and lactate dehydrogenase assays were performed to evaluate cell viability. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis were employed to assess the mRNA and protein expression levels of Prdx1-4 and Prdx-SO3. Furthermore, a dual luciferase reporter assay was used to assess the interaction between activator protein-1 (AP-1) and Trx1. The present study demonstrated that OGD/R decreased the cell viability and increased cellular damage, which was more marked following Trx1 knockdown. The expression of Prdx1-4 and Prdx-SO3 protein was higher in the cells subjected to OGD/R. Knockdown of Trx1 markedly decreased the levels of Prdx1-4 but increased Prdx-SO3 mRNA and protein levels. The results of the present study also suggested that AP-1 directly activated the expression of Trx1. The present study demonstrated that Trx1 exerts its neuroprotective effects by preventing oxidative stress in astrocytes via maintaining Prdx expression.
Keywords: thioredoxin 1, peroxiredoxin, activator protein-1, oxidative stress, neuroprotection
Introduction
Ischemic cerebrovascular disease has high rates of morbidity, mortality and recurrence, and may result in disability (1). A previous study has demonstrated that oxidative stress is key in pathophysiologic processes (2). Oxidative stress occurs due to an imbalance between production of reactive oxygen species and the cell's capacity to neutralize them via its intrinsic anti-oxidant defense (3). The endogenous thioredoxin (Trx)-peroxiredoxin (Prdx) system is important in fighting oxidative stress damage due to its high anti-oxidant capacity within the body (4).
Thioredoxin reductase (TrxR) catalyzes the reduction of thioredoxin with simultaneous oxidation of NADPH. Trx1 is crucial for maintaining and increasing the anti-oxidant activity of the Prdxs. The 2-Cys Prdxs are the predominant Prdx sub-family and comprise Prdx1, -2, -3 and -4 (5). All 2-Cys Prdxs contain a cysteine residue that is oxidized to cysteine sulfenic acid (-SOH) by peroxides. Cys-SOH forms a disulfide bond with the remaining cysteine, which is, in turn, reduced by Trx1 (6). Previous studies have demonstrated that Trx1 returns Prdxs to their reduced forms to maintain their normal biological functions (7,8). Increasing the transcriptional activity of Trx1 may be an effective way to reduce cell damage following ischemia/reperfusion. In 2009, Soriano et al (6) reported that the transcription factor activator protein-1 (AP-1) may be associated with the expression of Trx1.
AP-1 is a redox-sensitive intracellular transcription factor composed of proteins belonging to the c-FBJ murine osteosarcoma viral oncogene homolog, c-Jun proto-oncogene and activating transcription factor families (9). AP-1, a cytosolic protein, responds to a variety of stimuli by regulating gene expression. Activated AP-1 translocates to the nucleus, recognizes 12-O-tetradecanoylphorbol-13-acetate response elements (TRE; 5′-TGAG/CTCA-3′) and induces the expression of a variety of genes (10). A previous study observed TRE sequences in the promoter areas of the Trx1 gene, while the mechanism of intracellular signaling remains to be elucidated (11).
The present study focused on the interaction between Trx1 and Prdxs to determine the underlying mechanisms of the neuroprotective effects of Trx1 following stroke. For this purpose, an in vitro cell model of oxygen glucose deprivation/reperfusion (OGD/R) was used. Furthermore, the association between AP-1 and Trx1 expression was investigated using a luciferase reporter assay to determine whether AP-1 directly upregulates Trx1 expression.
Materials and methods
Reagents and cell culture
A total of 72 neonatal Sprague-Dawley rats (weight, male, 300–600 g; female, 250–500 g) were obtained from the Animal Experiment Center of Chongqing Medical University (Chongqing, China) and sacrificed by decapitation 1 day after birth. Six rats were used per experimental group. They were maintained in an atmosphere of 40–70% humidity at 18–26°C. The animal protocol was approved by the Institutional Animal Care and Use Committee of Chongqing Medical University (Chongqing, China) and all experimental procedures were approved by Chongqing Medical University Biomedical Ethics Committee (Chongqing, China). The procedures complied with the current national and international laws and recommendations.
High-glucose Dulbecco's modified Eagle's Medium (DMEM) and glucose-free DMEM were purchased from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Fetal bovine serum (FBS), Dulbecco's phosphate-buffered saline (PBS), 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxyme thoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS), lactate dehydrogenase (LDH) as well as penicillin and streptomycin were purchased from Gibco (Thermo Fisher Scientific, Inc.). Dimethylsulfoxide was purchased from Merck & Co., Inc. (Whitehouse Station, NJ, USA).
Primary astrocytes were isolated from one-day-old Sprague-Dawley rats as described previously () and cultured in 10% FBS/DMEM medium. Sub-culturing was performed after 6–7 days, in a humidified atmosphere containing 5% CO2, the cells were then cultured until they reached 90% confluence.
