Abstract
Prenatal forms of autosomal dominant polycystic kidney disease (ADPKD) are rare but can be recurrent in some families, suggesting a common genetic modifying background. Few patients have been reported carrying, in addition to the familial mutation, variation(s) in polycystic kidney disease 1 (PKD1) or HNF1 homeobox B (HNF1B), inherited from the unaffected parent, or biallelic polycystic kidney and hepatic disease 1 (PKHD1) mutations. To assess the frequency of additional variations in PKD1, PKD2, HNF1B, and PKHD1 associated with the familial PKD mutation in early ADPKD, these four genes were screened in 42 patients with early ADPKD in 41 families. Two patients were associated with de novo PKD1 mutations. Forty patients occurred in 39 families with known ADPKD and were associated with PKD1 mutation in 36 families and with PKD2 mutation in two families (no mutation identified in one family). Additional PKD variation(s) (inherited from the unaffected parent when tested) were identified in 15 of 42 patients (37.2%), whereas these variations were observed in 25 of 174 (14.4%, P=0.001) patients with adult ADPKD. No HNF1B variations or PKHD1 biallelic mutations were identified. These results suggest that, at least in some patients, the severity of the cystic disease is inversely correlated with the level of polycystin 1 function.
Keywords: pediatrics, polycystic kidney disease, human genetics, cystic kidney
Autosomal dominant polycystic kidney disease (ADPKD; MIM# 173900 and 613095) is the most common inherited kidney disorder and occurs in 1 in 400 to 1 in 1000 individuals worldwide.1 ADPKD is typically an adult-onset disease characterized by progressive, bilateral cyst development often resulting in ESRD. The disease is genetically heterogeneous with two genes identified: polycystic kidney disease 1 (PKD1) (MIM# 601313; located on chromosome 16p13.3), responsible for approximately 85% of patients,2 and polycystic kidney disease 2 (PKD2) (MIM# 173910; located on chromosome 4q21),3 responsible for approximately 15% of patients. Variability in the phenotype is greatly influenced by the mutated gene (ESRD occurs earlier in patients with PKD1 mutations than in patients with PKD2 mutations)4 and the type of mutation (truncating mutations in PKD1 leading to earlier ESRD than nontruncating mutations).5 However, there is some intrafamilial variability, which may be extreme in a small proportion of patients presenting with fetal enlarged hyperechogenic kidneys resembling the phenotype observed in autosomal recessive PKD. Prognosis of these prenatal ADPKD patients was initially reported as poor,6–8 but a more recent series reported a favorable prognosis in most patients, at least in childhood.9 Although prenatal ADPKD is a rare condition, recurrence of patients in the same families suggested the involvement of genetic factors.
In the last 5 years, a few patients with prenatal ADPKD have been shown to carry, in addition to the PKD mutation that segregate in the family, other DNA variation(s) affecting either the second PKD allele or another gene involved in renal cystic diseases, such as polycystic kidney and hepatic disease 1 (PKHD1) or HNF1 homeobox B (HNF1B).10,11 However, because these findings were only reported as case reports, we have currently no idea regarding the proportion of patients of early ADPKD associated with the coinheritance of the familial mutation and of additional variation(s) in PKD1 or PKD2, or of additional HNF1B or PKHD1 mutation. To address this question, we analyzed the entire coding sequence of PKD1, PKD2, HNF1B, and PKHD1 genes in a series of 42 patients with ADPKD belonging to 41 families, diagnosed before birth (n=40) or at birth (n=2).
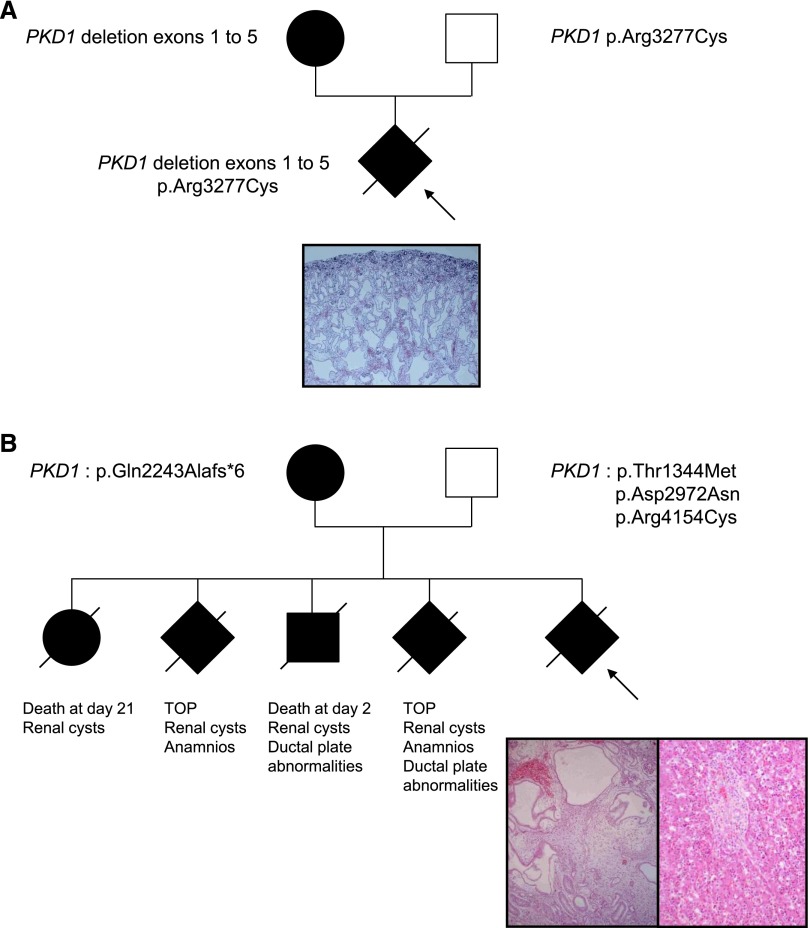
