Abstract
Objective
Accumulating evidence links the intestinal microbiota and colorectal carcinogenesis. Fusobacterium nucleatum may promote colorectal tumour growth and inhibit T-cell-mediated immune responses against colorectal tumours. Thus, we hypothesized that the amount of Fusobacterium nucleatum in colorectal carcinoma might be associated with worse clinical outcome.
Design
We utilised molecular pathological epidemiology database of 1,069 rectal and colon cancer cases in the Nurses’ Health Study and the Health Professionals Follow-up Study, and measured Fusobacterium nucleatum DNA in carcinoma tissue. Cox proportional hazards model was used to compute hazard ratio (HR), controlling for potential confounders, including microsatellite instability (MSI, mismatch repair deficiency), CpG island methylator phenotype (CIMP), KRAS, BRAF, and PIK3CA mutations, and LINE-1 hypomethylation (low-level methylation).
Results
Compared to Fusobacterium nucleatum-negative cases, multivariable HRs (95% confidence interval) for colorectal cancer-specific mortality in Fusobacterium nucleatum-low cases and Fusobacterium nucleatum-high cases were 1.25 (0.82 to 1.92) and 1.58 (1.04 to 2.39), respectively (p for trend = 0.020). The amount of Fusobacterium nucleatum was associated with MSI-high (multivariable odds ratio, 5.22; 95% CI, 2.86 to 9.55) independent of CIMP and BRAF mutation status, whereas CIMP and BRAF mutation were associated with Fusobacterium nucleatum only in univariate analyses (p < 0.001) but not in multivariate analysis that adjusted for MSI status.
Conclusions
The amount of Fusobacterium nucleatum DNA in colorectal cancer tissue is associated with shorter survival, and may potentially serve as a prognostic biomarker. Our data may have implications in developing cancer prevention and treatment strategies through targeting gastrointestinal microflora by diet, probiotics, and antibiotics.
Keywords: bacteria, colorectum, gut microbiome, adaptive immunity
INTRODUCTION
More than 100 trillion (1014) microorganisms inhabit the human intestinal tract and play an important role in health and disease conditions, including cancer.1–4 A growing body of evidence suggests a potential link between the microbiota and colorectal carcinogenesis.5–13 Proportions of colorectal cancers with specific molecular features including microsatellite instability (MSI), CpG island methylator phenotype (CIMP)-high, and BRAF mutation have been shown to decrease continuously from ascending colon to rectum, supporting a gradual change in pathogenic influence of intestinal microbiota and luminal contents along the proximal-distal axis.14
Studies have demonstrated an enrichment of Fusobacterium nucleatum in human colorectal adenomas and carcinomas compared to adjacent normal tissue.15–17 Experimental studies have shown that Fusobacterium nucleatum activates the WNT signaling pathway in colorectal carcinoma cells and may promote colorectal tumour growth,18 and that Fusobacterium nucleatum may inhibit T-cell-mediated immune responses against colorectal tumours.16, 19 Consistent with these lines of experimental evidence, a higher amount of tissue Fusobacterium nucleatum DNA has been associated with advanced disease stage6, 7, 20 and a lower density of T-cells in human colorectal carcinoma tissue.21 However, the prognostic significance of Fusobacterium nucleatum DNA in colorectal cancer tissue, controlling for clinical, pathological, and tumour molecular features, remains uncertain. We hypothesized that a higher amount of tissue Fusobacterium nucleatum DNA might be associated with worse clinical outcome in colorectal cancer.
To test this hypothesis, we utilised over 1,000 colorectal carcinoma cases in two U.S. nationwide prospective cohort studies (the Nurses’ Health Study and the Health Professionals Follow-up Study), and examined the amount of tissue Fusobacterium nucleatum DNA in relation to colorectal cancer mortality. Use of our comprehensive database enabled us to examine its prognostic role, while controlling for potential confounders including statuses of MSI, CIMP, and BRAF mutation.
METHODS
Study population
We utilised the database of two U.S. nationwide prospective cohort studies, the Nurses’ Health Study (NHS, with 121,701 women enrolled in 1976) and the Health Professionals Follow-up Study (HPFS, with 51,529 men enrolled in 1986).22, 23 Every 2 years, participants were sent follow-up questionnaires to gather information on health and lifestyle factors, and asked whether they had received diagnoses of major disease including cancers. The National Death Index was used to ascertain deaths of study participants and identify fatal colorectal carcinoma cases. Follow-up has exceeded 90% for each two year questionnaire. Study physicians reviewed medical records for all colorectal cancer cases, and assigned the cause of death for all deceased cases. Formalin-fixed paraffin-embedded (FFPE) tissue blocks were collected from hospitals where participants with colorectal carcinoma had undergone tumour resection. We included both colon and rectal carcinoma cases, considering the colorectal continuum model.24 A single pathologist (S.O.), who was unaware of other data, conducted a centralized review of hematoxylin and eosin-stained tissue sections from all colorectal carcinoma cases, and recorded pathological features. Tumour differentiation was categorised as well to moderate or poor (>50% vs.≤50% glandular area). We analysed available data on tissue Fusobacterium nucleatum DNA and patient survival in 1,069 colorectal carcinoma cases diagnosed up to 2008. Written informed consent was obtained from all study participants. The institutional review boards at the Harvard T.H. Chan School of Public Health and the Brigham and Women’s Hospital (Boston, MA, USA) approved the cohort studies.