Groups and cell treatment
Astrocytes were divided into three groups: i) Control group, untreated astrocytes; ii) negative control group, astrocytes transfected with negative lentivirus (LV3-NC); and iii) siTrx1 group, astrocytes transfected with the most effective of four lentiviruses designed to carry small interfering RNA (siRNA) targeting Trx1 LV3-54: 5′-GGGAGACAAGCTTGTGGTAGT-3′, LV3-135: 5′-CTGTGACAAGTATTCCAATGT-3′, LV3-222: 5′-GCCGACCTTCCAGTTCTATAA-3′, LV3-288:5′-GCTCGAAGCCACTATTACGGA-3′ and LV3-NC: 5′-TTCTCCGAACGTGTCACGT-3′, as indicated by Trx1 knockdown.
SiTrx1 lentivirus construction and cell transfection
The lentiviruses were constructed by Shanghai GenePharma Co., Ltd. (Shanghai, China). These Trx1 constructs were LV3-54, LV3-135, LV3-222 and LV3-288. LV3-NC served as the negative control.
The astrocytes were seeded into six-well plates at a concentration of 2×105 cells/well. When the astrocytes approached 80–90% confluency, they were transfected with 10 µl LV3-54, LV3-135, LV3-222, LV3-288 and LV3-NC (multiplicity of infection, 50). The media was changed on the second day. After 72 h, the efficiency of transfection was observed using a fluorescent microscope. Trx1 siRNA knockdown efficiency was confirmed by reverse-transcription quantitative polymerase chain reaction (RT-qPCR).
Establishment of oxygen glucose deprivation/reperfusion (OGD/R model
At three days following transfection, astrocytes were rinsed twice with PBS and cultured in glucose-free DMEM in a hypoxic atmosphere using a hypoxic chamber (8000DH, Thermo Fisher Scientific, Inc.) containing 5% O2, 94.5% N2 and 0.5% CO2 at 37°C for 4 h. Following OGD, the cells were placed in normal medium and immediately transferred to an incubator containing 5% CO2 in air for 24 h of recovery at 37°C. The cells in the control group were treated identically except for the OGD treatment.
Cell viability assay
An MTS assay was used to determine the number of surviving cells following OGD/R. Astrocytes were seeded in 96-well plates at 1×103 cells/well with glucose-free media. Following OGD/R, cells were incubated in normal fresh medium containing 1 mg/ml MTS at 37°C for 3 h in the dark. The absorption of the wells was then directly measured at a wavelength of 490 nm after 20 min using a microplate reader (Multiskan FC; Thermo Fisher Scientific, Inc.). A blank control was established for each treatment group by adding by adding 200 µl PBS.
LDH assay
LDH activity was quantitatively detected by measuring the LDH release from the damaged cells into the culture medium using a LDH assay kit (Nanjing Jancheng Bioengineering Institute, Nanjing, China). Astrocytes were seeded in six-well culture plates at 5×103/well prior to the experiment. The supernatant was collected and transferred to a 1.5-ml centrifuge tube following OGD/R. Cellular release of LDH was determined according to the manufacturer's instructions (Nanjing Jiancheng Bioengineering Institute). The absorbance (A) was measured using a Multiskan FC microplate reader at a wavelength of 450 nm and LDH release was calculated as follows: LDH release (%)=[(Aassay group)−(Acontrol group)]/[(Astandard group)−(Ablank group)]×200.
RNA isolation and RT-qPCR
The expression levels of Trx1 and Prdx genes were examined using RT-qPCR. RNA was isolated following cell lysis using an RNAiso Plus kit (Takara Biotechnology Co., Ltd., Dalian, China). Total RNA was reverse-transcribed into cDNA using a cDNA reverse transcription kit (Takara Biotechnology Co., Ltd.) at 37°C for 15 min, 85°C for 5 sec, and stored at −20°C. The products were used for two-step qPCR. The primer sequences are listed in Table I. PCR reactions were performed with iTaq™ Universal SYBR Green (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The thermocycling program was set as follows: Initial denaturation at 94°C for 10 min; 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 15 sec and extension at 70°C for 15 sec; and a final extension at 70°C for 5 min. The quantification cycle (Cq) data were collected using a CFX manager (Bio-Rad Laboratories, Inc.). Expression of Trx1 was normalized to β-actin. The relative quantification of gene expression was analyzed using the 2−ΔΔCq method (12,13). The fold change in target gene cDNA relative to the internal control β-actin was calculated as follows: Fold change = 2−ΔΔCq, ΔΔCq = (CqSample − Cqβ-actin) − (CqControl − Cqβ-actin).
Table I.