The sex ratio was 0.48 (20 men, 22 women). Forty patients occurred in 39 families with a known history of ADPKD. Two occurred sporadically but were included in the study because they presented as prenatal enlarged hyperechogenic kidneys compatible with ADPKD and were associated with a de novo PKD1 mutation. For the familial patients, the affected parent was the father in 20 patients and the mother in 19 patients. Average age of the affected parent was 36.7 years (range, 18–55). On average, four relatives (range, 0–11) per case were known to be affected (but data were incomplete in five families), and one (range, 0–3) was known to have reached end stage renal failure at a mean age of 49 years (range, 30–74) (however, this information was unknown in seven families). Renal ultrasound of the unaffected parent, who was 36 year old on average (range, 30–44), was performed in 14 patients. It was normal in 13 patients and showed one cyst in each kidney (6 and 11 mm) in one 37-year-old unaffected father. However, these ultrasounds were performed in 14 different centers with different apparatus. None of the unaffected parents underwent magnetic resonance imaging, which may have detected microcysts not identified by standard ultrasound. Table 1 shows prenatal features of the 40 patients diagnosed before birth and postnatal renal function at last follow-up, when known, in all patients. The mean term of discovery of renal anomalies was 23.2 weeks of gestation (range, 15–36; unknown in three patients), and mean kidneys’ size before birth was +3.9SD (range, 0–+14). Amniotic fluid volume was unknown in six patients, normal in 27 patients, and decreased in six patients; an anamnios was observed in one patient. Four pregnancies were terminated after parents’ request and case-by-case evaluation by the multidisciplinary prenatal committee in each center. Autopsy was performed after informed consent in all patients. Renal histology of patients 3 (Figure 1A) and 39 (Figure 1B) is shown in Figure 1. Patient 39 displayed severe renal cystic disease involving all nephronic structures, including glomeruli and abnormal liver histology with persistence of the ductal plate (Figure 1B). Patients 4 and 6 displayed complete disorganization of the renal architecture with a lack of corticomedullary differentiation and glomerular and tubular cysts, without renal dysplasia. Thirty-eight patients were born. Average term of birth was 39.2 weeks of gestation (range, 32–41) for the 29 patients for which this information was available. One presented with neonatal respiratory symptoms caused by moderate lung hypoplasia. All patients had postnatal ultrasound, and the last examination showed mean kidney sizes of +3SD (range, 0–+12) at an average age of 44 months (range, 0–201). Mean creatinine level (eGFR was frequently not available) was 33.7 μM/L (range, 7–150) at an average age of 50.3 months (range, 1–240). The two patients diagnosed at birth presented with palpable nephromegaly confirmed by renal ultrasound. Serum creatinine in these two patients was 28 and 150 μM/L at age 2 and 20 years, respectively.
Table 1.
Prenatal features in the 41 patients diagnosed before birth (all but patients 23 and 26) and renal function at the last follow-up
| Patient No. | Term of Discovery | Kidney Sizes (SD) | Hyperechogenicity (Y/N) | Serum Creatinine (μM) (Age in mo) |
|---|---|---|---|---|
| 1 | Unknown | Unknown | Y | 35 (79) |
| 2 | 22 | +4 | Y | 24 (35) |
| 3 | 22 | Unknown | Y | TOP |
| 4 | 22 | +6 | Y | TOP |
| 5 | 22 | Unknown | Y | 38 (57) |
| 6 | 15 | +14 | Y | TOP |
| 7 | 22 | +2 | Y | 16 (27) |
| 8 | 32 | +3 | Y | 34 (2) |
| 9 | 22 | +4 | Y | 31 (48) |
| 10 | 25 | Unknown | Unknown | 13 (5) |
| 11 | 22 | +2 | Unknown | Unknown |
| 12 | 32 | 0 | Unknown | 51 (132) |
| 13 | 17 | +3 | Y | 38 (92) |
| 14 | 22 | +3 | Y | 36 (47) |
| 15 | 22 | Unknown | Y | 7 (15) |
| 16 | 27 | +3 | Y | 33 (48) |
| 17 | 24 | +1 | Unknown | 27 (42) |
| 18 | 22 | +3 | Unknown | 23 (14) |
| 19 | 26 | +4 | Y | 25 (12) |
| 20 | 22 | +1 | Y | Unknown |
| 21 | 25 | +2 | Y | 26 (24) |
| 22 | 18 | +3 | Y | 13 (6) |
| 23 | At birth | +14 | Y | 150 (240) |
| 24 | 20 | Unknown | Unknown | 34 (102) |
| 25 | 22 | Unknown | Y | 32 (13) |
| 26 | At birth | Unknown | Unknown | 28 (30) |
| 27 | 20 | Unknown | Unknown | 17 (6) |
| 28 | 36 | Unknown | Unknown | 36 (83) |
| 29 | 22 | +10 | Y | 62 (52) |
| 30 | 22 | +4 | Unknown | 17 (4) |
| 31 | 22 | +3 | Y | 24 (1) |
| 32 | 33 | +3 | Y | 20 (3) |
| 33 | 22 | +3 | Y | 17 (10) |
| 34 | 22 | 0 | Y | 20 (12) |
| 35 | 22 | +3 | Y | 27 (20) |
| 36 | 23 | +2 | Unknown | 30 (49) |
| 37 | 25 | +3 | Unknown | 17 (8) |
| 38 | 22 | +6 | Y | 95 (173) |
| 39 | 22 | Unknown | Y | TOP |
| 40 | Unknown | Unknown | Y | 60 (201) |
| 41 | Unknown | Unknown | Y | Unknown |
| 42 | 22 | Unknown | Y | 25 (68) |
Kidney size was evaluated according to Chitty and Altman.29 Y, yes; n, no; TOP, termination of pregnancy.
