Abstract
Thermodynamic integration (TI) can provide accurate binding free energy insights in a lead optimization program, but its high computational expense has limited its usage. In the effort of developing an efficient and accurate TI protocol for FabI inhibitors lead optimization program, we carefully compared TI with different Amber molecular dynamics (MD) engines (sander and pmemd), MD simulation lengths, the number of intermediate states and transformation steps, and the Lennard-Jones and Coulomb Softcore potentials parameters in the one-step TI, using eleven benzimidazole inhibitors in complex with Francisella tularensis enoyl acyl reductase (FtFabI). To our knowledge, this is the first study to extensively test the new AMBER MD engine, pmemd, on TI and compare the parameters of the Softcore potentials in the one-step TI in a protein-ligand binding system. The best performing model, the one-step pmemd TI, using 6 intermediate states and 1 ns MD simulations, provides better agreement with experimental results (RMSD = 0.52 kcal/mol) than the best performing implicit solvent method, QM/MM-GBSA from our previous study (RMSD = 3.00 kcal/mol), while maintaining similar efficiency. Briefly, we show the optimized TI protocol to be highly accurate and affordable for the FtFabI system. This approach can be implemented in a larger scale benzimidazole scaffold lead optimization against FtFabI. Lastly, the TI results here also provide structure-activity relationship insights, and suggest the para-halogen in benzimidazole compounds might form a weak halogen bond with FabI, which is a well-known halogen bond favoring enzyme.
Keywords: thermodynamic integration (TI), enoyl acyl reductase (FabI), explicit solvent models, free energy calculation, implicit solvent models, halogen bond, benzimidazole-based inhibitors
Introduction
F. tularensis is the bacterial pathogen that causes tularemia and is a potential bioweapon. Current tularemia treatments are limited due to their toxicity or the requirement for intravenous administration.1 Therefore, a safe and orally available small molecule is desirable in the event of a potential tularemia outbreak. Among various antibacterial targets, enoyl acyl reductase (FabI) has been proven essential in F. tularensis, and differs structurally from the human fatty acid synthesis complex. Hence, FabI is a promising antibacterial F. tularensis target, and FabI inhibitors can potentially avoid human toxicity.
Free energy calculations potentially offer a valuable ligand binding free energy estimate and could prioritize compounds for synthesis and experimental testing. There are two major categories of free energy calculations. The first category includes implicit solvent methods such as Molecular Mechanics - Poisson-Boltzmann Surface Area (MM-PBSA), Molecular Mechanics - Generalized Born Surface Area (MM-GBSA), and Linear Interaction Energy (LIE). These relatively inexpensive methods offer accurate compound affinity ranking, but do not provide accurate binding free energy values.2–11 The second category includes explicit solvent methods such as thermodynamic integration (TI), free energy perturbation (FEP), Bennett’s acceptance ratio method (BAR),12 lambda dynamics,13,14 and weighted histogram analysis method (WHAM),15 which provide more accurate binding free energy values than implicit solvent methods. These explicit solvent methods have shown excellent accuracy in ligand binding affinity prediction across various therapeutic targets, including HIV protease,16,17 HIV reverse transcriptase,18 factor Xa,7 fructose 1,6-biphosphatase,19–21 and others.22–24 Among these explicit solvent methods, TI is probably the most common due to its simplicity in post processing (no molecular dynamics trajectory required), straightforward insertion of extra intermediate states for more accurate estimation, and intuitive integrand shape visualization to identify abnormal intermediate state transitions. It is also the best-supported explicit solvent method in the Amber Molecular Dynamics suite.
