Abstract
Genetic background effects have long been recognized, and in some cases studied, but they are often viewed as a nuisance by molecular biologists. We suggest that genetic variation represents a critical frontier for molecular studies today. Human genetics has seen a surge of interest in genetic variation and its contributions to disease, but insights into disease mechanisms are difficult since information about gene function is lacking. In contrast, model organism genetics has excelled at revealing molecular mechanisms of cellular processes, but often de-emphasizes genetic variation and its functional consequences. We argue that model organism biology would benefit from incorporating natural variation, both to capture how well laboratory lines exemplify the species they represent and to inform on molecular processes and their variability. Such a synthesis would also greatly expand the relevance of model systems for studies of complex trait variation, including disease.
Keywords: natural variation, model organism, genetic background, laboratory strain, phenotypic diversity, genetic mechanism
Model organisms provide powerful experimental platforms to elucidate the basic biological functions of cells. With the implicit understanding that fundamental mechanisms and processes are likely conserved across species, experimentally facile organisms emerged as choice tools for biological exploration. The intense focus on representative models, including bacteria, budding yeast, fruit flies, round worms, zebrafish, mustard weed, and house mice, has provided mechanistic insights across the kingdoms of life. These insights include, for example, a basic understanding of cellular processes (such as genome replication, transcription, and translation), organismal functions (including reproduction, development, and cell-cell communication), and environmental interactions (spanning immunity, stress responses, and circadian relationships).
The initial power of model-organism genetics emerged in part from the early acceptance of common – often inbred – lines of laboratory models, such that the knowledge generated in one lab could be readily incorporated by other researchers studying the same lines. Focusing on a limited set of lines minimized the impact of genetic background, so that mechanisms underlying fundamental processes could be worked out. But while the focus on the same laboratory lines fostered a deep understanding of those models, it has limited the scope of biological knowledge –not just to a limited set of model species, but in reality to only a small number of individual genotypes.
On first principles, individuals within a species are expected to vary in their genetic properties –indeed, this variation is often the subject of genetic and evolutionary investigation. But the potential for functional differences across genetic backgrounds is routinely ignored in molecular studies. Here, we define functional genetic differences as DNA variants that confer molecular or organismal phenotypic differences among individuals, regardless of the effects on evolutionary fitness. Understanding the phenotypic consequences of naturally occurring genetic variation remains a major challenge in biology today. This is acutely clear in the case of personalized medicine, where the hope is that an individual’s genotype will inform on disease predisposition, prognosis, and treatment. Although genetic variation has been documented extensively in humans, the ability to make predictions about phenotypic variation remains limited.
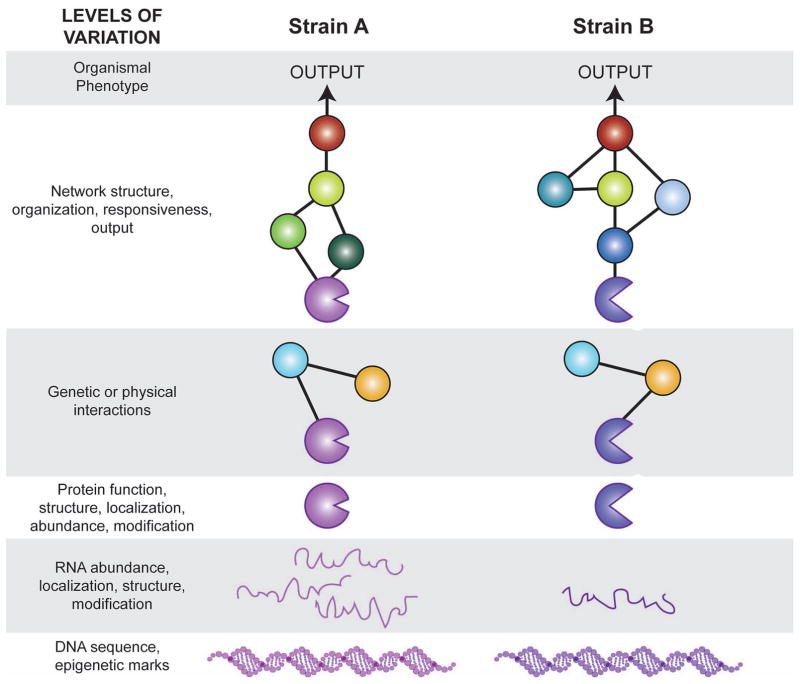
It is in this light that the experimental tools of model organisms offer tremendous potential to amplify our knowledge of how genetic variation modulates molecular processes and phenotypic outcomes. In this perspective, we argue that natural variation is a critical parameter that should be incorporated into models of molecular and cellular biology. Variation in gene expression, protein function, molecular interactions, and network organization can be thought of as an orthogonal dimension of biology that could be incorporated into molecular and cellular models (Figure 1). We believe that expanding model organism biology to incorporate a view of natural variation will cement the importance of model organisms for understanding both basic and applied genetics, including the causes of complex trait variation.