Figure 1.
Pedigrees of patients 3 and 39 with genotypes and renal/liver histology. (A) Patient 3: renal histology shows cystic dilations involving tubules and glomeruli, regularly distributed within the parenchyma. The superficial cortex is respected. (B) Patient 39: family history shows four previous instances of early death or termination of pregnancy (TOP) for renal cystic disease (with ductal plate abnormalities in the two patients with autopsy). Renal histology of the fetus (left panel) shows large and irregularly distributed cysts, involving tubules and glomeruli. Liver histology (right panel) shows the persistence of the ductal plate. Although we were not able to test fetal DNA, the unaffected father carries three PKD1 variants, all predicted to be deleterious. Original magnification, ×10.
The mutation inherited from the affected parent was identified in all but one (patient 5) of the 39 familial patients (Supplemental Table 1, Table 2). It was affecting PKD1 in 36 families and PKD2 in two families. Therefore, the overall mutation detection rate achieved in familial patients was 97.4% (38/39). PKD1 mutations consist in 22% nonsense (n=8), 30% frameshift (n=11), 11% splicing (n=4), 2.7% frame insertion (n=1), 2.7% large rearrangement (n=1), and 30.5% missense (n=11), confirming the high level of allelic heterogeneity. In addition, two de novo PKD1 missense mutations were identified in the two patients that occurred sporadically (Table 2). Average age at ESRD in families with nontruncating PKD1 mutation was 54.3 years (range, 34–71), and it was 49.2 years (range, 30–74) in families with truncating mutations.
Table 2.
PKD1 and PKD2 variations identified in index cases and affected and unaffected parents
| Patient No. | Affected Parent | Proband | Affected Parent | Unaffected Parent | Reference | |||
|---|---|---|---|---|---|---|---|---|
| Nucleotide Change(s) (Gene Involved) | Amino Acid Change(s) (Gene Involved) | Nucleotide Change (Gene Involved) | Amino Acid Change (Gene Involved) | Nucleotide Change (Gene Involved) | Amino Acid Change (Gene Involved) | |||
| 1 | Father | c.4804del (PKD1) | p.Arg1602Alafs*31 (PKD1) | c.4804del (PKD1) | p.Arg1602Alafs*31 (PKD1) | This study | ||
| c.9815G>A (PKD1) | p.Arg3272His (PKD1) | c | c | This study | ||||
| 2 | Mother | c.964C>G (PKD2) | p.Arg322Gly (PKD2) | c.964C>G (PKD2) | p.Arg322Gly (PKD2) | This study | ||
| 3 | Mother | c.1- ?_1201+?del (PKD1) | c.1-?_1201+?del (PKD1) | This study | ||||
| c.9829C>T (PKD1) | p.Arg3277Cys (PKD1) | c.9829C>T (PKD1) | p.Arg3277Cys (PKD1) | This study | ||||
| 4 | None | c.2534T>C (PKD1) | p.Leu845Ser (PKD1) | d | 30 | |||
| 5 | Mother | d | ||||||
| 6 | Father | c.2180T>C (PKD1) | p.Leu727Pro (PKD1) p.Thr3945Met (PKD1) | c.2180T>C (PKD1) | p.Leu727Pro (PKD1) | 20 | ||
| c.11834C>T (PKD1) | c.11834C>T (PKD1) | p.Thr3945Met (PKD1)d | This study | |||||
| 7 | Mother | c.6472C>T (PKD1) | pGln2158* (PKD1) | c.6472C>T (PKD1) | p.Gln2158* (PKD1) | d | 31 | |
| 8 | Father | c.7108T>A (PKD1) | p.Cys2370Ser (PKD1) p.Arg4154Cys (PKD1) | c.7108T>A (PKD1) | p.Cys2370Ser (PKD1) | 20 | ||
| c.12460C>T (PKD1) | c.12460C>T (PKD1) | p.Arg4154Cys (PKD1)d | 12 | |||||
| 9 | None | c.8536A>C (PKD1) | p.Thr2846Pro (PKD1) | d | This study | |||
| 10 | Mother | c.9568G>A (PKD1) | c.9568G>A (PKD1) | c | This study | |||
| 11 | Mother | c.11969T>G (PKD1) | p.Leu3990Arg (PKD1) | c.11969T>G (PKD1) | p.Leu3990Arg (PKD1) | This study | ||
| 12 | Mother | c.9563A>T (PKD1) | p.Asn3188Ile (PKD1) | c.9563A>T (PKD1) | p.Asn3188Ile (PKD1) | d | This study | |
| 13 | Father | c.1010_1013dup (PKD1) | p.Leu339Glyfs*33 (PKD1) | c.1010_1013dup (PKD1) | p.Leu339Glyfs*33 (PKD1) | This study | ||
| c.8129C>A (PKD1) | p.Thr2710Asn (PKD1) | c.8129C>A (PKD1) | p.Thr2710Asn (PKD1) | This study | ||||
| 14 | Mother | c.12503dup (PKD1) | p.Ser4169Leufs*41 (PKD1) | c.12503dup (PKD1) | p.Ser4169Leufs*41 (PKD1) | This study | ||