Primer sequences used for quantitative polymerase chain reaction.
| Gene | Gene ID | Primer sequence (5′-3′) | Product length (bp) |
|---|---|---|---|
| Trx1 | NM_053800.3 | F: CCTTCTTTCATTCCCTCTGTGA R: CCCAACCTTTTGACCCTTTTTA |
143 |
| Prdx1 | NM_057114.1 | F: CATTGCTCAGGATTATGGAGTC R: CATTTATTGTTATCTGGCGAAGG |
104 |
| Prdx2 | NM_017169.1 | F: CGTGGTCCTCTTTTTCTATCCA R: CTTTTAGTCACATCAGCAAGCA |
219 |
| Prdx3 | NM_022540.1 | F: TGCTTTTCTTCTACCCTTTGGA R: CATTCTTTCTTGGCGTGTTGAT |
165 |
| Prdx4 | NM_053512.2 | F: CCTCTGCTGCTGTTCCTGTTAC R: AAATCTTGGCTTTGCTTAGGTG |
175 |
| β-actin | NM_031144 | F: CACCCGCGAGTACAACCTTC R: CCCATACCCACCATCACACC |
207 |
Trx1, thioredoxin 1; Prdx, peroxiredoxin; F, forward; R, reverse.
Western blot analysis
Cultured cells were washed twice using PBS and the proteins were extracted and collected in lysis buffer containing a protease inhibitor cocktail (Beyotime Institute of Biotechnology, Shanghai, China). Equal quantities of total cellular protein extracts (40 µg) in each group were separated using 16% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Beyontime Institute of Biotechnology). The proteins were transferred to 0.22-µm polyvinylidene fluoride membranes (Beijing Dingguo Changsheng Biotechnology Co., Ltd., Beijing, China). These membranes were blocked with 5% non-fat milk in Tris-buffered saline with Tween 20 (TBST; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 2 h. Blots were incubated overnight at 4°C with the following primary antibodies obtained from Abcam (Cambridge, MA, USA): polyclonal rabbit anti-rat Prdx1 (1:2,000; cat. no. ab59539), polyclonal rabbit anti-rat Prdx2 (1:2,000; cat. no. ab59539), polyclonal rabbit anti-rat Prdx3 (1:2,000; cat. no. ab73349), monoclonal mouse anit-rat Prdx4 (1:2,000; cat. no. 16943), polyclonal rabbit anti-rat Prdx-SO3 (1:2,000; cat. no. ab16830) and β-actin (1:2,000; cat. no. A5441; Sigma-Aldrich). Following three washes in TBST, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (goat anti-rabbit IgG; 1:3,000; cat. no. BA1054; rabbit anti-rat IgG, 1:3,000; cat. no. BA1058), obtained from Wuhan Boster Biological Technology, Ltd. (Wuhan, China), at room temperature for 2 h, followed by another three washes. The protein concentration was determined using an Enhanced BCA Protein Assay kit (Beyotime Institute of Biotechnology). The bands were scanned using an imaging densitometer (ChemDoc XRS; Bio-Rad Laboratories, Inc.), and the results were quantified using Quantity One software (version 24.0; Bio-Rad Laboratories, Inc.).
Dual-luciferase reporter assays
The binding sites of AP-1 and Trx1 were analyzed by National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov/genome). A luciferase reporter vector driven by a fragment from the promoter of Trx1 (pGLO-Trx1-Luc) was subjected to site-directed mutagenesis of its TRE (Biomed Gene Technology Co., Ltd., Wuhan, China). Exponentially growing HEK293 cells (Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China) were seeded in 24-well plates at a density of 3×104/well, and the cells were divided into three groups: i) Control group; ii) Trx1 (WtTRE)-Luc group; and iii) Trx1(MtTRE)-Luc group. Each group was transfected with 0.8 µg pGLO-Trx1-Luc reporter and 0.8 µg AP-1 overexpression plasmid (Biomed Gene Technology Co., Ltd.) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The control group remained untreated. The medium was replaced after 4–6 h of transfection. Following treatment (24 h), the cells were lysed using 100 µl of Passive Lysis Buffer (Promega Corporation, Madison, WI, USA) and the lysates were analyzed using a Dual-Luciferase Reporter Assay kit according to the manufacturer's protocol (Promega Corporation). Firefly-based reporter gene activity was normalized to the Renilla control in all cases.
Statistical analysis
All experiments were performed at least three times and values are expressed as the mean ± standard error of the mean. Data were analyzed using one-way analysis of variance, followed by a post-hoc Tukey's test. For comparative analysis between two groups, a Student's t-test was used. Statistical analysis was performed using SPSS, version 17.0 (SPSS, Inc., Chicago, IL, USA) and P<0.05 was considered to indicate a statistically significant difference.