Quantitative polymerase chain reaction (PCR) for Fusobacterium nucleatum
DNA was extracted from colorectal carcinoma tissue in whole-tissue sections of FFPE tissue blocks using QIAamp DNA FFPE Tissue Kit (Qiagen). We performed a quantitative PCR assay to measure the amount of tissue Fusobacterium nucleatum DNA, after assay validation as previously described.21 Custom TaqMan primer/probe sets (Applied Biosystems) for the nusG gene of Fusobacterium nucleatum and for the reference human gene, SLCO2A1 were used as previously described.7 Each reaction contained 80 ng of genomic DNA and was assayed in 20 μL reactions containing 1× final concentration TaqMan Environmental Master Mix 2.0 (Applied Biosystems) and each TaqMan Gene Expression Assay (Applied Biosystems), in a 96-well optical PCR plate. Amplification and detection of DNA was performed with the StepOnePlus Real-Time PCR Systems (Applied Biosystems) using the following reaction conditions: 10 min at 95ºC and 45 cycles of 15 sec at 95ºC and 1 min at 60ºC.
Our validation study has previously shown that in colorectal carcinoma cases with detectable Fusobacterium nucleatum DNA, the cycle threshold (Ct) values in the quantitative PCR for Fusobacterium nucleatum and SLCO2A1 decreased linearly with the log-transformed amount of input DNA from the same specimen (r2 > 0.99), and that the inter-assay coefficient of variation of Ct values from the same specimen in five different batches was 1% or less for all targets.21 Each specimen was analysed in duplicate for each target in a single batch, and we used the mean of the two Ct values for each target. The amount of tissue Fusobacterium nucleatum DNA in each specimen was calculated as a relative unitless value normalized with SLCO2A1 using the 2− ΔCt method (where ΔCt = “the mean Ct value of Fusobacterium nucleatum” - “the mean Ct value of SLCO2A1”).21
Analyses of MSI, DNA methylation, and KRAS, BRAF, and PIK3CA mutations
DNA was extracted from colorectal carcinoma tissue in whole-tissue sections from FFPE tissue blocks. MSI status was analysed with the use of 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487) as previously described.25 We defined MSI-high as the presence of instability in ≥30% of the markers, and MSI-low/microsatellite stable (MSS) as instability in <30% of the markers. Methylation analyses of long interspersed nucleotide element-1 (LINE-1)26, 27 and eight promoter CpG islands specific for CIMP (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1)28, 29 were performed. PCR reaction and pyrosequencing were performed for KRAS (codons 12, 13, 61, and 146),30, 31 BRAF (codon 600),25 and PIK3CA (exons 9 and 20).32, 33
Statistical analysis
All statistical analyses were conducted using SAS (version 9.3, SAS Institute, Cary, NC) and all p values were two-sided. Our primary hypothesis testing was a linear trend test in Cox proportional hazards regression model to assess an association of the amount of tissue Fusobacterium nucleatum DNA with colorectal cancer-specific mortality. Overall mortality was a secondary outcome. Cases with detectable Fusobacterium nucleatum DNA were categorised as low versus high based on the median cut point amount of Fusobacterium nucleatum DNA, while cases without detectable Fusobacterium nucleatum DNA were categorised as negative. Test for a linear trend was conducted across the ordinal categories (negative [0], low [1], and high [2]) of the amount of tissue Fusobacterium nucleatum DNA as a continuous variable in the Cox proportional hazards regression model. A two-sided α level was set at 0.05 for our primary hypothesis testing.
For analyses of colorectal cancer-specific mortality, deaths as a result of other causes were censored. To control for confounders, we used multivariable Cox proportional hazards regression models. Multivariable models included disease stage as a stratifying variable using strata function in the SAS proc phreg command. In addition to the amount of tissue Fusobacterium nucleatum DNA, the multivariable model initially included sex, age at diagnosis (continuous), year of diagnosis (continuous), family history of colorectal cancer in a first-degree relative (present vs. absent), tumour location (proximal colon vs. distal colon vs. rectum), MSI (high vs. MSI-low/MSS), CIMP (high vs. low/negative), KRAS (mutant vs. wild-type), BRAF (mutant vs. wild-type), PIK3CA (mutant vs. wild-type), and tumour LINE-1 methylation level (continuous). A backward stepwise elimination with a threshold of p = 0.05 was used to select variables in the final models. For cases with missing information on LINE-1 methylation level (5.1%), we assign a separate indicator variable. For cases with missing information in any of the categorical covariates (family history of colorectal cancer in a first-degree relative [0.4%], tumour location [0.3%], MSI [4.3%], CIMP [8.5%], KRAS [7.5%], BRAF [3.6%], and PIK3CA [9.3%]), we included these cases in the majority category of a given covariate to minimize the number of variables in multivariable Cox models. We confirmed that excluding the cases with missing information in any of the covariates did not substantially alter results (data not shown). Previous experimental studies provide evidence for potentiating effects of Fusobacterium nucleatum on colorectal tumour progression.16, 18 If the hypothesis that tissue Fusobacterium nucleatum is associated with shorter survival is true, high disease stage and poor tumour differentiation (both of which are associated with tissue Fusobacterium nucleatum in the current study) are likely mediators on the causal pathway from the amount of tissue Fusobacterium nucleatum DNA to shorter survival. Thus, we did not include disease stage or tumour differentiation in multivariable Cox proportional hazards regression models in our secondary analysis. The proportionality of hazards assumption was assessed by a time-varying covariate, using an interaction term of colorectal cancer-specific survival term and the amount of Fusobacterium nucleatum DNA (p = 0.45). The Kaplan-Meier method was used to describe the distribution of colorectal cancer-specific and overall survival, and the log-rank test for trend was performed to assess a linear trend in survival probability across the ordinal categories (negative [0], low [1], and high [2]) of the relative amount of tissue Fusobacterium nucleatum DNA.
All cross-sectional univariable analyses for clinical, pathological, and tumour molecular associations were secondary exploratory analyses, with an adjusted two-sided α level of 0.003 (= 0.05/16) for multiple hypothesis testing. To assess associations between the ordinal categories of the amount of tissue Fusobacterium nucleatum DNA and other categorical variables, Fisher’s exact test was performed. To compare mean age and mean LINE-1 methylation levels, an analysis of variance assuming equal variances was performed.
