Abstract
The aim of the present study was to investigate the genetic etiology of patients with nonsyndromic hearing impairment through gene analysis, and provide accurate genetic counseling and prenatal diagnosis for deaf patients and families with deaf children. Previous molecular etiological studies have demonstrated that the most common molecular changes in Chinese patients with nonsyndromic hearing loss (NSHL) involved gap junction protein β 2, solute carrier family 26, member 4 (SLC26A4), and mitochondrial DNA 12S rRNA. A total of 117 unrelated NSHL patients were included. Mutation screening was performed by Sanger sequencing in GJB2, 12S rRNA, and the hot-spot regions of SLC26A4. In addition, patients with a single mutation of SLC26A4 in the hot-spot regions underwent complete exon sequencing to identify a mutation in the other allele. A total of 36 of the 117 deaf patients were confirmed to have two pathogenic mutations, which included 4 deaf couples, husband or wife in 11 deaf couples and 17 deaf individuals. In addition, prenatal diagnoses was performed in 7 pregnant women at 18–21 weeks gestation who had previously given birth to a deaf child, and the results showed that two fetal genotypes were the same as the proband's genotypes, four fetuses carried one pathogenic gene from their parents, and one fetus was identified to have no mutations. Taken together, the genetic testing of deaf patients can provide reasonable guidance to deaf patients and families with deaf children.
Keywords: nonsyndromic hearing impairment, gap junction protein β 2, mitochondrial DNA, solute carrier family 26, member 4, gene diagnosis, genetic counseling
Introduction
Hearing loss is one of the most common birth defects. According to the World Health Organization, 360,000,000 individuals worldwide have disabling hearing loss (www.who.int/), including 328,000,000 adults and 32,000,000 children. Deafness is a multi-factorial disease, predominantly induced by genetic or environmental causes or their interactions (1). It is suggested that ~50% of deafness is hereditary, which includes syndromic hearing loss and nonsyndromic hearing loss (NSHL), which comprises ~70% of cases of hereditary deafness (2). Of NSHL, almost 80% of cases is due to autosomal recessive inheritance, and it has been demonstrated that autosomal recessive NSHL is associated with >50 genes (3) (http://hereditaryhearingloss.org/), including the gap junction protein β 2 (GJB2), solute carrier family 26, member 4 (SLC26A4) and mitochondrial DNA 12S rRNA genes. In NSHL, ~50% of deafness is caused by mutation of the GJB2 gene (4,5), and in the GJB2 gene, >90 mutations have been identified. GJB2 35delC is the most common mutation in the European population (6), while 235delC is common in the Asian population (7). Mutation of the SLC26A4 gene is the second leading cause of NSHL, which contains 21 exons and has been identified to contain ~200 mutations, with IVS7-2A>G and 2168A>G being the two most common mutations in Asian population (8–10). The 1,555A>G mutation in the mitochondrial DNA 12S rRNA gene was the first identified genetic cause for aminoglycoside-induced NSHL (11). Previous molecular etiological studies have shown that GJB2, SLC26A4 and 12S rRNA are three common deafness-associated genes in NSHL among the Chinese population (12–15). Therefore, mutational screening in these three genes may be effective and beneficial for diagnosing deafness in this population.
It is common for deaf couples to have children together; certain couples may have the same disease-causing genetic mutation. Thus, confirmation of the pathogenic genotype of deaf patients may assist in determining the genotype of their children. In families with children affected by NSHL, it is important to confirm the genotypes of the couples and their deaf children, in order to determine the risk in their next pregnancy. In the present study, the GJB2 gene, SLC26A4 gene and 12S rRNA were examined by Sanger sequencing in 117 deaf patients, including 39 deaf couples and 39 deaf individuals. Furthermore, seven pregnant women were offered prenatal diagnosis via amniocentesis at 18–21 weeks gestation. The aim of the present study was to analyze the genetics of deafness and provide appropriate genetic counseling about hearing loss to our patients.
