Abstract
Enterococci are Gram positive and catalase- negative cocci that are found in the gastrointestinal tract of mammals and birds, and are readily isolated from soil, surface and waters. The aim of this study was to discriminate between Enterococcus isolates based on repetitive element sequence based –PCR (Rep-PCR) with the BOXA2R primer and their antibiotics profile. Enterococci isolates were obtained from 180 fecal samples. The isolates were identified by biochemical reaction and specific identification was confirmed by PCR with species specific primers. All isolates were subjected to Rep typing and antimicrobial susceptibility tests. Rep-PCR analysis of 180 isolates revealed 93 REP types with forty-five single types (ST1 to ST45) and forty-eight common types (CT1 to 48). Antibiotic susceptibility tests exhibited that 53 (29.4%), 43 (23.8%), 11 (6.1%) and 9 (5%) were resistant to erythromycin, tetracycline, gentamicin and ciprofloxacin respectively but among the isolates, sixteen were multi drug resistant (MDR). These MDR isolates showed 11 Rep types with seven single types and four common types. In addition, 81.2% of MDR isolates were from male subjects and the average age of these persons was more than fifty years. This study showed that 56.2% of MDR isolates were homogeneous with 95 % similarity, and high rate of resistance to tetracycline and erythromycin (81.2%) were observed in these isolates. The concern about these normal flora isolates are the pathogenic potential of these bacteria through the horizontal transfer of antibiotic resistance and virulence genes.
Key Words: Enterococcus, Rep- PCR, antibiotic profile, normal flora
Enterococci are Gram positive, aerotolerant fermentative and catalase-negative cocci that are found in the gastrointestinal (GI) tract of mammals, birds and are readily isolated from soil, surface waters and sediments. The ability of this organism to survive in various host environments provides great temptation to be selected as a fecal contaminant indicator (1).
On the other hand, they are becoming significant pathogens worldwide, especially with respect to nosocomial infections (2). Multiple antibiotic exposure in hospital settings, promotes a high level of resistance to glycopeptides and aminoglycosides, which are of great clinical importance, providing them with an evolutionary pressure for selective advantage (3).
Among enterococcus species, E. faecalis and E. Faecium are important and have been isolated from healthy persons and patients. These species are causative agents of a variety of human infections such as bloodstream, heart, abdomen and urinary tract infections (4).
Differentiation of enterococci at the species level is mostly made by phenotypic methods, such as detecting the utilization of some carbohydrates and other substrates (5). In recent years, molecular techniques such as ribotyping, pulsed-field gel electrophoresis, Phene Plate (PhP) typing and repetitive element sequence based –PCR (Rep-PCR) have been used as effective methods for detecting and discriminating of pathogenic as well as non pathogenic isolates (1).
Among these methods, Rep-PCR is a DNA fingerprint technique that uses repetitive intergenic DNA sequences to differentiate between sources of fecal pollution. In this technique, the DNA present between adjacent repetitive extragenic elements is amplified by PCR and various DNA fragment sizes are produced. The PCR products are separated based on their size by agarose-gel electrophoresis to yield fingerprint patterns. In the final step, these patterns can be analyzed with the pattern recognition computer software (6,7).
Rep- PCR method is more accurate, reproducible and efficient than some other typing methods such as ribotyping, PFGE and PhP typing (6). On the other hand, this method is suitable for microbial ecology and subtype analysis investigations (8).
In this study, Rep-PCR with the BOXA2R primers was used to discriminate between enterococcus isolates and their antibiotics profile was assessed.
Materials and methods
Sample collection and species identification
Enterococci isolates were obtained from 180 fecal samples (persons between 20 to 70 years old) during 2014 in Tehran, Iran. The isolates were identified by their growth on m-enterococcus agar (ME agar), Gram staining, catalase reaction and streaking on bile aesculin azid agar plates (9). Colonies with a black zone of aesculin hydrolysis were considered as presumptive enterococci. Final identification was confirmed by PCR with species specific primers (10).
Antimicrobial susceptibility tests
Resistance phenotypes were determined by disk diffusion method on Mueller Hinton agar according to the guidelines of the clinical and laboratory standards institute (CLSI) (11). Antibio-tic disc test, included vancomycin, ciprofloxacin, tetracycline, erythromycin, chloramphenicol, ampi-cillin, linezolid, teicoplanin, gentamycin and syner-cid (quinupristin and dalfopristin). E. faecalis ATCC 29212 was used as reference strain for antimicrobial susceptibility testing.