We conducted logistic regression analyses to assess associations of the amount of tissue Fusobacterium nucleatum DNA (an ordinal predictor variable [negative, low, and high]) with each component of the American Joint Committee on Cancer (AJCC) staging system, including pT stage (an ordinal outcome variable [pT1 vs. pT2 vs. pT3 vs. pT4]), pN stage (an ordinal outcome variable [pN0 vs. pN1 vs. pN2]), and M stage (a binary outcome variable [M0 vs. M1]). Test for a linear trend was conducted across the ordinal categories (negative [0], low [1], and high [2]) of the amount of tissue Fusobacterium nucleatum DNA as a continuous variable in logistic regression models. The multivariable logistic regression model initially included age (continuous), sex, year of diagnosis (continuous), family history of colorectal carcinoma in a first-degree relative (present vs. absent), tumour location (proximal colon vs. distal colon vs. rectum), MSI (high vs. MSI-low/MSS), CIMP (high vs. low/negative), KRAS (mutant vs. wild-type), BRAF (mutant vs. wild-type), PIK3CA (mutant vs. wild-type), and LINE-1 methylation level (continuous). For cases with missing information in any of the covariates, we assigned a separate (“missing”) indicator variable. A backward stepwise elimination with a threshold of p = 0.05 was used to select variables in the final models. To assess independent associations of MSI, CIMP, and BRAF mutation status (predictor variables) with the amount of tissue Fusobacterium nucleatum DNA (an ordinal outcome variable [negative vs. low vs. high]), we performed multivariable ordinal logistic regression analysis. In addition to MSI, CIMP, and BRAF mutation status, the multivariable ordinal logistic regression model initially included age (continuous), sex, year of diagnosis (continuous), family history of colorectal carcinoma in a first-degree relative (present vs. absent), tumour location (proximal colon vs. distal colon vs. rectum), KRAS (mutant vs. wild-type), PIK3CA (mutant vs. wild-type), and LINE-1 methylation level (continuous). For cases with missing information in any of the covariates, we assigned a separate (“missing”) indicator variable. A backward stepwise elimination was performed with a threshold of p = 0.05 to select covariates in the final model. We assessed the proportional odds assumption in the ordinal logistic regression model, which was generally satisfied (p ≥ 0.06).
RESULTS
Fusobacterium nucleatum in colorectal cancer tissue and patient mortality
We measured the relative amount of Fusobacterium nucleatum DNA in tumour tissue of 1,069 colorectal carcinoma cases within the Nurses’ Health Study and the Health Professionals Follow-up Study, using the quantitative PCR assay as previously described.21 Table 1 shows clinical, pathological, and tumour molecular features of the 1,069 cases. Fusobacterium nucleatum DNA was detected in colorectal carcinoma tissue in 134 (13%) of the 1,069 cases. We equally dichotomised the cases with detectable Fusobacterium nucleatum DNA into low versus high.
Table 1.
Characteristics according to the amount of Fusobacterium nucleatum DNA in colorectal cancer tissue
| Characteristic* | All patients (n = 1,069) | The amount of Fusobacterium nucleatum DNA in colorectal cancer tissue
|
p Value† | ||
|---|---|---|---|---|---|
| Negative (n = 935) | Low (n = 67) | High (n = 67) | |||
| Mean age ± SD (year) | 69.3 ± 8.8 | 69.2 ± 8.8 | 71.1 ± 9.0 | 68.8 ± 8.3 | 0.21 |
| Sex | 0.36 | ||||
| Men | 449 (42%) | 400 (43%) | 26 (39%) | 23 (34%) | |
| Women | 620 (58%) | 535 (57%) | 41 (61%) | 44 (66%) | |
| Year of diagnosis | 0.016 | ||||
| Prior to 1995 | 351 (33%) | 322 (34%) | 12 (18%) | 17 (25%) | |
| 1996 to 2000 | 298 (28%) | 260 (28%) | 18 (27%) | 20 (30%) | |
| 2001 to 2008 | 420 (39%) | 353 (38%) | 37 (55%) | 30 (45%) | |
| Family history of colorectal carcinoma in a first-degree relative | 0.27 | ||||
| Absent | 857 (80%) | 745 (80%) | 59 (88%) | 53 (80%) | |
| Present | 208 (20%) | 187 (20%) | 8 (12%) | 13 (20%) | |
| Tumour location | 0.001 | ||||
| Caecum | 178 (17%) | 145 (15%) | 13 (20%) | 20 (30%) | |
| Ascending to transverse colon | 346 (32%) | 296 (32%) | 23 (34%) | 27 (41%) | |
| Splenic flexure to sigmoid | 311 (29%) | 287 (31%) | 12 (18%) | 12 (18%) | |