Figure 1. Natural variation at multiple levels.
The diagram depicts natural variation emerging for a given gene and its mRNA/protein product in one strain (left column) and for the orthologous gene and products in a second genetic background (right column). Circles and lines represent proteins and the interactions between them, respectively.
The pluses and perils of laboratory lines
The amount of information that has been collected from laboratory lines is remarkable. Decades of classical breeding, forward- and reverse-genetic screens, and molecular dissection have produced detailed descriptions of cellular and organismal function. Many model organisms were among the first in their clades to be sequenced at the genome scale, including Saccharomyces cerevisiae [1], Escherichia coli [2], Caenorhabditis elegans [3], Drosophila melanogaster [4], Arabidopsis thaliana [5], and Mus musculus [6]. Subsequent technological innovations, including high-throughput sequencing, methods to quantify and locate proteins and metabolites, and robotic and molecular tools to rapidly generate and phenotype mutants, have enabled unprecedented genetic characterization of a few strains of model organisms. While the technology exists to generate these data in a wider range of strains and species, it is really only in the context of the extensive background knowledge for model genetic lines that such high-throughput data can be integrated in a unifying framework. Each discovery is interpreted in light of considerable existing knowledge for that strain, often substantially accelerating understanding. This is especially important for systems-biology models that integrate diverse large-scale datasets, preferably generated from the same genetic background. Hence, there remains an inherent advantage in generating more knowledge on the same laboratory lines.
However, the strategy of focusing on particular lines has important drawbacks that should be recognized. It is becoming increasingly realized that laboratory lines of model organisms are often outliers of their species. These individuals have been selected, deliberately or inadvertently, for their ease of manipulation. Several recent reviews present the unusual features of model genetic backgrounds [7–12] and so we highlight only a few cases here. For example, common lab strains of S. cerevisiae are unusual in that they exist as stable haploids, lack the natural flocculence common to many wild isolates, harbor a variety of auxotrophies, and are easy to manipulate genetically [9]. Laboratory stocks of D. melanogaster – including the sequenced reference strain – are less active than wild flies, often have impaired senses [13, 14], and mostly descend from North American populations that have a complex, genetically admixed history [15, 16]. Commonly used classical inbred strains of house mice, which are in fact hybrids of multiple subspecies [17], grow faster, reproduce earlier, generate larger litters, gain weight more easily, and are much less aggressive than wild mice [18–20]. More fundamentally, specific aspects of molecular biology can be unusual in laboratory lines, perhaps obscuring natural processes from view. For example, laboratory lines of D. melanogaster lack transposable P elements as well as the regulators that suppress their transposition [21]; had it not been for studies crossing natural and laboratory fly lines, the discovery and subsequent exploitation of P elements may not have occurred.
