Abstract
In our previous study, the upregulation of microRNA (miR)-29c was identified in the mother of a fetus with a congenital heart defect. However, the functional and regulatory mechanisms of miR-29c in the development of the heart remain to be elucidated. In the present study, the role and mechanism of miR-29c inhibition in heart development were investigated in an embryonic carcinoma cell model. Inhibition of miR-29c promoted proliferation, and suppressed the apoptosis and differentiation of P19 cells. It was also demonstrated that Wingless-related MMTV integration site 4 (Wnt4) was a target of miR-29c, determined using bioinformatic analysis combined with luciferase assays. The inhibition of miR-29c stimulated the WNT4/β-catenin pathway, promoting proliferation of the P19 cells, but suppressing their differentiation into cardiomyocytes. Furthermore, the inhibition of miR-29c promoted the expression of B cell lymphoma-2 and inhibited cell apoptosis. These results demonstrate the significance of miR-29c in the process of cardiac development and suggest that miR-29c dysregulation may be associated with the occurrence of CHD. Thus, miR-29c may have therapeutic potential in the future.
Keywords: microRNA-29c, proliferation, P19 embryonic carcinoma cell, apoptosis, differentiation, Wnt4
Introduction
Congenital heart defects (CHDs) represent the most common category of birth defect, with a prevalence of 8–12/1,000 births (1,2). This number can be substantially higher if the 30% of embryos or fetuses lost prior to birth due to heart malformations are included (3). The causes of CHDs are complex, with environmental and genetic factors being of importance (4). In twins, higher levels of concordance are observed in monozygotic twins, compared with dizygotic twins, suggesting a more significant etiological contribution by genetic, rather than environmental factors (5). The process of heart development requires precise temporal-spatial regulation of gene expression, in which the highly conserved regulatory networks of transcription factors and genes accurately control the signaling pathways required for normal heart morphogenesis.
MicroRNAs (miRNAs) are a class of small non-coding RNAs, which critically modulate gene expression at a post-transcriptional level by interacting with the 3′ untranslated regions (UTRs) of specific mRNA targets, inhibiting protein translation or promoting the degradation of cognate target mRNAs (6). miRNA profiling studies have demonstrated their important roles in biological processes, including cell proliferation and death (7), differentiation (8), intracellular signaling and cell movement (9,10). Previous investigations have implicated miRNAs as having a ʻfine tuningʼ effect in cardiac development (11). The miR-1/miR-133 clusters are the most abundantly expressed miRNAs in the heart (12), and were the first and are the most extensively investigated miRNAs among all cardiac development-specific miRNAs (13). The overexpression of miR-1 leads to a thin ventricular wall and embryonic fatality at E13.5 (14), and the deletion of miR-1–2 results in embryonic fatality at E15.5 (12), each, in part, due to defective ventricular myocyte proliferation. Similarly, gain- and loss-of function studies of miR-133 a indicate that miR-133 a negatively regulates cardiomyocyte proliferation during heart development (15,16). By contrast, in vitro studies have suggested that miR-1 and miR-133 promote cardiomyocyte differentiation from embryonic stem (ES) cells. Although other miRNAs, which exert importance effects on cardiac development are rapidly emerging, current understanding remains limited. The identification of additional regulatory components may provide a more detailed and complete understanding, and efforts to identify additional cardiac-specific miRNAs is required.
Our previous studies revealed that miR-29c is significantly upregulated in pregnant women with fetal CHD (17). This finding indicates that miR-29c may act as another heart morphogenesis-specific miRNA. However, investigations on miR-29c have focused predominantly on tumors. Nguyen et al (18) found that low levels of miR-29c were associated with the progression of melanoma, and Wang et al (19) identified an inhibitory role in hepatocellular carcinoma. Other investigations have revealed that miR-29c is involved in leukemia (20), glioma (21), bladder cancer (22) and nasopharyngeal (23) and gastric carcinoma (24). Taken together, it has become apparent that a common feature of tumorigenesis is the downregulation of miR-29c, and that the presence of miR-29c inhibits tumor cell growth, which may be indicative of improved prognosis. In addition, the downregulation of miR-29c may be associated with aberrant cardiac morphogenesis, although reports on the involvement of miR-29c in heart development are limited. The present study investigated the effects of miR-29c on cardiac development by manipulating the expression levels of miR-29c in murine P19 embryonic carcinoma cells (ECCs), which can be induced to differentiate into cardiomyocytes with dimethylsulfoxide (DMSO). The present study also used an miRNA 'sponge' technique to establish prolonged miR-29c loss of function. The aim of the present study was to examine the role and mechanism of miR-29c inhibition in the regulation of heart development.
Materials and methods
'Sponge' design and construction of miR-29c inhibition plasmids
The present study utilized a previously reported miRNA 'sponge' technique, using duplex oligonucleotides (25). Sense and antisense oligonucleotides, containing two miR-29c binding sites (MBSs), were separated by a four nucleotide spacer sequence. Each MBS had a central mismatch between positions 9–12 of miR-29c, to create a bulge to prevent rapid turnover of the sponge by endonucleolytic cleavage (26) and to achieve sustained inhibition of miR-29c. A negative control sponge was used, which had a similar design, but with a randomly-scrambled sequence between nucleotides 2–8. The two sponges were separately digested using endonuclease SanDI, and a pGLV3 vector containing the SanDI recognition site was introduced, resulting in miR-29c-sequestering and negative control plasmids. To identify and acquire the miR-29c-inhibited cells for use in subsequent experiments, green fluorescent protein (GFP) and puromycin-coding sequences were cloned downstream of the MBS. The pGLV3 vector contained the GFP gene and puromycin-resistance gene and was designed by GenePharma (Shanghai, China). Oligonucleotides that contained the endonuclease SanD site (GGGT/ACCC) were also chemically synthesized by GenePharma.