| Rectosigmoid and rectum | 231 (22%) | 205 (22%) | 19 (28%) | 7 (11%) | |
| pT stage (depth of tumour invasion) | 0.0008 | ||||
| pT1 (submucosa) | 99 (10%) | 93 (11%) | 1 (1.6%) | 5 (8.2%) | |
| pT2 (muscularis propria) | 205 (21%) | 189 (22%) | 12 (19%) | 4 (6.6%) | |
| pT3 (subserosa) | 620 (63%) | 532 (62%) | 41 (66%) | 47 (77%) | |
| pT4 (serosa or other organs) | 57 (5.8%) | 44 (5.1%) | 8 (13%) | 5 (8.2%) | |
| pN stage (number of positive lymph nodes) | 0.84 | ||||
| pN0 (0) | 602 (63%) | 531 (64%) | 35 (58%) | 36 (61%) | |
| pN1 (1–3) | 211 (22%) | 182 (22%) | 16 (27%) | 13 (22%) | |
| pN2 (≥4) | 138 (15%) | 119 (14%) | 9 (15%) | 10 (17%) | |
| AJCC disease stage | 0.003 | ||||
| I | 241 (25%) | 225 (26%) | 9 (15%) | 7 (11%) | |
| II | 325 (33%) | 274 (32%) | 23 (37%) | 28 (45%) | |
| III | 278 (28%) | 239 (28%) | 25 (40%) | 14 (23%) | |
| IV | 133 (14%) | 115 (14%) | 5 (8.1%) | 13 (21%) | |
| Tumour differentiation | < 0.0001 | ||||
| Well to moderate | 965 (90%) | 862 (92%) | 55 (83%) | 48 (72%) | |
| Poor | 102 (9.6%) | 72 (7.7%) | 11 (17%) | 19 (28%) | |
| MSI status | < 0.0001 | ||||
| MSI-low/MSS | 858 (84%) | 780 (87%) | 42 (66%) | 36 (55%) | |
| MSI-high | 165 (16%) | 114 (13%) | 22 (34%) | 29 (45%) | |
| MLH1 hypermethylation | < 0.0001 | ||||
| Absent | 844 (86%) | 759 (89%) | 48 (79%) | 37 (60%) | |
| Present | 134 (14%) | 96 (11%) | 13 (21%) | 25 (40%) | |
| CIMP status | < 0.0001 | ||||
| Low/negative | 800 (82%) | 716 (84%) | 48 (79%) | 36 (58%) | |
| High | 178 (18%) | 139 (16%) | 13 (21%) | 26 (42%) | |
| BRAF mutation | 0.0009 | ||||
| Wild-type | 866 (84%) | 771 (86%) | 50 (78%) | 45 (68%) | |
| Mutant | 165 (16%) | 130 (14%) | 14 (22%) | 21 (32%) | |
| KRAS mutation | 0.44 | ||||
| Wild-type | 566 (57%) | 499 (57%) | 29 (50%) | 38 (61%) | |
| Mutant | 423 (43%) | 370 (43%) | 29 (50%) | 24 (39%) | |
| PIK3CA mutation | 0.88 | ||||
| Wild-type | 813 (84%) | 713 (84%) | 48 (83%) | 52 (83%) | |
| Mutant | 157 (16%) | 136 (16%) | 10 (17%) | 11 (17%) | |
| Mean LINE-1 methylation level (%) ± SD | 63.3 ± 10.0 | 63.1 ± 10.0 | 64.8 ± 10.7 | 65.2 ± 8.9 | 0.11 |
AJCC, American Joint Committee on Cancer; CIMP, CpG island methylator phenotype; LINE-1, long interspersed nucleotide element-1; MSI, microsatellite instability; MSS, microsatellite stable; SD, standard deviation.
Percentage indicates the proportion of cases with a specific clinical, pathological, or tumour molecular feature according to the amount of Fusobacterium nucleatum DNA in colorectal cancer tissue. There were cases which had missing values for any of the characteristics except for age, sex, and year of diagnosis.
To assess associations between the ordinal categories (negative, low, and high) of the amount of Fusobacterium nucleatum DNA in colorectal cancer tissue and categorical variables, Fisher’s exact test was performed. To compare mean age and mean LINE-1 methylation levels, an analysis of variance was performed. We adjusted two-sided α level to 0.003 (= 0.05/16) by simple Bonferroni correction for multiple hypothesis testing.
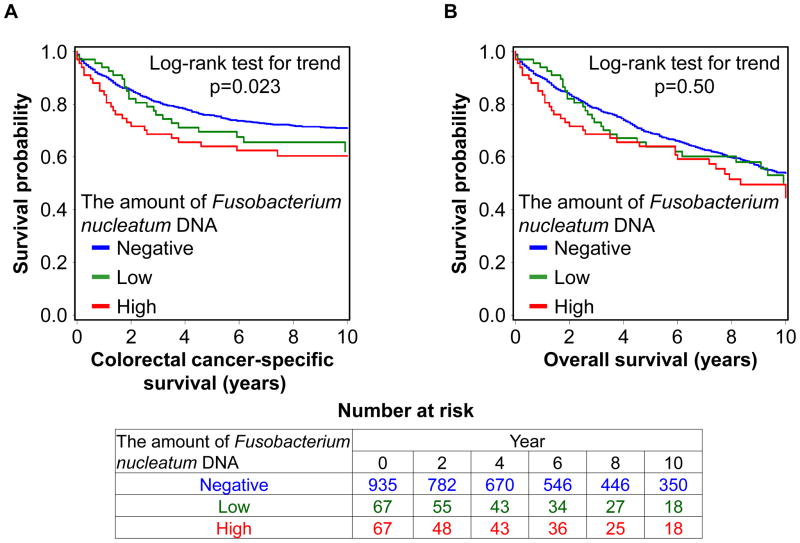
To test our primary hypothesis, we examined the relationship between the relative amount of tissue Fusobacterium nucleatum DNA and patient mortality (table 2). In the 1,069 colorectal cancer cases, there were 578 deaths, including 315 colorectal cancer-specific deaths, during a median patient follow-up of 10.7 years (interquartile range: 7.0 to 15.8) for censored cases. The amount of tissue Fusobacterium nucleatum DNA was associated with higher colorectal cancer-specific mortality in univariable (p for trend = 0.023) and multivariable Cox regression analyses (p for trend = 0.020). Compared to Fusobacterium nucleatum-negative cases, multivariable hazard ratios (HRs) for colorectal cancer-specific mortality in Fusobacterium nucleatum-low cases and Fusobacterium nucleatum-high cases were 1.25 (95% confidence interval [CI], 0.82 to 1.92) and 1.58 (95% CI, 1.04 to 2.39), respectively. In Kaplan-Meier analysis, a higher amount of tissue Fusobacterium nucleatum DNA was associated with shorter colorectal cancer-specific survival (p = 0.023 by the log-rank test for trend; figure 1A).
Table 2.
