Abstract
Five putative novel Pseudomonas species shown to be pathogenic to citrus have been characterized in a screening of 126 Pseudomonas strains isolated from diseased citrus leaves and stems in northern Iran. The 126 strains were studied using a polyphasic approach that included phenotypic characterizations and phylogenetic multilocus sequence analysis. The pathogenicity of these strains against 3 cultivars of citrus is demonstrated in greenhouse and field studies. The strains were initially grouped phenotypically and by their partial rpoD gene sequences into 11 coherent groups in the Pseudomonas fluorescens phylogenetic lineage. Fifty-three strains that are representatives of the 11 groups were selected and analyzed by partial sequencing of their 16S rRNA and gyrB genes. The individual and concatenated partial sequences of the three genes were used to construct the corresponding phylogenetic trees. The majority of the strains were identified at the species level: P. lurida (5 strains), P. monteilii (2 strains), P. moraviensis (1 strain), P. orientalis (16 strains), P. simiae (7 strains), P. syringae (46 strains, distributed phylogenetically in at least 5 pathovars), and P. viridiflava (2 strains). This is the first report of pathogenicity on citrus of P. orientalis, P. simiae, P. lurida, P. moraviensis and P. monteilii strains. The remaining 47 strains that could not be identified at the species level are considered representatives of at least 5 putative novel Pseudomonas species that are not yet described.
Introduction
The plant pathogenic Pseudomonas species that belong to the Pseudomonas syringae species complex include P. cannabina, P. avellanae, P. amygdali, P. ficuserectae, P. savastanoi, P. tremae, P. meliae, P. caricapapayae and P. syringae [(ISPP Taxonomy of Plant Pathogenic Bacteria Committee; http://www.isppweb.org/names_bacterial.asp); [1]]. P. syringae is the first species in the Top 10 plant pathogenic bacteria [2]. Traditionally, these pathogens have been differentiated from other Pseudomonas species according to their colony morphology, ability to induce a hypersensitivity response in non-host plants, and the presence or absence of pectinase and arginine dihydrolase [3]. Some authors have also included the pectinolytic species P. viridiflava and the oxidase-positive species P. cichorii within this group [4]. Both species groups are monophyletic in the rpoD gene’s phylogenetic tree and in multilocus sequence analysis (MLSA) and are considered members of the P. syringae phylogenetic group as defined by Mulet and collaborators [5].
P. syringae and P. viridiflava, which cause citrus blast and black pit disease, are pathogenic species to citrus plants. Blast is a disease of the leaves and twigs, and black pit is a disease of the fruit. P. syringae and P. viridiflava are widespread on citrus foliage, although their presence does not always lead to disease development. In general, the occurrence of disease symptoms is influenced by several factors, such as temperature, humidity, oxygen depletion, varietal susceptibility and the virulence of the bacterial strains. Under favorable conditions, the disease develops very quickly and could cause substantial economic losses [2]. These diseases are widespread in Iran under cool and wet conditions. Citrus blast is one of the most important citrus diseases in the northern citrus growing provinces of Iran, which represents 60% of the total citrus growing area in Iran (approximately 100,000 ha). Damage is mainly caused by the prevailing climatic conditions in this area of the Caspian Sea belt [6, 7, 8]. The disease has caused considerable damage to citrus orchards in recent years in this area, although it has not caused much damage in the other orchards in Iran. No exact crop loss data are available. Farmers use bactericidal compounds to control the disease (e.g., copper oxychloride); however, this practice could cause serious damage to the environment and human health and also promotes the selection of pathogenic strains with increased tolerance to copper [9, 10]. Symptoms of the disease, initially water-soaked lesions turning to brown to black necrotic areas, most commonly begin on young leaves and twigs. Leaf lesions extend through the petiole to the stem and expand in both directions. Expansion of the lesions often leads to girdling of the affected branch and withering of the portions distal to the lesion [11].
Differentiation of species in the P. syringae species complex by phenotypic characteristics, 16S rDNA phylogenetic analysis and cell wall fatty acid composition lacked the required resolution for reliable differentiation of the taxa [12, 13, 14]. Recently, the convenience of sequence-based analysis for rapid and precise identification of plant pathogenic Pseudomonas has been proposed by several authors [15, 16, 17]. Partial sequences of the rpoD gene have been proposed for the differentiation of Pseudomonas species, with a species cut-off at 95% sequence similarity [18, 5].
More than 1,000 strains were isolated in an initial screening from samples of citrus leaves and twigs with blast disease symptoms collected from different regions of Iran (Gilan, Mazandaran and Golestan provinces). One hundred and forty strains were positive in the pathogenicity tests conducted, and the strains that were phenotypically related to Pseudomonas strains (126) were included in the present study. The collection of 126 Pseudomonas strains has been identified and characterized taxonomically by extensive numerical analyses of phenotypic features and by a multilocus sequencing approach, which includes analyses of partial sequences of the 16S rRNA, gyrB and rpoD genes. Strains of Pseudomonas species previously not considered as pathogenic to plants (P. simiae, P. orientalis, P. moraviensis and P. monteilii) and at least five putative novel species were isolated and characterized taxonomically by means of a polyphasic approach; the pathogenic properties of these strains to citrus were demonstrated in the present study.
Materials and Methods
Sampling sites
Citrus orchards in Northern Iran cover an area located in the Caspian Sea belt (in Golestan, Mazandaran and Gilan provinces); this area extends in the north over 400 km from east to west and lies between the shore of the Caspian Sea and the first slopes of the Alborz mountain range. These provinces are geographically divided into two parts: the coastal plains and the mountainous areas. The Alborz Mountain Range surrounds the coastal strip and plains of the Caspian Sea like a huge barrier. The Gilan province has a humid subtropical climate with the heaviest rainfall in Iran (1,900 mm rainfall, humidity reaches up to 90% in summer with temperatures of over 26°C); the Mandazaran province has a moderate subtropical climate (1,200 mm average rainfall and 25°C in summer and 8°C in winter); the Golestan province has a moderate and humid climate (average annual temperature 18.2°C and annual rainfall 556 mm).
Bacterial strains
One hundred twenty-six Pseudomonas strains were isolated from diseased citrus leaves and stems. No specific permission from any organization in Iran was required for the isolation of these bacterial strains because the research was for a PhD thesis. The field studies did not involve endangered or protected species. A segment of the leaf or twig lesion was surface sterilized in 0.5% sodium hypochlorite for two minutes and washed three times with sterile distilled water (SDW). The segment was cut into small pieces in drops of SDW, left for 10–20 min in a sterile laminar air flow cabinet and drops of the suspension were streaked on plates of nutrient agar (NA) medium (Merck, Germany). The streaked plates were incubated at 25°C. Single colonies were cultured on NA containing 1% sucrose (NAS), and after two days of growth, the plates were maintained at 4–6°C for short-term storage and routine use. Suspensions in SDW were stored at room temperature. For long storage periods, the cells were maintained in 25% glycerol at -70°C. The origins and geographical locations of the strains are indicated in S1 Table.