The phenotypic differences between research lines and wild individuals emerge for a variety of reasons. Some features are the result of artificial selection for adaptation to laboratory environments; others reflect the historical bottleneck of choosing a few strains to serve as the common references for the species. Many of the accumulated differences in commonly used lab strains may in fact be deleterious in nature (even if they are beneficial to life in the lab). For example, mice from one widely used laboratory strain go blind within the first few months of life, a characteristic that would almost certainly be detrimental in their natural habitat [22]. Despite such unusual phenotypes, lab lines are often taken as sole representatives of their species and even of their clades. We argue that for the vast majority of biological processes, the extent to which mechanistic details vary within species is only beginning to emerge.
How much variation is there?
DNA sequencing has revealed tremendous genetic variation within and among natural populations of model-organism species. On average, a random pair of homologous, nuclear, noncoding sequences differs by 0.3–0.8% in S. cerevisiae [23–25], 0.5–1% in D. melanogaster [26], 0.1–0.4% in C. elegans [27, 28], and 0.1–0.2% in M. musculus [29]. A large number of variants have the potential to affect phenotypes of interest. For example, surprising numbers of nonsense mutations are segregating in natural populations of both model organisms [30–32] and humans. The average human genome harbors predicted loss-of-function variants in 250–300 genes, according to the 1,000 Genomes Project [33]. Yet the functional consequences of genetic variation in model organisms (and in other species) remain mostly uncharacterized.
The common assumption is that “important” features of the organism will be conserved within and even between species, and indeed conservation can be a signature of evolutionary constraint (but can also emerge from low mutation rates). However, a lack of conservation does not necessarily imply an absence of importance to the organism. In fact, a low-conservation signature can be produced by rapid evolution in one or more lineages, specifically because of genetic changes that are adaptively significant [34–37]. Adaptive variation within model species (e.g. in populations from contrasting environments) could be particularly informative in studies dissecting molecular variation, since this type of variation is less likely to emerge clearly from mutagenesis in the laboratory. The lack of conservation can also arise if one genetic change is compensated by other mutations through epistasis. This phenomenon is clearly seen in the evolution of transcription-factor binding sites across closely related yeast species, where the disappearance of a binding element at one promoter position is compensated by the appearance of a novel instance of that element elsewhere in the regulatory region [38].
Another example of the complex interplay between molecular mechanism and constraint is the wholesale rewiring of transcriptional regulatory networks over long evolutionary time frames in fungi. The co-regulation of ribosomal protein (RP) genes is highly conserved across kingdoms, and the acute response of RP genes to environmental insults persists over a billion years of fungal evolution [39]. Yet regulation of RP genes is perhaps the clearest case where the upstream regulatory mechanisms are completely different across fungal species. Concerted evolution in the network required changes in both upstream regulatory connections and in downstream transcriptional elements controlling each of the more than a hundred genes [40–43]. In this example, regulatory mechanisms have diverged substantially across species even as tight co-regulation and environmental responsiveness of the genes has been highly conserved – and it is likely the co-regulated response, rather than the mechanism producing it, that is important for fitness. While it might be expected that long evolutionary timeframes are required to enable wholesale network rewiring, evidence for within-species regulatory rewiring exists: regulation of yeast cell adhesion requires the filamentous MAP kinase pathway in one strain but an entirely different upstream regulatory process in another [44]. These examples illustrate the potential for mechanistic variation in producing the same cellular output.
Although the mechanistic underpinnings in many cases remain murky, high-throughput experiments are now quantifying the prevalence of genetic background effects on functional variation within a species [45]. Comparing gene knockout libraries from two strains of S. cerevisiae revealed that nearly 5% of genes scored essential in one strain are dispensable for survival in the other [46]. How this disparity is tolerated across strains is largely unknown but could be informative from a mechanistic perspective. In C. elegans, the reduced expression of ~20% of interrogated genes produced a phenotype that varied considerably across two different lines [47]. Genetic interactions can also vary at a high rate. One study screened two D. melanogaster lines for genetic modifiers of a hypomorphic allele of scalloped, a transcription factor involved in wing development: nearly 74% of all modifiers produced different effects in the two lines. While most of these differences related to the severity of the phenotype, several modifiers produced opposite effects in the two strain backgrounds. Dissecting the genetic basis of such phenotypic differences has been a longstanding interest of many biologists; we contend that leveraging such differences specifically to understand cellular mechanisms presents an exciting opportunity.
