Abstract
Cytokines and their intercellular signals regulate the multipotency of mesenchymal stem cells (MSCs). The present study established the MSC lines SG-2, -3, and -5 from the bone marrow of green fluorescent protein (GFP)-transgenic mice. These cell lines clearly expressed mouse MSC markers Sca-1 and CD44, and SG-2 and -5 cells retained the potential for osteogenic and adipogenic differentiation in the absence of members of the transforming growth factor (TGF)-β superfamily. By contrast, SG-3 cells only retained adipogenic differentiation potential. Analysis of cytokine and cytokine receptor expression in these SG cell lines showed that bone morphogenetic protein (BMP) receptor 1B was most highly expressed in the SG-3 cells, which underwent osteogenesis in response to BMP, while TGF-β receptor II was most highly expressed in SG-3 and -5 cells. However, it was unexpectedly noted that phosphorylation of Smad 2, a major transcription factor, was induced by TGF-β1 in SG-2 cells but not in SG-3 or -5 cells. Furthermore, TGF-β1 clearly induced the expression of Smad-interacting transcription factor CCAAT/enhancer binding protein-β in SG-2 but not in SG-3 or -5 cells. These results demonstrated the establishment of TGF-β-responsive SG-2 MSCs, BMP-responsive SG-3 MSCs and TGF-β/BMP-unresponsive SG-5 MSCs, each of which was able to be traced by GFP fluorescence after transplantation into in vivo experimental models. In conclusion, the present study suggested that these cell lines may be used to explore how the TGF-β superfamily affects the proliferation and differentiation status of MSCs in vivo.
Keywords: green fluorescent protein-transgenic mouse, mesenchymal stem cells, bone morphogenetic protein-2, transforming growth factor-β, CCAAT/enhancer binding protein-β
Introduction
Mesenchymal stem cells (MSCs), which were first derived from bone marrow, have self-renewal properties and are able to differentiate into a variety of mesenchymal tissue types (1–3). In stem cell therapy, human bone marrow-derived MSCs (BM-MSCs) are expanded in vitro and subsequently autoimplanted, which eliminates the risk of immune rejection. BM-MSCs are able to differentiate into osteoblasts, chondrocytes and adipocytes (4), and are a major source of bone regeneration and remodeling during homeostasis (5–8). In addition, immunophenotype evaluation demonstrated that mouse BM-MSCs express Sca-1 and CD44, but not CD11b or CD45 (9).
The transforming growth factor (TGF)-β superfamily includes the TGF-β/activin/Nodal family and the bone morphogenetic protein (BMP)/growth and differentiation factor (GDF)/Mullerian inhibiting substance (MIS) family (10). On the cell surface, binding of ligands to receptors triggers the formation of a tetrameric complex of type I and II receptors. Type II receptor kinase activates type I receptor kinase, which transduces the signal through phosphorylation of receptor-activated Smads (R-Smads) (11–14). Smad proteins are the central mediators of TGF-β superfamily signaling. R-Smads, including Smad 1, Smad 5 and Smad 8, are primarily activated by BMP-specific type I receptors, whereas Smad 2 and Smad 3 are activated by the TGF-β-specific type I receptors. Activated R-Smads form complexes with the common mediator Smads (Co-Smads; e.g., Smad 4), which translocate into the nucleus, where they and their partner proteins regulate the transcription of specific target genes. Abnormal intensity of Smad-mediated TGF-β/BMP signals is associated with various human diseases, including bone and immune disorders, fibrosis, and cancer progression or metastasis (15). Of note, TGF-β superfamily-induced intracellular signals affect osteogenesis and adipogenesis of MSCs; for instance, BMP has been observed to potentiate osteogenic and adipogenic differentiation of undifferentiated mesenchymal cells (16). By contrast, TGF-β potentiates osteogenic differentiation of BM-MSCs (17,18), although none of these results have been confirmed in vivo.
Recent studies have focused on controlling TGF-β/BMP signals for the discovery of pharmacotherapeutics; however, the detection of therapeutic molecular targets in these pathways has not been successful, probably because most trials are performed in vitro, not in vivo (19,20). Therefore, it is important to establish appropriate in vivo experimental models to evaluate the role of TGF-β/BMP signaling in disease development or healing. The present study aimed to establish MSC cell lines derived from bone marrow of green fluorescent protein (GFP)-transgenic mice; the cells and their diverse, intracellular BMP and TGF-β signals can be tracked after transplantation into in vivo experimental models. These cell lines are available for in vivo molecular studies that aim to determine how the TGF-β superfamily affects MSC proliferation and differentiation in diseases including fibrosis and cancer progression or metastasis (21,22), and in tissue repair processes, including tissue reconstruction and anti-inflammatory responses (23).
Materials and methods
Bone marrow-derived cells from GFP-transgenic mice
All experimental procedures were performed in accordance with the guidelines established by the Animal Studies Committee at Iwate Medical University (Iwate, Japan). A total of four GFP-transgenic mice (24) were obtained from the Center for in vivo Science, Iwate Medical University (Iwate, Japan). The mice were sacrificed by excessive inhalation of CO2. Cells were flushed from the tibia of three-week-old GFP-transgenic mice with phosphate-buffered saline (PBS) containing 0.5% fetal bovine serum (FBS; PAA Laboratories, GE Healthcare, Piscataway, NJ, USA) and 2 mM EDTA, and then seeded into plastic cell culture dishes (Nunc; Thermo Fisher Scientific, Waltham, MA, USA) with Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA) containing 10% FBS. The cells were cultured for 1 week under hypoxic conditions (5% O2, 5% CO2 and 90% N2). Cells were re-plated upon reaching 80% confluence.
Co-transfection of hTERT and SV40 large T antigen (SV40LT) genes
The expanded cells were transfected with pBABE-neo-hTERT and pBABE-pur-SV40LT plasmids encoding neomycin and puromycin resistance (provided by Addgene, Cambridge, MA, USA) with Lipofectamine LTX (Invitrogen Life Technologies, Carlsbad, CA, USA) according to manufacturer instructions. Cells were then incubated in DMEM containing 10% FBS, 150 µg/ml G418 (Gibco-BRL, Thermo Fisher Scientific) or 1 µg/ml puromycin (Gibco-BRL) under hypoxic conditions for 12–15 days. The surviving cells were trypsinized and allowed to grow in 90-mm culture dishes.