Physiological and biochemical tests
Biochemical characteristics and carbon source utilization were tested using API 20NE strips (bioMérieux, Marcy l’Etoile, France) and the Biolog GN2 microplates MicroLog System (Biolog Inc. Hayward, California, U.S.A.) according to the manufacturer’s instructions. Fluorescent pigment production was tested on King B medium (Merck, Germany) [19]. Numerical analyses of the phenotypic tests were performed using the computer program MVSP (Multi-Variate Statistical Package, version 3.22, Kovach Computing Services, Anglesey, UK). A similarity matrix was generated using the Simple Matching Coefficient, and a dendrogram was constructed using the unweighted pair group method with arithmetic averages (UPGMA).
Pathogenicity tests on citrus plants
Three different citrus cultivars were used in the pathogenicity tests: Alemow (Citrus macrophylla, the most sensitive cultivar in an extensive natural infection in 1998 in northern Iran) and the two most abundant cultivars in the region, Washington navel (Citrus sinensis) and Sour Orange (Citrus aurantium). Other plants were not tested for susceptibility. Five month old seedlings were maintained in a greenhouse (15°C and high relative humidity) and used for the pathogenicity tests. Forty-one strains were also tested under field conditions on new twigs or leaves at temperatures between 8–20°C and prolonged wetting by rain or fog. The strains were grown on NA at 25°C for 24 h, and the cells were suspended in sterile distilled water. Bacterial suspensions were adjusted to an optical density of 0.2 at 620 nm, corresponding to approximately 1 × 108 CFU/ml, as determined by dilution plating. One hundred microliters of bacterial suspension was injected into the intercellular space of orange leaves with a 0.5 mm needle and syringe [20]. All pathogenicity tests were conducted at least twice. Control plants were treated with sterile distilled water. The inoculated plants were examined after two weeks. Disease severity was measured in mm2 of the necrotic lesion. Re-isolation from the representative isolates on plates of NA was performed 15–21 days after inoculation. The results were statistically analyzed with Sigma Plot version 11 and plotted as boxes and whiskers. This plot provided summary statistics for five values: the minimum, the maximum, the median, the 25th percentile, and the 75th percentile. LOPAT tests were performed as previously described: levan production [20], oxidase test [21], potato rot symptoms [22], arginine dihydrolase test and tobacco hypersensitivity reaction [22]. The reaction of the isolates in the LOPAT tests and the utilization of selected carbon compounds were determined to verify the identity of the isolates recovered compared with the inoculum, proving Koch postulates.
DNA extraction, PCR amplification and DNA sequencing conditions
For DNA extraction, a bacterial suspension was prepared in 500 μl of 0.2 mM EDTA, 30 μl of 1 M NaOH was added, and the sample was boiled for 5 min. After 2 min centrifugation at 16.000 x g, the supernatant was recovered. PCR amplification was performed with a DNA thermocycler (Eppendorf). Each reaction mixture contained 5 μl of PCR buffer (EG Healthcare) and 5 μl of each of the four deoxynucleoside triphosphates (Roche) at a final concentration of 200 μM each. A total of 2.5 μl of each primer was used at a concentration of 10 μM, with 5 U of Taq DNA polymerase (EG Healthcare), in a total volume of 50 μl. The cycling conditions for the rpoD (PsEG30F/PsEG790R) [18], 16S rRNA (16F27/16R1492) [23] and gyrB (BAUP2/APrU) genes [24] included a denaturation period at 94°C for 5 min, followed by 30 cycles of amplification (denaturation at 94°C for 1 min, primer annealing at 55°C (48°C for rpoD) for 1 min, and primer extension at 72°C for 1.5 min). A final elongation step was carried out at 72°C for 10 min. The amplified products were purified with MultiScreen HTS PCR 96-well filter plates (Millipore) according to the manufacturer’s instructions. Sequencing reactions were performed using ABI Prism BigDye Terminator version 3.1, and the sequences were read with an automatic sequence analyzer (3130 genetic analyzer; Applied Biosystems).
Phylogenetic analysis
The sequence analysis procedures were performed as previously described by Mulet and collaborators [18]. Individual trees were generated for the 16S rRNA, gyrB, and rpoD partial gene alignments. An analysis of three concatenated genes (16S rRNA, gyrB, rpoD; 2,818 nucleotides in total) was performed as described by Mulet et al. [25]. A concatenated gene tree was constructed with individual alignments in the following order: 16S rRNA, gyrB and rpoD. The length and nucleotide positions are in reference to P. aeruginosa type strain DSM 50071T: the 16S rRNA gene (X06684) nucleotide positions from 98 to 1372; the gyrB gene (AB039386) nucleotide positions from 326 to 1122; and the rpoD gene (AB039607) nucleotide positions from 94 to 743.
Nucleotide sequence accession numbers
The nucleotide sequences determined in this study have been deposited in the EMBL database under the following accession numbers: the 16S rRNA gene from HG805683 to HG805808; the gyrB gene from HG805628 to HG805682; and the rpoD gene from HG805502 to HG805627. All sequence accession numbers used in this article are shown in S1 Table.
Results
Strain isolation and pathogenicity tests
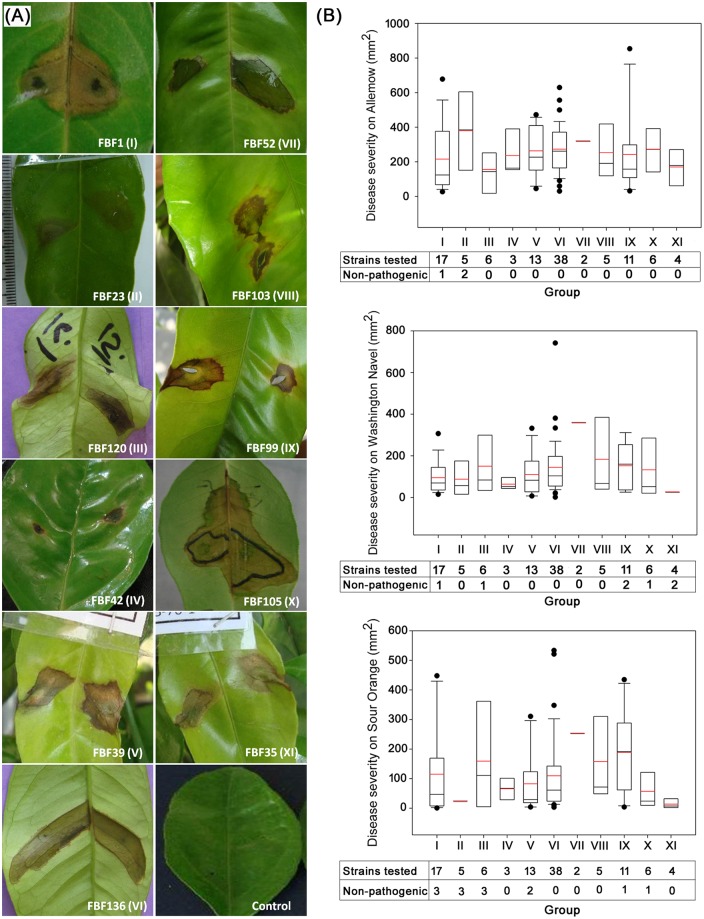
More than 1,000 strains were isolated in an initial screening from samples of citrus leaves and twigs with blast disease symptoms collected from different regions of Iran (Gilan, Mazandaran and Golestan provinces) in 2009–2010. One hundred and forty strains were positive in the pathogenicity tests conducted, and the strains phenotypically related to Pseudomonas strains (126) were studied further (Table 1, S1, S2 and S3 Tables). The 126 strains were able to induce lesions in citrus leaves under greenhouse conditions and were selected for characterization. Forty-one of the isolates were tested and produced disease symptoms under natural field conditions with a monthly average temperature of 21°C. The symptoms were less severe under field conditions. There was a considerable difference in virulence among strains of the same group as depicted by the diameter of the lesion and the rate of expansion of necrosis. Disease severities in greenhouse experiments of the 126 strains distributed in 11 phenotypic and phylogenetic groups (I to XI, see below) are depicted in Fig 1 and S3 Table. The highest median severity, measured in mm2 of the lesion, was found among strains in group VII of the P. syringae phylogenetic group. In the majority of cases (84%), the Alemow cultivar was less resistant than the other two cultivars tested (S3 Table). As examples, results of the tests of 11 strains, one of each group in the greenhouse experiments are shown in Fig 1.