The amount of Fusobacterium nucleatum DNA in colorectal cancer tissue and patient mortality
| The amount of Fusobacterium nucleatum DNA | No. of cases | Colorectal cancer-specific mortality
|
Overall mortality
|
||||
|---|---|---|---|---|---|---|---|
| No. of events | Univariable HR (95% CI) | Multivariable stage-stratified HR (95% CI)* | No. of events | Univariable HR (95% CI) | Multivariable stage-stratified HR (95% CI)* | ||
| Negative | 935 | 265 | 1 (reference) | 1 (reference) | 511 | 1 (reference) | 1 (reference) |
| Low | 67 | 24 | 1.31 (0.86–2.00) | 1.25 (0.82–1.92) | 32 | 1.01 (0.70–1.44) | 0.84 (0.59–1.21) |
| High | 67 | 26 | 1.51 (1.01–2.26) | 1.58 (1.04–2.39) | 35 | 1.14 (0.81–1.61) | 1.08 (0.76–1.52) |
| p for trend† | 0.023 | 0.020 | 0.50 | 0.99 | |||
CI, confidence interval; HR, hazard ratio.
The multivariable stage-stratified Cox regression model initially included sex, age, year of diagnosis, family history of colorectal cancer in parent or sibling, tumour location, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and long interspersed nucleotide element-1 (LINE-1) methylation level. A backward elimination with a threshold of p = 0.05 was used to select variables in the final models.
Test for a linear trend was conducted across the ordinal categories (negative [0], low [1], and high [2]) of the amount of Fusobacterium nucleatum DNA in colorectal cancer tissue as a continuous variable in the Cox regression model.
Figure 1.
Kaplan-Meier curves for colorectal cancer-specific survival (A) and overall survival (B) according to the amount of Fusobacterium nucleatum DNA in colorectal cancer tissue. The p value was calculated by the log-rank test for trend (two-sided). The tables (bottom) show the number of patients who remained alive and at risk of death at each time point after the diagnosis of colorectal cancer.
In a secondary analysis of overall mortality as an outcome, the amount of tissue Fusobacterium nucleatum DNA was not significantly associated with overall mortality (p for trend = 0.99; table 2: p = 0.50 by the log-rank test for trend; figure 1B).
Considering that disease stage and tumour differentiation may be on the causal pathway from the amount of tissue Fusobacterium nucleatum DNA to shorter survival, we also used the multivariable Cox regression model that did not include disease stage or tumour differentiation in a further secondary analysis, and observed a significant association of the amount of tissue Fusobacterium nucleatum DNA with higher colorectal cancer-specific mortality (p for trend = 0.0001; supplementary table S1).
Tissue Fusobacterium nucleatum in relation to other features in colorectal cancer
As shown in Table 1, a higher amount of tissue Fusobacterium nucleatum DNA was associated with proximal tumour location, higher pT stage, poor tumour differentiation, MSI-high, MLH1 hypermethylation, CIMP-high, and BRAF mutation (p≤0.001 with the adjusted α level of 0.003 for multiple hypothesis testing).
As an exploratory analysis, we examined associations of tissue Fusobacterium nucleatum DNA with pT stage, pN stage, and M stage (table 3). The amount of tissue Fusobacterium nucleatum DNA was associated with higher pT stage in univariable (p for trend = 0.0003) and multivariable ordinal logistic regression analyses (p for trend = 0.0007). The association of tissue Fusobacterium nucleatum DNA with pN or M stage was not statistically significant (p for trend ≥ 0.029 with the adjusted α level of 0.003; table 3).
Table 3.
Association of the amount of Fusobacterium nucleatum DNA in colorectal cancer tissue with each component of the American Joint Committee on Cancer staging system
| Univariable OR (95% CI) | Multivariable OR (95% CI)* | |||
|---|---|---|---|---|
| Model for pT stage (n = 981, as an ordinal outcome variable [pT1 vs. pT2 vs. pT3 vs. pT4]) | ||||
| The amount of Fusobacterium nucleatum DNA |
|
Negative | 1 (reference) | 1 (reference) |
| Low | 2.22 (1.27–3.90) | 2.41 (1.37–4.24) | ||
| High | 2.24 (1.27–3.95) | 2.02 (1.14–3.56) | ||
| p for trend† | 0.0003 | 0.0007 | ||
| Model for pN stage (n = 951, as an ordinal outcome variable [pN0 vs. pN1 vs. pN2]) | ||||
| The amount of Fusobacterium nucleatum DNA |
|
Negative | 1 (reference) | 1 (reference) |
| Low | 1.21 (0.72–2.04) | 1.48 (0.86–2.52) | ||
| High | 1.15 (0.68–1.94) | 1.68 (0.96–2.92) | ||
| p for trend† | 0.45 | 0.029 | ||
| Model for M stage (n = 977, as a binary outcome variable [M0 vs. M1]) | ||||
| The amount of Fusobacterium nucleatum DNA |
|
Negative | 1 (reference) | 1 (reference) |
| Low | 0.56 (0.22–1.43) | 0.81 (0.31–2.11) | ||
| High | 1.70 (0.90–3.24) | 2.33 (1.16–4.69) | ||
| p for trend† | 0.32 | 0.045 | ||
CI, confidence interval; OR, odds ratio.
The logistic regression analysis model initially included age, sex, year of diagnosis, family history of colorectal carcinoma in parent or sibling, tumour location, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and long interspersed nucleotide element-1 (LINE-1) methylation level. A backward stepwise elimination with a threshold of p = 0.05 was used to select variables in the final models.
Test for a linear trend was conducted across the ordinal categories (negative [0], low [1], and high [2]) of the amount of Fusobacterium nucleatum DNA in colorectal cancer tissue as a continuous variable in the logistic regression model for pT stage (an ordinal outcome variable), pN stage (an ordinal outcome variable), or M stage (a binary outcome variable). We adjusted two-sided α level to 0.003 for multiple hypothesis testing.