Single-cell cloning
Single-cell clones were obtained using the limited dilution method. After hTERT and SV40LT transfection and selection with G418 and puromycin, the surviving cells were seeded on a 96-well plate (Nunc) at 0.5% cells per well, and then cultured under hypoxic conditions. After 10 days, the cells were sub-cultured in 24-well plates (Nunc). This was repeated until confluence was reached at 20 days after single cell cloning. Population doubling (PD) was defined as the number of doublings required for a single cell to reach confluence in a 60-mm culture dish (Nunc) under hypoxic conditions. PD was estimated for clones SG-2, -3, -5 and -6.
Telomeric repeat amplification protocol
Telomerase activity in bone marrow-derived cell lines was assayed by the stretch PCR method using the Quantitative Telomerase Detection kit (Allied Biotech, Vallejo, CA, USA) according to manufacturer's instructions. The PCR mixture contained QTD premixed buffer and SYBR green one dye (cat. no. MT3010; Allied Biotech, Inc., St. Benicia, CA, USA). Quantification of telomerase activity was performed under the following amplification conditions using a Thermal Cycler Dice Real Time system (Takara Bio, Otsu, Japan) according to manufacturer's instructions: 25°C for 20 min, initial activation at 95°C for 10 min, denaturation at 90°C for 30 s, annealing at 60°C for 30 s, and a final extension of 40 cycles at 72°C for 30 s. The PCR products were separated by 10–20% polyacrylamide gel electrophoresis and stained with ethidium bromide.
Detection of SV40LT by immunocytochemistry
Bone marrow-derived cell lines were seeded onto eight-well culture slides (BD Biosciences, Franklin Lakes, NJ, USA). After 24 h, the cells were fixed with 4% paraformaldehyde and washed five times with PBS. For detection of SV40LT, cells were incubated with mouse monoclonal anti-SV40LT (1:100; cat. no. ab16879; Abcam, Cambridge, UK) antibody for 1 h at room temperature. The cells were then incubated with Alexa Fluor 594 goat polyclonal anti-mouse secondary antibodies (1:500; cat. no. A11005; Thermo Fisher Scientific) and DAPI (1:500; cat. no. D9542; Sigma-Aldrich) for 30 min at room temperature. Fluorescence was examined by using a fluorescence microscope (Olympus ix70; Olympus Corporation, Tokyo, Japan).
Detection of MSC markers by flow cytometry
A total of 1.0×105 bone marrow-derived cell lines (SG-2, -3, -5 and -6) were suspended in PBS containing 0.5 FBS and 2 mM EDTA and incubated with phycoerythrin-conjugated monoclonal anti-mouse Sca-1 (1:10; cat. no. 130-093-224), monoclonal anti-mouse CD44 (1:10; cat. no. 130-096-838), monoclonal anti-mouse CD11b (1:10; cat. no. 130-091-240) or monoclonal anti-mouse CD45 (1:10; cat. no. 130-091-610) antibody for 1 h at 4°C in the dark. All antibodies were purchased from Miltenyi Biotec (Bergisch Gladbach, Germany). Acquisition was performed using an EPICS XL ADC system (Beckman Coulter, Brea, CA, USA).
Adipogenic and osteogenic differentiation
In vitro differentiation was performed according to a previous study by our group (25). To induce osteogenic differentiation, confluent cells were incubated in osteogenic differentiation medium (ODM) under hypoxic conditions for two weeks. Bone matrix mineralization was evaluated by Alizarin red S (Sigma-Aldrich) staining. To induce adipogenic differentiation, cells were cultured to near confluence and cultured in adipogenic differentiation medium (ADM) under hypoxic conditions for two weeks. At the end of the differentiation period, lipid droplets were stained with Oil Red O (Sigma-Aldrich).
Expression profiling of cytokines and cytokine receptors
Gene expression profiling was performed using a PrimerArray of mouse cytokine-cytokine receptor interaction (Takara Bio) in combination with a Thermal Cycler Dice Real Time System (Takara Bio) according to manufacturer's instructions. This PrimerArray is a set of real-time reverse transcription-polymerase chain reaction (RT-PCR) primers used for the analysis of RNA expression. The array contains a mixture of 96 primer pairs for 88 target genes and eight housekeeping genes. Quantification of gene expression was performed using a PrimerArray Analysis Tool version 2.0 (Takara Bio).
Western blot analysis
SG-2, -3 and -5 cells were serum-starved overnight and stimulated with 50 ng/ml BMP-2 (R&D Systems, Minneapolis, MN, USA) or 50 ng/ml TGF-β1 (R&D Systems). The cells were washed twice with ice-cold PBS and then lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.2, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing protease and phosphatase inhibitor cocktails (Sigma-Aldrich). The protein content was quantified using the bicinchoninic acid method (Pierce; Thermo Fisher Scientific). Samples containing equal amounts of protein were separated by 12.5% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Merck Millipore, Darmstadt, Germany). After blocking with 5% nonfat dry milk in 50 mM Tris-HCl, pH 7.2, containing 150 mM NaCl and 0.1% Tween-20 for 2 h at room temperature, the membrane was incubated with primary rabbit monoclonal anti-phospho-Smad 5 (1:1,000; cat. no. ab92698; Abcam), anti-Smad 1/5/8 (1:1,000, cat. no. 12656; Cell Signaling Technology, Danvers, MA, USA), rabbit monoclonal anti-phospho-Smad 2 (1:1,000; cat. no. 04-953; Merck Millipore), or anti-Smad 2/3 (1:1,000; cat. no. 610843; BD Biosciences) antibody overnight at 4°C, with mouse monoclonal anti-β-actin (1:1,000; cat. no. sc-47778; Santa Cruz Biotechnology, Dallas, TX, USA) antibody as a loading control. The blots were incubated with alkaline phosphatase-conjugated secondary antibody and developed using the 5-bromo-4-chloro-3′-indolyphosphate/nitro-blue tetrazolium membrane phosphatase substrate system (cat. no. 50-81-00; Kirkegaard & Perry Laboratories, Gaithersburg, MD, USA).