Table 1. Strains used in this study and their assignation to phenotypic clusters and phylogenetic groups in the rpoD gene tree and in the MLSA analysis.
| Strain | Phenotypic cluster and rpoD group | rpoD gene closest type strain | Similarity (%) | Phylogenetic group (3 genes) | Closest type strain (3 genes) | Similarity (%) |
|---|---|---|---|---|---|---|
| FBF1, FBF9, FBF10, FBF11, FBF17, FBF34, FBF43, FBF48, FBF51, FBF64, FBF66, FBF80, FBF81, FBF84, FBF86, FBF95 | I | P. orientalis DSM 17489T | 97.8 | I | P. orientalis DSM 17489T | 97 |
| FBF7, FBF8, FBF23, FBF41, FBF56, FBF140, FBF141, FBF142, FBF143, FBF144 | II | P. synxantha LMG 2335T/ P. veronii LMG 17761T/ P. libanensis CIP105460T | 92–93.7 | II | P. synxantha LMG 2335T | 96 |
| FBF110, FBF112, FBF120, FBF121, FBF126, FBF128, FBF130 | III | P. simiae OLIT | 99.7–99.8 | III | P. simiae OLIT | 99.96 |
| FBF5, FBF20, FBF31, FBF42 | IV | P. lurida P513/18T | 99.5 | IV | P. lurida P513/18T | 99.6–99.7 |
| FBF25 | I and IVa | P. lurida P513/18T | 99.5 | IV | P. lurida P513/18T | 99.7 |
| FBF21, FBF22, FBF28, FBF30, FBF38, FBF39, FBF53, FBF54, FBF55, FBF59, FBF73, FBF75, FBF93 | V | P. rhodesiae LMG 17764T | 91.7 | V | P. marginalis ATCC 10844T/ P. grimontii CIP 106645T | 94.8–94.9 |
| FBF12, FBF32, FBF61, FBF62, FBF69, FBF104, FBF119 | VI-A | P. syringae ATCC 19310T | 97.1–97.8 | nd | nd | |
| FBF16, FBF33, FBF36, FBF40, FBF60, FBF68, FBF77, FBF78, FBF79, FBF83, FBF97, FBF106, FBF113, FBF115, FBF116, FBF139 | VI-B | P. syringae ATCC 19310T | 96.8–98.2 | VI | P. tremae LMG 22121T | 97.1–97.3 |
| FBF124, FBF136 | VI-B and VI-Ca | P. syringae ATCC 19310T | 97.8 | VI | P. tremae LMG 22121T | 97.3 |
| FBF13, FBF71, FBF72, FBF74, FBF98, FBF107, FBF108, FBF118, FBF134 | VI-C | P. syringae ATCC 19310T | 97.1–97.5 | VI | P. tremae LMG 22121T / P. syringae ATCC 19310T | 97.1 |
| FBF27, FBF46, FBF47, FBF63, FBF111, FBF138 | VI-D | P. syringae ATCC 19310T | 99.7–99.8 | VI | P. syringae ATCC 19310T | 99.3–99.4 |
| FBF109, FBF125, FBF135 | VI-E | P. syringae ATCC 19310T | 97.2–98.0 | VI | P. syringae ATCC 19310T | 98.6 |
| FBF49, FBF82, FBF91 | VI-F and VI-Ba | P. syringae ATCC 19310T | 98.2–99.5 | nd | nd | |
| FBF52, FBF100, FBF117 | VII | P. viridiflava ATCC 13223T | 91.8–97.8 | VII | P. viridiflava ATCC 13223T | 95.6–99.3 |
| FBF24, FBF58, FBF102, FBF103, FBF122 | VIII | P. syringae ATCC 19310T | 85–85.4 | VIII | P. meliae CCUG 51503T / P. tremae LMG 22121T | 91.6–91.7 |
| FBF2, FBF65, FBF67, FBF85, FBF87, FBF88, FBF89, FBF90, FBF92, FBF96, FBF99 | IX | P. moraviensis DSM 16007T | 93.6–97.2 | IX | P. moraviensis DSM 16007T/ P. koreensis LMG 21318T | 96.2–97.2 |
| FBF15, FBF50, FBF57, FBF101, FBF105, FBF114 | X | P. monteilii ATCC 700476T | 94–99 | X | P. monteilii ATCC 700476T | 95.8–98.9 |
| FBF18, FBF19, FBF35, FBF44 | XI | P. japonica JCM 21532T | 81.2 | XI | P. japonica JCM 21532T | 91.9 |
aStrains FBF124 and FBF136 were included in phenotypic cluster VI-B and in group VI-C in the rpoD and MLSA analysis; strains FBF49, FBF82 and FBF91 were included in phenotypic cluster VI-F and in the rpoD gene group VI-B; strain FBF25 was included in phenotypic cluster I and in the rpoD gene and MLSA group IV.
Fig 1. Pathogenicity tests and disease severity comparison for all Pseudomonas strains analyzed.
A. Pathogenicity test in Sour Orange citrus leaves for each group of strains. B. Disease severity comparison of Pseudomonas strains of each group tested in three plant genotypes, measured by the lesion area expressed in mm2 two weeks after inoculation. The number of strains and the number of non-pathogenic strains for each group tested are indicated below each graph. The black horizontal line represents the median and the red horizontal line represents the mean values (if only 2 values are considered, mean and median values are identical); the boundary of the box closest to zero indicates the 25th percentile and the boundary farthest from zero indicates 75th percentile. Whiskers (error bars) above and below the box indicate the 90th and 10th percentiles. The black dots indicate outlier values.
Phenotypic characterization and identification
The following preliminary phenotypic tests were used to identify the strains as possible members of the genus Pseudomonas: cell morphology, Gram staining, motility, oxidase, catalase, oxidation/fermentation of glucose and production of a fluorescent pigment on King B medium (S3 Table). All isolates were identified initially as P. syringae and P. viridiflava by the reactions in the LOPAT tests.