Materials and methods
Subjects
In the present study 117 deaf patients, aged between 3 months and 35 years were recruited. These patients were diagnosed with nonsyndromic neurosensory deafness at Nanjing Maternity and Child Health Care Hospital (Nanjing, China) between 2009 and 2013, with hearing loss of >70 dB. Any patients with systemic disease, dysgnosia, syndromic deafness or a history of meningitis, otitis media or trauma of the ear were excluded from the investigations. The present study was performed in accordance with the declaration of Helsinki, and was performed following approval from the ethics committee of Nanjing Maternity and Child Health Care Hospital (Nanjing Medical University, Nanjing, China). Written informed consent was obtained from the patient.
DNA extraction
To obtain DNA, 2 ml peripheral blood was collected and then placed into tubes with ethylene diamine tetraacetic acid (EDTA; Greiner Bio-One International GmbH). For the pregnant women offered prenatal diagnosis, 10 ml amniotic fluid was collected at 18–21 weeks gestation, and DNA was extracted using a DNA extraction kit (Xiamen Zhishan Biological Company, Xiamen, China), according to manufacturer's protocol, followed by storage at −20°C.
Primer design
Primers were designed, according to those previously described (13–15), which were used to perform polymerase chain reaction (PCR) amplification and sequencing. The primer sequences are listed in Table I, and were synthesized by Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The coding sequences of GJB2 were amplified, following which GJB2 genetic mutations were detected by Sanger sequencing (13,16,17). Furthermore, mitochondrial DNA from 1,428 to 1,890 were also amplified and 1,555A>G mutation of the mitochondrial 12S rRNA gene were detected (15,18,19). The hot-spot regions of the SLC26A4 gene, including exons 5, 7, 8, 10, 17 and 19, were also amplified, of which exons 7 and 8 were amplified in one amplicon. Therefore, five fragments were amplified to cover the six exons and flanking sequence, following which the SLC26A4 gene mutation was detected. For patients with one allelic variant in the hot-spot regions, the other exons were sequenced one by one, until two mutant alleles were identified, as previously described (14,20,21).
Table I.
PCR primers for the GJB2 gene, SLC26A4 gene and mitochondrial DNA 12S rRNA.
| Gene | Exon | PCR primer (5′–3′) | Product length (bp) |
|---|---|---|---|
| GJB2 | 2 | F-TTGGTGTTTGCTCAGGAAGA | 872 |
| R-GGTTGCCTCATCCCTCTCAT | |||
| SLC26A4 | 5 | F-CCTATGCAGACACATTGAACATTTG | 442 |
| R-TGAGCCTTAATAAGTGGGGTCTTG | |||
| 7+8 | F-CATGGTTTTTCATGTGGGAAGATTC | 502 | |
| R-AGACTGACTTACTGACTTAATGT | |||
| 10 | F-AAATACTCAGCGAAGGTCTTGC | 250 | |
| R-CGAGCCTTCCTCTGTTGC | |||
| 17 | F-CCAAGGAACAGTGTGTAGGTC | 423 | |
| R-CCCATGTATTTGCCCTGTTGC | |||
| 19 | F-TCACTTGAACTTGGGACGCGGA | 383 | |
| R-CAACAGCTAGACTAGACTTGTG | |||
| Mitochondrial 1 allelic | 1,555 | F-GCAGTAAACTAAGAGTAGAGT | 463 |
| DNA 12S rRNA | R-GGCTCTCCTTGCAAAGTTAT |
GJB2, gap junction protein β 2; SLC26A4, solute carrier family 26, member 4; PCR, polymerase chain reaction; F, forward; R, reverse.