RNA isolation and RT-quantitative (q)PCR
SG-2, -3 and -5 cells were stimulated with TGF-β1 (1–50 ng/ml; R&D Systems) for 24 h. Total RNA was isolated using ISOGEN reagent (Nippongene, Tokyo, Japan) according to the manufacturer's instructions. First-strand cDNA was synthesized from total RNA with the PrimeScript RT Reagent kit (Takara Bio). RT-qPCR was performed on a Thermal Cycler Dice Real Time System (Takara Bio) with SYBR Premix Ex Taq II (Takara Bio). Expression of CCAAT/enhancer binding protein-β (C/EBPβ) was normalized to β-actin and relative expression levels were calculated as a fold-increase or -decrease relative to the control. Transcripts were detected with primers (designed using a Perfect Real Time Support system; Takara Bio) for C/EBPβ (sense, 5′-GACAAGCTGAGCGACGAGTA-3′; and anti-sense, 5′-AGCTGCTCCACCTTCTTCTG-3′) and β-actin (sense, 5′-CATCCGTAAAGACCTCTATGCCAAC-3′ and anti-sense, 5′-ATGGAGCCACCGATCCACA-3′). For each PCR run, cDNA derived from 50 ng total RNA was used. Following initial denaturation at 95°C for 30 s, a two-step cycle procedure was used (denaturation at 95°C for 5 s and annealing and extension at 60°C for 30 s) for 40 cycles. The relative mRNA expression levels in each sample were calculated using the 2−ΔΔCT method (26).
Statistical analysis
All experiments were repeated at least three times. Representative images or data are shown. Values are expressed as the mean ± standard deviation. Differences between control and test samples were analyzed using paired two-tailed Student's t-tests. P<0.05 was considered to indicate a statistically significant difference between values.
Results
Establishment of cell lines from the bone marrow of GFP-transgenic mice
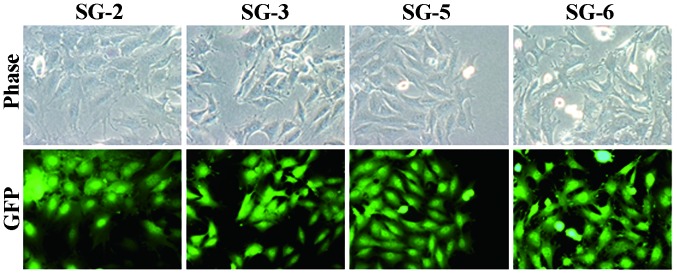
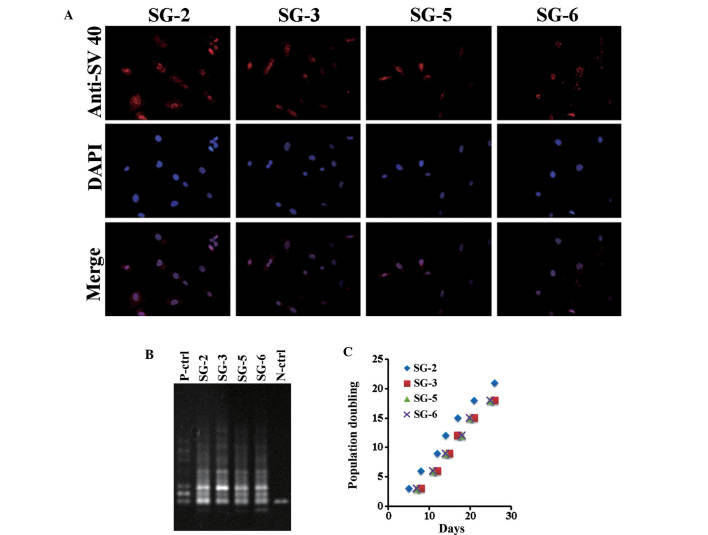
Bone marrow cells were flushed from the tibia of GFP mice and cultured under hypoxic conditions. Adherent cells were transformed with hTERT and SV40LT vectors, yielding four single cell-derived cell lines SG-2, -3, -5 and -6. As shown in Fig. 1, the cell lines exhibited fibroblastic morphology (Fig. 1, upper panel) and GFP fluorescence (Fig. 1, lower panel). All cell lines exhibited nuclear SV40LT expression (Fig. 2A). At PD 20, a stretch PCR assay indicated telomerase activity in all cell lines, but not in the negative control (Fig. 2B). Thus, the bone marrow-derived cell lines exhibited telomerase activity and SV40LT expression. All cell lines grew at a similar rate of ~1 PD every two days (Fig. 2C). The cells divided at least 60 times and were passaged >30 times, thus demonstrating successful immortalization.
Figure 1.
SG-2, -3, -5, and -6 cell lines derived from the bone marrow of tibia from GFP-transgenic mice imaged by phase-contrast (upper panel) and fluores-cence (blue filter; lower panel) microscopy. GFP, green fluorescence protein.
Figure 2.
Immortalizing gene expression in SG-2, -3, -5, and -6 cells. (A) SV40 large T antigen immunofluorescence imaging with Alexa Fluor 594 and DAPI (nuclei). (B) Telomerase activity assessed by stretch polymerase chain reaction assay using 10–20% polyacrylamide gels and visualization with ethidium bromide, and are representative of three independent experiments. (C) Population doubling under hypoxic conditions. Population doubling and the incubation day were considered zero when single-cell cloning was performed. P/N-ctrl, positive/negative control.