Incorporating natural variation as an orthogonal variable in cellular networks
Recently, the genetic tools of laboratory lines have been exploited to understand the consequences of variation found in nature. We submit that the reverse can also be true: natural variation could significantly illuminate our understanding of molecular mechanisms. Natural phenotypic variation can be conceptualized as an orthogonal dimension of biology. Just as an individual’s DNA sequence can be mapped onto a reference genome, we argue that natural variation in gene expression, protein function, and molecular interactions can be mapped onto cellular networks elucidated in reference lines of model organisms (Figure 1). Adding this orthogonal level of information could reveal profound insights into how cells function, by pointing to individual genes, proteins, or other elements that vary functionally, by revealing mechanistic plasticity across genetic backgrounds, and by suggesting the features of the cellular network (or its output) that are actually under constraint.
Several case studies highlight the potential for genetic variation to elucidate underlying molecular or physiological mechanisms, particularly those relevant to human health. One recent study by our colleagues incorporated genetic variation from the Drosophila Genetic Reference Panel (DGRP) [48, 49] into a D. melanogaster model of traumatic brain injury (TBI) [50]. The DGRP consists of >200 inbred lines of wild fly isolates, selected to represent common polymorphisms in a natural population, and has served as a powerful community resource for mapping complex traits [48, 49]. Mapping TBI sensitivity implicated genes linked to intestinal barrier function; this link, coupled with subsequent molecular studies in the laboratory strain, revealed that a low-sugar diet following TBI protects against death in multiple different genetic backgrounds. This example highlights the ability of natural variation to reveal physiological connections that determine and affect phenotypes, including disease. Another example is provided by a series of experiments exploring differences in pathogen susceptibility in two different commonly used strains of C. elegans. Genetic mapping of differential pathogen responses, using recombinant inbred lines emerging from a cross of the two parental strains, implicated a neuronal peptide receptor and, through subsequent mapping, an upstream ubiquitin ligase that regulates a behavioral response to pathogen recognition [51, 52]. Combining this information with molecular, structural, and physiological experiments in the lab strain not only implicated the genetic basis for the variable phenotype but also revealed a new molecular model for pathogen avoidance and contributed to a broader understanding of mechanosensory neuronal circuits [53].
There are those who would argue that genetic and phenotypic variation segregating in nature is less informative for molecular studies than deep laboratory mutagenesis in a single pure-bred line. We recognize that laboratory mutagenesis is highly effective at dissecting gene and protein function in a single genetic background. We argue, however, that the outcome and interpretation of such experiments could be significantly different if done in a different genetic background, as highlighted by several examples above. At issue then is the question being asked: if the goal is to elucidate molecular mechanisms in a single individual, saturating laboratory mutagenesis may indeed be the best approach. But if the aim is to dissect a molecular mechanism to serve as a model for other organisms, and especially for human disease that is likely to be complex, then the additional step of at least considering natural variation may significantly expand the scope.
Capturing natural functional variation in model organisms
Realizing the power of natural variation for model organism biology will require changes in perspective and in practice. Ultimately, different labs may choose to incorporate natural variation to different extents, from simply scanning allele frequencies of mechanistically implicated genes to performing detailed experiments in multiple backgrounds. While it is impractical to expect a deep understanding of all genetic backgrounds found in nature, we envisage several realistic avenues through which natural variation can be incorporated into molecular studies.