PCR amplification
The GoTaq® PCR system was purchased from Promega Corporation (Madison, WI, USA). The total volume of the PCR reaction mixture was 50 µl, including 10 µl of 5X Green GoTaq® Reaction Buffer, 2 µl of 10 mmol/l dNTP, 1 µl of each (10 µmol/l) primer, 0.25 µl of 5 U/µl GoTaq® DNA polymerase, 5 µl of the 20 ng/µl DNA template and 30.75 µl distilled water. The amplification reaction conditions for exons 7 and 8 of the SLC26A4 gene were as follows: 95°C for 5 min; followed by 35 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec, and a final step of 72°C for 7 min. For the remaining six exons, the reaction conditions were as follows: 95°C for 5 min; followed by 35 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 1 min, and a final step of 72°C for 7 min. The amplified products were stored at 4°C.
Sequence analysis
Following PCR, agarose gel electrophoresis was performed, and the products were purified according to TIANgel Midi DNA Purification kits (Tiangen Biotech Co., Ltd., Beijing, China), following which the purified products were sequenced. Sequencing kits were obtained from Life Technologies (Grand Island, NY, USA). The reaction system contained 1 µl Bigdye, 3 µl buffer, 2 µl primers and 5 µl purified products in a total volume of 20 µl. The reaction conditions were as follows: 95°C for 1 min; followed by 35 cycles of 95°C for 10 sec, 50°C for 5 sec and 60°C for 4 min. The amplified products were stored at 4°C. The sequences of the primers are listed in Table I. In addition, the sequencing products were purified with 100 mM EDTA solution and alcohol solution. Following purification, the products were dried and were dissolved in 100% HiDi Formamide (Gibco; Thermo Fisher Scientific, Inc.). Subsequently, all products were sequenced (forward and reverse) using an ABI3130 analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.). The sequence results were aligned with standard sequences using SeqBuilder procedures on Lasergene software, version 7.1 (DNASTAR, Inc., Madison, WI, USA). The GJB2, SLC26A4 and mitochondrial DNA 12S rRNA genetic standards were NM_004004 [GenBank, National Center for Biotechnology Information (NCBI), www.ncbi.nlm.nih.gov/nuccore/NM_004004], ENSG00000091137 (Ensembl, European Bioinformatics Institute, www.ensembl.org/Homo_sapiens/Gene/Sequence?g=ENSG00000091137) and the Cambridge reference sequence, NC_001807 (GenBank, NCBI, www.ncbi.nlm.nih.gov/nuccore/NC_001807.4), respectively.
Results
Genetic sequencing of 117 deaf patients
In the present study, genetic detection was performed in 117 deaf patients, of which 36 cases were confirmed to carry two pathogenic mutations. The positive rate was 30.77% (Table II), including 19 cases with GJB2 gene mutations (16.24%), 12 cases with SLC26A4 gene mutations (10.26%) and five cases with the 1,555A>G allelic mutation of mitochondrial DNA 12S rRNA (4.27%). In total, four GJB2 gene mutations (Fig. 1), and eight SLC26A4 gene mutations (Fig. 2) were identified.
Table II.
Mutant alleles of deafness-associated genes in 36 deaf patients.
| Gene | Mutant allele | Patients (n) | Total (n) | Rate (%) |
|---|---|---|---|---|
| GJB2 | 235delC/235delC | 9 | 19 | 16.24 |
| 235delC/299_300delAT | 5 | |||
| 235delC/176_191del16bp | 2 | |||
| 299_300delAT/299_300delAT | 1 | |||
| 176_191del16bp/176_191del16bp | 1 | |||
| 235delC/511_512insAACG | 1 | |||
| SLC26A4 | IVS7-2A>G/IVS7-2A>G | 4 | 12 | 10.26 |
| IVS7-2A>G/1226 G>A | 1 | |||
| IVS7-2A>G/1174A>T | 1 | |||
| IVS7-2A>G/2027T>A | 1 | |||
| IVS7-2A>G/2168 A>G | 1 | |||
| 589G>A/589G>A | 1 | |||
| 1174A>T/1975 G>C | 1 | |||
| 1226G>A/1226G>A | 1 | |||
| 1079C>T/2168A>G | 1 | |||
| Mitochondrial DNA | 1555A>G | 5 | 5 | 4.27 |
| 12S rRNA | ||||
| Total | 36 | 30.77 |
GJB2, gap junction protein β 2; SLC26A4, solute carrier family 26, member 4.