Table 4 shows the distribution of colorectal cancer cases according to combined MSI/CIMP/BRAF status. As an exploratory analysis, we performed multivariable ordinal logistic regression analysis to assess independent associations of MSI, CIMP, and BRAF mutation status with the amount of tissue Fusobacterium nucleatum DNA (table 5). The amount of tissue Fusobacterium nucleatum DNA was associated with MSI-high (multivariable odds ratio, 5.22; 95% CI, 2.86 to 9.55), independent of CIMP and BRAF mutation status. In contrast, CIMP or BRAF mutation was not associated with Fusobacterium nucleatum after adjusting for MSI status.
Table 4.
The amount of Fusobacterium nucleatum DNA in colorectal cancer tissue in relation to combined MSI/CIMP/BRAF status
| Combined MSI/CIMP/BRAF status
|
The amount of Fusobacterium nucleatum DNA
|
|||||
|---|---|---|---|---|---|---|
| MSI status | CIMP status | BRAF mutation | All patients (n = 953) | Negative (n = 833) | Low (n = 59) | High (n = 61) |
| High | High | Mutant | 83 | 56 (67%) | 10 (12%) | 17 (21%) |
| High | High | Wild-type | 37 | 29 (78%) | 2 (5.4%) | 6 (16%) |
| High | Low/negative | Mutant | 3 | 1 (33%) | 1 (33%) | 1 (33%) |
| High | Low/negative | Wild-type | 32 | 21 (66%) | 6 (19%) | 5 (15%) |
| MSI-low/MSS | High | Mutant | 29 | 27 (93%) | 0 | 2 (6.9%) |
| MSI-low/MSS | High | Wild-type | 27 | 25 (93%) | 1 (3.7%) | 1 (3.7%) |
| MSI-low/MSS | Low/negative | Mutant | 40 | 38 (95%) | 1 (2.5%) | 1 (2.5%) |
| MSI-low/MSS | Low/negative | Wild-type | 702 | 636 (91%) | 38 (5.4%) | 28 (4.0%) |
CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stable. Percentage indicates the proportion of Fusobacterium nucleatum-negative, Fusobacterium nucleatum-low, or Fusobacterium nucleatum-high cases among all cases with a given specific combined MSI/CIMP/BRAF status.
Table 5.
Ordinal logistic regression analysis to assess independent associations of MSI, CIMP, and BRAF mutation status with the amount of Fusobacterium nucleatum DNA in colorectal cancer tissue
| Model for the amount of Fusobacterium nucleatum DNA (n = 953, as an ordinal outcome variable [negative vs. low vs. high]) | Univariable OR (95% CI) | Multivariable OR (95% CI)* |
|---|---|---|
| MSI-high (vs. MSI-low/MSS) | 4.53 (3.04–6.75) | 5.22 (2.86–9.55) |
| CIMP-high (vs. CIMP-low/negative) | 2.51 (1.65–3.80) | 0.71 (0.35–1.44) |
| BRAF mutant (vs. wild-type) | 2.23 (1.46–3.42) | 1.14 (0.62–2.10) |
CI, confidence interval; CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stable; OR, odds ratio.
In addition to MSI, CIMP, and BRAF mutation status, the multivariable ordinal logistic regression analysis model initially included age, sex, year of diagnosis, family history of colorectal carcinoma in parent or sibling, tumour location, KRAS and PIK3CA mutations, and LINE-1 methylation level. A backward stepwise elimination with a threshold of p = 0.05 was used to select variables in the final models, and the “year of diagnosis” variable remained in the final model.
DISCUSSION
We conducted this study to test the hypothesis that a higher amount of tissue Fusobacterium nucleatum might be associated with worse clinical outcome in colorectal cancer. Utilising the database of the 1,069 colorectal carcinoma cases in the two U.S. nationwide prospective cohort studies, we observed the association between the amount of tissue Fusobacterium nucleatum DNA and higher colorectal cancer-specific mortality.
Recent studies have provided mechanistic insights into the relationship between Fusobacterium nucleatum and colorectal tumour progression. Fusobacterium nucleatum expresses the virulence factor FadA on the bacterial cell surface, which has been shown to activate the WNT signaling pathway in colorectal carcinoma cells and promote colorectal tumour growth.18 Fusobacterium nucleatum may inhibit T-cell-mediated immune responses against colorectal tumours in the ApcMin/+ mouse model.16, 19 Our recent study has shown an inverse association between the amount of tissue Fusobacterium nucleatum DNA and CD3+ T-cell density in colorectal cancer.21 In the present study, a higher amount of tissue Fusobacterium nucleatum DNA was associated with a higher pT stage and worse clinical outcome. These lines of evidence together with the findings from our current study support the hypothesis that Fusobacterium nucleatum-high colorectal cancers may represent a more biologically aggressive cancer subtype. In light of possible roles of Fusobacterium nucleatum in down-regulating T-cell-mediated antitumour immune responses and in promoting colorectal tumour progression, future investigations may be warranted to explore potential influence of tissue Fusobacterium nucleatum on efficacy of the T-cell-based immunotherapies for colorectal cancer.
Colorectal cancers develop through the accumulation of genetic and epigenetic alterations, influenced by microbial and other environmental exposures and host responses to the exposures.34–38 In the current study, a higher amount of tissue Fusobacterium nucleatum DNA was associated with key tumour molecular features of colorectal cancer, including MSI-high, CIMP-high, LINE-1 hypomethylation, and BRAF mutation, which have been associated with clinical outcome in colorectal cancer.39–46 By utilising over 1,000 human colorectal carcinoma cases, to our knowledge we provided the first evidence that supports the prognostic significance of the amount of Fusobacterium nucleatum DNA in colorectal cancer tissue, independent of clinical, pathological, and major tumour molecular features. In addition, our current study could demonstrate that tissue Fusobacterium nucleatum was associated with MSI-high, but not with CIMP-high or BRAF mutation in multivariate analysis that adjusted for each other.