1) Understand the phenotypic distribution of natural strains and determine where laboratory lines fall along that spectrum
Surveying the distribution of a particular phenotype is now often feasible, thanks to medium- and high-throughput techniques and broader availability of natural lines to test. Knowledge of the phenotypic distribution would inform our perspective on the potential diversity of underlying mechanisms and may help prioritize follow-up experiments. For example, comparing phenotypic differences with sequence variation in genes of interest could suggest alternate genetic backgrounds whose subsequent study may be particularly informative from a mechanistic perspective. Knowledge of the phenotypic distribution will also clarify whether lab-strain phenotypes are representative of segregating variation or are unusual compared to other individuals in the species. Even when laboratory lines lie at the outer ranges of the phenotypic distribution, understanding the mechanistic underpinnings in lab strains can still be useful; in fact, studying unusual strains in the context of the broader population may be a powerful model for human disease, in which specific genetic backgrounds are particularly susceptible to disease.
2) Expand detailed molecular studies to a broader set of diverse genetic backgrounds
To expand our understanding of mechanistic possibilities, model-organism communities should consider establishing diverse sets of representative lines, chosen to capture the genetic and phenotypic variation of the species. We advocate a two-step framework: 1) a broad survey of phenotypic diversity as outlined above and 2) detailed molecular dissection, when informative, on strategically chosen genetic backgrounds that could be particularly illuminating from a mechanistic perspective. The first step may be more realistically facilitated by a community-wide approach to establish strain panels, which would enable targeted investigation of specific lines by individual labs, to validate detailed molecular studies done in laboratory strains. Additional reference lines should be chosen to maximize variation, e.g. geographically diverse isolates including representatives from the species’ ancestral range and adaptively unique natural populations, such as those from extreme environments or distinct substrates. A promising example of a community-wide effort is the Arabidopsis 1001 Genomes Project, in which ecologically diverse A. thaliana lines sampled from across the ancestral species range are being sequenced to catalyze functional dissection of genetic variation [54]. This and other mapping resources for model organism communities [55, 54, 28, 56, 48, 49, 57] not only enable mapping of phenotypic differences in parental lines, but can unmask cryptic variation that provides a powerful complement to laboratory mutagenesis (e.g. [58, 59]).
3) Represent the potential for natural variation when presenting mechanistic studies
Finally, it is important to recognize that variation exists, even when it is not the main point of a study. Results should be described in terms of the strain or genetic background used and not as the sole representation of the species (unless broader information in other lines exists). This is especially important for cross-species comparisons, which frequently consider only one strain – in many cases a laboratory line – as a single representative of each species. We also encourage model organism databases to organize our knowledge of molecular, genomic, and phenotypic variation; a nice example is the Mouse Phenome Database that captures various phenotypes of diverse mouse lines [60] .
Concluding remarks
Major advances in biology have often come from combining established perspectives. A synthesis incorporating natural variation into molecular and cellular biological studies could accelerate our understanding of life, leveraging the depth achieved by studying laboratory lines of model organisms and the breadth afforded by incorporating natural variation. This synthesis would also reinforce the relevance of model organism genetics for understanding disease biology and its interaction with genetic variation.
Trends Box.
“Please provide as a separate Word .doc file 3 – 5 bullet points with recent advances in the area, including emerging concepts and/or distinctions, that constitute a main motivation for the discussion developed in the article. The Trends box may not exceed 900 characters including spaces.”
Laboratory lines of model organisms have provided a wealth of information about biological functions. Many of these lines are derived from species with significant genetic and functional variation in nature.
The long-standing focus on particular genetic backgrounds has enabled the elucidation of core biological functions, but it has limited our understanding of the levels and impacts of natural variation found in wild populations.
A broader synthesis incorporating natural variation into model-organism research would illuminate biological mechanisms, reveal the genetic basis for phenotypic variation, and solidify the importance of model organisms for understanding both basic and personalized genetics.