Figure 1.
GJB2 mutations, detected using direct DNA sequencing. The black arrows indicate single nucleotide mutations. (A) A heterozygous mutation of c.235delC. (B) A heterozygous mutation of c.299_300delAT. (C) A heterozygous mutation of c.176_191del16bp. (D) A heterozygous mutation of c.511_512insAACG.
Figure 2.
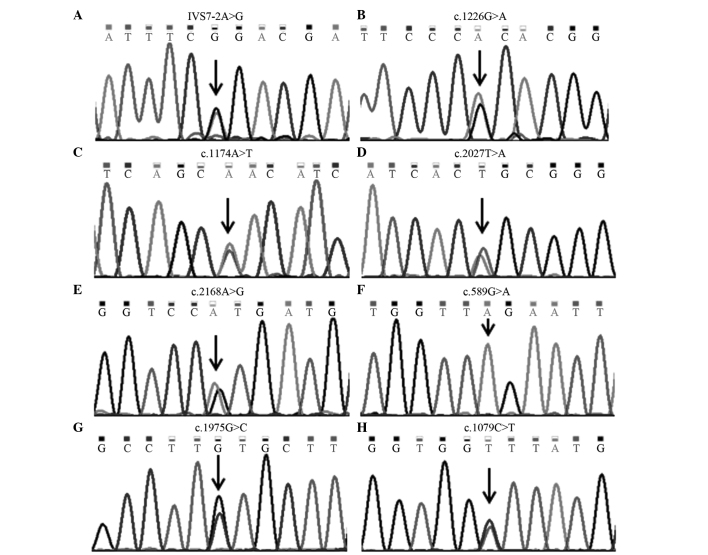
SLC26A4 mutations, detected using direct DNA sequencing. The black arrows indicate single nucleotide mutations. (A) A heterozygous mutation of IVS7-2A>G. (B) A heterozygous mutation of c.1226G>A. (C) A heterozygous mutation of c.1174A>T. (D) A heterozygous mutation of c.2027T>A. (E) A heterozygous mutation of c.2168A>G. (F) A homozygous mutation of c.589G>A. (G) A heterozygous mutation of c.1975G>C. (H) A heterozygous mutation of c.1079C>T.
Genetic sequencing of 39 deaf couples
The 117 deaf patients recruited in the present study included 39 pairs of deaf couples, of which four couples were confirmed to have two deafness-causing mutations (10.26%). Among these, deafness in one couple was caused by SLC26A4 gene mutations, whose children will be deaf. In one deaf couple, deafness was caused by GJB2 and SLC26A4 gene mutations, respectively. The present study hypothesized that their children will have a low risk of deafness with no need for prenatal diagnosis as the parents pathogenic mutations are in different genes, the child will be heterozygous for these two genes which are associated with autosomal recessive hearing loss. Furthermore, in one couple, the husband had two GJB2 gene mutations, whereas the wife had 1,555A>G mutation of mitochondrial DNA 12S rRNA. Their children would carry a GJB2 gene heterozygous mutation and mitochondrial DNA mutation, and may be advised to avoid taking aminoglycoside antibiotics as the mutation 1,555A>G is important in aminoglycoside-induced nonsyndromic deafness (18). In addition, the husband of one couple had 1,555A>G mutation of mitochondrial DNA 12S rRNA, whereas the wife carried two GJB2 gene mutation. The present study hypothesized that their children would be at low risk of deafness due to their child carrying a heterozygous GJB2 gene mutation. The results of genetic sequencing indicated that, in 11 couples, only one of each couple (28.21%) was confirmed to have deafness-causing mutations, which indicated that their offspring were at low risk of deafness. Table III shows the results of genetic analyses.
Table III.