Cell culture and differentiation
Murine P19 ECCs (5×106 cells/ml) were obtained from American Type Culture Collection (Manassas, VA, USA). The P19 cells (2×106 cells/ml) were cultured in α-mimimal essential medium (α-MEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (complete medium; (HyClone; GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, UK). For the induction of P19 cell differentiation, DMSO (Sigma-Aldrich, St. Louis, MO, USA) was diluted in the culture medium to a final concentration of 1.0%. The P19 cells aggregated over the following 4 days to form embryoid bodies (EBs), during which time the medium was replenished every 24 h. On day 4, the EBs were transferred to a 6-well plate and cultured in DMSO-free medium to allow adherent growth of the P19 cells. The culture medium was refreshed every 48 h during the ensuing course of differentiation. Images of the morphologies of the derived EBs and differentiated cell patches were captured microscopically (Primovert-ZM300FL; Carl Zeiss, Göttingen, Germany).
Establishment of a stable cell line of miR-29c inhibition
The miR-29c inhibition and negative control plasmids were transfected into P19 cells at 70–80% confluence using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 2×106 cells were transfected with 4 µg plasmid in serum-free α-MEM, according to the manufacture's protocol. Complete medium replaced the serum-free α-MEM, and images were captured under fluorescent microscopy to identify transfected cells exhibiting GFP luminescence. Subsequently, the cells were filtered in medium containing 2 µg/ml puromycin (Sigma-Aldrich) and the expression levels of miR-29c were quantified by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Cells were stably miR-29c inhibited and every experiment was performed within 2 weeks
Cell proliferation and cell cycle
A Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc., Rockville, MD, USA) assay was used to evaluate the proliferation rate of the P19 cells. Briefly, 2,000 cells/well were seeded into 96-well plates and monitored every 24 over 4 days. According to the manufacturer's protocol, 10 µl of CCK-8 reagent (1 mg/ml) was added to the medium, and the cells were maintained for 3 h at 37°C in an incubator containing 5% CO2. The numbers of cells were assessed by measuring the optical density (OD)450 using a microplate reader (Synergy H4 Hybrid; Bio-Tek Instruments, Inc., Winooski, VT, USA).
For cell cycle analysis, the cells were synchronized by serum-starvation for 24 h and placed in complete medium for the subsequent 24 h, during which cells were harvested at 6 h intervals. The adherent and non-adherent cells were centrifuged at 4°C, 100 × g for 5 min. The sedimented cells were washed twice with phosphate-buffered saline (PBS) at 4°C and were resuspended in 70% ethanol at −20°C overnight. The cells were then washed once with PBS and resuspended in 500 µl propidium iodide/RNase staining buffer (BD Biosciences, San Diego, CA, USA) for 15 min at room temperature. Subsequently, 1×104 cells were analyzed by flow cytometry using a FACSCalibur (BD Biosciences) instrument, and the proportion of cells in each cell-cycle phase were determined according to DNA content using FlowJo software (version 7.6.1; Tree Star, Inc., Ashland, OR, USA).
Cell apoptosis and Hoechst staining
Molecular and cellular techniques were used to evaluate the apoptotic status of the P19 cells. The cells were cultured in 6-well plates at a density of 2×105 cells/well. Complete medium was then replaced with serum-free α-MEM for 24 h, when the cells reached 70% confluence, to induce apoptosis. The adherent cells were liberated by trypsin digestion (1 mg/ml; Gibco; Thermo Fisher Scientific, Inc.) and were pooled with the non-adherent cells. Collectively, 1×105 cells were centrifuged at 4°C, 100 × g for 5 min. Following aspiration of the supernatant, 100 µl of 1X assay buffer and 5 µl Annexin V-APC/7-AAD (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) were added to the cells, which were resuspended. The samples were analyzed using flow cytometry, as described above. For each sample, 20,000 gated events were analyzed.
Hoechst staining was used to observe the apoptotic cells, characterized by pyknotic and fragmented nuclei, emitting intense fluorescence. Apoptosis of the cells was induced, as described above, and the cells were stained with Hoechst 33258 dye (Beyotime Institute of Biotechnology, Shanghai, China), which fluoresces blue on interaction with double-stranded DNA. Analysis of fluorescence was performed using microscopy under ultraviolet (UV) excitation at 348 nm and emission at 480 nm. A total of five microscopic fields were randomly-selected, and images were captured of each experimental group of cells.
RNA extraction and RT-qPCR
For miRNA extraction, an miRNeasy Mini kit (Qiagen, Austin, TX, USA) was used, according to the manufacturer's protocol. RNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Rockland, DE, USA) cDNA synthesis was performed using a TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). RT-qPCR was performed using an Applied Biosystems 7500 Fast Real-time PCR cycler (Applied Biosystems; Thermo Fisher Scientific, Inc.). U6 was used as an internal control. The primers for miR-29c and U6 were purchased from Applied Biosystems (Thermo Fisher Scientific, Inc.). TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used for the extraction of total RNA. Aliquots of 1 µg RNA were used to synthesize cDNA using a Thermo Scientific RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). qPCR, using SYBR Green (Invitrogen; Thermo Fisher Scientific, Inc.), was performed, according to the manufacturer's protocol. Briefly, the samples were incubated at 95°C for 10 min for initial denaturation, and then subjected to 40 PCR cycles, each consisting of 95°C for 15 sec and 60°C for 60 sec. Gapdh served as an internal mRNA control. The relative mRNA expression levels were calculated using the 2−ΔΔCq method (27). The sequences of the primers used are presented in Table I.