Results
Effects of Trx1 knockdown in astrocytes
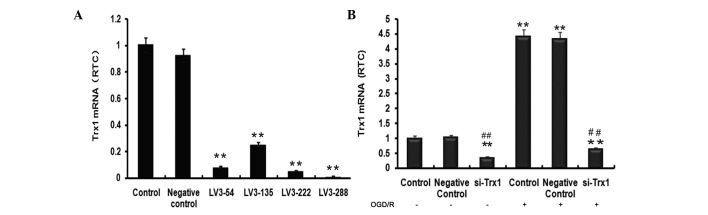
To select the most effective lentivirus fragment for knockdown of Trx1, the efficiency of lentiviral transfection was examined by fluorescent microscopy (results not shown). In order to detect the level of Trx1 knockdown, RT-qPCR was performed to evaluate Trx1 mRNA levels (Fig. 1A). Trx1 mRNA levels indicated a significant reduction of Trx1 following treatment with the four siRNAs compared with the control group (P<0.01). The largest decrease was observed in the LV3-288 group (P<0.01), approaching a knockdown efficiency of 90±2.21%. LV3-288 (target sequence, 5′-GCTCGAAGCCACTATTACGGA-3′) was therefore selected for use in all subsequent experiments.
Figure 1.
Trx1 mRNA expression levels following knockdown or OGD/R. (A) The expression of Trx1 was significantly knocked down by LV3-288. (B) Trx1 mRNA expression demonstrated a significant decrease following Trx1 knockdown but a significant increase following OGD/R. Values are expressed as the mean ± standard error of the mean. **P<0.01 vs. the controls, ##P<0.01 vs. si-Trx1 group. RTC, relative to control; Trx1, thioredoxin 1; LV, lentivirus; si, small interfering; OGD/R, oxygen glucose deprivation/reperfusion.
OGD-induced oxidative stress damage increases Trx1 mRNA expression levels
To investigate the impact of OGD/R on Trx1 mRNA expression levels and the efficiency of the siRNA used in inhibiting the upregulation of Trx1 under OGD/R conditions in astrocytes, RT-qPCR analysis was performed. As presented in Fig. 1B, Trx1 mRNA expression in the si-Trx1 group was reduced by 66±3.61% and 86±2.93% compared with the controls, respectively (P<0.01). Of note, Trx1 mRNA levels in astrocytes subjected to OGD/R were significantly elevated compared with those in the control groups (P<0.01). However, treatment with LV3-288 completely abrogated these increases (P<0.01). These results indicated that OGD/R markedly increased Trx1 mRNA expression levels, which was efficiently blocked by LV3-288.
Trx1 knockdown decreases astrocyte viability following OGD/R
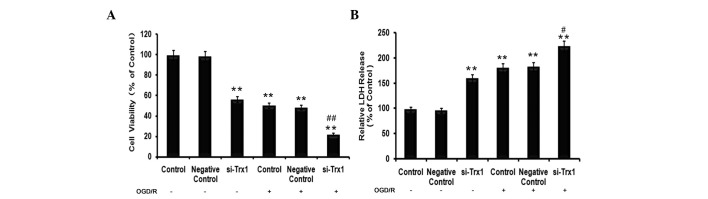
To determine the effects of Trx1 on cell viability and cell damage in astrocytes following OGD/R, MTS and LDH assays were performed. The results of the MTS assay demonstrated that, compared with the control group, the cell viability of astrocytes decreased by 49±1.64% (P<0.05) in the OGD/R group (Fig. 2A). However, the viability of cells pre-treated with siRNA against Trx1 in the OGD/R group decreased to 77±1.83% of that of the control cells (P<0.01). LDH is a stable cytoplasmic enzyme present in the majority of cells. It is rapidly released into the cell culture supernatant upon damage to the plasma membrane (14). As presented in Fig. 2B, treatment of astrocytes with siRNA against Trx1 significantly increased their LDH release (P<0.01). Furthermore, cells in the OGD/R groups released significantly more LDH than the control cells (P<0.01), which was further enhanced by knockdown of Trx1 (P<0.05 vs. OGD/R group).
Figure 2.
Effect of Trx1 on cell viability and cell damage following exposure to OGD/R. (A) Cell viability was measured using the MTS assay. (B) Cell damage was evaluated with LDH levels. Values are expressed as the mean ± standard error of the mean (n=6 from 3 independent experiments). *P<0.05, **P<0.01 vs. the control; #P<0.05, ##P<0.01 vs. the si-Trx1 group. Trx1, thioredoxin 1; LDH, lactate dehydrogenase; si, small interfering; OGD/R, oxygen glucose deprivation/reperfusion.