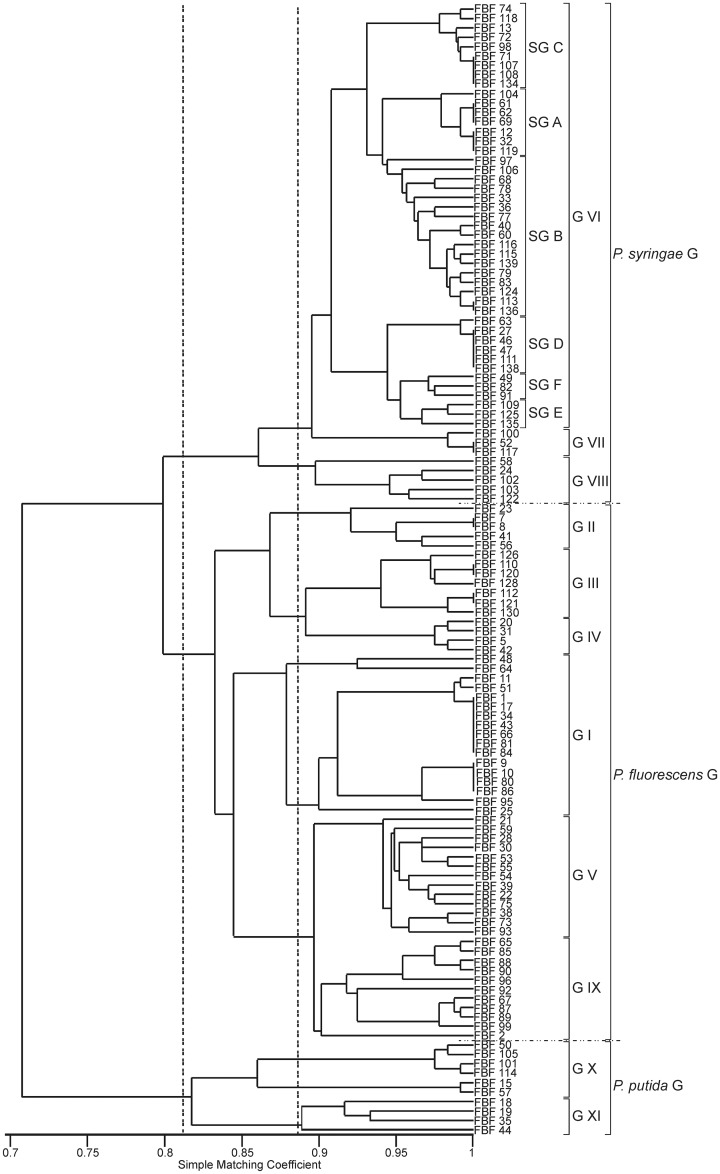
The results of the biochemical and physiological tests based on API 20 NE strips and Biolog GN2 microplates are given in S3 Table. A similarity matrix was generated using a simple matching coefficient, and the results are depicted in a dendrogram constructed by the UPGMA algorithm. Three main clusters were observed at 81% similarity, and these clusters corresponded to the P. putida, P. syringae and P. fluorescens phenogroups (Fig 2). Eleven phenotypic groupings were detected with 89–90% similarity; 2 clusters corresponded to the P. putida phenotype, 6 corresponded to the P. fluorescens phenotype, and 3 corresponded to the P. syringae phenotype. Exopolysaccharides (EPS) were only produced by strains belonging to the P. fluorescens phenotypic clusters I, II, and V and the P. syringae clusters VI and VII (Fig 2).
Fig 2. Phenotypic clustering of the studied strains.
A similarity matrix based on the phenotypic traits was generated using a simple matching coefficient and the dendrogram constructed by UPGMA.
The rpoD gene phylogenetic groups
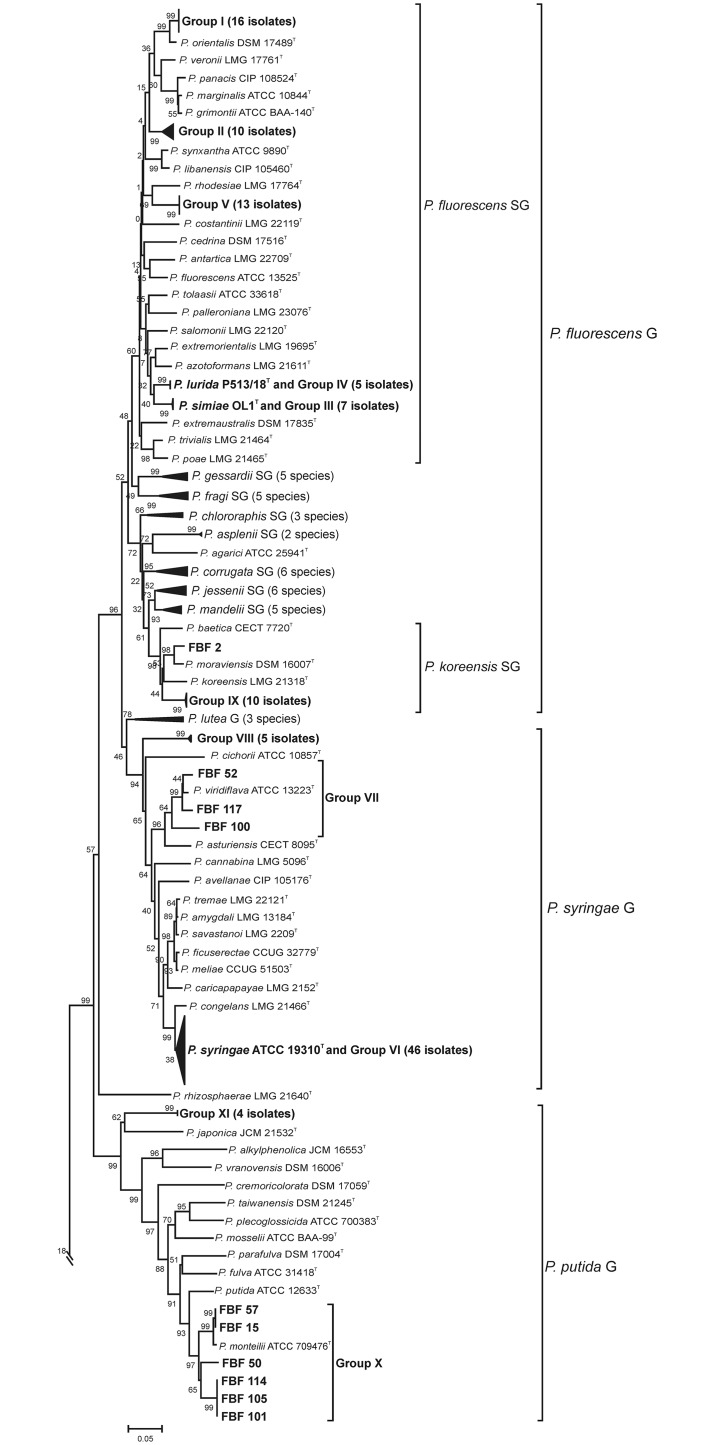
In the majority of cases, the similarity of the isolates to the species type strains and the branching order in the rpoD phylogenetic tree (Fig 3 and Table 1) allowed for the identification of the strains at the species level. Eleven rpoD gene sequence groups were identified (Table 2). Forty-six isolates belonged to phenotypic cluster VI in the P. syringae species complex. Three isolates of phenotypic cluster VII were close to the P. viridiflava type strain. However, the remaining isolates were related to other Pseudomonas species. Five strains of phenotypic cluster VIII were located in an independent branch between the phylogenetic groups of P. syringae and P. lutea. Sixteen strains of phenotypic cluster I were located in the P. fluorescens phylogenetic subgroup, close to P. orientalis. The remaining isolates in the P. fluorescens phylogenetic subgroup clustered as follows: 7 strains of phenotypic cluster III were close to P. simiae; 5 strains of phenotypic cluster IV were close to P. lurida, 13 strains of phenotypic cluster V were close to P. rhodesiae; 11 strains of phenotypic cluster IX were close to P. koreensis and P. moraviensis; 10 strains of phenotypic cluster II formed an independent branch in the P. fluorescens phylogenetic subgroup, close to P. libanensis, P. synxantha and P. veronii. Some isolates were affiliated with species from the P. putida phylogenetic group; 6 strains of phenotypic cluster X were placed close to P. monteilii, and 4 strains of phenotypic cluster XI were close to P. japonica.