Table I.
Primer sets used for reverse transcription-quantitative polymerase chain reaction analysis.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Bcl-2 | CAGACATGCACCTACCCAGC | GTCGCTACCGTCGTGACTTC |
| Bax | CCAGCCCATGATGGTTCTGAT | CCGGCGAATTGGAGATGAACT |
| Gata4 | CCAACTGCCAGACTACCAC | GGACCAGGCTGTTCCAAGA |
| cTnT | GGAGTACGAGGAGGAACAGG | GTCCACTCTCTCTCCATCGG |
| Mef2c | CAGCACTGACATGGATAAGG | CTGCCAGGTGGGATAAGAACG |
| Wnt4 | TGTGCAAACGGAACCTTGAG | ATGCCCTTGTCACTGCAAAG |
| β-catenin | AGACAGCTCGTTGTACTGCT | GTGTCGTGATGGCGTAGAAC |
| Gapdh | AACTTTGGCATTGTGGAAGG | CACATTGGGGGTAGGAACAC |
Bcl-2, B cell lymphoma-2; Bax, Bcl-2-associated X protein; Gata4, GATA binding protein 4; cTnT, cardiac troponin T; Mef2c, myocyte enhancer factor 2c; Wnt4, Wingless-related MMTV integration site 4.
Western blot analysis
The proteins were extracted from frozen P19 cell samples using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology) and protein concentrations were quantified using a Bicinchoninic Acid Protein Assay kit (Beyotime Institute of Biotechnology). Subsequently, 20 µg of denatured protein was loaded onto 10% SDS-PAGE gels, electrophoretically resolved and transferred onto polyvinylidene-fluoride membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membranes were then blocked at room temperature with 5% non-fat milk for 2 h, washed in Tris-buffered saline containing 0.1% Tween® 20 (TBST), and then incubated at 4°C overnight with monoclonal rabbit anti-mouse Bcl-2 antibody (cat. no. ab32124; Abcam, Cambrige, UK), monoclonal rabbit anti-mouse Bax antibody (cat. no. ab32503; Abcam), monoclonal rabbit anti-mouse GAPDH antibody (cat. no. ab37168; Abcam), monoclonal rabbit anti-mouse cTnT antibody (cat. no. ab92546; Abcam), polyclonal rabbit anti-mouse Mef2c antibody (cat. no. ab78888; Abcam) and polyclonal rabbit anti-mouse GATA4 antibody (cat. no. sc-9053; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). The membranes were then washed with TBST and incubated with horseradish peroxidase-conjugated polyclonal goat anti-rabbit IgG secondary antibody (1:5,000 dilution; cat. no. ab97200; Abcam) for 2 h at room temperature. Following a further three washes with TBST, the blots were visualized using ECLPlus reagents (Beyotime Institute of Biotechnology). For Western blot analysis, GAPDH was used as an internal control. Protein expression levels were quantified using ImageJ2x software (version 2.1.4.5; Rawak Software, Inc., Stuttgart, Germany).
miR-29c target prediction and luciferase assays
To further examine the miR-29c regulatory mechanisms in cardiac development, the present study accessed online resources to investigate the potential gene network targets. In brief, miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/index.php) was used to predict potential target genes. The outcome of this process identified Wingless-related MMTV integration site 4 (Wnt4) as a likely target of miR-29c. To further validate this prediction, functional Luciferase assays were performed. To determine whether miR-29c can bind to the 3′UTR of Wnt4, wild-type (wt), Wnt4 3′UTR sequence (pGL3-Wnt4 3′UTR-wt) and pGL3-Wnt4 3′UTR-mutant (mut) sequence (comprising the Wnt4 3′UTR sequence with a number of mutations), Luciferase reporter plasmids were used (Promega Corporation, Madison, WI, USA). A total of 1×105 cells were seeded, in triplicate, in 48-well plates for 24 h, and then transfected with 50 ng of the Luciferase reporter plasmids and 10 nM miRNA using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h post-transfection, the cells were lysed and Luciferase activity was assayed using a Dual-Luciferase Reporter Assay system (Promega Corporation).
Statistical analysis
Data are expressed as the mean ± standard deviation. Differences among groups were analyzed using one-way analysis of variance, and the Student-Newman-Keuls method was used for further multiple comparisons. P-values in the present study were two-sided, and P<0.05 was considered to indicate a statistically significant difference. Analyses were performed using SPSS software (v. 13.0, SPSS, Inc., Chicago, IL, USA).