Trx1 knockdown inhibits increases in Prdx expression following OGD/R
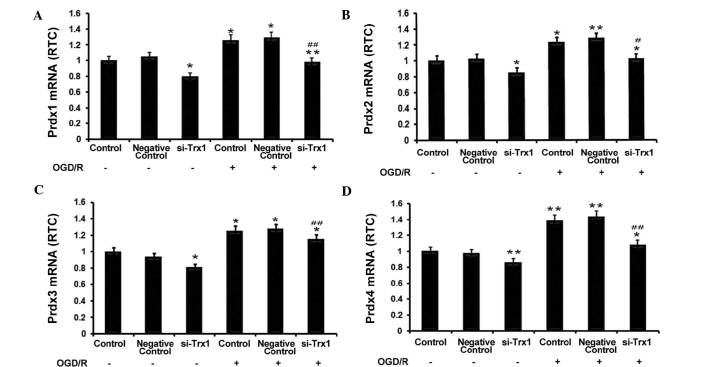
RT-qPCR was performed to investigate the association between Trx1 and Prdxs. The results showed that Trx-1 knockdown significantly reduced Prdx1-4 expression in astrocytes (P<0.05 or P<0.01) (Fig. 3). Furthermore, OGD/R significantly enhanced the expression of Prdx1-4 (P<0.05 or P<0.01 vs. no OGD/R), which was partly abrogated by knockdown of Trx1 (P<0.05 or P<0.01 vs. OGD/R control groups). These observations indicated that enhanced expression of Trx1 during OGD/R increases the expression of the anti-oxidant Prdxs.
Figure 3.
Trx1 knockdown suppresses 2-Cys Prdxs expression. (A) Prdx1, (B) Prdx2, (C) Prdx3 and (D) Prdx4 were determined by reverse-transcription quantitative polymerase chain reaction. Prdx1-4 mRNA expression increased following OGD/R, which was inhibited by Trx1 knockdown. Values are expressed as the mean ± standard error of the mean (n=4). *P<0.05, **P<0.01 vs. the controls; #P<0.05, ##P<0.01 vs. the si-Trx1 group. Trx1, thioredoxin 1; si, small interfering; OGD/R, oxygen glucose deprivation/reperfusion; Prdx, peroxiredoxin.
Trx1 knockdown decreases Prdx protein expression but increases Prdx-SO3 protein expression
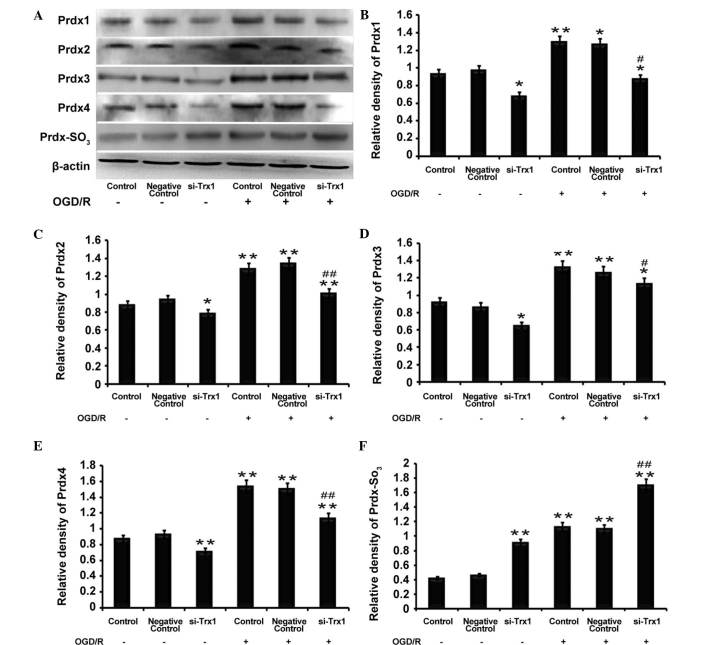
Western blot analysis demonstrated that the protein expression of Prdx1-4 and Prdx-SO3 was significantly elevated following OGD/R (P<0.05 or P<0.01) (Fig. 4), which was consistent with the RT-qPCR results. However, the levels of Prdx1-4 protein were reduced following Trx1 knockdown regardless of OGD/R. Furthermore, Prdx-SO3 protein expression was increased following Trx1 knockdown (P<0.01) (Fig. 4). The results demonstrated that, under oxidative stress, the decrease in Prdx expression levels was accompanied by an increase in Prdx-SO3. This results in an imbalance of the redox equilibrium and leads to damaged cells and tissue. These results indicated that Trx1 enhances Prdx activity and drives the response to oxidative stress via increasing the expression of Prdxs and inhibiting Prdx-SO3.
Figure 4.
Trx1 knockdown attenuates 2-Cys Prdxs protein expression and promotes Prdx-SO3 formation. (A) Representative western blot of Prdx1-4 and Prdx-SO3 in astrocytes following Trx1 knockdown. Quantification of (B) Prdx1, (C) Prdx2, (D) Prdx3, (E) Prdx4 and (F) Prdx-SO3 was performed by densitometric analysis. Following Trx1 knockdown, expression levels of Prdx1-4 were decreased, while PRDX-SO3 was elevated. In addition, PRDXs1-4 and PRDX-SO3 protein expression was increased following OGD/R treatment. Values are expressed as the mean ± standard error of the mean (n=4). *P<0.05, **P<0.01 vs. the control; #P<0.05, ##P<0.01 vs. the si-Trx1 group. Trx1, thioredoxin 1; si, small interfering; OGD/R, oxygen glucose deprivation/reperfusion; Prdx, peroxiredoxin.