Acknowledgments
We thank our colleagues David Wassarman and Barry Ganetzky and two reviewers for insightful comments. The authors were supported by grants from the NIH (R01GM083989 to APG, R01GM100426 to BAP, and R01GM111797 to JEP) and NSF (DEB1353737 to BAP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274(5287):546, 563–7. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 2.Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–62. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Consortium CeS. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–8. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 4.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 5.Arabidopsis Genome I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 6.Mouse Genome Sequencing C. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 7.Alfred J, Baldwin IT. New opportunities at the wild frontier. Elife. 2015;4 doi: 10.7554/eLife.06956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frezal L, Felix MA. C. elegans outside the Petri dish. Elife. 2015;4 doi: 10.7554/eLife.05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liti G. The fascinating and secret wild life of the budding yeast S. cerevisiae. Elife. 2015;4 doi: 10.7554/eLife.05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markow TA. The secret lives of Drosophila flies. Elife. 2015;4 doi: 10.7554/eLife.06793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phifer-Rixey M, Nachman MW. Insights into mammalian biology from the wild house mouse Mus musculus. Elife. 2015;4 doi: 10.7554/eLife.05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sterken MG, Snoek LB, Kammenga JE, Andersen EC. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 2015;31(5):224–31. doi: 10.1016/j.tig.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kain JS, Stokes C, de Bivort BL. Phototactic personality in fruit flies and its suppression by serotonin and white. Proc Natl Acad Sci U S A. 2012;109(48):19834–9. doi: 10.1073/pnas.1211988109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senthilan PR, Piepenbrock D, Ovezmyradov G, Nadrowski B, Bechstedt S, Pauls S, Winkler M, Mobius W, Howard J, Gopfert MC. Drosophila auditory organ genes and genetic hearing defects. Cell. 2012;150(5):1042–54. doi: 10.1016/j.cell.2012.06.043. [DOI] [PubMed] [Google Scholar]
- 15.Kao JY, Zubair A, Salomon MP, Nuzhdin SV, Campo D. Population genomic analysis uncovers African and European admixture in Drosophila melanogaster populations from the south-eastern United States and Caribbean Islands. Mol Ecol. 2015;24(7):1499–509. doi: 10.1111/mec.13137. [DOI] [PubMed] [Google Scholar]
- 16.Pool JE. The Mosaic Ancestry of the Drosophila Genetic Reference Panel and the D. melanogaster Reference Genome Reveals a Network of Epistatic Fitness Interactions. Mol Biol Evol. 2015 doi: 10.1093/molbev/msv194. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, Tucker P, Boursot P, McMillan L, Churchill GA, de Villena FP. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 2011;43(7):648–55. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bronson FH. Energy allocation and reproductive development in wild and domestic house mice. Biol Reprod. 1984;31(1):83–8. doi: 10.1095/biolreprod31.1.83. [DOI] [PubMed] [Google Scholar]
- 19.Miller RA, Dysko R, Chrisp C, Seguin R, Linsalata L, Buehner G, Harper JM, Austad S. Mouse (Mus musculus) stocks derived from tropical islands: new models for genetic analysis of life-history traits. J Zool. 2000;250:95–104. doi: 10.1111/j.1469-7998.2000.tb00580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harper JM. Wild-derived mouse stocks: an underappreciated tool for aging research. Age (Dordr) 2008;30(2–3):135–45. doi: 10.1007/s11357-008-9057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engels WR. The P family of transposable elements in Drosophila. Annu Rev Genet. 1983;17:315–44. doi: 10.1146/annurev.ge.17.120183.001531. [DOI] [PubMed] [Google Scholar]