Genetic sequencing of 15 couples.
| Couple | Gender/age (years) | GJB2 mutation | SLC26A4 mutation | 12S rRNA mutation | |
|---|---|---|---|---|---|
| 1 | Female/20 | 299_300delAT/299_300delAT | – | – | |
| Male/28 | – | – | – | ||
| 2 | Female/29 | 235delC/511_512insAACG | – | – | |
| Male/30 | – | – | – | ||
| 3 | Female/22 | – | – | – | |
| Male/29 | – | – | 1555A>G | ||
| 4 | Female/28 | – | – | 1555A>G | |
| Male/29 | 235delC/235delC | – | – | ||
| 5 | Female/24 | – | 1226G>A/1226G>A | – | |
| Male/25 | – | IVS7-2A>G/2027T>A | – | ||
| 6 | Female/24 | – | – | 1555A>G | |
| Male/24 | – | – | – | ||
| 7 | Female/28 | 235delC/235delC | – | – | |
| Male/28 | – | – | – | ||
| 8 | Female/23 | 235delC/235delC | – | – | |
| Male/24 | – | – | – | ||
| 9 | Female/26 | – | – | – | |
| Male/27 | – | IVS7-2A>G/1226 G>A | – | ||
| 10 | Female/28 | – | – | – | |
| Male/30 | – | 1555A>G | |||
| 11 | Female/25 | 235delC/176_191del16bp | – | – | |
| Male/25 | – | – | – | ||
| 12 | Female/25 | 235delC/235delC | – | – | |
| Male/26 | – | – | – | ||
| 13 | Female/26 | – | – | ||
| Male/31 | – | 589G>A/589G>A | – | ||
| 14 | Female/28 | 235delC/299_300delAT | – | – | |
| Male/29 | – | IVS7-2A>G/IVS7-2A>G | – | ||
| 15 | Female/29 | 235delC/176_191del16bp | – | – | |
| Male/34 | – | – | 1555A>G | ||
GJB2, gap junction protein β 2; SLC26A4, solute carrier family 26, member 4.
Genetic sequencing of families with deaf children and prenatal diagnosis
In the 39 deaf patients, 17 cases were identified with GJB2 gene or SLC26A4 gene mutations. Subsequently, their parents received genetic analysis to confirm whether they were carriers (Table IV). In 7/17 families, prenatal diagnosis was performed during their next pregnancy. The results showed that two fetuses had the same genotype as the proband. These two families decided to terminate the pregnancy. No mutations were identified in one fetus, and the genotypes of the other four fetuses were the same as one parent. The result of clinical follow-up demonstrated that they had normal hearing following birth.
Table IV.
Genetic sequencing of 17 couples with deaf children.
| Couple | Mutation
|
Follow-up | |||
|---|---|---|---|---|---|
| Child (proband) | Father | Mother | Fetus | ||
| 1a | 235delC/299_300delAT | 299_300delAT | 235delC | 235delC/299_300delAT | Ending pregnancy |
| 2a | 235delC/299_300delAT | 235delC | 299_300delAT | 299_300delAT/WT | Normal hearing |
| 3a | 235delC/235delC | 235delC | 235delC | 235delC/WT | Normal hearing |
| 4a | 176_191del16bp/176_191del16bp | 176_191del16bp | 176_191del16bp | 176_191del16bp/WT | Normal hearing |
| 5b | IVS7-2A>G/IVS7-2A>G | IVS7-2A>G | IVS7-2A>G | WT/WT | Normal hearing |
| 6b | IVS7-2A>G/IVS7-2A>G | IVS7-2A>G | IVS7-2A>G | IVS7-2A>G/IVS7-2A>G | Ending pregnancy |
| 7b | IVS7-2A>G/2168A>G | IVS7-2A>G | 2168A>G | IVS7-2A>G/WT | Normal hearing |
| 8a | 235delC/299_300delAT | 299_300delAT | 235delC | ||
| 9a | 235delC/299_300delAT | 299_300delAT | 235delC | ||
| 10a | 235delC/235delC | 235delC | 235delC | ||
| 11a | 235delC/235delC | 235delC | 235delC | ||
| 12a | 235delC/235delC | 235delC | 235delC | ||
| 13a | 235delC/235delC | 235delC | 235delC | ||
| 14b | IVS7-2A>G/1174A>T | 1174A>T | IVS7-2A>G | ||
| 15b | IVS7-2A>G/IVS7-2A>G | IVS7-2A>G | IVS7-2A>G | ||
| 16b | 1174A>T/1975G>C | 1174A>T | 1975G>C | ||
| 17b | 1079C>T/2168A>G | 2168A>G | 1079C>T | ||
GJB2 gene mutation;
SLC26A4 gene mutation; WT, wile-type allele.