AP-1 mediates Trx1 expression
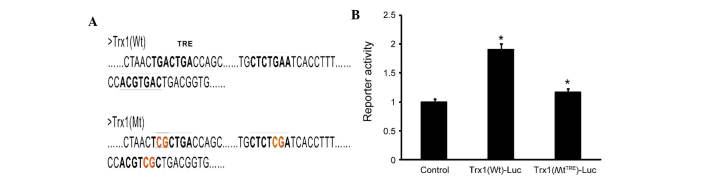
To determine whether AP-1 directly regulates the expression of Trx1, three tandem AP-1 binding sites were predicted in the Trx1 promoter using bioinformatics (Fig. 5A). The sequences were verified using the NCBI database. The AP-1 plasmid as well as the luciferase reporter plasmids PGLO-Trx1(Wt) and PGLO-Trx1(MtTRE) were constructed. The luciferase activity in cells co-transfected with PGLO-Trx1(Wt)-Luc and AP-1-transfected was significantly increased by 60±4.35% compared with the control group and the PGLO-Trx1(MtTRE)-Luc group(Fig. 5B). These results demonstrated that site-directed mutagenesis of the TRE sequences inhibited the specific binding between AP-1 and TRE (P<0.05), leading to a marked reduction of luciferase activity. These results demonstrated that the expression of Trx1 is directly and positively regulated by AP-1.
Figure 5.
Expression of Trx1 is partly dependent on AP-1. (A) Three tandem AP-1 binding sites are present in the Trx1 promoter. (B) The combining capacity of AP-1 and Trx1 was detected by dual luciferase activity assay. Astrocytes were transfected with luciferase reporter constructs driven by a Wt or TRE-mutated fragment from the Trx1 promoter as well as an AP-1 plasmid. At 24 h post-transfection, luciferase activity of the Wt plasmid was shown to be increased by AP-1, while the mutant vector was not affected by Ap-1. Values are expressed as the mean ± standard error of the mean (n=6). *P<0.05 vs. the control. Trx1, thioredoxin 1; si, small interfering; AP-1, activator protein-1; Luc, luciferase; Wt, wild-type; MtTRE, TRE mutant; TRE, 12-O-tetradecanoylphorbol-13-acetate response element.
Discussion
The Trx system is composed of Trx, Trx receptor (TrxR) and NADPH, and exerts vital anti-oxidant effects depending on the activity of TrxR (15,16). A previous study has demonstrated that in thioredoxin transgenic mice, cell damage and the outbreak index of cerebral ischemia/reperfusion injury were markedly attenuated in the presence of high expression levels of Trx1 (17). Other previous studies further indicated that Trx1 was induced in response to oxidative stress resulting from cerebral ischemia in anaerobic environments (18–20). The present study also suggested that the Trx1 expression was significantly increased following OGD/R, and the MTS and LDH results demonstrated that OGD/R decreased the viability of astrocytes, which was markedly aggravated by Trx1 knockdown. These findings suggested that Trx1 may protect astrocytes from damage following OGD/R and that it protects from oxidative stress damage by exerting an anti-oxidant effects.
In addition to its contribution to redox processes by reducing inter- and intra-chain protein disulfide bonds, the Trx1 system also maintains the activity of important anti-oxidant enzymes, such as peroxiredoxins (Prdxs) (20–23). In 2010, Hwang et al (24) demonstrated that Trx1 markedly improves the neuroprotection of Prdxs in Mongolian gerbils. In the present study, the expression of Prdx1-4 was observed to decrease following Trx1 knockdown, while Prdx-SO3 expression increased. The expression of Prdxs was demonstrated to be closely associated with Trx1. These results suggested that Trx1 is the cofactor of Prdxs and that Trx1 maintains Prdx activity in response to oxidative stress damage.
According to a previous study, the upregulation of Trx1 expression is an important step of neuronal responses to oxidative stress (25). A series of transcription factor binding sites have been identified in the promoter region of Trx1, including SP-1 transcription factor, AP-1, nuclear factor-κB and nuclear factor erythroid 2-like 2 (26–28). Among these, AP-1 has received increasing attention in recent years. Previous studies have demonstrated that sulfiredoxin expression is regulated by the transcription factor AP-1, which mediates its upregulation by synaptic activity in neurons (29–31). In the present study, AP-1 was observed to be directly activate the expression of Trx1 by specific binding to the TRE. Thus, increasing AP-1 levels may be an effective strategy for enhancing Trx-1 expression and achieving anti-oxidant effects for the treatment of stroke.