| c.4831G>A (PKD1) | p.Val1611Ile (PKD1) | c.4831G>A (PKD1) | p.Val1611Ile (PKD1) | This study | ||||
| 15 | Mother | c.974G>C (PKD2) | p.Arg325Pro (PKD2) | c.974G>C (PKD2) | p.Arg325Pro (PKD2) | This study | ||
| 16 | Mother | c.9089_9096del (PKD1) | p.Leu3030Profs*36 (PKD1) | c.9089_9096del (PKD1) | p.Leu3030Profs*36 (PKD1) | cd | This study | |
| 17 | Father | c.2833_2834del (PKD1) | p.Val945Thrfs*155 (PKD1) | c.2833_2834del (PKD1) | p.Val945Thrfs*155 (PKD1) | d | 16 | |
| 18 | Father | c.6548_6551del (PKD1) | p.Thr2183Serfs*28 (PKD1) p.Asp1332Asn (PKD1) | c.6548_6551del (PKD1) | p.Thr2183Serfs*28 (PKD1) | This study | ||
| c.3994G>A (PKD1) | c.3994G>A (PKD1) | p.Asp1332Asn (PKD1) | 15 | |||||
| 19 | Mother | c.7978G>T (PKD1) | p.Asp2660Tyr (PKD1) p.Ser4054Phe (PKD1) | c.7978G>T (PKD1) | p.Asp2660Tyr (PKD1) | c | c | This study |
| c.12161C>T (PKD1) | This study | |||||||
| 20 | Father | c.3202del (PKD1) | p.Gln1068Serfs*36 (PKD1) | c.3202del (PKD1) | p.Gln1068Serfs*36 (PKD1) | This study | ||
| 21 | Mother | c.5517G>T (PKD1) | p.Trp1839Cys (PKD1) | c.5517G>T (PKD1) | p.Trp1839Cys (PKD1) | This study | ||
| c.5830G>A (PKD1) | p.Gly1944Arg (PKD1) | c.5830G>A (PKD1) | p.Gly1944Arg (PKD1)d | This study | ||||
| 22 | Father | c.5764C>T (PKD1) | p.Gln1922* (PKD1) | c.5764C>T (PKD1) | p.Gln1922* (PKD1) | d | 30 | |
| 23 | Father | c.9562A>G (PKD1) | p.Asn3188Asp (PKD1) | c.9562A>G (PKD1) | p.Asn3188Asp (PKD1) | This study | ||
| c.6173A>G (PKD1) | p.Gln2058Arg (PKD1) | c.6173A>G (PKD1) | p.Gln2058Arg (PKD1)d | This study | ||||
| 24 | Father | c.8497C>T (PKD1) | p.Pro2833Ser (PKD1) | c.8497C>T (PKD1) | p.Pro2833Ser (PKD1) | cd | This study | |
| 25 | Father | c.12440dup (PKD1) | p.Glu4148Glyfs*9 (PKD1) | c.12440dup (PKD1) | p.Glu4148Glyfs*9 (PKD1) | This study | ||
| 26 | Mother | c.12003+1G>A (PKD1) | c.12003+1G>A (PKD1) | cd | 16 | |||
| 27 | Father | c.11713–2A>G (PKD1) | c.11713–2A>G (PKD1) | This study | ||||
| 28 | Father | c.11538–2A>G (PKD1) | c.11538–2A>G (PKD1) | 16 | ||||
| 29 | Father | c.2582G>A (PKD1) | p.Trp861* (PKD1) | c.2582G>A (PKD1) | p.Trp861* (PKD1) | 16 | ||
| c.12074A>G (PKD1) | p.Glu4025Gly (PKD1) | c.12074A>G (PKD1) | p.Glu4025Gly (PKD1) | This study | ||||
| 30 | Father | c.11451_11453dup (PKD1) | p.Gly3818dup (PKD1) | c.11451_11453dup (PKD1) | p.Gly3818dup (PKD1) | d | This study | |
| 31 | Mother | c.5873G>A (PKD1) | p.Trp1958* (PKD1) | c.5873G>A (PKD1) | p.Trp1958* (PKD1) | |||
| c.12460C>T (PKD1) | p.Arg4154Cys (PKD1) | c.12460C>T (PKD1) | p.Arg4154Cys (PKD1) | 12 | ||||
| 32 | Mother | c.11884C>T (PKD1) | p.Gln3962* (PKD1) | c.11884C>T (PKD1) | p.Gln3962* (PKD1) | d | This study | |
| 33 | Father | c.8017–3C>G (PKD1) | c.8017–3C>G (PKD1) | This study | ||||
| 34 | Mother | c.8998del (PKD1) | p.Arg3000Alafs*74 (PKD1) | c.8998del (PKD1) | p.Arg3000Alafs*74 (PKD1) | This study | ||
| 35 | Mother | c.6472C>T (PKD1) | p.Gln2158* (PKD1) | c.6472C>T (PKD1) | p.Gln2158* (PKD1) | 32 | ||
| 36 | Father | c.7115C>G (PKD1) | p.Ser2372Cys (PKD1) | c.7115C>G (PKD1) | p.Ser2372Cys (PKD1) | 16 | ||
| 37 | Mother | c.689G>C (PKD1) | p.Cys230Ser (PKD1) | c.689G>C (PKD1) | p.Cys230Ser (PKD1) | 20 | ||
| 38 | Father | c.11614G>T (PKD1) | p.Glu3872* (PKD1) | c.11614G>T (PKD1) | p.Glu3872* (PKD1) | c.9548G>A (PKD1) | p.Arg3183Gln (PKD1) | 31 |
| c.9548G>A (PKD1) | p.Arg3183Gln (PKD1) | |||||||
| 39 | Mother | a | a | c.6727_6730del (PKD1) | p.Gln2243Alafs*6 (PKD1) | c.4031C>T (PKD1) | p.Thr1344Met (PKD1) p.Asp2972Asn (PKD1) | This study |
| c.8914G>A (PKD1) | p.Arg4154Cys (PKD1) | 13 | ||||||
| c.12460C>T (PKD1) | 12 | |||||||
| 40 | Father | c.10343del (PKD1) | p.Pro3448Glnfs*25 (PKD1) | b | b | c | 16 | |
| 41 | Father | c.10343del (PKD1) | p.Pro3448Glnfs*25 (PKD1) | b | b | 16 | ||
| 42 | Father | c.10232G>A (PKD1) | p.Trp3411* (PKD1), p.Asn3295Ser (PKD1) | c.10232G>A (PKD1) | p.Trp3411* (PKD1) | c.9884A>G (PKD1) | p.Asn3295Ser (PKD1) | This study |
| c.9884A>G (PKD1) | ||||||||