Discussion
The provision of appropriate genetic counseling for patients with hearing loss remains a challenge in clinical practice. Deafness exhibits marked genetic heterogeneity and phenotypic variability (1,2). At present, hundreds of genes have been identified to cause hereditary hearing loss, including the GJB2 gene, SLC26A4 gene and the mitochondrial DNA 12S rRNA 1,555A>G mutation, which have been confirmed to be closely associated with NSHL (1,16–23).
The present study used Sanger sequencing to analyze mutations of GJB2, SLC26A4 and the mitochondrial DNA 12S rRNA 1,555A>G in 117 patients with NSHL. The results revealed that 36 of the 117 patients (30.77%) carried two deafness-causing mutations, including the GJB2 gene mutations (16.24%), SLC26A4 gene mutations (10.26%) and mitochondrial DNA 12SrRNA 1555 locus mutation (4.27%). The 235delC mutation in the GJB2 gene mutation has been identified at the highest rate in patients in the present study and is also the most common in the Asian population (24). The present study identified the mitochondrial DNA 12S rRNA 1,555A>G locus mutation in ~4.27% of the patients, which follows the pattern of maternal inheritance (11,18). It may be the situation that there are healthy individuals with normal hearing carrying the 1,555A>G mutation. Thus, once this mutation is identified, the family members can avoid the use of aminoglycoside antibiotics, preventing drug-induced deafness (25). The SLC26A4 gene contains 21 exons. The present study selected certain hot-spot regions, SLC26A4 exons 5, 7, 8, 10, 17 and 19, for sequencing. The IVS7-2A>G mutation is the most common mutation in the SLC26A4 gene (10). For patients with one allelic variant in the hot-spot regions, the remaining exons were sequenced one by one until two mutant alleles had been identified. The results found the second mutant allele, the missense mutation 1079C>T, in exon 9 of one patient. In addition, 11 patients carrying SLC26A4 mutations were detected using hot-spot region screening, whereas only one case was identified by complete exon sequencing. According to the SLC26A4 gene mutation spectrum, the Sanger sequencing method detecting hot-spot regions of the SLC26A4 gene has been found to have clinical value in identifying mutations associated with deafness (14).
In the present study, there were 39 deaf couples. Traditionally, it is thought that there is a high possibility of deaf couples having hearing-impaired offspring, however, the majority of couples with hearing impairment have deafness induced by environmental causes or due to different pathogenic genes, thus, the risk of their children being deaf is low (12). Additionally, in couples with deafness due to GJB2 and SLC26A4 gene mutation, their children were predicted to be carriers with normal hearing. In one couple, in which the mother had hearing loss caused by the mitochondrial DNA, 12S rRNA mutation, their children were predicted to be a carrier with normal hearing, in the absence of use with ototoxic drugs. However, they cannot be a carrier of the mutational gene if the father has hearing loss caused by the mitochondrial DNA 12S rRNA mutation, the children would be predicted to have normal hearing. Therefore, genetic analysis of deafness-associated genes can reduce anxiety in deaf couples, and determine the risk in their next pregnancy.
In the present study, 3 deafness-associated genes in 39 deaf patients and their parents were sequenced, and the results revealed 17 cases carried two confirmed pathogenic mutations, whose parents were both carriers of the same genetic mutation. In addition, seven pregnant women were offered prenatal diagnosis. The genetic analysis revealed that the genotypes of two fetuses were the same as the probands. Of the remaining fetuses, four were carriers, and one was identified without mutations in the genes analyzed.