Results
miR-29c suppression in P19 cells
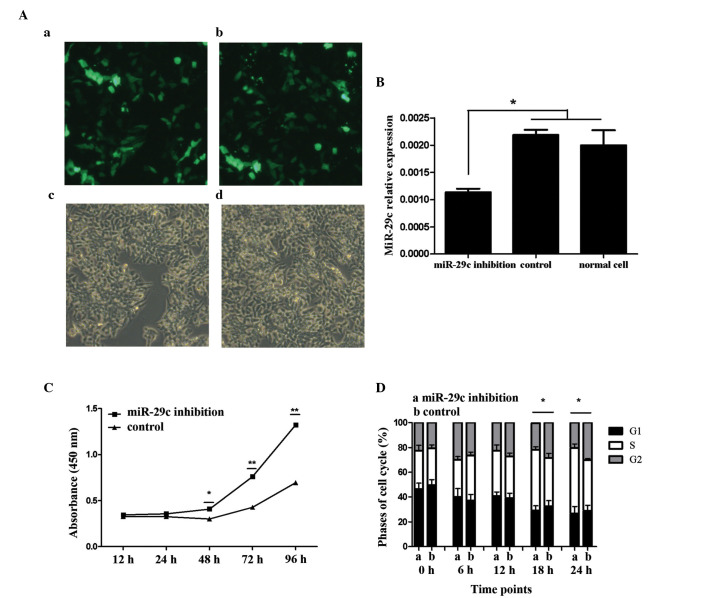
Continuous low expression of miR-29c was a precondition of the present study. At 2 days following transfection of with the miR-29c-inhibitory or negative-control plasmids, the P19 cells were observed using fluorescent microscopy. Under bright field illumination, the transfected cells were culture-vessel adherent, and their morphological characteristics exhibited few differences from the normal P19 cells. Under dark-field imaging, the majority of the cells exhibited GFP fluorescence when subjected to UV light, indicative of successful plasmid transfection (Fig. 1A). Subsequently, the present study examined whether the plasmids functioned as intended in suppressing the activity of miR-29c. Using RT-qPCR, the expression levels of miR-29c were quantified. In the miR-29c-inhibited cells, the levels of target miRNA were significantly reduced, whereas no statistical difference was observed between the control and the non-targeted groups (Fig. 1B; P<0.05).
Figure 1.
Expression of miR-29c in P19 cells and promotion of P19 cell proliferation by miR-29c. Transfection efficiency was determined by the expression of green fluorescent protein using fluorescence microscopy. (A) (a) miR-29c inhibition and (b) control plasmids were successfully transfected and expressed. No significant differences in transfection efficiency were found between the (c) miR-29c-inhibited and (d) control groups (magnification, x100). (B) Confirmation of the expression of miR-29c by reverse transcription-quantitative polymerase chain reaction analysis. Data are presented as the mean ± standard deviation of three experiments. miR-29c was significantly downregulated in the miR-29c inhibition group, compared with the control- and no-vector P19 cell group (*P<0.05). Expression levels of miR-29c were similar between the control- and no-vector P19 cell groups, indicating the vector itself exerted no effect. (C) Cell proliferation was monitored over 4 days consecutively using a Cell Counting Kit-8 assay, performed three times with six replicates in each group. Representative proliferation curves are shown (*P<0.05 and **P<0.01). (D) Cell cycle analysis of the P19 cells was performed using flow cytometry to detect proliferation. Data are presented as the mean ± standard deviation of three experiments (*P<0.05). Results from the two assays indicated that miR-29c inhibition promoted P19 cell proliferation. miR, microRNA.
Inhibition of miR-29c promotes P19 cell proliferation
For each treatment group in the presents tudy, six replicates were used to estimate the variance caused by random factors. From the OD values, it was found that the level of random error was so minor that error bars were almost indiscernible (Fig. 1C). As culture duration increased, the numbers of cells in the miR-29c inhibition group and the control vector group increased exponentially, and differences in proliferation rate became apparent at 48 h, and further diverged at 72 and 96 h. The OD values of the miR-29c inhibition group were significantly higher, compared with those of the control group at 48, 72 and 96 h (Fig. 1C; P<0.05 and P<0.01). Cell cycle analysis reinforced the results obtained from cell proliferation analysis. The number of cells in each of the three cell cycle stages were counted using flow cytometry. No difference was observed initially, however, 18–24 h following cell seeding, a high number of cells in the miR-29c inhibition group were in the S phase (Fig. 1D; P<0.05). This result suggested that cell proliferation in the miR-29c inhibition group was stimulated to a greater extent, compared with the control group.
Inhibition of miR-29c suppresses P19 cell apoptosis
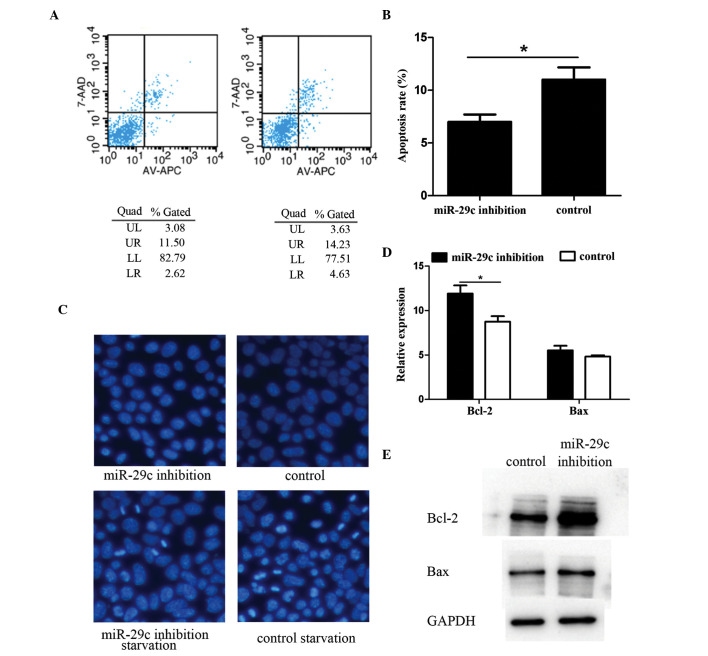
Phosphatidylserine is usually only present on the inner surface of the cell membrane bilayer. At the onset of apoptosis, phosphatidylserine translocates to the outer layer, and can be detected using Annexin V and quantified using flow cytometry. The results of the present study showed that the miR-29c inhibition group had a lower level of apoptosis, compared with the control group (Fig. 2A and B). To further verify these flow cytometry results, Hoechst staining was performed to visualize apoptosis. Following serum starvation, a small proportion of cells commenced apoptosis and emitted intense fluorescence, indicative of pyknotic and fragmented nuclei. An increased number of apoptotic cells were observed in the miR-29c inhibition group, compared with the control (Fig. 2C). Additionally, the activity of apoptosis-associated genes in the two groups of cells using RT-qPCR and Western blotting. Data indicated that the gene expression of pro-apoptotic gene B cell lymphoma-2 (Bcl-2)-associated X protein (Bax) was almost the same in the two groups, however, the anti-apoptotic gene, Bcl-2, was differentially expressed. The expression level in the miR-29c-inhibited P19 cells was significantly higher, compared with that in the control group (Fig. 2D and E; P<0.05).