Not analyzed (fetal DNA was degraded).
DNA from affected parent was not available.
DNA from unaffected parent was not available.
Unaffected parent had renal ultrasound.
Among the 38 probands for whom the familial mutation was identified, 15 were found to carry additional PKD1 variant(s) scored as likely pathogenic (see Methods) and/or located at invariant positions in polycystin 1 (PC-1) orthologs to Xenopus (Supplemental Figure 1, Table 2), shown for 13 of them to be inherited from the unaffected parent (DNA from healthy parent was not available in two patients; Table 2). The two probands with de novo PKD1 misense mutations were not found to carry additional variation in PKD1 or PKD2. One proband (patient 39), displaying a severe renal and liver phenotype, could not be tested because of the poor quality of the fetal DNA. However, his unaffected father was shown to carry three PKD1 variations. Paternal grandparents’ DNA was not available. There were 15 different variations identified in addition to the familial mutations, one of them (c.12460C>T p.Arg4154Cys) being found three times. Ten out of the 15 variations were located in invariant positions in PC-1 orthologs to Xenopus, and four were in highly conserved positions. They are shown in Supplemental Table 2, with the scores established to evaluate their putative pathogenicity by different programs (see Methods). Six were previously reported,10,12–15 and three were present in our in-house database: p.Thr2710N and p.Arg3183Gln were associated with another PKD1 or PKD2 mutation in patients diagnosed in the fourth decade, and the p.R3277C mutation was identified at the homozygous state in a woman transplanted at 50 years. No additional PKD1 or PKD2 variations were identified in the other patients. We did not identify biallelic mutations in PKHD1. In addition to known polymorphisms and silent variations, four heterozygous PKHD1 changes were detected. One heterozygous frame shift mutation (c.9689del [p.Asp3230Valfs*34]) was identified in patient 20, an 8-year-old boy with unknown renal function. That mutation was inherited from his affected father, who was therefore also carrying both the PKD1 and PKHD1 mutations, and had an eGFR of 81 ml/min per 1.73 m2 at the age of 30 years. One unreported heterozygous missense variation (p.Val1087Met [c.3259G>A]) was identified in the affected mother of patient 39, who had a normal renal function at the age of 41 years. Two other missense variations (p.His491Gln [c.1473C>A] and p.Ser3210Cys [c.9629C>G]), known as rs 368613297 and rs141081295, were identified in patients 29 (a 4-year-old girl with a stage 2 chronic renal failure and carrying the p.Glu4025Gly PKD1 variation in addition to a truncating PKD1 mutation) and 4 (a fetus presenting with very large hyperechogenic kidneys and carrying a single p.Leu845Ser de novo mutation in PKD1), respectively. Finally, no HNF1B mutation was detected in this series. To evaluate the frequency of additional variations in PKD1 in a population of patients affected with classic (with first symptoms in adulthood) ADPKD, we went back to 174 patients of an in-house adult cohort5,16 for whom the complete exploration of the PKD1 gene was completed. In addition to the polymorphic variants known to be frequent in PKD1 (9.05 per patient on average), we identified other rare variants, not described in the common databases (the Single Nucleotide Polymorphism Database and the Exome Sequencing Project) and predicted to be pathogenic, in only 25 (14.4%) out of these 174 patients, including the patients carrying the three variations previously mentioned (Supplemental Table 3). That proportion is very significantly (P=0.001) smaller than the 16 of 43 (37.2%) of our series of early ADPKD. In addition, the p.Arg2765Cys variant was identified in nine patients. However, the p.Arg2765Cys variant, previously reported as a likely incompletely penetrant allele,10 was not considered as such because it is frequently observed in the Genkyst cohort,5,16 without significant effect on the phenotype (A. Marie-Pierre, unpublished data).