In conclusion, the gene sequencing performed in the present study was convenient and reliable for the genetic testing of NSHL. In addition, the method could be applied in clinical laboratories due to its simple process and low cost, providing easier genetic assessment. In addition, results from the present study provided accurate information for genetic counseling and prenatal diagnosis.
Acknowledgments
The present study was supported by the Program for Leading Talents and Medical Innovative Team of Jiangsu Province (grant no. LJ201109), the Special Funds of Clinical Medicine Science and Technology of Jiangsu Province (grant no. BL2012039), the Science and Technology Research and Development Program of Health Bureau of Nanjing City (grant no. YKK11059) and the Training Program for Young Talents of Nanjing City.
References
- 1.Shearer AE, Smith RJ. Genetics: Advances in genetic testing for deafness. Curr Opin Pediatr. 2012;24:679–686. doi: 10.1097/MOP.0b013e3283588f5e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cryns K, Van Camp G. Deafness genes and their diagnostic applications. Audiol Neurootol. 2004;9:2–22. doi: 10.1159/000074183. [DOI] [PubMed] [Google Scholar]
- 3.Hoefsloot LH, Feenstra I, Kunst HP, Kremer H. Genotype phenotype correlations for hearing impairment: Approaches to management. Clin Genet. 2014;85:514–523. doi: 10.1111/cge.12339. [DOI] [PubMed] [Google Scholar]
- 4.Rabionet R, Gasparini P, Estivill X. Molecular genetics of hearing impairment due to mutations in gap junction genes encoding beta connexins. Hum Mutat. 2000;16:190–202. doi: 10.1002/1098-1004(200009)16:3<190::AID-HUMU2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 5.Kenneson A, Van Naarden Braun K, Boyle C. GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: A HuGE review. Genet Med. 2002;4:258–274. doi: 10.1097/00125817-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Snoeckx RL, Huygen PL, Feldmann D, Marlin S, Denoyelle F, Waligora J, Mueller-Malesinska M, Pollak A, Ploski R, Murgia A, et al. GJB2 mutations and degree of hearing loss: A multicenter study. Am J Hum Genet. 2005;77:945–957. doi: 10.1086/497996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohtsuka A, Yuge I, Kimura S, Namba A, Abe S, Van Laer L, Van Camp G, Usami S. GJB2 deafness gene shows a specific spectrum of mutations in Japan, including a frequent founder mutation. Hum Genet. 2003;112:329–333. doi: 10.1007/s00439-002-0889-x. [DOI] [PubMed] [Google Scholar]
- 8.Hilgert N, Smith RJ, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: Which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681:189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang QJ, Zhao YL, Rao SQ, Guo YF, Yuan H, Zong L, Guan J, Xu BC, Wang DY, Han MK, et al. A distinct spectrum of SLC26A4 mutations in patients with enlarged vestibular aqueduct in China. Clin Genet. 2007;72:245–254. doi: 10.1111/j.1399-0004.2007.00862.x. [DOI] [PubMed] [Google Scholar]
- 10.Du W, Guo Y, Wang C, Wang Y, Liu X. A systematic review and meta-analysis of common mutations of SLC26A4 gene in Asian populations. Int J Pediatr Otorhinolaryngol. 2013;77:1670–1676. doi: 10.1016/j.ijporl.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 11.Kokotas H, Petersen MB, Willems PJ. Mitochondrial deafness. Clin Genet. 2007;71:379–391. doi: 10.1111/j.1399-0004.2007.00800.x. [DOI] [PubMed] [Google Scholar]