Figure 2.
miR-29c inhibition suppresses P19 cell apoptosis. Apoptosis was detected using flow cytometry in three independent experiments. Representative results are shown in the (A) (a) miR-29c inhibition group and (b) control group. (B) Apoptotic cell frequency is shown, and the magnitude of the inhibition of apoptosis by miR-29c was significant, comparison with that of the control group. Hoechst staining was performed to evaluate apoptosis in terms of cell morphology. (C) Following serum starvation, apoptosis of the cells began and the apoptotic rate was lower in the miR-29c inhibition group, compared with the control group (magnification, ×200). Bcl-2 gene family members were detected using (D) reverse transcription-quantitative polymerase chain reaction and (E) Western blot analyses. The results showed that the expression levels of survival-promoting Bcl-2 were significantly higher in the miR-29c inhibited group. Protein expression levels of cell death-promoting Bax were not affected (*P<0.05). miR, microRNA; Bcl-2, B cell lymphoma 2; Bax, Bcl-2-associated X protein; Quad, quadrant.
Inhibition of miR-29c suppresses P19 cell differentiation
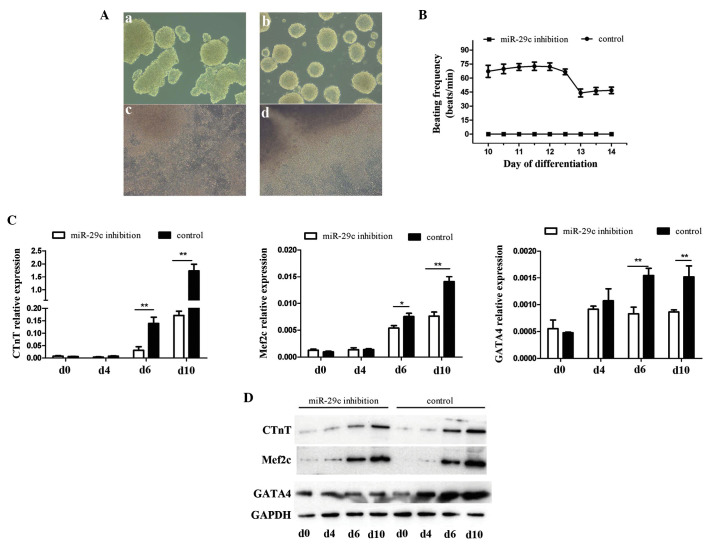
EB formation is a key step for P19 cell differentiation into beating cardiomyocytes. The results of the present study showed that cells in the miR-29c inhibition group formed more irregular EBs (Fig. 3Aa and b) when cultured under the same conditions as the control group. The cells were also more readily disaggregated when transferred to 6-well culture plates. Furthermore, the control cells exhibited more uniform outward proliferation, centered on the EBs, whereas the miR-29c-inhibited cells appeared more disordered (Fig. 3Ac and d). On day 10, beating cells were observed in the control group, whereas the cells exhibiting miR-29c inhibition were quiescent in comparison. Cells were also collected at different time points, in order to detect the expression levels of the cardiogenesis-associated genes, cardiac troponin T (Ctnt), myocyte enhancer factor 2c. (Mef2c) and GATA binding protein 4 (Gata4) using RT-qPCR (Fig. 3C) and Western blot (Fig. 3D) analyses. The results from these two methods were largely concordant. Gene expression levels were significantly elevated during the course of differentiation, peaking at day 6 (Gata4) and day 10 (Ctnt, Mef2c), indicative of the success of differentiation, which was also identified in the above-mentioned morphological analysis. Comparisons were made between the two groups, which revealed that cardiogenic gene expression levels in miR-29c-inhibited cells were significantly lower than those in the control group on days 6 and 10 (P<0.05 and P<0.01). These results suggested that miR-29c inhibition suppressed DMSO-induced myocardial cell differentiation in P19 cells.
Figure 3.
Inhibition of miR-29c-suppresses P19 cell differentiation. (A) Comparison of cell morphology at day 4 between the (a) miR-29c-inhibited and (b) control group, and at day 10 in the (c) miR-29c-inhibited and (d) control group. (B) Characteristics of beating P19 cell clusters between days 10 and 14. (C) Expression levels of cardiogenesis-associated genes, determined using reverse transcription-quantitative polymerase chain reaction analysis (*P<0.05 and **P<0.01). (D) Expression levels of cardiogenesis-associated proteins, determined using Western blot analysis. miR, microRNA; Bcl-2, B cell lymphoma-2; Bax, Bcl-2-associated X protein; Gata4, GATA binding protein 4; cTnT, cardiac troponin T; Mef2c, myocyte enhancer factor 2c; Wnt4, Wingless-related MMTV integration site 4.