Bone marrow-derived SG cell lines have MSC-like features
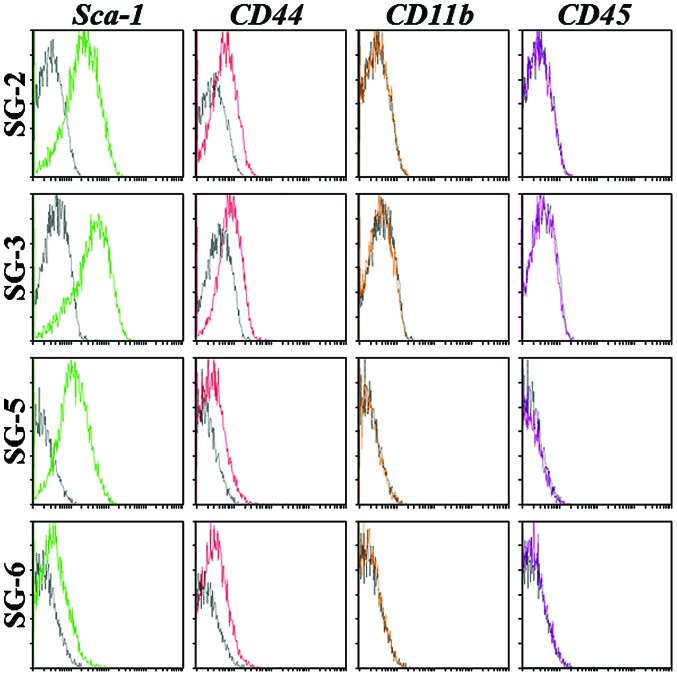
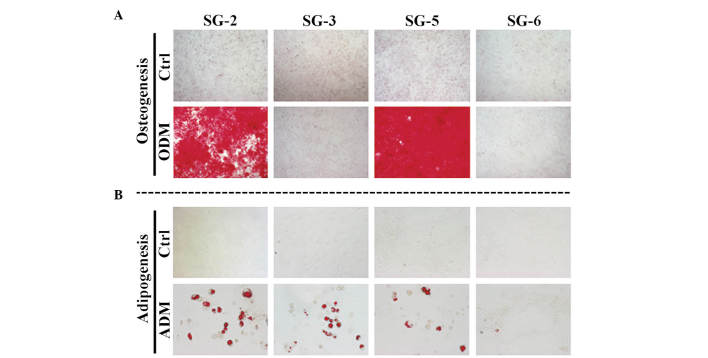
To determine their MSC character, the SG-2, -3, 5, and -6 cell lines were analyzed for the expression of mouse MSC markers and differentiation potential. Sca-1 is the most reliable MSC marker in mice and was strongly expressed in SG-2, -3 and -5 cells, but only weakly expressed in SG-6 cells (Fig. 3). CD44 was detected at similar levels in all cell lines, although the hematopoietic stem cell markers CD11b and CD45 were not detected. The expression patterns of MSC markers suggested that the SG-2, -3 and -5 cell lines were MSCs. Next, the present study evaluated the osteogenic and adipogenic differentiation potentials of these cell lines. Bone matrix mineralization indicated by Alizarin red staining was highest in SG-5 cells (Fig. 4A). Although miner-alization was observed in SG-2 cells, it was markedly lower than that in SG-5 and was not detected in SG-3 and SG-6 cells. Thus, SG-2 and -5 cells retained their osteogenic differentiation potential, although at different levels of efficiency. Lipid droplet formation indicated by Oil Red O staining was more intense in SG-2 cells than in SG-3 and -5 cells (Fig. 4B). Thus, SG-2, -3, and -5 retained their adipogenic differentiation potential to various degrees.
Figure 3.
Identification of mouse mesenchymal stem cell markers in SG-2, -3, -5, and -6 cell by flow cytometry. The cell lines were incubated with phycoerythrin-conjugated control immunoglobulin G (black), anti-Sca-1 (green), anti-CD44 (red), anti-CD11b (yellow), or anti-CD45 (purple) antibody and acquisition was performed on a EPICS XL ADC system.
Figure 4.
Differentiation potential of SG-2, -3, -5, and -6. The cell lines were cultured in (A) ODM or (B) ADM. After 2 weeks, the cells were evaluated for (A) extracellular matrix mineralization by alizarin red staining and (B) for adipogenic differentiation by Oil-Red O staining. Ctrl, control; ODM, osteogenic differentiation medium; ADM, adipogenic differentiation medium.
Intercellular signaling by TGF-β and BMP in SG cells
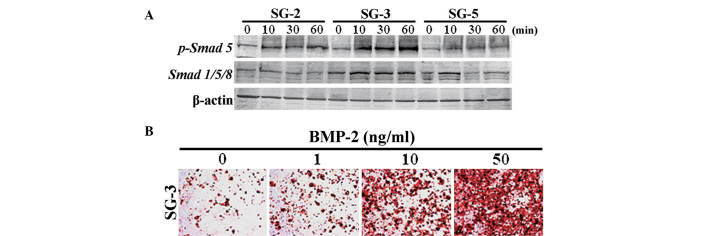
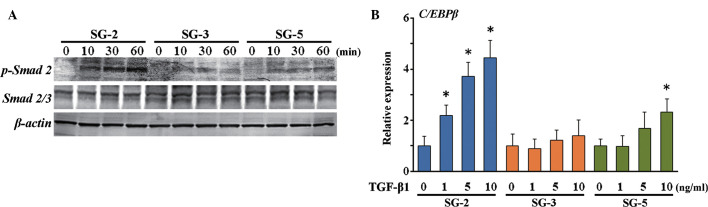
In order to identify the expression profiles of cytokines and cytokine receptors in SG cells, the present study performed PrimerArray analyses and compared the results derived from SG-2, -3 and -5 cells. Differentially expressed genes are shown in Table I. BMP receptor 1B (Bmpr1b) was most highly expressed in SG-3 cells, while TGF-β receptor II (Tgfbr2) was most highly expressed in SG-3 and -5 cells, indicating differential sensitivities to BMP-2 and TGF-β. Smad 5, which is a major signaling factor activated by BMP, was most significantly phosphorylated in BMP 2-stimulated SG-3 cells (Fig. 5A). In addition, osteogenic differentiation in SG-3 cells was induced by stimulation with BMP-2 in a dose-dependent manner (Fig. 5B), suggesting these cells retained the capacity to differentiate into adipogenic (Fig. 4B) and osteogenic lineages. However, phosphorylation of Smad 2, which is activated by TGF-β, was highest in the MSC-like SG-2 cells in response to TGF-β1 (Fig. 6A). Of note, mRNA expression of C/EBPβ, which is an immune- and inflammatory response-associated as well as Smad-interacting transcription factor, was induced in SG-2 cells by TGF-β1 in a dose-dependent manner (Fig. 6B). Thus, the SG-3 cells were BMP-responsive and the SG-2 cells were TGF-β-responsive, while SG-5 cells were BMP/TGF-β-unresponsive MSCs.