Somatic second hit mutation, similar to the carcinogenesis Knudson’s model, was the first proposed mechanism for cysts formation in ADPKD, but it is now accepted that genetics or environmental additional factors can influence the severity of the disease. Recent data have shown that the extent of cyst formation is inversely correlated with the level of PC-1 function.17 Accordingly, the association of an additional variation to the disease-causing mutation is expected to lead to a more severe (and earlier) disease.10,18,19 However, in 26 out of 41 patients of our series with an identified mutation, no additional PKD1-PKD2 variation was identified. These patients might be associated with a reduced level of PC-1 because of variation(s) modifying the expression of PKD1 at the transcriptional or post-transcriptional level or because of epigenetic factors. Alternatively these patients may be associated with mutations in other genes involved in the cilia-dependent pathway; however, the role of polycystins in signaling within primary cilia and the relationship of polycystins with other cilia proteins is currently not well understood. In contrast with previously reported data,11 we did not identify additional HNF1B variations or biallelic PKHD1 mutations in our series. Nevertheless, a possible role of the heterozygous PKHD1 variations (shown in supplemental Table 4) that we identified in modifying the phenotype cannot be ruled out.
In our series, the proportion (95%) of prenatal instances of ADPKD carrying a PKD1 mutation is more important than the 85% of patients observed in a large series of classic ADPKD.16,20,21 In addition, the average age at ESRD in the affected relatives in our series is earlier than in a larger cohort of ADPKD patients carrying a PKD1 mutation.5 In particular, the age at ESRD in affected ancestors carrying a nontruncating mutation was 54.3 years (vs 67.9 years as reported by Cornec-Le Gall and colleagues5). This may indicate that the level of reduction of PC-1 function associated with these familial mutations is important. Only two instances of prenatal ADPKD occurred in families carrying a PKD2 missense mutation. Interestingly, all variations inherited from the unaffected parent involved the PKD1 gene. All were located in invariant and/or highly conserved regions and/or were predicted as pathogenic in at least four of five tools; however, the parents carrying these variants were all a priori unaffected (some having a normal kidney ultrasound after the age of 30 years). This likely illustrates the fact that there is a threshold of PC-1 function above which no cysts formation . When the level of PC-1 activity goes below that threshold, cysts initiation and progression ensues. The improvement in ultrasound imaging in the last years may have contributed to the detection of some recent instances of prenatal ADPKD. However, the rate (35.7%) of additional PKD1 variation(s) that we report here, which is much more frequent than in a cohort of patients affected with classic (adult) ADPKD, confirms our current understanding that the rate of cyst progression and growth is a regulated trait that is earliest and quickest as the function of PC-1 is altered.
Concise Methods
Patients
This was a retrospective study that included 42 patients from 41 families sent from pediatric nephrology, obstetrics, and genetics departments from France and Tunisia between December 2011 and February 2014. DNA or blood sample from all patients were received for molecular screening in the laboratory of molecular genetics in Brest, France, with written informed consents and clinical information. This study was conducted with the approval of the Comité de Protection des Personnes pour la recherche biomédicale Ile de France 2.