Fig 3. Phylogenetic tree of the Pseudomonas strains based on the nucleotide sequences of the rpoD gene (717 nt).
The scale bar represents the number of substitutions per site. The number shown next to each node indicates the percentage bootstrap values of 1000 replicates. Cellvibrio japonicus was used as the outgroup.
Table 2. Assignation to pathovars of strains identified as members of the P. syringae phylogenetic group based on rpoD gene sequences.
| Strain | P. syringae group closest pathovar reference strain | Similarity (%) |
|---|---|---|
| FBF27, FBF47 | P. syringae PDDCC 3023T | 100 |
| FBF16, FBF36, FBF40, FBF49, FBF60, FBF68, FBF78, FBF79, FBF82, FBF83, FBF97, FBF106, FBF113, FBF115, FBF116, FBF139 | P. syringae pv. dysoxyli N255 | 99.1–99.5 |
| FBF24, FBF46, FBF58, FBF102, FBF103, FBF111, FBF122, FBF138 | P. syringae pv. lapsa N2096 / P. syringae pv. atrofaciens N2612 | 83.5–100 |
| FBF13, FBF71, FBF72, FBF74, FBF98, FBF107, FBF108, FBF118, FBF124, FBF134, FBF136 | P. syringae pv. papulans N2848 | 99.1–99.7 |
| FBF12, FBF32, FBF33, FBF61, FBF62, FBF69, FBF77, FBF91, FBF104, FBF109, FBF119, FBF125, FBF135 | P. syringae pv. solidagae ICMP 16925 | 97.7–98.4 |
| FBF63 | P. syringae pv. syringae | 100 |
| FBF52, FBF100, FBF117 | P. viridiflava PDDCC 2848 / P. syringae pv. ribicola N963 | 90.7–97.9 |
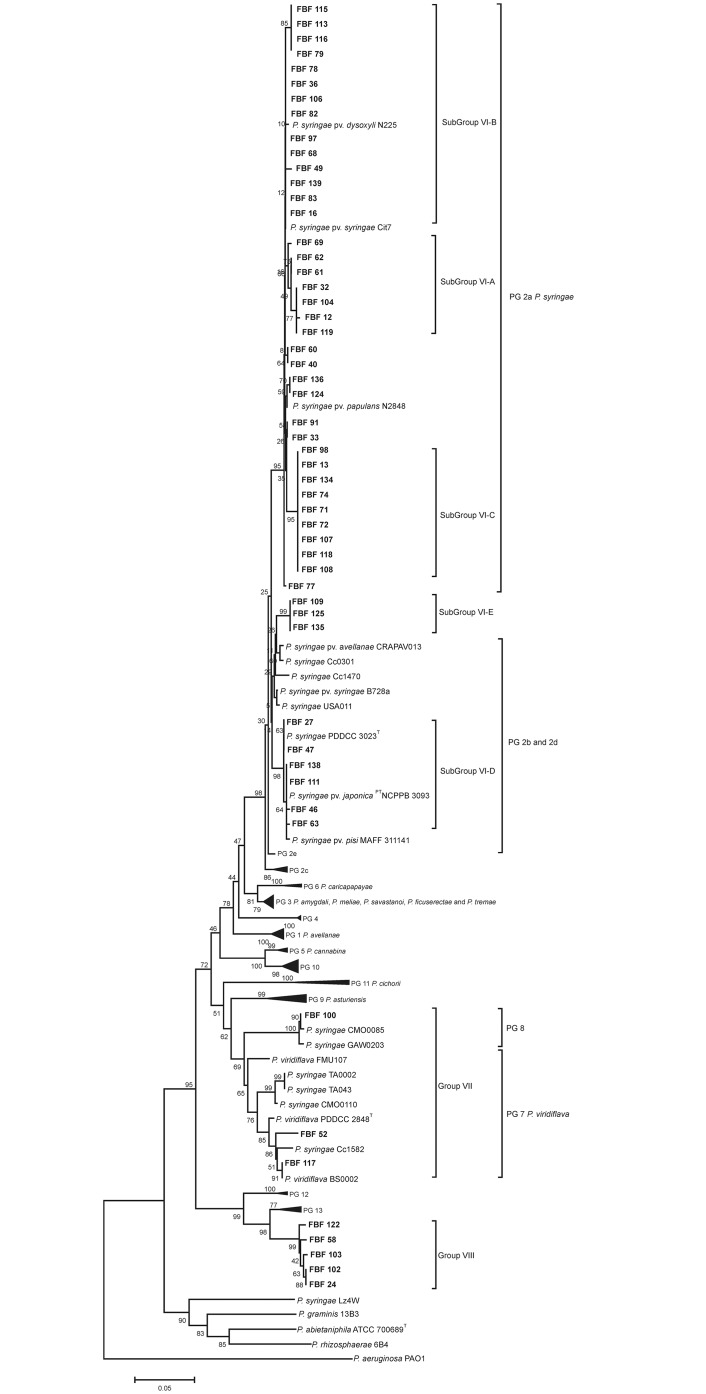
To further identify the 54 strains affiliated with the P. syringae species complex, a phylogenetic tree was constructed using the partial sequences of the rpoD gene (455 nt) published by Parkinson et al. [12] and Berge et al. [26]. As depicted in Fig 4 and Table 2, the majority of the strains in our study were closely affiliated in the rpoD sequence with strains of known pathovars of P. syringae (pv. dysoxyli, 16 strains; pv. solidagae, 10 strains; pv. lapsa/pv. atrofaciens, 3 strains; pv. papulans, 11 strains; and pv. syringae, 1 strain) and with strains classified in 3 of the 13 phylogroups (PG) defined by Berge and collaborators (PG 2a, 37 strains; PG 2b, 6 strains, together with P. syringae type strain; PG 2, but distinct to the subgroups already defined, 3 strains; PG7, 2 strains, together with P. viridiflava type strain) [26]. However, the 5 following strains appeared in a separate phylogroup in the species complex and were not affiliated with any known pathovar or phylogroup: strains FBF24, 58, 102, 103 and 122.
Fig 4. Phylogenetic tree based on the rpoD gene sequence (455 nt) of the strains assigned to the P. syringae species complex including the pathovar reference strains and strains of the 13 phylogroups (PG) defined by Berge et al. [26].