Table I.
Genes for which expression changed by >2-fold in SG-2 vs. SG-3 and SG-5.
| Gene symbol | Gene name | Fold change with SG-2
|
|
|---|---|---|---|
| vs. SG-3 | vs. SG-5 | ||
| Bmpr1b | Bone morphogenetic protein receptor, type 1B | 45.89 | −2.362 |
| Egfr | Epidermal growth factor receptor | 4.228 | 6.453 |
| Ifngr2 | Interferon γ receptor 2 | 1.181 | −3.534 |
| Il17ra | Interleukin 17 receptor A | 2.266 | 1.790 |
| Il18r1 | Interleukin 18 receptor 1 | 3.811 | 1.007 |
| Pdgfrb | Platelet derived growth factor receptor, beta polypeptide | 2.514 | 1.625 |
| Cx3cl1 | Chemokine (C-X3-C motif) ligand 1 | 5.315 | 2.346 |
| Tgfbr1 | Transforming growth factor, beta receptor I | 2.056 | 1.778 |
| Tgfbr2 | Transforming growth factor, beta receptor II | 2320 | 2702 |
| Hgf | Hepatocyte growth factor | 14.83 | 5.098 |
| Kdr | Kinase insert domain protein receptor | 8.574 | 2.888 |
| Lepr | Leptin receptor | 2.144 | 1.495 |
| Pdgfb | Platelet-derived growth factor, B polypeptide | 4.823 | 4.993 |
| Pdgfra | Platelet-derived growth factor receptor, alpha polypeptide | 2.329 | 1.444 |
| Prlr | Prolactin receptor | −2.071 | −1.014 |
| Ccl9 | Chemokine (C-C motif) ligand 9 | −2.174 | 1.050 |
| Tnfrsf1b | Tumor necrosis factor receptor superfamily, member 1b | 2.129 | 1.879 |
| Cd40 | CD40 antigen | −1.319 | −27.48 |
| Il2rg | Interleukin 2 receptor, γ chain | 7.160 | 2.789 |
| Lifr | Leukemia inhibitory factor receptor | 1.602 | 4.112 |
| Kitl | Kit ligand | 2.000 | 1.283 |
| Cntfr | Ciliary neurotrophic factor receptor | 5.938 | 5.242 |
| Cxcl14 | Chemokine (C-X-C motif) ligand 14 | 7.835 | 5.205 |
| Il17rb | Interleukin 17 receptor B | −2.250 | 3.074 |
| Ccl8 | Chemokine (C-C motif) ligand 8 | 1.919 | 2.346 |
Figure 5.
Osteogenic differentiation of SG-3 cells was induced by BMP-2. (A) Phosphorylation status was analyzed by western blotting, and the blots are representative of three independent experiments. (B) After 2 weeks of culture in osteogenic differentiation medium containing BMP-2, bone matrix mineralization was evaluated by Alizarin red S staining. BMP, bone morphogenetic protein.
Figure 6.
Expression of C/EBPβ induced by TGF-β in SG-2 cells. (A) The phosphorylation status was analyzed by western blotting, and the blots are representative of three independent experiments. (B) Transcript expression of C/EBPβ was assessed after 24 h, normalized to β-actin and expressed as a fold-increase or -decrease relative to the control (0 ng/ml TGF-β). Values are expressed as the mean ± standard deviation. *P<0.05 vs. untreated group. C/EBPβ, CCAAT/enhancer binding protein-β; TGF, transforming growth factor; p, phosphorylated.
Discussion
In the present study, the bone marrow of GFP mice was used to establish three MSC lines immortalized by transfection with SV40LT and hTERT. SV40LT-transformed cells are not tumorigenic (27–30); therefore, SV40LT is commonly used to immortalize primary mammalian cells. Another commonly used gene for immortalization is TERT, which maintains telomere length to enable cells to indefinitely proliferate. TERT expression is high in stem cells, while it is reduced upon differentiation. Restoration of TERT activity in normal somatic cells can lead to their immortalization and may be associated with the acquisition of characteristics associated with cellular transformation (31). It has been indicated that ectopic expression of the mouse TERT catalytic subunit does not affect embryonic stem cell proliferation or differentiation in vitro, but protects them from cell death during differentiation (32). Therefore, the cell lines generated in the present study may be used to study stem cell proliferation and differentiation.
The expression of MSC markers Sca-1+ and CD44+ and the absence of hematopoietic stem cell markers CD11b− and CD45− were confirmed in SG-2, -3 and -5 cells, which exhibited osteogenic and adipogenic differentiation potential. These results strongly suggested that the MSC-like potential of the cells was preserved. The present study focused on cytokines and cytokine receptors that are expressed specifically in each cell line to clarify the differentiation mechanism of MSCs: Bmpr1b was most highly expressed in SG-3 but not in SG-2 or -5 cells, whereas Tgfbr2 was most highly expressed in SG-3 and -5 but not in SG-2 cells. BMP-2 induced phosphorylation of Smad 5 in SG-3 but not in SG-2 and -5 cells. Furthermore, BMP-2 induced osteogenic differentiation of SG-3 cells but did not affect osteogenic differentiation in SG-2 and -5 cells (data not shown). By contrast, TGF-β unexpectedly but clearly induced Smad 2 phosphorylation in SG-2 cells, in which expression of TGF-β receptors I and II was lower than in SG-3 and -5 cells, suggesting that Smad 2 itself or signal transduction molecules upstream of Smad 2 may have been inactivated in SG-3 and -5 cells. Thus, the present study established TGF-β-responsive SG-2 cells, BMP-responsive SG-3 cells and TGF-β/BMP-unresponsive SG-5 cells that can be traced by GFP fluorescence after transplantation into in vivo experimental models.