- 22.Chang B, Hawes NL, Hurd RE, Davisson MT, Nusinowitz S, Heckenlively JR. Retinal degeneration mutants in the mouse. Vision Res. 2002;42(4):517–25. doi: 10.1016/s0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- 23.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O'Kelly MJ, van Oudenaarden A, Barton DB, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. Population genomics of domestic and wild yeasts. Nature. 2009;458(7236):337–41. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458(7236):342–5. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang QM, Liu WQ, Liti G, Wang SA, Bai FY. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol. 2012;21(22):5404–17. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- 26.Lack JB, Cardeno CM, Crepeau MW, Taylor W, Corbett-Detig RB, Stevens KA, Langley CH, Pool JE. The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics. 2015;199(4):1229–41. doi: 10.1534/genetics.115.174664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cutter AD, Dey A, Murray RL. Evolution of the Caenorhabditis elegans genome. Mol Biol Evol. 2009;26(6):1199–234. doi: 10.1093/molbev/msp048. [DOI] [PubMed] [Google Scholar]
- 28.Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Felix MA, Kruglyak L. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012;44(3):285–90. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geraldes A, Basset P, Smith KL, Nachman MW. Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Mol Ecol. 2011;20(22):4722–36. doi: 10.1111/j.1365-294X.2011.05285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jelier R, Semple JI, Garcia-Verdugo R, Lehner B. Predicting phenotypic variation in yeast from individual genome sequences. Nat Genet. 2011;43(12):1270–4. doi: 10.1038/ng.1007. [DOI] [PubMed] [Google Scholar]
- 31.Wohlbach DJ, Rovinskiy N, Lewis JA, Sardi M, Schackwitz WS, Martin JA, Deshpande S, Daum CG, Lipzen A, Sato TK, Gasch AP. Comparative genomics of Saccharomyces cerevisiae natural isolates for bioenergy production. Genome Biol Evol. 2014;6(9):2557–66. doi: 10.1093/gbe/evu199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Jiang L, Chen Y, Haelterman NA, Bellen HJ, Chen R. FlyVar: a database for genetic variation in Drosophila melanogaster. Database (Oxford) 2015;2015 doi: 10.1093/database/bav079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genomes Project C. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bishop JG, Dean AM, Mitchell-Olds T. Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc Natl Acad Sci U S A. 2000;97(10):5322–7. doi: 10.1073/pnas.97.10.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292(5525):2281–5. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- 36.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3(2):137–44. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 37.Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 2006;16(6):580–5. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 38.Doniger SW, Fay JC. Frequent gain and loss of functional transcription factor binding sites. PLoS Comput Biol. 2007;3(5):e99. doi: 10.1371/journal.pcbi.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasch AP. Comparative genomics of the environmental stress response in ascomycete fungi. Yeast. 2007;24(11):961–76. doi: 10.1002/yea.1512. [DOI] [PubMed] [Google Scholar]
- 40.Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2(12):e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci U S A. 2005;102(20):7203–8. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hogues H, Lavoie H, Sellam A, Mangos M, Roemer T, Purisima E, Nantel A, Whiteway M. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell. 2008;29(5):552–62. doi: 10.1016/j.molcel.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8(3):e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chin BL, Ryan O, Lewitter F, Boone C, Fink GR. Genetic variation in Saccharomyces cerevisiae: circuit diversification in a signal transduction network. Genetics. 2012;192(4):1523–32. doi: 10.1534/genetics.112.145573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandler CH, Chari S, Dworkin I. Does your gene need a background check? How genetic background impacts the analysis of mutations, genes, and evolution. Trends Genet. 2013;29(6):358–66. doi: 10.1016/j.tig.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, Bernstein DA, Rolfe PA, Heisler LE, Chin B, Nislow C, Giaever G, Phillips PC, Fink GR, Gifford DK, Boone C. Genotype to phenotype: a complex problem. Science. 2010;328(5977):469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vu V, Verster AJ, Schertzberg M, Chuluunbaatar T, Spensley M, Pajkic D, Hart GT, Moffat J, Fraser AG. Natural Variation in Gene Expression Modulates the Severity of Mutant Phenotypes. Cell. 2015;162(2):391–402. doi: 10.1016/j.cell.2015.06.037. [DOI] [PubMed] [Google Scholar]