- 12.Yuan Y, You Y, Huang D, Cui J, Wang Y, Wang Q, Yu F, Kang D, Yuan H, Han D, Dai P. Comprehensive molecular etiology analysis of nonsyndromic hearing impairment from typical areas in China. J Transl Med. 2009;7:79. doi: 10.1186/1479-5876-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai P, Yu F, Han B, Liu X, Wang G, Li Q, Yuan Y, Liu X, Huang D, Kang D, et al. GJB2 mutation spectrum in 2,063 Chinese patients with nonsyndromic hearing impairment. J Transl Med. 2009;7:26. doi: 10.1186/1479-5876-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Y, Guo W, Tang J, Zhang G, Wang G, Han M, Zhang X, Yang S, He DZ, Dai P. Molecular epidemiology and functional assessment of novel allelic variants of SLC26A4 in non-syndromic hearing loss patients with enlarged vestibular aqueduct in China. PLoS One. 2012;7:e49984. doi: 10.1371/journal.pone.0049984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Li R, Chen J, Liao Z, Zhu Y, Qian Y, Xiong S, Heman-Ackah S, Wu J, Choo DI, Guan MX. Mutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside-induced and non-syndromic hearing loss. Hum Genet. 2005;117:9–15. doi: 10.1007/s00439-005-1276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estivill X, Fortina P, Surrey S, Rabionet R, Melchionda S, D'Agruma L, Mansfield E, Rappaport E, Govea N, Milà M, et al. Connexin-26 mutations in sporadic and inherited sensorineural deafness. Lancet. 1998;351:394–398. doi: 10.1016/S0140-6736(97)11124-2. [DOI] [PubMed] [Google Scholar]
- 17.Morell RJ, Kim HJ, Hood LJ, Goforth L, Friderici K, Fisher R, Van Camp G, Berlin CI, Oddoux C, Ostrer H, et al. Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N Engl J Med. 1998;339:1500–1505. doi: 10.1056/NEJM199811193392103. [DOI] [PubMed] [Google Scholar]
- 18.Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, Bai Y, Young WY, Guan MX. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pryor SP, Madeo AC, Reynolds JC, Sarlis NJ, Arnos KS, Nance WE, Yang Y, Zalewski CK, Brewer CC, Butman JA, Griffith AJ. SLC26A4/PDS genotype-phenotype correlation in hearing loss with enlargement of the vestibular aqueduct (EVA): Evidence that Pendred syndrome and non-syndromic EVA are distinct clinical and genetic entities. J Med Genet. 2005;42:159–165. doi: 10.1136/jmg.2004.024208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Zhu QW, Yuan YY, Huang SS, Han DY, Huang DL, Dai P. Identification of SLC26A4 c.919-2A>G compound heterozygosity in hearing-impaired patients to improve genetic counseling. J Transl Med. 2012;10:225. doi: 10.1186/1479-5876-10-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun SC, Liu YX, Peng YS, Li HF, Xie CY. Analysis of GJB2 gene coding sequence in patients with nonsyndromic hearing loss. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2011;28:409–413. doi: 10.3760/cma.j.issn.1003-9406.2011.04.011. In Chinese. [DOI] [PubMed] [Google Scholar]
- 23.Dai P, Stewart AK, Chebib F, Hsu A, Rozenfeld J, Huang D, Kang D, Lip V, Fang H, Shao H, et al. Distinct and novel SLC26A4/Pendrin mutations in Chinese and U.S. patients with nonsyndromic hearing loss. Physiol Genomics. 2009;38:281–290. doi: 10.1152/physiolgenomics.00047.2009. [DOI] [PubMed] [Google Scholar]
- 24.Chun JY, Shin SK, Min KT, Cho W, Kim J, Kim SO, Hong SP. Performance evaluation of the TheraTyper-GJB2 assay for detection of GJB2 gene mutations. J Mol Diagn. 2014;16:573–583. doi: 10.1016/j.jmoldx.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Kent A, Turner MA, Sharland M, Heath PT. Aminoglycoside toxicity in neonates: Something to worry about? Expert Rev Anti Infect Ther. 2014;12:319–331. doi: 10.1586/14787210.2014.878648. [DOI] [PubMed] [Google Scholar]