Of note, TGF-β stimulation of SG-2 induced expression of C/EBPβ, a Smad-interacting transcription factor (33). C/EBPβ is a member of the C/EBP family of transcription factors (C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ, C/EBPε and C/EBPξ) (34–36). C/EBPβ was first described in 1990 as a basic leucine zipper-structured factor that binds to the interleukin (IL)-1-responsive element in the IL-6 promoter (37). C/EBPβ is highly expressed in myelomonocytic cells and macrophages (38–41). Extracellular signals, including differentiation- or proliferation-inducing agents, hormones, cytokines and inflammatory substances, as well as bacterial and other microbial products can activate or inhibit C/EBPβ via distinct signal transduction pathways. The expression and/or activation of C/EBPβ is regulated by transcriptional mechanisms, mammalian target of rapamycin (mTOR)-mediated alternative translation, post-translational modifications and protein-protein interactions (35,42,43). Upon its activation, C/EBPβ induces or represses a variety of genes, including cytokines, chemokines and their receptors, other pro-inflammatory genes and pro-proliferative or differentiation-associated markers, as well as metabolic enzymes (34). C/EBPβ thereby affects associated cellular functions, including proliferation (40,42), differentiation (39,41,44), metabolic regulation (45,46) and orchestration of the immune response (47–49). Furthermore, C/EBPβ is implicated in the pathogenesis of various common diseases, including cancer, hyper-/hypo-inflammation and bacterial/viral infections (34,50). Expression and activation of C/EBPβ induces the production of monocyte chemotactic protein-1 (MCP-1) (51–54), a member of the C-C motif chemokine ligand-2, which induces leukocyte migration to inflamed tissues and organs (55,56). In addition, MCP-1 is secreted by primary breast tumors and stimulates migration of MSCs to tumor lesions (57). MCP-1, stromal cell-derived factor-1, macrophage inflammatory protein-1α and monocyte chemotactic protein-3 are the most widely reported MSC homing factors (58–61). Stem cell therapy relies on the appropriate homing and engraftment capacity of stem cells (62). Therefore, SG-2 can be used for in vivo studies of TGF-β-dependent anti-inflammation and stem cell homing.
Abnormal intensity of Smad-mediated TGF-β/BMP signals in MSCs is associated with various human diseases, including bone and immune disorders, fibrosis, and cancer progression or metastasis. However, the detection of therapeutic molecular targets for these diseases in the TGF-β/BMP signaling pathways has not been successful as most studies have been performed in vitro. The present study established MSC lines, including TGF-β-responsive SG-2, BMP-responsive SG-3, and TGF-β/BMP-unresponsive SG-5 cells, which can be traced by GFP fluorescence after transplantation into in vivo experimental models. These cell lines can be used to explore how TGF-β/BMP-induced Smad-mediated signals affect proliferation and differentiation of MSCs in vivo, providing insight into various human diseases, including bone and immune disorders, fibrosis and cancer progression or metastasis.
Acknowledgments
The present study was supported in part by KAKENHI grants-in-aid from the Japan Society for the Promotion of Science (grant nos. 25893221 to S.S., 25463053 to N.C., 26893248 to J.Y., 26462932 to H.K. and 26670852 to A.I.), a grant-in-aid for Strategic Medical Science Research Centre from the Ministry of Education, Culture, Sports, Science and Technology of Japan (2010–2014), a grant from the Keiryokai Research Foundation (no. 120 to S.S., 2013) and the Fund of Academic Society for Research in Otolaryngology (Kansai Medical University, Hirakata, Japan).
References
- 1.Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 2.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Docheva D, Popov C, Mutschler W, Schieker M. Human mesenchymal stem cells in contact with their environment: Surface characteristics and the integrin system. J Cell Mol Med. 2007;11:21–38. doi: 10.1111/j.1582-4934.2007.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baksh D, Song L, Tuan RS. Adult mesenchymal stem cells: Characterization, differentiation and application in cell and gene therapy. J Cell Mol Med. 2004;8:301–316. doi: 10.1111/j.1582-4934.2004.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kassem M, Abdallah BM, Saeed H. Osteoblastic cells: Differentiation and transdifferentiation. Arch Biochem Biophys. 2008;473:183–187. doi: 10.1016/j.abb.2008.03.028. [DOI] [PubMed] [Google Scholar]
- 6.Jones E, Yang X. Mesenchymal stem cells and bone regeneration: Current status. Injury. 2011;42:562–568. doi: 10.1016/j.injury.2011.03.030. [DOI] [PubMed] [Google Scholar]
- 7.Proff P, Römer P. The molecular mechanism behind bone remodelling: A review. Clin Oral Investig. 2009;13:355–362. doi: 10.1007/s00784-009-0268-2. [DOI] [PubMed] [Google Scholar]
- 8.Lazar-Karsten P, Dorn I, Meyer G, Lindner U, Driller B, Schlenke P. The influence of extracellular matrix proteins and mesenchymal stem cells on erythropoietic cell maturation. Vox Sang. 2011;101:65–76. doi: 10.1111/j.1423-0410.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- 9.Yew TL, Chang MC, Hsu YT, Weng WH, Tsai CC, Chiu FY, Hung SC, He FY. Efficient expansion of mesenchymal stem cells from mouse bone marrow under hypoxic conditions. J Tissue Eng Regen Med. 2013;7:984–993. doi: 10.1002/term.1491. [DOI] [PubMed] [Google Scholar]
- 10.Weiss A, Attisano L. The TGF beta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol. 2013;2:47–63. doi: 10.1002/wdev.86. [DOI] [PubMed] [Google Scholar]
- 11.Miyazawa K, Shinozaki M, Hara T, Furuya T, Miyazono K. Two major Smad pathways in TGF-beta superfamily signalling. Genes Cells. 2002;7:1191–1204. doi: 10.1046/j.1365-2443.2002.00599.x. [DOI] [PubMed] [Google Scholar]