We acknowledge limitations of our study. First, the data on cancer recurrence were limited in the two cohorts. However, colorectal cancer-specific mortality is a reasonable cancer-specific outcome in the current study, which utilised the population-based data of long-term patient follow-up, since median survival for recurrent (local or metastatic) colorectal cancer was approximately 10 to 20 months during much of the time period of this study.47 Second, the data on cancer treatment were limited. However, it is unlikely that the distribution of chemotherapy use could substantially differ according to the amount of tissue Fusobacterium nucleatum DNA, because the data on tissue Fusobacterium nucleatum DNA were not available for treatment decisions.
Strengths of this study include the use of our molecular pathological epidemiology48-51 database (of over 1,000 colorectal carcinoma cases in the two U.S. nationwide, prospective cohort studies), which integrated epidemiologic exposures, clinicopathologic features, key tumour molecular features, and tissue Fusobacterium nucleatum DNA in colorectal carcinoma. Importantly, our colorectal cancer specimens were derived from a large number of hospitals in diverse settings across the U.S., which increases the generalizability of our findings. In addition, the sample size and comprehensiveness of the colorectal cancer database enabled us to assess the prognostic significance of tissue Fusobacterium nucleatum DNA, controlling for potential confounders.
In conclusion, the amount of tissue Fusobacterium nucleatum DNA was associated with higher colorectal cancer-specific mortality. These findings need to be validated in additional populations, as analytical and clinical validations are important to implement clinical use of tumour tissue biomarkers.52 Upon validation, Fusobacterium nucleatum DNA in colorectal carcinoma tissue may serve as a prognostic biomarker. In addition, our population-based data may provide insights for future studies to develop strategies for colorectal cancer prevention and treatment through targeting the microbiota.
Supplementary Material
Significance of this study.
What is already known about this subject?
Microorganisms play an important role in health and disease conditions, including cancer.
Fusobacterium nucleatum has been shown to promote colorectal tumour growth and inhibit antitumour immune responses in animal models.
Fusobacterium nucleatum DNA is detectable in a subset of human colorectal neoplasias.
What are the new findings?
The amount of Fusobacterium nucleatum DNA in colorectal cancer tissue is positively associated with colorectal cancer-specific mortality, independent of clinical, pathological, and major tumour molecular features.
The amount of Fusobacterium nucleatum DNA in colorectal cancer tissue is positively associated with pT stage.
The amount of Fusobacterium nucleatum in colorectal cancer tissue is associated with MSI-high both in univariable and multivariable analyses (independent of CIMP and BRAF mutation status), whereas CIMP and BRAF mutation are associated with Fusobacterium nucleatum only in univariate analyses but not after adjusting for MSI status.
How might it impact on clinical practice in the foreseeable future?
Fusobacterium nucleatum DNA in colorectal carcinoma tissue may serve as a potential prognostic biomarker.
Our population-based data can provide insights for the development of new colorectal cancer prevention and treatment strategies through targeting the microbiota.
Acknowledgments
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Funding: This work was supported by U.S. National Institutes of Health (NIH) grants [P01 CA87969 to S.E. Hankinson; UM1 CA186107 to M.J. Stampfer; P01 CA55075 and UM1 CA167552 to W.C. Willett; P50 CA127003 to C.S.F.; R01 CA137178 to A.T.C.; R01 CA151993 and R35 CA197735 to S.O.; and K07 CA190673 to R.N.]; and by grants from The Paula and Russell Agrusa Fund for Colorectal Cancer Research, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. K.M. is supported by a fellowship grant from Uehara Memorial Foundation and a grant from Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers from Japanese Society for the Promotion of Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- Ct
cycle threshold
- FFPE
formalin-fixed paraffin-embedded
- HPFS
Health Professionals Follow-up Study
- HR
hazard ratios
- LINE-1
long interspersed nucleotide element-1
- MSI
microsatellite instability
- MSS
microsatellite stable
- NHS
Nurses’ Health Study
- PCR
polymerase chain reaction
- SD
standard deviation
Footnotes
Author contributions: Kosuke Mima, Reiko Nishihara, Zhi Rong Qian, and Yin Cao contributed equally as co-first authors. Jeffrey A. Meyerhardt, Andrew T. Chan, Charles S. Fuchs, and Shuji Ogino contributed equally as co-last authors. All authors contributed to review and revision. Danny A. Milner, Edward L. Giovannucci, Levi A. Garraway, Gordon J. Freeman, Glenn Dranoff, Wendy S. Garrett, Curtis Huttenhower, Matthew Meyerson, Jeffrey A. Meyerhardt, Andrew T. Chan, Charles S. Fuchs, and Shuji Ogino developed the main concept and designed the study. Andrew T. Chan, Charles S. Fuchs, and Shuji Ogino wrote grant applications. Kosuke Mima, Reiko Nishihara, Zhi Rong Qian, Yin Cao, Yasutaka Sukawa, Juhong Yang, Ruoxu Dou, Yohei Masugi, Mingyang Song, Jeffrey A. Meyerhardt, Andrew T. Chan, Charles S. Fuchs, and Shuji Ogino were responsible for collection of tumour tissue, and acquisition of epidemiologic, clinical and tumour tissue data, including histopathological and immunohistochemical characteristics. Kosuke Mima, Yasutaka Sukawa, Aleksandar D. Kostic, Marios Giannakis, Susan Bullman, Danny A. Milner, Wendy S. Garrett, Curtis Huttenhower, Matthew Meyerson, Charles S. Fuchs, and Shuji Ogino performed data analysis and interpretation. Kosuke Mima, Reiko Nishihara, and Shuji Ogino drafted the manuscript. Jonathan A. Nowak, Mingyang Song, Marios Giannakis, Hideo Baba, Edward L. Giovannucci, Gordon J. Freeman, Glenn Dranoff, Wendy S. Garrett, Matthew Meyerson, Jeffrey A. Meyerhardt, Andrew T. Chan, Charles S. Fuchs, and Shuji Ogino contributed to editing and critical revision for important intellectual contents.