Wnt4 is a target of miR-29c
The involvement of miR-29c as an important regulatory factor in the intricate process of cardiac development has been reported, but remains to be fully elucidated. The present study aimed to identify potential target genes through which miR-29c may exert regulatory effects on the interactive gene network/s involved in cardiomyocyte differentiation. Bioinformatic analysis indicated that Wnt4 may be a likely target of miR-29c. Luciferase assays were also performed, and revealed that luminescent activity was significantly reduced by transfection of miR-29c with a wt vector carrying the 3′-UTR of Wnt4. Transfection with a mutant vector showed no decrease in Luciferase activity (Fig. 4A). These results indicated that the 3′-UTR of Wnt4 either contains miR-29c binding sites or that Wnt4 is a target of miR-29c.
Figure 4.
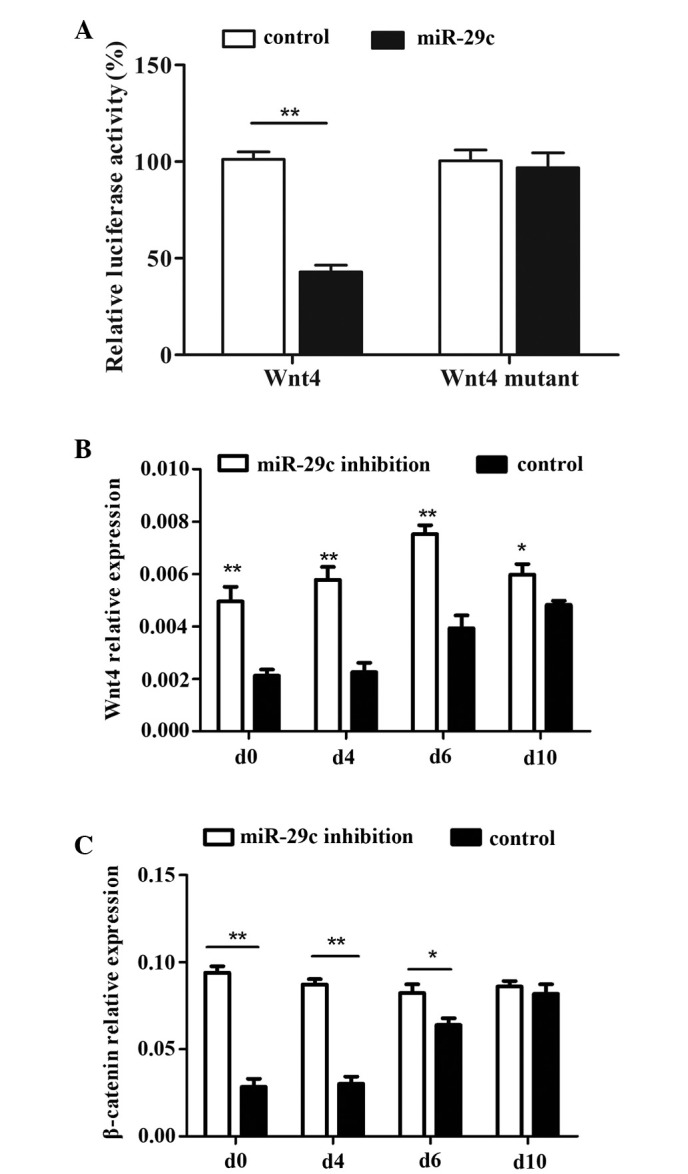
miR-29c targets Wnt4 and regulates cardiac differentiation through the WNT/β-catenin pathway. (A) A Luciferase assay revealed Wnt4 as a potential target of miR-29c. Expression levels of (B) Wnt4 and (C) β-catenin during P19 cell differentiation were detected using reverse transcription-quantitative polymerase chain reaction (*P<0.05 and **P<0.01). The results indicated that miR-29c inhibition stimulated the WNT/β-catenin pathway and inhibited P19 cell differentiation. miR, microRNA; Wnt4, Wingless-related MMTV integration site 4.
The well-known canonical WNT signaling pathway, acting via its major transducer, β-catenin, was shown to exert significant effects on cardiac development. The present study detected the protein expression levels of Wnt4 and β-catenin during P19 cell differentiation, and found that β-catenin exhibited concordance with Wnt4. The expression levels at each time point were significantly higher in the miR-29c inhibited group, compared with the control group (Fig. 4B and C; P<0.05 and P<0.01). The expression levels of Wnt4 and β-catenin remained relatively stable in the miR-29c inhibition group, but were increased in the control cells during the course of P19 cell differentiation. These results suggested that the expression levels of Wnt4 are downregulated during early differentiation, and that maintenance of the elevated protein levels of WNT4 may interfere with normal differentiation. The expression levels of Wnt4 were modulated by miR-29c, and were involved in the regulation of P19 cell differentiation into cardiomyocytes.