Mutation Analysis
Molecular testing of PKD1, PKD2, and HNF1B genes was performed by direct sequencing, followed by quantitative fluorescent multiplex PCR, Multiplex Ligation-Dependent Probe Amplification (MRC-Holland), and array-comparative genomic hybridization, as previously described16 in all but one index patients (as fetal DNA was degraded). When variations were identified, both parents were tested for the variations. However, DNA from the unaffected parent was unavailable in seven patients (including two patients with a PKD1 variation identified in addition to the familial mutation), and DNA from the deceased affected parent was unavailable once. The PKHD1 gene was tested as previously described.22
Scoring Missense Mutations
The scoring for pathogenicity prediction for missense mutations was performed using a multisequence alignment of PC-1 orthologs of human, macaque, mouse, rat, dog, opossum, and Xenopus and with the combination of five different methods, including the Grantham matrix scoring system, Align-GVGD,23 PolyPhen (http://genetics.bwh.harvard.edu/pph/),24,25 SIFT,26,27 and MutationTaster.28 From the results of these analyses, the missense variations were considered as pathogenic for the following: (1) a Grantham distance >60 (corresponding to a nonconservative substitution); (2) a score ≥C15 with Align-GVGD; (3) a possibly or probably damaging prediction with PolyPhen; (4) a deleterious prediction with SIFT; and (5) a disease-causing prediction with MutationTaster. Variations located on invariable or highly conserved positions and/or scored as pathogenic by at least four tools were taken into account. Effect of changes on splice site was checked by Alamut software (Interactive Biosoftware, Rouen, France).
Statistical Analyses
Data were analyzed using chi-squared and Fisher exact tests. All of the tests were two-sided. P values <0.05 were considered significant.
Disclosures
None.
Supplementary Material
Acknowledgments
Part of this work was funded by the French Société de Néphrologie. We thank the families for their participation. We are grateful to Dr. Marie-Claire Gubler for helpful discussions and analysis of fetal kidneys’ pathology. We are grateful to Christine Bole-Feysot and Patrick Nitschke, from the Plateforme Génomique Institut Imagine, for help with PKHD1 analysis. We thank Olivier Gribouval for help with the figure.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014101051/-/DCSupplemental.
References
- 1.Wilson PD: Polycystic kidney disease. N Engl J Med 350: 151–164, 2004 [DOI] [PubMed] [Google Scholar]
- 2.The European Polycystic Kidney Disease Consortium : The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell 78: 725, 1994 [PubMed] [Google Scholar]
- 3.Mochizuki T, Wu G, Hayashi T, Xenophontos SL, Veldhuisen B, Saris JJ, Reynolds DM, Cai Y, Gabow PA, Pierides A, Kimberling WJ, Breuning MH, Deltas CC, Peters DJ, Somlo S: PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272: 1339–1342, 1996 [DOI] [PubMed] [Google Scholar]
- 4.Hateboer N, v Dijk MA, Bogdanova N, Coto E, Saggar-Malik AK, San Millan JL, Torra R, Breuning M, Ravine D, European PKD1-PKD2 Study Group : Comparison of phenotypes of polycystic kidney disease types 1 and 2. Lancet 353: 103–107, 1999 [DOI] [PubMed] [Google Scholar]
- 5.Cornec-Le Gall E, Audrézet MP, Chen JM, Hourmant M, Morin MP, Perrichot R, Charasse C, Whebe B, Renaudineau E, Jousset P, Guillodo MP, Grall-Jezequel A, Saliou P, Férec C, Le Meur Y: Type of PKD1 mutation influences renal outcome in ADPKD. J Am Soc Nephrol 24: 1006–1013, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blyth H, Ockenden BG: Polycystic disease of kidney and liver presenting in childhood. J Med Genet 8: 257–284, 1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross DG, Travers H: Infantile presentation of adult-type polycystic kidney disease in a large kindred. J Pediatr 87: 760–763, 1975 [DOI] [PubMed] [Google Scholar]
- 8.Proesmans W, Van Damme B, Casaer P, Marchal G: Autosomal dominant polycystic kidney disease in the neonatal period: Association with a cerebral arteriovenous malformation. Pediatrics 70: 971–975, 1982 [PubMed] [Google Scholar]
- 9.Boyer O, Gagnadoux MF, Guest G, Biebuyck N, Charbit M, Salomon R, Niaudet P: Prognosis of autosomal dominant polycystic kidney disease diagnosed in utero or at birth. Pediatr Nephrol 22: 380–388, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Rossetti S, Kubly VJ, Consugar MB, Hopp K, Roy S, Horsley SW, Chauveau D, Rees L, Barratt TM, van’t Hoff WG, Niaudet P, Torres VE, Harris PC: Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney Int 75: 848–855, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergmann C, von Bothmer J, Ortiz Brüchle N, Venghaus A, Frank V, Fehrenbach H, Hampel T, Pape L, Buske A, Jonsson J, Sarioglu N, Santos A, Ferreira JC, Becker JU, Cremer R, Hoefele J, Benz MR, Weber LT, Buettner R, Zerres K: Mutations in multiple PKD genes may explain early and severe polycystic kidney disease. J Am Soc Nephrol 22: 2047–2056, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrichot RA, Mercier B, Simon PM, Whebe B, Cledes J, Ferec C: DGGE screening of PKD1 gene reveals novel mutations in a large cohort of 146 unrelated patients. Hum Genet 105: 231–239, 1999 [DOI] [PubMed] [Google Scholar]