Multilocus sequence analysis
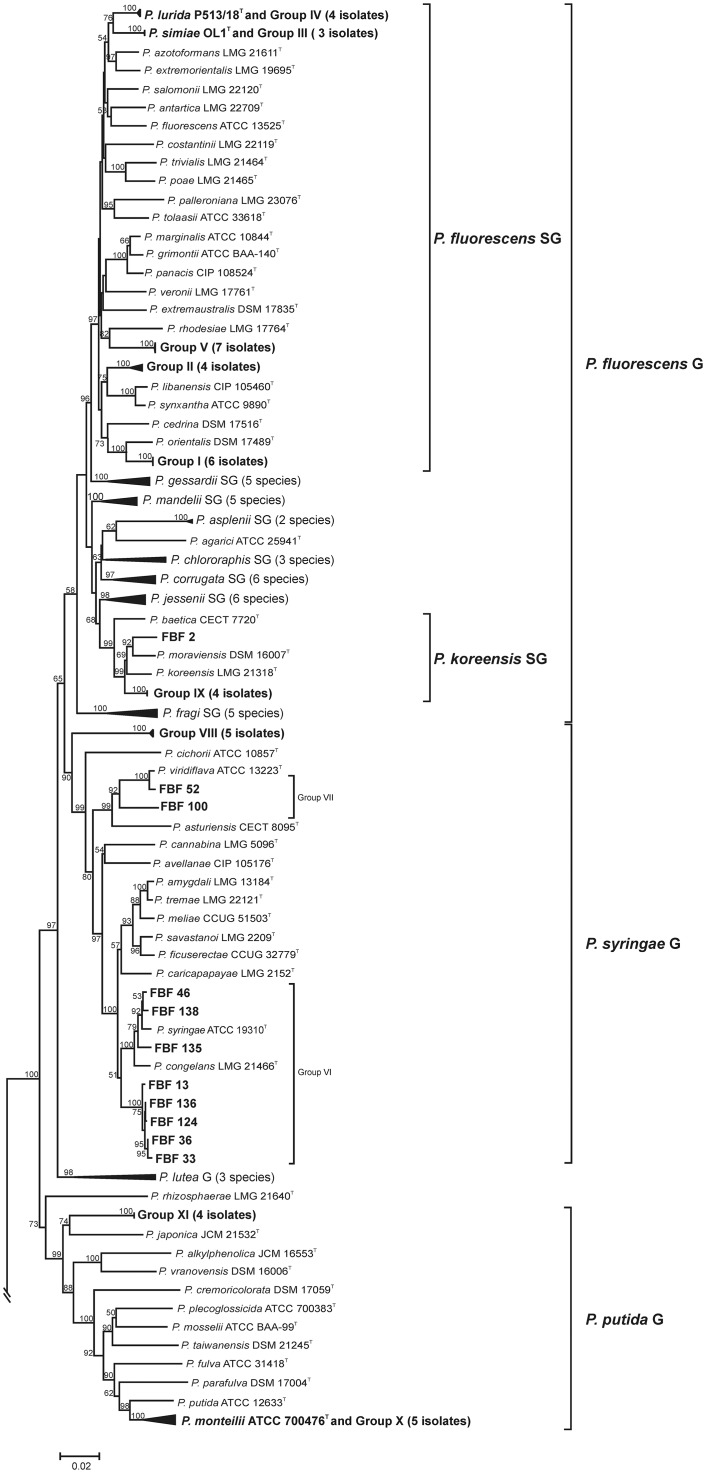
To determine the phylogenetic affiliation of the rpoD gene groupings in the genus Pseudomonas, 53 strains were selected as representatives of the 11 rpoD gene groupings for a multilocus sequence analysis (MLSA) according to the following criterion: at least 2 strains per rpoD phylogenetic group were selected to have representative strains of each phylogroup and to assess the phylogenetic diversity within each group. Phylogenetic analyses based on the concatenated sequences (2,818 nt) of the 16S rRNA (1,296 nt), gyrB (805 nt) and rpoD genes (717 nt) confirmed the distribution of the isolates in the 11 phylogenetic and phenotypic groupings (Figs 2, 3 and 4). The robustness of the concatenated tree was demonstrated by the high bootstrap values at all branches. The MLSA groups (Fig 5) were congruent with the groups in the rpoD analysis. The 53 representative strains were located in the P. fluorescens phylogenetic group (5 in the P. koreensis SG; 24 in the P. fluorescens SG), the P. syringae group (15 strains), and the P. putida group (9 strains) (Fig 5).
Fig 5. Phylogenetic consensus tree of the Pseudomonas strains based on the nucleotide sequences of the 16S rRNA, rpoD, and gyrB genes of 53 selected strains.
The scale bar represents the number of substitutions per site. The number shown next to each node indicates the percentage bootstrap values of 1000 replicates. Cellvibrio japonicus was used as the outgroup.
Assignment of the strains to species
Following the criteria proposed by Mulet and collaborators [5], the strains were assigned to known species in the MLSA analysis when the similarity to a type strain was higher than 97% and both strains were located in the same phylogenetic branch (Fig 5). Assignments are indicated in Table 1 and S2 Table. Group VI was the largest among the 11 groups, and the eight strains comprising the group were members of the P. syringae group. These eight strains were located in two independent branches; one branch had three isolates that were close to P. syringae and P. congelans, and there were five strains in the other independent branch. Group VII (two strains) consisted of isolates identified as P. viridiflava. Group VIII (5 strains) included isolates that have a similarity lower than 92% with the type strain of any other species in the group and are, therefore, considered members of a putative new species in the P. syringae phylogenetic group (Fig 5).
The other 6 groups (I, II, III, IV, V and IX) are affiliated with the P. fluorescens phylogenetic group as defined by Mulet and collaborators [27, 5] and are closely related to species that have not previously been described as plant pathogens. The highest similarity detected for the isolates of Group I was with P. orientalis (96.9–97.0%). The isolates of Group II were close to the type strains of P. libanensis and P. synxantha (95.9–96.0% for isolate FBF23; 95.9–96.0% for isolate FBF56).
The 3 strains of Group III are phylogenetically close to P. simiae (99.9%). The 4 strains of Group IV are phylogenetically close to P. lurida (99.7% for strains FBF25, 31 and 42 and 97.7% for strain FBF5). The 7 isolates of Group V showed a similarity of 94.8–94.9% with P. marginalis and P. grimontii. The isolates of Group IX are related to the two species of the P. koreensis subgroup, P. koreensis (96.7% similarity) and P. moraviensis (96.8% similarity).
Members of the two remaining groups (X and XI) belonged to the P. putida phylogenetic group. The isolates of Group X are related to P. monteilii (95.8–98.9% similarity), which is a species isolated from clinical specimens [28] and environmental samples [29]. The closest type strain to the isolates of Group XI was P. japonica, although this strain exhibited a low level of similarity (91.9%).
The distribution of the strains in groups and the geographical regions of isolation are indicated in S1 and S2 Tables and in S1 Fig. Representative strains of all groups were detected in the Mazandaran region, groups II, III, IV, VII, VIII and X were not detected in the Gilan region and groups II, V, VII and XI were not detected in the Golestan region; groups I, VI and IX were present in all 3 regions. There was no correlation between cultivars and groups. Strains representatives of all groups were isolated in more than one cultivar.
Discussion
The physiological and biochemical characteristics of the isolates were consistent with the characteristics previously described for the genus Pseudomonas, although discrepancies with known features of the classical citrus pathogens existed in the LOPAT tests (levan production, oxidase activity, potato soft rot, arginine dihydrolase activity, tobacco hypersensitivity). The phytopathogenic, oxidase-negative fluorescent Pseudomonas species have been traditionally identified as either P. syringae or P. viridiflava. Several strains in our study (71 strains) showed a LOPAT pattern different from the patterns corresponding to P. syringae or P. viridiflava. The polyphasic taxonomic study of the 126 strains analyzed demonstrated that only strains in group VI (P. syringae, 46 strains) and group VII (P. viridiflava, 2 strains) could be identified as members of these two species. Strains belonging to P. orientalis, P. synxantha, P. simiae, P. lurida, and P. monteilii were also identified; furthermore, at least 5 putative novel species were identified.