- 12.Heldin CH, Landström M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21:166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Meulmeester E, Ten Dijke P. The dynamic roles of TGF-β in cancer. J Pathol. 2011;223:205–218. doi: 10.1002/path.2785. [DOI] [PubMed] [Google Scholar]
- 14.Imamura T, Oshima Y, Hikita A. Regulation of TGF-β family signaling by ubiquitination and de ubiquitination. J Biochem. 2013;154:481–489. doi: 10.1093/jb/mvt097. [DOI] [PubMed] [Google Scholar]
- 15.Herhaus L, Sapkota GP. The emerging roles of deubiquitylating enzymes (DUBs) in the TGFβ and BMP pathways. Cell Signal. 2014;26:2186–2192. doi: 10.1016/j.cellsig.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SY, Lee JH, Kim JY, Bae YC, Suh KT, Jung JS. BMP2 increases adipogenic differentiation in the presence of dexamethasone, which is inhibited by the treatment of TNF-α in human adipose tissue-derived stromal cells. Cell Physiol Biochem. 2014;34:1339–1350. doi: 10.1159/000366341. [DOI] [PubMed] [Google Scholar]
- 17.Pountos I, Georgouli T, Henshaw K, Bird H, Jones E, Giannoudis PV. The effect of bone morphogenetic protein-2, bone morphogenetic protein-7, parathyroid hormone and platelet-derived growth factor on the proliferation and osteogenic differentiation of mesenchymal stem cells derived from osteoporotic bone. J Orthop Trauma. 2010;24:552–556. doi: 10.1097/BOT.0b013e3181efa8fe. [DOI] [PubMed] [Google Scholar]
- 18.Yokota J, Chosa N, Sawada S, Okubo N, Takahashi N, Hasegawa T, Kondo H, Ishisaki A. PDGF-induced PI3K-mediated signal enhances TGF-β-induced osteogenic differentiation of human mesenchymal stem cells in the TGF-β-activated MEK-dependent manner. Int J Mol Med. 2014;33:534–542. doi: 10.3892/ijmm.2013.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray P, Chapman SC. Cytoskeletal reorganaization drives mesenchymal condensation and regulates downstream molecular signaling. PLoS One. 2015;10:e0134702. doi: 10.1371/journal.pone.0134702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sengle G, Carlberg V, Tufa SF, Charbonneau NL, Smaldone S, Carlson EJ, Ramirez F, Keene DR, Sakai LY. Abnormal activation of BMP signaling causes myopathy in Fbn2 null mice. PLoS Genet. 2015;11:e1005340. doi: 10.1371/journal.pgen.1005340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramman R, Humphreys BD. Kidney pericytes: Roles in regeneration and fibrosis. Semin Nephrol. 2014;34:374–383. doi: 10.1016/j.semnephrol.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Touboul C, Vidal F, Pasquier J, Lis R, Rafii A. Role of mesenchymal cells in the natural history of ovarian cancer: A review. J Transl Med. 2014;12:271. doi: 10.1186/s12967-014-0271-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wise AF, Ricardo SD. Mesenchymal stem cells in kidney inflammation and repair. Nephrology (Carlton) 2012;17:1–10. doi: 10.1111/j.1440-1797.2011.01501.x. [DOI] [PubMed] [Google Scholar]
- 24.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. 'Green mice' as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/S0014-5793(97)00313-X. [DOI] [PubMed] [Google Scholar]
- 25.Aomatsu E, Takahashi N, Sawada S, Okubo N, Hasegawa T, Taira M, Miura H, Ishisaki A, Chosa N. Novel SCRG1/BST1 axis regulates self-renewal, migration and osteogenic differentiation potential in mesenchymal stem cells. Sci Rep. 2014;4:3652. doi: 10.1038/srep03652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Gee CJ, Harris H. Tumorigenicity of cells transformed by Simian virus 40 and of hybrids between such cells and normal diploid cells. J Cell Sci. 1979;36:223–240. doi: 10.1242/jcs.36.1.223. [DOI] [PubMed] [Google Scholar]
- 28.Howell N. Suppression of transformation and tumorigenicity in interspecies hybrids of human SV40-transformed and mouse 3T3 cell lines. Cytogenet Cell Genet. 1982;34:215–229. doi: 10.1159/000131809. [DOI] [PubMed] [Google Scholar]
- 29.Kahn P, Topp WC, Shin S. Tumorigenicity of SV40-transformed human and monkey cells in immunodeficient mice. Virology. 1983;126:348–360. doi: 10.1016/0042-6822(83)90484-1. [DOI] [PubMed] [Google Scholar]
- 30.Nitta M, Katabuchi H, Ohtake H, Tashiro H, Yamaizumi M, Okamura H. Characterization and tumorigenicity of human ovarian surface epithelial cells immortalized by SV40 large T antigen. Gynecol Oncol. 2001;81:10–17. doi: 10.1006/gyno.2000.6084. [DOI] [PubMed] [Google Scholar]
- 31.Kang MK, Park NH. Extension of cell life span using exogenous telomerase. Methods Mol Biol. 2007;371:151–165. doi: 10.1007/978-1-59745-361-5_12. [DOI] [PubMed] [Google Scholar]
- 32.Lee MK, Hande MP, Sabapathy K. Ectopic mTERT expression in mouse embryonic stem cells does not affect differentiation but confers resistance to differentiation- and stress-induced p53-dependent apoptosis. J Cell Sci. 2005;118:819–829. doi: 10.1242/jcs.01673. [DOI] [PubMed] [Google Scholar]
- 33.Abraham S, Sweet T, Khalili K, Sawaya BE, Amini S. Evidence for activation of the TGF-beta1 promoter by C/EBP beta and its modulation by Smads. J Interferon Cytokine Res. 2009;29:1–7. doi: 10.1089/jir.2008.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: Structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/bj20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zahnow CA. CCAAT/enhancer-binding protein beta: Its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med. 2009;11:e12. doi: 10.1017/S1462399409001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 37.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams SC, Cantwell CA, Johnson PF. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 39.Katz S, Kowenz-Leutz E, Müller C, Meese K, Ness SA, Leutz A. The NF-M transcription factor is related to C/EBP beta and plays a role in signal transduction, differentiation and leukemogenesis of avian myelomonocytic cells. EMBO J. 1993;12:1321–1332. doi: 10.1002/j.1460-2075.1993.tb05777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haas SC, Huber R, Gutsch R, Kandemir JD, Cappello C, Krauter J, Duyster J, Ganser A, Brand K. ITD- and FL-induced FLT3 signal transduction leads to increased C/EBP beta-LIP expression and LIP/LAP ratio by different signalling modules. Br J Haematol. 2010;148:777–790. doi: 10.1111/j.1365-2141.2009.08012.x. [DOI] [PubMed] [Google Scholar]