- 13.Bouba I, Koptides M, Mean R, Costi CE, Demetriou K, Georgiou I, Pierides A, Siamopoulos K, Deltas CC: Novel PKD1 deletions and missense variants in a cohort of Hellenic polycystic kidney disease families. Eur J Hum Genet 9: 677–684, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Yu C, Yang Y, Zou L, Hu Z, Li J, Liu Y, Ma Y, Ma M, Su D, Zhang S: Identification of novel mutations in Chinese Hans with autosomal dominant polycystic kidney disease. BMC Med Genet 12: 164, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossetti S, Hopp K, Sikkink RA, Sundsbak JL, Lee YK, Kubly V, Eckloff BW, Ward CJ, Winearls CG, Torres VE, Harris PC: Identification of gene mutations in autosomal dominant polycystic kidney disease through targeted resequencing. J Am Soc Nephrol 23: 915–933, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Audrézet MP, Cornec-Le Gall E, Chen JM, Redon S, Quéré I, Creff J, Bénech C, Maestri S, Le Meur Y, Férec C: Autosomal dominant polycystic kidney disease: Comprehensive mutation analysis of PKD1 and PKD2 in 700 unrelated patients. Hum Mutat 33: 1239–1250, 2012 [DOI] [PubMed] [Google Scholar]
- 17.Fedeles SV, Tian X, Gallagher AR, Mitobe M, Nishio S, Lee SH, Cai Y, Geng L, Crews CM, Somlo S: A genetic interaction network of five genes for human polycystic kidney and liver diseases defines polycystin-1 as the central determinant of cyst formation. Nat Genet 43: 639–647, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopp K, Ward CJ, Hommerding CJ, Nasr SH, Tuan HF, Gainullin VG, Rossetti S, Torres VE, Harris PC: Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J Clin Invest 122: 4257–4273, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vujic M, Heyer CM, Ars E, Hopp K, Markoff A, Orndal C, Rudenhed B, Nasr SH, Torres VE, Torra R, Bogdanova N, Harris PC: Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J Am Soc Nephrol 21: 1097–1102, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossetti S, Consugar MB, Chapman AB, Torres VE, Guay-Woodford LM, Grantham JJ, Bennett WM, Meyers CM, Walker DL, Bae K, Zhang QJ, Thompson PA, Miller JP, Harris PC, CRISP Consortium : Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 18: 2143–2160, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Torra R, Badenas C, Darnell A, Nicolau C, Volpini V, Revert L, Estivill X: Linkage, clinical features, and prognosis of autosomal dominant polycystic kidney disease types 1 and 2. J Am Soc Nephrol 7: 2142–2151, 1996 [DOI] [PubMed] [Google Scholar]
- 22.Failler M, Gee HY, Krug P, Joo K, Halbritter J, Belkacem L, Filhol E, Porath JD, Braun DA, Schueler M, Frigo A, Alibeu O, Masson C, Brochard K, Hurault de Ligny B, Novo R, Pietrement C, Kayserili H, Salomon R, Gubler MC, Otto EA, Antignac C, Kim J, Benmerah A, Hildebrandt F, Saunier S: Mutations of CEP83 cause infantile nephronophthisis and intellectual disability. Am J Hum Genet 94: 905–914, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavtigian SV, Greenblatt MS, Lesueur F, Byrnes GB, IARC Unclassified Genetic Variants Working Group : In silico analysis of missense substitutions using sequence-alignment based methods. Hum Mutat 29: 1327–1336, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramensky V, Bork P, Sunyaev S: Human non-synonymous SNPs: Server and survey. Nucleic Acids Res 30: 3894–3900, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sunyaev SR, Eisenhaber F, Rodchenkov IV, Eisenhaber B, Tumanyan VG, Kuznetsov EN: PSIC: Profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng 12: 387–394, 1999 [DOI] [PubMed] [Google Scholar]
- 26.Ng PC, Henikoff S: Predicting deleterious amino acid substitutions. Genome Res 11: 863–874, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng PC, Henikoff S: Accounting for human polymorphisms predicted to affect protein function. Genome Res 12: 436–446, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D: MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7: 575–576, 2010 [DOI] [PubMed] [Google Scholar]
- 29.Chitty LS, Altman DG: Charts of fetal size: Kidney and renal pelvis measurements. Prenat Diagn 23: 891–897, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Thomas R, McConnell R, Whittacker J, Kirkpatrick P, Bradley J, Sandford R: Identification of mutations in the repeated part of the autosomal dominant polycystic kidney disease type 1 gene, PKD1, by long-range PCR. Am J Hum Genet 65: 39–49, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan YC, Blumenfeld JD, Anghel R, Donahue S, Belenkaya R, Balina M, Parker T, Levine D, Leonard DG, Rennert H: Novel method for genomic analysis of PKD1 and PKD2 mutations in autosomal dominant polycystic kidney disease. Hum Mutat 30: 264–273, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Rossetti S, Chauveau D, Kubly V, Slezak JM, Saggar-Malik AK, Pei Y, Ong AC, Stewart F, Watson ML, Bergstralh EJ, Winearls CG, Torres VE, Harris PC: Association of mutation position in polycystic kidney disease 1 (PKD1) gene and development of a vascular phenotype. Lancet 361: 2196–2201, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.