The molecular data confirmed that the partial sequence analysis of the rpoD gene is sufficient for rapid isolate classification. The same strain groupings were obtained using the rpoD sequence alone and in the analysis based on the concatenated 3 genes (16S rRNA, gyrB and rpoD), as was previously described for Pseudomonas species [27] and for the pathovars in P. syringae [12]. The MLSA phylogroups defined by Parkinson et al. [12] and Berge et al. [26] are maintained in this study. The groupings of strains obtained by the molecular methods in the MLSA study were similar to the groupings obtained by the biochemical and physiological tests. The combined results showed a high diversity among Pseudomonas strains pathogenic to citrus in Iran. No clear distribution of species by region was detected, and only members of group I assigned to P. orientalis, group VI assigned to P. syringae and group IX that could not be assigned to a known species were found in the three geographical regions. The severity of the lesions induced by the strains assigned to species not yet described as pathogenic and the putative novel species were of a similar order of magnitude as the P. syringae and P. viridiflava isolates.
Plant pathogenic Pseudomonas spp. have been described in the P. fluorescens phylogenetic lineage rather than in the P. aeruginosa lineage [16]. The 21 Pseudomonas species described by Bull and collaborators [16] belong mainly to the P. syringae phylogenetic group (14 species), with 5 species associated with the P. fluorescens subgroup and 2 species associated with the P. corrugata subgroup. The majority of the strains used in the present study belonged to the P. syringae phylogenetic group (54 strains) and the P. fluorescens subgroup (51 strains). Several strains were assigned to species that have not been described as plant pathogens and have been isolated previously from diverse habitats. P. orientalis (group I, 16 strains) has been isolated from spring waters in Lebanon [30]; P. simiae (group III, 7 strains) has been isolated from a monkey [31] and from Antarctic samples [32]; P. lurida (group IV, 5 strains) has been found as a plant growth promoting bacterium in the phyllosphere of grasses [33] and in high altitude rhizospheric soil from the Uttarakh and Himalayas [34, 35]; P. monteilii (group X, 6 strains) has been isolated from clinical specimens [28] and environmental samples [29]. Strains in groups II (10 strains), V (13 strains), VII (1 strain), VIII (5 strains), IX (10 strains), X (4 strains) and XI (4 strains), which represented 37% of the isolates, could not be assigned to a known species and are considered representatives of at least 5 putative novel Pseudomonas species because each group is phylogenetically and phenotypically homogeneous and distinct from known species type strains.
The results of the present study showed that citrus blast disease in the northern citrus producing provinces is caused by a diverse population of Pseudomonas strains belonging to species and pathovars including the two universally known species P. syringae pv. syringae [36] and P. viridiflava [7]. We demonstrated, for the first time, that the P. lurida, P. orientalis and P. simiae strains are pathogenic to plants. The probable differences between these species in characteristics such as host range with and outside Rutaceae and over summering are new issues in need of resolution. The analysis of the rpoD gene partial sequences is a fast and reliable method to differentiate Pseudomonas plant pathogenic species, and the Pseudomonas species diversity is higher than previously thought for citrus plants. At least 5 putative novel species have been detected, and their formal proposal is underway. These results are in accordance with the proposal of Scotta and collaborators [37], who recommended the use of the rpoD gene for routine and epidemiological studies of Pseudomonas clinical strains. This method is well suited for routine assessments of citrus plant lots for quality control to limit the chance of increasing the genetic diversity of Pseudomonas populations through the importation of foreign plants.
Supporting Information
(PDF)
(PDF)
(PDF)
++, strongly positive; +, positive; w+, weakly positive; -, negative; ND, not determined.
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files. The data will be available without restriction after the acceptance for publication. The authors have provided all sequences in case you need them, and accession number have been added to the text (Lines 208-210). Sequence accession numbers are indicated for each strain in supplementary S1 Table. All sequences will be available, after acceptance of the manuscript publication, from the EMBL database (accession numbers HG805502-HG805808).
Funding Statement
The work was supported by BOS2011 CGL24318 and Consolider CSD2009-00006 from Spanish Ministry of Economy and Competitiveness - MINECO.
References
- 1.Young JM. Taxonomy of Pseudomonas syringae. J Plant Pathol. 2010; 92,S1.5–S1.14 [Google Scholar]
- 2.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Path. 2012; 13, 614–629. 10.1111/j.1364-3703.2012.00804.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lelliott RA, Billing E, Hayward AC. A determinative scheme for the fluorescent plant pathogenic Pseudomonads. J Appl Bacteriol. 1966; 29, 470–489. 10.1111/j.1365-2672.1966.tb03499.x [DOI] [PubMed] [Google Scholar]
- 4.Hojo H, Koyanagi M, Tanaka M, Kajihara S, Ohnishi K, Kiba A, Hikichi Y. The hrp genes of Pseudomonas cichorii are essential for pathogenicity on eggplant but not on lettuce. Microbiology 2008; 154, 2920–2928. 10.1099/mic.0.2008/021097-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulet M, Gomila M, Scotta C, Sánchez D, Lalucat J, García-Valdés E. Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analysis approaches in species discrimination within the genus Pseudomonas. Syst Appl Microbiol. 2012; 35, 455–464. 10.1016/j.syapm.2012.08.007 [DOI] [PubMed] [Google Scholar]
- 6.Razinataj M, Taghavi SM. A comparison of Pseudomonas viridiflava isolates from different hosts by phenotypic characteristics and pathogenicity in Fars Province. Iranian J Agricul Sci. 2004; 35, 253–263. [Google Scholar]
- 7.Shams-Bakhsh M, Rahimian H. Comparative study on agents of citrus blast and bacterial canker of stone fruits in Mazandaran. Iranian J Plant Pathol. 1997; 33, 132–143. [Google Scholar]
- 8.Taghavi SM, Ziaee M. Comparison of Pseudomonas syringae pv. syringae Van Hall isolates from different hosts based on phenotypic characteristics, serological properties and pathogenicity. J Sci Technol Agricul Nat Res. 2003; 7, 199–213. [Google Scholar]
- 9.Brent KJ, Hollomon DW, Federation GCP. Fungicide resistance: the assessment of risk. Global Crop Protection Federation Brussels, Belgium: 1998. [Google Scholar]