DNA extraction and REP-PCR
Bacterial DNA was prepared with DNA extraction kit (Qiagen, Hilden, Germany). BOX-PCR analysis was ideally performed using a BOXA2R oligonucleotide sequence (5′-ACG TGGTTTGAAGAGATTTTCG-3′) as described by Pangallo et al.(1). Briefly, PCR assay was performed in a total volume of 25 μl containing 50 ng DNA, 1.25 U Taq DNA polymerase, 1xPCR buffer, 2.5 mM MgCl2 ,200 μM dNTP, 30 pmol BOXA2 primer and using the following conditions: initial denaturation at 95 °C for 7 min, 35 cycles of denaturation at 90 °C for 30 s ,annealing at 40 °C for 1 min and elongation at 60 °C for 6 min with final extension at 65 °C for 16 min. Electrophoresis of the PCR products was performed in 1% agarose gel for 150 min at 80 V followed by visualization in UV transilluminator after staining with ethidium bromide.
Results
Prevalence of enterococci species
A total of 180 enterococci isolates were obtained from fecal samples. 96 (53.3%) and 84 (46.7%) of these isolates were from male and female subjects, respectively. Species identification showed that 158 (87.7%) and 22 (12.3%) of isolates were E. faecium and E. faecalis, respectively.
Antibiotic resistance
All of the 180 isolates (100%) were susceptible to chloramphenicol, ampicillin, linezolid, teicoplanin, vancomycin and synercid (quinupristin and dalfopristin) but antibiotic susceptibility tests exhibited that 53 (29.4%), 43 (23.8%), 11 (6.1%) and 9 (5%) of isolates were resistant to erythromycin, tetracycline, gentamicin and ciprofloxacin, respectively. Among these isolates, sixteen were multi drug resistant (MDR) with resistance to erythromycin and tetracycline (13 isolates), erythromycin and ciprofloxacin (2 isolates) and ciprofloxacin and tetracycline (1 isolate). Of the total MDR isolates, 14 (81.5%) were E. faecium and 2 (12.5%) were E. faecalis.
Thirteen (81.2%) and three (18.8%) of MDR isolates were obtained from male and female subjects, respectively, and 62.5% of isolates were from persons aged more than fifty years.
REP-PCR typing of MDR isolates
REP-PCR analysis of 180 isolates showed 93 REP types with forty-five (25%) single types (ST1 to ST45) and forty-eight common types (CT1 to 48). Forty-eight common types included 135 (%75) isolates. Each common type was observed in 2 to 4 isolates. The diversity analysis of Simpson’s index showed REP-PCR with Di =0.96.
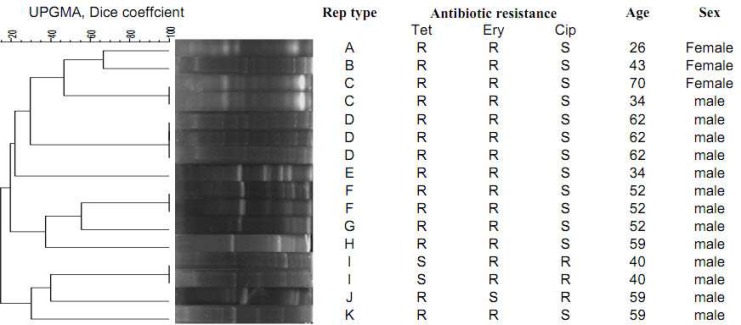
On the other hand, the analysis of sixteen MDR isolates showed 11 Rep types with four common types covering 9 (56.2%) isolates with 95 % similarity. The remaining 7 isolates (43.8%) were highly divergent belonging to seven single types (ST). All REP types, except for CT I and ST J, were resistant to tetracycline and erytromycine. On the other hand, CT I was resistant to ciprofloxacin and erytromycine. In addition, ST J was resistant to ciprofloxacin and tetracycline (Figure 1).
Fig. 1.
Dendrogram cluster analysis of REP-PCR data for 16 MDR Enterococcus isolates. Age, sex, Rep type and patterns are indicated. Cip: ciprofloxacin; Ery: erythromycin; Tet: tetracycline
Discussion
The enterococci are members of the intestinal microflora and include more than 20 different species, but E. faecalis and E. faecium are dominant in clinical isolates, environment and foods (12).
In the present study, all of the isolates (100%) were susceptible to chloramphenicol, ampicillin, linezolid, teicoplanin, synercid (quinupristin and dalfopristin) and vancomycin. Susceptibility to vancomycin was also observed in other studies (13, 14). On the other hand, inconsistent with the results of Ali et al. in 2013 in Pakistan (15) and Kim et al. in 2013 in Korea (13), our results showed the resistance to erythromycin, tetracycline and ciprofloxacin. This may result from the choice of antibiotics and consumption dose that had an effect on antibiotic resistance worldwide.
Moreover, colonization and infection with MDR enterecocci- those strains with significant resistance to two or more antibiotics- occur worldwide. Acquiring plasmid- encoded antibiotic resistance genes and virulence factors are the concern for this MDR normal flora, because they can become pathogenic and may be very important in public health (16).
On the other hand, sixteen MDR isolates were obtained from all of the samples. High rate of resistance to tetracycline and erythromycin (81.2%) was observed in our MDR isolates, which is similar to a report from Pakistan (15). In addition, 81.2% of MDR isolates were from male subjects and the average age of these persons was more than fifty years.