- 48.Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, Richardson MF, Anholt RR, Barron M, Bess C, Blankenburg KP, Carbone MA, Castellano D, Chaboub L, Duncan L, Harris Z, Javaid M, Jayaseelan JC, Jhangiani SN, Jordan KW, Lara F, Lawrence F, Lee SL, Librado P, Linheiro RS, Lyman RF, Mackey AJ, Munidasa M, Muzny DM, Nazareth L, Newsham I, Perales L, Pu LL, Qu C, Ramia M, Reid JG, Rollmann SM, Rozas J, Saada N, Turlapati L, Worley KC, Wu YQ, Yamamoto A, Zhu Y, Bergman CM, Thornton KR, Mittelman D, Gibbs RA. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482(7384):173–8. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang W, Massouras A, Inoue Y, Peiffer J, Ramia M, Tarone AM, Turlapati L, Zichner T, Zhu D, Lyman RF, Magwire MM, Blankenburg K, Carbone MA, Chang K, Ellis LL, Fernandez S, Han Y, Highnam G, Hjelmen CE, Jack JR, Javaid M, Jayaseelan J, Kalra D, Lee S, Lewis L, Munidasa M, Ongeri F, Patel S, Perales L, Perez A, Pu L, Rollmann SM, Ruth R, Saada N, Warner C, Williams A, Wu YQ, Yamamoto A, Zhang Y, Zhu Y, Anholt RR, Korbel JO, Mittelman D, Muzny DM, Gibbs RA, Barbadilla A, Johnston JS, Stone EA, Richards S, Deplancke B, Mackay TF. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014;24(7):1193–208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katzenberger RJ, Chtarbanova S, Rimkus SA, Fischer JA, Kaur G, Seppala JM, Swanson LC, Zajac JE, Ganetzky B, Wassarman DA. Death following traumatic brain injury in Drosophila is associated with intestinal barrier dysfunction. Elife. 2015;4 doi: 10.7554/eLife.04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323(5912):382–4. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang HC, Paek J, Kim DH. Natural polymorphisms in C. elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature. 2011;480(7378):525–9. doi: 10.1038/nature10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schafer WR. Mechanosensory molecules and circuits in C. elegans. Pflugers Arch. 2015;467(1):39–48. doi: 10.1007/s00424-014-1574-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, Wang X, Ott F, Muller J, Alonso-Blanco C, Borgwardt K, Schmid KJ, Weigel D. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43(10):956–63. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 55.Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O'Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, Tarantino LM, Threadgill D, Toth LA, Valdar W, de Villena FP, Warden C, Whatley S, Williams RW, Wiltshire T, Yi N, Zhang D, Zhang M, Zou F Complex Trait C. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36(11):1133–7. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 56.King EG, Merkes CM, McNeil CL, Hoofer SR, Sen S, Broman KW, Long AD, Macdonald SJ. Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 2012;22(8):1558–66. doi: 10.1101/gr.134031.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long AD, Macdonald SJ, King EG. Dissecting complex traits using the Drosophila Synthetic Population Resource. Trends Genet. 2014;30(11):488–95. doi: 10.1016/j.tig.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell M, Ganetzky B. Identification of Mob2, a novel regulator of larval neuromuscular junction morphology, in natural populations of Drosophila melanogaster. Genetics. 2013;195(3):915–26. doi: 10.1534/genetics.113.156562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paaby AB, White AG, Riccardi DD, Gunsalus KC, Piano F, Rockman MV. Wild worm embryogenesis harbors ubiquitous polygenic modifier variation. Elife. 2015;4 doi: 10.7554/eLife.09178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grubb SC, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2014;42(Database issue):D825–34. doi: 10.1093/nar/gkt1159. [DOI] [PMC free article] [PubMed] [Google Scholar]