- 10.Edwards C. The Impact of Pesticides on the Environment In: Pimentel D, Lehman H. (Eds.). The Pesticide Question. Springer; US: 1993; 2011. pp. 13–46. [Google Scholar]
- 11.Timmer LW, Garnsey SM, Graham JH. Compendium of citrus diseases. American Phytopathological Society (APS Press) 2000 [Google Scholar]
- 12.Parkinson N, Bryant R, Bew J, Elphinstone J. Rapid phylogenetic identification of members of the Pseudomonas syringae species complex using the rpoD locus. Plant Pathol. 2011; 60, 338–344. 10.1111/j.1365-3059.2010.02366.x [DOI] [Google Scholar]
- 13.Stead DE. Grouping of plant-pathogenic and some other Pseudomonas spp. by using cellular fatty acid profiles. Int J Syst Evol Microbiol. 1992; 42, 281–295. [Google Scholar]
- 14.Moore ERB, Mua M, Arnscheidt A, Böttger EC, Hutson RA, Collins MD et al. The determination and comparison of the 16S rDNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships. Syst Appl Microbiol. 1996; 19, 476–492. 10.1016/S0723-2020(96)80021-X [DOI] [Google Scholar]
- 15.Bull CT, De Boer SH, Denny TP, Firrao G, Fischer-Le Saux M, Saddler GS, et al. Comprehensive list of names of plant pathogenic bacteria, 1980–2007. J Plant Pathol. 2010; 92, 551–592. [Google Scholar]
- 16.Bull CT, De Boer SH, Denny TP, Firrao G, Fischer-Le Saux M, Saddler GS, et al. List of new names of plant pathogenic bacteria (2008–2010). J Plant Pathol. 2012; 94, 21–27. 10.4454/jpp.fa.2011.003 [DOI] [Google Scholar]
- 17.Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, Harayama S. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology. 2000; 146, 2385–2394. [DOI] [PubMed] [Google Scholar]
- 18.Mulet M, Bennasar A, Lalucat J, García-Valdés E. An rpoD-based PCR procedure for the identification of Pseudomonas species and for their detection in environmental samples. Mol Cell Probes. 2009; 23, 140–147. 10.1016/j.mcp.2009.02.001 [DOI] [PubMed] [Google Scholar]
- 19.King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fuorescin. J Lab Clin Med. 1954; 44, 301–307. [PubMed] [Google Scholar]
- 20.Schaad NW, Jones JB, Chun W. Laboratory guide for identification of plant pathogenic bacteria (3th ed.). 2001; p 373 APS Press, Minnesota. [Google Scholar]
- 21.Ewing JH, Johnson JG. The differentiation of Aeromonas and C27 cultures from Enterobacteriaceae. Syst Evol Microbiol. 1960; 10, 223–230. [Google Scholar]
- 22.Klement Z, Farkas GL, Lovreicovich L. Hypersensitive reaction induced by phytopathogenic bacteria in the tobacco leaf. Phytopathology. 1964; 54, 474–477. [Google Scholar]
- 23.Lane DJ. 16S/23S rRNA sequencing, p 115–175. In Stackebrand E., Goodfellow M. (ed). Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons, New York: 1991 [Google Scholar]
- 24.Cladera AM, Bennasar A, Barceló M, Lalucat J, García-Valdés E. Comparative genetic diversity of Pseudomonas stutzeri genomovars, clonal structure, and phylogeny of the species. J Bacteriol. 2004; 186, 5239–5248. 10.1128/JB.186.16.5239-5248.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulet M, Gomila M, Gruffaz C, Meyer JM, Palleroni NJ, Lalucat J, et al. Phylogenetic analysis and siderotyping as useful tools in the taxonomy of Pseudomonas stutzeri: description of a novel genomovar. Int J Syst Evol Microbiol. 2008; 58, 2309–2315. 10.1099/ijs.0.65797-0 [DOI] [PubMed] [Google Scholar]
- 26.Berge O, Monteil CL, Bartoli C, Chandeysson C, Guilbaud C, Sands C. et al. A user’s guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS ONE. 2014; 9(9): e105547 10.1371/journal.pone.0105547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulet M, Lalucat J, García-Valdés E. DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol. 2010; 12, 1513–1530. 10.1111/j.1462-2920.2010.02181.x [DOI] [PubMed] [Google Scholar]
- 28.Elomari M, Coroler L, Verhille S, Izard D, Leclerc H. Pseudomonas monteilii sp. nov., isolated from clinical specimens. Int J Syst Evol Microbiol. 1997; 47, 846–852. [DOI] [PubMed] [Google Scholar]
- 29.Masuda M, Yamasaki Y, Ueno S, Inoue A. Isolation of bisphenol A-tolerant/degrading Pseudomonas monteilii strain N-502. Extremophiles 2007; 11, 355–362. 10.1007/s00792-006-0047-9 [DOI] [PubMed] [Google Scholar]
- 30.Dabboussi F, Hamze M, Elomari M, Verhille S, Baida N, Izard D, Leclerc H. Taxonomic study of bacteria isolated from Lebanese spring waters: proposal for Pseudomonas cedrella sp. nov. and P. orientalis sp. nov. Res Microbiol. 1999; 150, 303–316. [DOI] [PubMed] [Google Scholar]
- 31.Vela AI, Gutiérrez MC, Falsen E, Rollán E, Simarro I, García P, Domínguez L, Ventosa A, Fernández-Garayzábal JF. Pseudomonas simiae sp. nov., isolated from clinical specimens from monkeys (Callithrix geoffroyi). Int J Syst Evol Microbiol. 2006; 56, 2671–2676. 10.1099/ijs.0.64378-0 [DOI] [PubMed] [Google Scholar]
- 32.Martínez-Rosales C, Castro-Sowinski S. Antarctic bacterial isolates that produce cold-active extracellular proteases at low temperature but are active and stable at high temperature. Polar Res. 2011; 30, 7123 10.3402/polar.v30i0.7123 [DOI] [Google Scholar]
- 33.Behrendt U, Ulrich A, Schumann P, Meyer JM, Spröer C. Pseudomonas lurida sp. nov., a fluorescent species associated with the phyllosphere of grasses. Int J Syst Evol Microbiol. 2007; 57, 979–985. 10.1099/ijs.0.64793-0 [DOI] [PubMed] [Google Scholar]
- 34.Mishra PK, Mishra S, Bisht SC, Selvakumar G, Kundu S, Bisht JK, Gupta HS. Isolation, molecular characterization and growth-promotion activities of a cold tolerant bacterium Pseudomonas sp. NARs9 (MTCC9002) from the Indian Himalayas. Biol Res. 2009; 42, 305–313. /S0716-97602009000300005. [PubMed] [Google Scholar]
- 35.Selvakumar G, Joshi P, Suyal P, Mishra PK, Joshi GK, Bisht JK, Bhatt JC, Gupta HS. Pseudomonas lurida M2RH3 (MTCC 9245), a psychrotolerant bacterium from the Uttarakhand Himalayas, solubilizes phosphate and promotes wheat seedling growth. World J Microbiol Biotechnol. 2011; 27, 1129–1135. 10.1007/s11274-010-0559-4 [DOI] [Google Scholar]
- 36.Whiteside JO, Garnsey SM, Timmer LW. Compendium of Citrus Diseases. (2 ed). American Phytopathology Society Press, Minnesota: 1989 [Google Scholar]
- 37.Scotta C, Mulet M, Sánchez D, Gomila M, Ramírez A, Bennasar A, García-Valdés E, Holmes B, Lalucat J. Identification and genomovar assignation of clinical strains of Pseudomonas stutzeri. Eur J Clin Microbiol Infect Dis. 2012; 31:2133–2139. 10.1007/s10096-012-1547-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
++, strongly positive; +, positive; w+, weakly positive; -, negative; ND, not determined.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. The data will be available without restriction after the acceptance for publication. The authors have provided all sequences in case you need them, and accession number have been added to the text (Lines 208-210). Sequence accession numbers are indicated for each strain in supplementary S1 Table. All sequences will be available, after acceptance of the manuscript publication, from the EMBL database (accession numbers HG805502-HG805808).