In conclusion, the results of the present study indicated that Trx1 may positively regulate the expression of anti-oxidant Prdxs following OGD/R and that its upregulation may be a suitable method for the prevention and treatment of cerebral disease. Further exploration using in vitro and in vivo studies with siRNA and overexpression technology are required to verify the underlying mechanisms of action of Trx1 and the interaction between Trx1 and Prdxs.
Acknowledgments
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81171090 and 81271460), the Natural Science Youth Foundation of China (grant no. 81301125) and the Natural Science Foundation of Chongqing Education Committee, China (grant no. KJ110313).
References
- 1.Liu R, Liu H, Ha Y, Tilton RG, Zhang W. Oxidative stress induces endothelial cell senescence via downregulation of Sirt6. Biomed Res Int. 2014:902842. doi: 10.1155/2014/902842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva DG, Belini Junior E, de Almeida EA, Bonini-Domingos CR. Oxidative stress in sickle cell disease: An overview of erythrocyte redox metabolism and current antioxidant therapeutic strategies. Free Radic Biol Med. 2013;65:1101–1109. doi: 10.1016/j.freeradbiomed.2013.08.181. [DOI] [PubMed] [Google Scholar]
- 3.Calabrese V, Cornelius C, Mancuso C, Pennisi G, Calafato S, Bellia F, Bates TE, Giuffrida Stella AM, Schapira T, Dinkova Kostova AT, Rizzarelli E. Cellular stress response: A novel target for chemoprevention and nutritional neuroprotection in aging, neurodegenerative disorders and longevity. Neurochem Res. 2008;33:2444–2471. doi: 10.1007/s11064-008-9775-9. [DOI] [PubMed] [Google Scholar]
- 4.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 5.Wood ZA, Schröder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/S0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 6.Soriano FX, Léveillé F, Papadia S, Higgins LG, Varley J, Baxter P, Hayes JD, Hardingham GE. Induction of sulfiredoxin expression and reduction of peroxiredoxin hyperoxidation by the neuroprotective Nrf2 activator 3H-1,2-dithiole-3-thione. J Neurochem. 2008;107:533–543. doi: 10.1111/j.1471-4159.2008.05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das KC, Das CK. Thioredoxin, a singlet oxygen quencher and hydroxyl radical scavenger: Redox independent functions. Biochem Biophys Res Commun. 2000;277:443–447. doi: 10.1006/bbrc.2000.3689. [DOI] [PubMed] [Google Scholar]
- 8.Lu J, Holmgren A. Thioredoxin system in cell death progression. Antioxid Redox Signal. 2012;17:1738–1747. doi: 10.1089/ars.2012.4650. [DOI] [PubMed] [Google Scholar]
- 9.Pronk TE, van der Veen JW, Vandebriel RJ, van Loveren H, de Vink EP, Pennings JL. Comparison of the molecular topologies of stress-activated transcription factors HSF1, AP-1, NRF2 and NF-κB in their induction kinetics of HMOX1. Biosystems. 2014;124:75–85. doi: 10.1016/j.biosystems.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Choi WJ. The Heterochromatin-1 phosphorylation contributes to TPA-Induced AP-1 expression. Biomol Ther (Seoul) 2014;22:308–313. doi: 10.4062/biomolther.2014.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen B, Guan D, Cui ZJ, Wang X, Shen X. Thioredoxin 1 downregulates MCP-1 secretion and expression in human endothelial cells by suppressing nucleartranslocation of activator protein 1 and redox factor-1. Am J Physiol Cell Physiol. 2010;298:C1170–1179. doi: 10.1152/ajpcell.00223.2009. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Xing G, Dong M, Li X, Zou Y, Fan L, Wang X, Cai D, Li C, Zhou L, Liu J, Niu Y. Neuroprotective effects of puerarin against beta-amyloid-induced neurotoxicity in PC12 cells via a PI3K-dependent signaling pathway. Brain Res Bull. 2011;85:212–218. doi: 10.1016/j.brainresbull.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 14.Guo H, Kong S, Chen W, Dai Z, Lin T, Su J, Li S, Xie Q, Su Z, Xu Y, Lai X. Apigenin mediated protection of OGD-evoked neuron-like injury in differentiated PC12 cells. Neurochem Res. 2014;39:2197–2210. doi: 10.1007/s11064-014-1421-0. [DOI] [PubMed] [Google Scholar]