Use of standardized official symbols: We use HUGO (Human Genome Organisation)-approved official symbols for genes and gene products, including APC, BRAF, CACNA1G, CDKN2A, CRABP1, IGF2, KRAS, MLH1, NEUROG1, PIK3CA, RUNX3, SLCO2A1, and SOCS1; all of which are described at www.genenames.org.
Competing interests: Dr. Chan previously served as a consultant for Bayer Healthcare, Millennium Pharmaceuticals, Pozen Inc, and Pfizer Inc. This study was not funded by Bayer Healthcare, Millennium Pharmaceuticals, Pozen Inc, or Pfizer Inc. Dr. Meyerson applies a patent on Fusobacterium in colorectal cancer diagnosis, and has ownership interest in and is a consultant and advisory board member for Foundation Medicine. The other authors declare that they have no competing interests.
The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
References
- 1.McLean MH, Dieguez D, Jr, Miller LM, et al. Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut. 2015;64:332–341. doi: 10.1136/gutjnl-2014-308514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 4.Arthur JC, Gharaibeh RZ, Muhlbauer M, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun. 2014;5:4724. doi: 10.1038/ncomms5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belcheva A, Irrazabal T, Robertson SJ, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 9.Prorok-Hamon M, Friswell MK, Alswied A, et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. 2014;63:761–770. doi: 10.1136/gutjnl-2013-304739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cougnoux A, Dalmasso G, Martinez R, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63:1932–1942. doi: 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Yang Y, Huycke MM. Commensal bacteria drive endogenous transformation and tumour stem cell marker expression through a bystander effect. Gut. 2015;64:459–468. doi: 10.1136/gutjnl-2014-307213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piazzi G, D'Argenio G, Prossomariti A, et al. Eicosapentaenoic acid free fatty acid prevents and suppresses colonic neoplasia in colitis-associated colorectal cancer acting on Notch signaling and gut microbiota. Int J Cancer. 2014;135:2004–2013. doi: 10.1002/ijc.28853. [DOI] [PubMed] [Google Scholar]
- 13.Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy AN, Araujo-Perez F, Azcarate-Peril A, et al. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito M, Kanno S, Nosho K, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 18.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 21.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015 Jun 4; doi: 10.1001/jamaoncol.2015.1377. Published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095–1105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamauchi M, Lochhead P, Morikawa T, et al. Colorectal cancer: a tale of two sides or a continuum? Gut. 2012;61:794–797. doi: 10.1136/gutjnl-2012-302014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irahara N, Nosho K, Baba Y, et al. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12:177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogino S, Kawasaki T, Kirkner GJ, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamura Y, Lochhead P, Yamauchi M, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18:2257–2268. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogino S, Lochhead P, Chan AT, et al. Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod Pathol. 2013;26:465–484. doi: 10.1038/modpathol.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Win AK, Buchanan DD, Rosty C, et al. Role of tumour molecular and pathology features to estimate colorectal cancer risk for first-degree relatives. Gut. 2015;64:101–110. doi: 10.1136/gutjnl-2013-306567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tillmans LS, Vierkant RA, Wang AH, et al. Associations between cigarette smoking, hormone therapy, and folate intake with incident colorectal cancer by TP53 protein expression level in a population-based cohort of older women. Cancer Epidemiol Biomarkers Prev. 2014;23:350–355. doi: 10.1158/1055-9965.EPI-13-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colussi D, Brandi G, Bazzoli F, et al. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148:77–87. e72. doi: 10.1053/j.gastro.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiovitz S, Bertagnolli MM, Renfro LA, et al. CpG island methylator phenotype is associated with response to adjuvant irinotecan-based therapy for stage III colon cancer. Gastroenterology. 2014;147:637–645. doi: 10.1053/j.gastro.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogino S, Shima K, Meyerhardt JA, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18:890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seppala TT, Bohm JP, Friman M, et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br J Cancer. 2015;112:1966–1975. doi: 10.1038/bjc.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manceau G, Marisa L, Boige V, et al. PIK3CA mutations predict recurrence in localized microsatellite stable colon cancer. Cancer Med. 2015;4:371–382. doi: 10.1002/cam4.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inamura K, Yamauchi M, Nishihara R, et al. Tumor LINE-1 methylation level and microsatellite instability in relation to colorectal cancer prognosis. J Natl Cancer Inst. 2014 Sep 4; doi: 10.1093/jnci/dju195. Published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn JB, Chung WB, Maeda O, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011;117:1847–1854. doi: 10.1002/cncr.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 48.Ogino S, Chan AT, Fuchs CS, et al. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bishehsari F, Mahdavinia M, Vacca M, et al. Epidemiological transition of colorectal cancer in developing countries: environmental factors, molecular pathways, and opportunities for prevention. World J Gastroenterol. 2014;20:6055–6072. doi: 10.3748/wjg.v20.i20.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Epplein M, Bostick RM, Mu L, et al. Challenges and opportunities in international molecular cancer prevention research: An ASPO Molecular Epidemiology and the Environment and International Cancer Prevention Interest Groups Report. Cancer Epidemiol Biomarkers Prev. 2014;23:2613–2617. doi: 10.1158/1055-9965.EPI-14-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campbell PT, Newton CC, Newcomb PA, et al. Association between Body Mass Index and Mortality for Colorectal Cancer Survivors: Overall and by Tumor Molecular Phenotype. Cancer Epidemiol Biomarkers Prev. 2015 Jun 2; doi: 10.1158/1055-9965.EPI-15-0094. Published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9 (Suppl 5):S1–32. doi: 10.6004/jnccn.2011.0137. quiz S33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.