Discussion
CHD is the most common type of birth defect and increases the burden on families and society, economically, emotionally and psychologically. Although it is difficult to discern which of the multitude of factors contributing to CHD have the most marked effects, for example genetic or environmental, it is apparent that disentangling genetic factors may be beneficial. An improved understanding of cardiac development may assist in elucidating the etiology of CHD and, thus, afford therapeutic or remedial interventions. In this regard, the EC cell culture system provides a suitable in vitro model system for the investigation of cardiac cell differentiation and development: EC cells are readily maintained in culture and are easily amenable to exogenous gene integration and genetic manipulation studies (28). Murine P19 EC cells have long been used in cardiomyocyte differentiation investigations due to their capacity to differentiate into beating cardiomyocytes under the induction of agents, including DMSO (10). In the past few decades, miRNAs have attracted substantial interest, and have prompted significant efforts as a consequence of their now-recognized roles in regulating heart development (25). Our previous study (15) found that miR-29c is upregulated during the gestation of a fetus with a CHD and, in the present study, the functions of miR-29c in cardiac development were further examined. In gain-and loss-of-function studies involving specific miRNAs, loss-of-function approaches appear superior as they reveal functions at physiological miRNA levels. By contrast, the delivery of exogenous miRNAs to biological systems can result in the repression of non-physiological target mRNAs, as miRNA-target interaction is typically concentration-dependent (29). To investigate the effect of miRNA loss-of-function, several approaches have been described in the literature, among which the miRNA 'sponge' approach was selected in the present study to inhibit miR-29c. Natural miRNA 'sponges' contain multiple high-affinity MBSs and can, thus, efficiently sequester specific miRNAs, thereby mini-mizing their interaction with endogenous mRNA targets, and establishing an endogenous regulatory mechanism (30,31).
In the present study, it was found that miR-29c inhibition promoted the proliferation, inhibited the apoptosis, and suppressed the differentiation of murine P19 cells into functional cardiomyocytes. Furthermore, Wnt4 mRNA was revealed as a target of miR-29c, thereby regulating the WNT/β-catenin signaling pathway and modulating cardiac development.
Cellular proliferation, for the purposes of cell renewal and differentiation into specific cell types, are two intrinsic features of EC cells. In this regard, the appropriate regulation of proliferation and timely differentiation are important for P19 EC cells to successfully differentiate into cardiomyocytes. The results of the present study suggested that miR-29c inhibition promoted the self-renewal of P19 cells, but inhibited the subsequent differentiation. The results of early studies of WNT signaling interactions have been clouded. Activation of the WNT/β-catenin pathway in chicken and frog embryos was initially reported to suppress cardiac cell specification (32), however, the opposite has been demonstrated in cultured ES cells (33). In addition, Ueno et al demonstrated a biphasic role for WNT/β-catenin signaling in cardiac specification, promoting cardiac differentiation prior to gastrulation and inhibition of heart formation during gastrulation, in vivo and in vitro (34). Qyang et al (35) reported that the WNT/β-catenin pathway promotes the renewal and expansion of Isl1(+) cardiovascular progenitors, but inhibits their differentiation into cardiomyocytes. Based on these findings, the present study hypothesized that inhibition of miR-29c leads to the overexpression of WNT4, which stimulates the canonical WNT/β-catenin pathway, promoting the proliferation (renewal) of P19 cells, but suppressing their differentiation into cardiomyocytes. The overwhelming majority of biological processes are likely to be regulated by more than one signaling pathway, in order for homeostatic stability to be maintained. Notch signaling, for example, is also involved in cardiac development, which is crucial and at least as important as those of the WNT/β-catenin pathway (36). However, the Notch and WNT signaling pathways can exert paradoxical effects on cell fate decisions, with each pathway promoting alternate and opposing outcomes (37). Studies of mammary stem cells, have demonstrate that WNT signaling promotes the maintenance of stem cell fate, whereas Notch signaling induces lineage commitment and differentiation (38). It may be that a delicate balance of WNT and Notch signaling is a prerequisite for appropriate and coordinated cardiac differentiation, if comparable contrary mechanisms exist during normal heart development. This requires further investigation.
The regulation of cell fate, including apoptosis, is crucial for embryonic development. Cardiac morphogenesis is a complex process, during which the nascent heart transforms from a single, simple tube into a fully septated heart with four chambers and a separated outflow tract (39). Apoptosis is indispensable in this process, as the morphology of the heart changes progressively, and specific structures emerge and dissipate temporally. Apoptosis is governed by two major pathways, an extrinsic pathway involving ῾death receptors᾿ and an intrinsic pathway involving mitochondria, which is regulated by members of the Bcl-2 gene family (40). Bcl-2 and Bax belong to the Bcl gene family, and have opposing effects on cell fate, with Bcl-2 enhancing cell survival and Bax promoting cell death. The present study revealed that apoptotic rates were significantly reduced in the miR-29c inhibition group, and that the survival-promoting expression levels of Bcl-2 were correspondingly elevated in the miR-29c-inhibited cells, with no difference in the expression levels of Bax.
In conclusion, the present study was a continuation of previous investigations, which examined the functions of miR-29c in cardiac development by inhibiting the expression of miR-29c in the P19 EC cell model, and evaluating the effects on cell proliferation, apoptosis and differentiation. Preliminary investigations of the mechanisms by which miR-29c regulates cardiac development were performed, which revealed that Wnt4 was a regulatory interaction target of miR-29c. Due to the clinical impact of CHD and other serious cardiac disorders, further investigations are warranted to clarify the gene network interactions involving miR-29c in mammalian cardiogenesis, to determine the potential therapeutic benefit.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (grant nos. 81370278 and 81300127). The manuscript was copyedited using a professional English editing service (Elixigen Co., Huntingdon Beach, CA, USA).