- 15.Sengupta R, Holmgren A. Thioredoxin and thioredoxin reductase in relation to reversible S-nitrosylation. Antioxid Redox Signal. 2013;18:259–269. doi: 10.1089/ars.2012.4716. [DOI] [PubMed] [Google Scholar]
- 16.Nickel C, Rahlfs S, Deponte M, Koncarevic S, Becker K. Thioredoxin networks in the malarial parasite Plasmodium falciparum. Antioxid Redox Signal. 2006;8:1227–1239. doi: 10.1089/ars.2006.8.1227. [DOI] [PubMed] [Google Scholar]
- 17.Das DK. Thioredoxin regulation of ischemic preconditioning. Antioxid Redox Signal. 2004;26:405–412. doi: 10.1089/152308604322899477. [DOI] [PubMed] [Google Scholar]
- 18.Taksgi Y, Hatori I, Nozaki K, Mitsui A, Ishikawa M, Hashimoto N, Yodoi J. Excitotoxic hippocampal injury is atenuated in thioredoxin transgenic mice. J Cereb Blood Flow Metab. 2000;20:829–833. doi: 10.1097/00004647-200005000-00009. [DOI] [PubMed] [Google Scholar]
- 19.Takagi Y, Horikawa F, Nozaki K, Sugino T, Hashimoto N, Yodoi J. Expression and distribution of redox regulatory protein, thioredoxin during transient focal brain ischemia in the rat. Neurosci Lett. 1998;251:25–28. doi: 10.1016/S0304-3940(98)00492-3. [DOI] [PubMed] [Google Scholar]
- 20.Mansur K, Iwashashi Y, Kiryu-Seo S, Su Q, Namikawa K, Yodoi J, Kiyama H. Up-regulation of thioredoxin expression in motor neurons after nerve injury. Brain Res Mol Brain Res. 1998;62:86–91. doi: 10.1016/S0169-328X(98)00244-7. [DOI] [PubMed] [Google Scholar]
- 21.Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Masutani H, Ueda S, Yodoi J. The thioredoxin system in retroviral infection and apoptosis. Cell Death Differ. 2005;12(Suppl 1):991–998. doi: 10.1038/sj.cdd.4401625. [DOI] [PubMed] [Google Scholar]
- 23.Kondo N, Nakamura H, Masutani H, Yodoi J. Redox regulation of human thioredoxin network. Antioxid Redox Signal. 2006;8:1881–1890. doi: 10.1089/ars.2006.8.1881. [DOI] [PubMed] [Google Scholar]
- 24.Hwang IK, Yoo KY, Lee CH, Kim DW, Choi JH, Kwon YG, Kim YM, Choi SY, Won MH. Changes in the expression of mitochondrial peroxiredoxin and thioredoxin in neurons and glia and their protective effects in experimental cerebral ischemic damage. Free Radic Biol Med. 2010;48:1242–1251. doi: 10.1016/j.freeradbiomed.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Liu M, Wang Y, Zheng L, Zheng W, Dong K, Chen S, Zhang B, Li Z. Fasudil reversed MCT-induced and chronic hypoxia-induced pulmonary hypertension by attenuating oxidative stress and inhibiting the expression of Trx1 and HIF-1α. Respir Physiol Neurobiol. 2014;201:38–46. doi: 10.1016/j.resp.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Bloomfield KL, Osborne SA, Kennedy DD, Clarke FM, Tonissen KF. Thioredoxin-mediated redox control of the transcription factor Sp1 and regulation of the thioredoxin gene promoter. Gene. 2003;319:107–116. doi: 10.1016/S0378-1119(03)00799-6. [DOI] [PubMed] [Google Scholar]
- 27.Kim YC, Yamaguchi Y, Kondo N, Masutani H, Yodoi J. Thioredoxin-dependent redox regulation of the antioxidant responsive element (ARE) in electrophile response. Oncogene. 2003;22:1860–1865. doi: 10.1038/sj.onc.1206369. [DOI] [PubMed] [Google Scholar]
- 28.Tanito M, Masutani H, Kim YC, Nishikawa M, Ohira A, Yodoi J. Sulforaphane induces thioredoxin through the antioxidant-responsive element and attenuates retinal light damage in mice. Invest Ophthalmol Vis Sci. 2005;46:979–987. doi: 10.1167/iovs.04-1120. [DOI] [PubMed] [Google Scholar]
- 29.Papadia S, Soriano FX, Léveillé F, Martel MA, Dakin KA, Hansen HH, Kaindl A, Sifringer M, Fowler J, Stefovska V, et al. Synaptic NMDA receptor activity boosts intrinsic antioxidant defenses. Nat Neurosci. 2008;11:476–487. doi: 10.1038/nn2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soriano FX, Baxter P, Murray LM, Sporn MB, Gillingwater TH, Hardingham GE. Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol Cells. 2009;27:279–282. doi: 10.1007/s10059-009-0050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei Q, Jiang H, Matthews CP, Colburn NH. Sulfiredoxin is an AP-1 target gene that is required for transformation and shows elevated expression in human skin malignancies. Proc Natl Acad Sci USA. 2008;105:19738–19743. doi: 10.1073/pnas.0810676105. [DOI] [PMC free article] [PubMed] [Google Scholar]