- 41.Gutsch R, Kandemir JD, Pietsch D, Cappello C, Meyer J, Simanowski K, Huber R, Brand K. CCAAT/enhancer-binding protein beta inhibits proliferation in monocytic cells by affecting the retinoblastoma protein/E2F/cyclin E pathway but is not directly required for macrophage morphology. J Biol Chem. 2011;286:22716–22729. doi: 10.1074/jbc.M110.152538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nerlov C. The C/EBP family of transcription factors: A paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–324. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Tsukada J, Yoshida Y, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54:6–19. doi: 10.1016/j.cyto.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Pham TH, Langmann S, Schwarzfischer L, El Chartouni C, Lichtinger M, Klug M, Krause SW, Rehli M. CCAAT enhancer-binding protein beta regulates constitutive gene expression during late stages of monocyte to macrophage differentiation. J Biol Chem. 2007;282:21924–21933. doi: 10.1074/jbc.M611618200. [DOI] [PubMed] [Google Scholar]
- 45.Liu S, Croniger C, Arizmendi C, Harada-Shiba M, Ren J, Poli V, Hanson RW, Friedman JE. Hypoglycemia and impaired hepatic glucose production in mice with a deletion of the C/EBP beta gene. J Clin Invest. 1999;103:207–213. doi: 10.1172/JCI4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Croniger CM, Millward C, Yang J, Kawai Y, Arinze IJ, Liu S, Harada-Shiba M, Chakravarty K, Friedman JE, Poli V, Hanson RW. Mice with a deletion in the gene for CCAAT/enhancer-binding protein beta have an attenuated response to cAMP and impaired carbohydrate metabolism. J Biol Chem. 2001;276:629–638. doi: 10.1074/jbc.M007576200. [DOI] [PubMed] [Google Scholar]
- 47.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273:29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- 48.Zwergal A, Quirling M, Saugel B, Huth KC, Sydlik C, Poli V, Neumeier D, Ziegler-Heitbrock HW, Brand K. C/EBP beta blocks p65 phosphorylation and thereby NF-kappa B-mediated transcription in TNF-tolerant cells. J Immunol. 2006;177:665–672. doi: 10.4049/jimmunol.177.1.665. [DOI] [PubMed] [Google Scholar]
- 49.Cappello C, Zwergal A, Kanclerski S, Haas SC, Kandemir JD, Huber R, Page S, Brand K. C/EBP beta enhances NF-kappaB-associated signalling by reducing the level of IkappaB-alpha. Cell Signal. 2009;21:1918–1924. doi: 10.1016/j.cellsig.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Nonne macher MR, Wigdahl B. CCAAT/enhancer-binding proteins and the pathogenesis of retrovirus infection. Future Microbiol. 2009;4:299–321. doi: 10.2217/fmb.09.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spooner CJ, Guo X, Johnson PF, Schwartz RC. Differential roles of C/EBP beta regulatory domains in specifying MCP-1 and IL-6 transcription. Mol Immunol. 2007;44:1384–1392. doi: 10.1016/j.molimm.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Abraham M, Shapiro S, Karni A, Weiner HL, Miller A. Gelatinases (MMP-2 and MMP-9) are preferentially expressed by Th1 vs. Th2 cells. J Neuroimmunol. 2005;163:157–164. doi: 10.1016/j.jneuroim.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 53.Guo J, Zhang H, Xiao J, Wu J, Ye Y, Li Z, Zou Y, Li X. Monocyte chemotactic protein-1 promotes the myocardial homing of mesenchymal stem cells in dilated cardiomyopathy. Int J Mol Sci. 2013;14:8164–8178. doi: 10.3390/ijms14048164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belema-Bedada F, Uchida S, Martire A, Kostin S, Braun T. Efficient homing of multipotent adult mesenchymal stem cells depends on FROUNT-mediated clustering of CCR2. Cell Stem Cell. 2008;2:566–575. doi: 10.1016/j.stem.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 55.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 56.Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: Basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 57.Dwyer RM, Potter-Beirne SM, Harrington KA, Lowery AJ, Hennessy E, Murphy JM, Barry FP, O'Brien T, Kerin MJ. Monocyte chemotactic protein-1 secreted by primary breast tumors stimulates migration of mesenchymal stem cells. Clin Cancer Res. 2007;13:5020–5027. doi: 10.1158/1078-0432.CCR-07-0731. [DOI] [PubMed] [Google Scholar]
- 58.Schenk S, Mal N, Finan A, Zhang M, Kiedrowski M, Popovic Z, McCarthy PM, Penn MS. Monocyte chemotactic protein-3 is a myocardial mesenchymal stem cell homing factor. Stem Cells. 2007;25:245–251. doi: 10.1634/stemcells.2006-0293. [DOI] [PubMed] [Google Scholar]
- 59.Zhou YL, Zhang HF, Li XL, Di RM, Yao WM, Li DF, Feng JL, Huang J, Cao KJ, Fu M. Increased stromal-cell-derived factor 1 enhances the homing of bone marrow derived mesenchymal stem cells in dilated cardiomyopathy in rats. Chin Med J (Engl) 2010;123:3282–3287. [PubMed] [Google Scholar]
- 60.Zhuang Y, Chen X, Xu M, Zhang LY, Xiang F. Chemokine stromal cell-derived factor 1/CXCL12 increases homing of mesenchymal stem cells to injured myocardium and neovascularization following myocardial infarction. Chin Med J (Engl) 2009;122:183–187. doi: 10.3901/jme.2009.07.183. [DOI] [PubMed] [Google Scholar]
- 61.Abbott JD, Huang Y, Liu D, Hickey R, Krause DS, Giordano FJ. Stromal cell-derived factor-1 alpha plays a critical role in stem cell recruitment to the heart after myocardial infarction but is not sufficient to induce homing in the absence of injury. Circulation. 2004;110:3300–3305. doi: 10.1161/01.CIR.0000147780.30124.CF. [DOI] [PubMed] [Google Scholar]
- 62.Mozid AM, Arnous S, Sammut EC, Mathur A. Stem cell therapy for heart diseases. Br Med Bull. 2011;98:143–159. doi: 10.1093/bmb/ldr014. [DOI] [PubMed] [Google Scholar]