References
- 1.Khoshnood B, Lelong N, Houyel L, Thieulin AC, Jouannic JM, Magnier S, Delezoide AL, Magny JF, Rambaud C, Bonnet D, et al. Prevalence, timing of diagnosis and mortality of newborns with congenital heart defects: A population-based study. Heart. 2012;98:1667–1673. doi: 10.1136/heartjnl-2012-302543. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman JIe. The global burden of congenital heart disease. Cardiovasc J Afr. 2013;24:141–145. doi: 10.5830/CVJA-2013-028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin CJ, Lin CY, Chen CH, Zhou B, Chang CP. Partitioning the heart: Mechanisms of cardiac septation and valve development. Development. 2012;139:3277–3299. doi: 10.1242/dev.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van der Bom T, Zomer AC, Zwinderman AH, Meijboom FJ, Bouma BJ, Mulder BJ. The changing epidemiology of congenital heart disease. Nat Rev Cardiol. 2011;8:50–60. doi: 10.1038/nrcardio.2010.166. [DOI] [PubMed] [Google Scholar]
- 5.Brennan P, Young ID. Congenital heart malformations: Aetiology and associations. Semin Neonatol. 2001;6:17–25. doi: 10.1053/siny.2000.0032. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Xiang R, Lei H, Chen M, Li Q, Sun H, Ai J, Chen T, Wang H, Fang Y, Zhou Q. The miR-17-92 cluster regulates FOG-2 expression and inhibits proliferation of mouse embryonic cardiomyocytes. Braz J Med Biol Res. 2012;45:131–138. doi: 10.1590/S0100-879X2012007500007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sluijter JP, van Mil A, van Vliet P, Metz CH, Liu J, Doevendans PA, Goumans MJ. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol. 2010;30:859–868. doi: 10.1161/ATVBAHA.109.197434. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics. 2012;10:246–253. doi: 10.1016/j.gpb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Wang DZ. microRNAs in cardiovascular development. J Mol Cell Cardiol. 2012;52:949–957. doi: 10.1016/j.yjmcc.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 13.Liu, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 15.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meder B, Katus HA, Rottbauer W. Right into the heart of microRNA-133a. Genes Dev. 2008;22:3227–3231. doi: 10.1101/gad.1753508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu S, Cao L, Zhu J, Kong L, Jin J, Qian L, Zhu C, Hu X, Li M, Guo X, et al. Identification of maternal serum microRNAs as novel non-invasive biomarkers for prenatal detection of fetal congenital heart defects. Clin Chim Acta. 2013;424:66–72. doi: 10.1016/j.cca.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen T, Kuo C, Nicholl MB, et al. Low miR-29c levels associated with TRG hypermethylation and melanoma progression. RNA Biology. 2011;8:357. [Google Scholar]
- 19.Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG, Jia JH. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun. 2011;411:586–592. doi: 10.1016/j.bbrc.2011.06.191. [DOI] [PubMed] [Google Scholar]
- 20.Zhou K, Yu Z, Yi S, Li Z, An G, Zou D, Qi J, Zhao Y, Qiu L. miR-29c down-regulation is associated with disease aggressiveness and poor survival in Chinese patients with chronic lymphocytic leukemia. Leuk Lymphoma. 2014;55:1544–1550. doi: 10.3109/10428194.2013.855308. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Li Y, Sun J, Wang Q, Sun C, Yan Y, Yu L, Cheng D, An T, Shi C, et al. Tumor-suppressive effects of miR-29c on gliomas. Neuroreport. 2013;24:637–645. doi: 10.1097/WNR.0b013e3283630126. [DOI] [PubMed] [Google Scholar]
- 22.Xu F, Zhang Q, Cheng W, Zhang Z, Wang J, Ge J. Effect of miR-29b-1* and miR-29c knockdown on cell growth of the bladder cancer cell line T24. J Int Med Res. 2013;41:1803–1810. doi: 10.1177/0300060513505266. [DOI] [PubMed] [Google Scholar]
- 23.Zeng X, Xiang J, Wu M, Xiong W, Tang H, Deng M, Li X, Liao Q, Su B, Luo Z. Circulating miR-17, miR-20a, miR-29c and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. Plos One. 2012;7:e46367. doi: 10.1371/journal.pone.0046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuo M, Nakada C, Tsukamoto Y, Noguchi T, Uchida T, Hijiya N, Matsuura K, Moriyama M. MiR-29c is down-regulated in gastric carcinomas and regulates cell proliferation by targeting RCC2. Mol Cancer. 2013;12:15. doi: 10.1186/1476-4598-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kluiver J, Slezak-Prochazka I, Smigielska-Czepiel K, Halsema N, Kroesen BJ, van den Berg A. Generation of miRNA sponge constructs. Methods. 2012;58:113–117. doi: 10.1016/j.ymeth.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 26.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Heyden MA, Defize LH. Twenty one years of P19 cells: What an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc Res. 2003;58:292–302. doi: 10.1016/S0008-6363(02)00771-X. [DOI] [PubMed] [Google Scholar]
- 29.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev. 2015;81:117–127. doi: 10.1016/j.addr.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Ebert MS, Sharp PA. MicroRNA sponges: Progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisenberg LM, Eisenberg CA. Wnt signal transduction and the formation of the myocardium. Dev Biol. 2006;293:305–315. doi: 10.1016/j.ydbio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 34.Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, Murry CE. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 36.de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22:244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caliceti C, Nigro P, Rizzo P, Ferrari R. ROS, Notch and Wnt signaling pathways: Crosstalk between three major regulators of cardiovascular biology. Biomed Res Int. 2014;2014:318714. doi: 10.1155/2014/318714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gittenberger-de Groot AC, Bartelings MM, Poelmann RE, Haak MC, Jongbloed MR. Embryology of the heart and its impact on understanding fetal and neonatal heart disease. Semin Fetal Neonatal Med. 2013;18:237–244. doi: 10.1016/j.siny.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Biala AK, Kirshenbaum LA. The interplay between cell death signaling pathways in the heart. Trends Cardiovasc Med. 2014;24:325–331. doi: 10.1016/j.tcm.2014.08.002. [DOI] [PubMed] [Google Scholar]