Molecular techniques used for typing of bacteria are variable in terms of standardization, cost, reproducibility, discriminatory power and interpretation. REP-PCR, is a simple procedure to distinguish closely related organisms. This method is less expensive and faster than many other methods such as PFGE and AFLP typing (17).
In the current study, the analyzes showed 93 REP types. 45 (25%) of these isolates were single types (ST) and the other 135 (75%) isolates belonged to Forty-eight common types with the biggest cluster containing four isolates. Furthermore, Rep PCR with MDR isolates detected 11 Rep types with four common types covering 9 (56.2%) isolates with 95 % similarity. Also, some strains with different Rep types were isolated from the same person, showing the heterogeneity of normal flora enterococci.
Although BOX-PCR genotyping have discriminatory power, is cheap and reproducible, but the incorrect grouping of some strains and difficulties of pattern analysis are some of the limitations of this method. The concern about these normal flora isolates are the pathogenic potential of these bacteria through the horizontal transfer of antibiotic resistance and virulence genes.
References
- 1.Pangallo D, Drahovska H, Harichova J, et al. Evaluation of different PCR-based approaches for the identification and typing of environmental enterococci. Antonie van Leeuwenhoek. 2008;93:193–203. doi: 10.1007/s10482-007-9193-z. [DOI] [PubMed] [Google Scholar]
- 2.Talebi M, Rahimi F, Katouli M, et al. Epidemiological link between wastewater and human vancomycin-resistant Enterococcus faecium isolates. Current microbiology. 2008;56:468–73. doi: 10.1007/s00284-008-9113-0. [DOI] [PubMed] [Google Scholar]
- 3.Lukášová J, Šustáčková A. Enterococci and antibiotic resistance. Acta VetBrno. 2003;72:315–23. [Google Scholar]
- 4.Ochoa SA, Escalona G, Cruz-Cordova A, et al. Molecular analysis and distribution of multidrug-resistant Enterococcus faecium isolates belonging to clonal complex 17 in a tertiary care center in Mexico City. BMC microbiology. 2013;13:291. doi: 10.1186/1471-2180-13-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russello GI, Farina C, Raglio A, et al. Typing of Enterococcus spp. strains isolated from patients with infective endocarditis by an automated repetitive-sequence-based PCR system. Microbiol Med. 2011;26:188–91. [Google Scholar]
- 6.Werner G. Molecular typing of Enterococci/VRE. J Bacteriol Parasitol. 2013;S:5:2155–9597. [Google Scholar]
- 7.Mohammed M, Abd El-Aziz H, Omran N, et al. Rep-PCR characterization and biochemical selection of lactic acid bacteria isolated from the Delta area of Egypt. International journal of food microbiology. 2009;128:417–23. doi: 10.1016/j.ijfoodmicro.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 8.Badgley BD, Nayak BS, Harwood VJ. The importance of sediment and submerged aquatic vegetation as potential habitats for persistent strains of enterococci in a subtropical watershed. Water research. 2010;44:5857–66. doi: 10.1016/j.watres.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. Journal of clinical microbiology. 1989;27:731–4. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kariyama R, Mitsuhata R, Chow JW, et al. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. Journal of clinical microbiology. 2000;38:3092–5. doi: 10.1128/jcm.38.8.3092-3095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Performance standards for antimicrobial susceptibility testing. Twentieth informational supplement. CLSI document M100-S20. Clinical and Laboratory Standards Institute (CLSI) 2014 [Google Scholar]
- 12.Meays CL, Broersma K, Nordin R, et al. Source tracking fecal bacteria in water: a critical review of current methods. Journal of environmental management. 2004;73:71–9. doi: 10.1016/j.jenvman.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Koo M, Cho A-R, Jeong A-R, et al. Antibiotic susceptibility and molecular typing of Enterococcus faecalis from retail pork meat products in Korea. J Kor Soc Appl Biol Chem. 2013;56:295–9. [Google Scholar]
- 14.Establishment of control system of antibiotics for livestock. Report of Korea Food and Drug Administration Project Int Veter Res Qua Serv. 2003 [Google Scholar]
- 15.Ali S, Hasan K, Bin Asif H, et al. Environmental enterococci: I. Prevalence of virulence, antibiotic resistance and species distribution in poultry and its related environment in Karachi, Pakistan. Lett Appl Microbio. 2014;58:423–32. [Google Scholar]
- 16.Vu J, Carvalho J. Enterococcus: review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. Front Biolo. 2011;6:357–66. [Google Scholar]
- 17.Deshpande LM, Fritsche TR, Moet GJ, et al. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagnostic microbiology and infectious disease. 2007;58:163–70. doi: 10.1016/j.diagmicrobio.2006.12.022. [DOI] [PubMed] [Google Scholar]