The explicit solvent methods, including TI, have been extremely computationally expensive, thus limiting their real world usage.25,26 There are several pioneering studies that have improved TI computational efficiency and accuracy by choosing the best performing integration method,27,28 optimizing the simulation length,29 sampling techniques,30–32 the number of transition steps,33–36 and the number of intermediate states7,37,38. Nonetheless, these studies have mostly focused on solvation energy but not ligand binding free energy in proteins on a large scale. Moreover, the Amber Molecular Dynamics suite recently enabled TI in pmemd, which has been proven to be a more efficient MD engine than the older and slower sander,39 but has not yet been extensively tested.40 Since recent alchemical free energy studies suggest that prediction performance is system dependent,41–43 it would be useful to study the effects of algorithms, simulation length, the number of transition steps and intermediate states on TI ligand binding free energy in F. tuleransis FabI. The optimized TI protocol here will assist in our lead optimization campaign against F. tuleransis FabI (FtFabI). 2,44,45–47
Methods
Experimental Enzymatic Activity
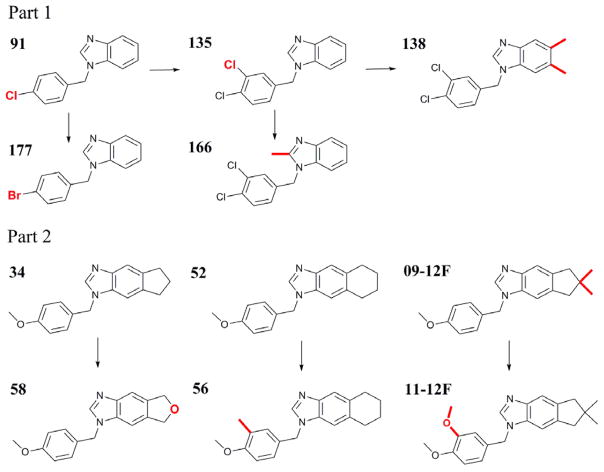
The benzimidazole scaffold FabI inhibitors in this study and their activities are listed in Figure 1. The details of their IC50 & Ki experimental determination have been previously described.47 The experimental binding free energy (ΔGbind) of these inhibitors was obtained from the experimentally determined Ki using Equation 1. T is room temperature (300K) and R is the ideal gas constant (1.9872×10−3 kcal K−1 mol−1).
Figure 1.
The eleven benzimidazole scaffold FtFabI inhibitors in this study. The TI transformation groups are highlighted in bold and indicated in arrows. Part 1 is the benchmark set and part 2 includes more compounds for the test.
| (1) |
Complex Preparation
The protein-ligand binding conformations were obtained from the PDB codes, 3UIC (FtFabI: compound 138)46, 4J3F (FtFabI: compound 56), and 4J1N.47 The starting conformations of the FtFabI complex with compound 91, 135, 166, 177, 34, 52, 58, 09–12F, and 11–12F were obtained by removing and/or adding corresponding functional groups (highlighted in red in Figure 1) in a benzimidazole inhibitor from the above three crystal structures. The RESP charges for the ligands and the cofactor, NADH, were derived from the HF/6-31G* theory level in Gaussian 0948 using the R.E.D. server.49,50 The General Amber force field (GAFF)51 was assigned to ligands and the FF14SB force field was assigned to the protein using antechamber in Amber v14.52 The ligand and protein-ligand complexes were solvated in a 10 Å octahedral TIP3P water box with counter ions (Na+ and Cl−) to neutralize the solvated systems.
Free Energy Calculation in the Three-Step Thermodynamic Integration
The TI simulations of pairs of similar ligands were separated into two parts: ligand transformation within the protein active site (ΔGbound) and ligand transformation in solvents (ΔGfree). The binding free energy difference between these two similar ligands (ΔΔG(A→B)) can be calculated from Equation 2:
| (2) |
Each ΔGbound and ΔGfree contains three steps. The first step is the charge removal of the ligand A region, which doesn’t exist on ligand B. The second step is the vdW (vdW) transformation of the unique regions between ligands A and B. The third step is to assign the charge back to the ligand B region, which doesn’t exist on ligand A. The Lennard-Jones Softcore potential (Equation 3) is activated in the second step to avoid any system instability as atoms disappear and appear,33 where λ is a coupling parameter, rab is the atomic distance between two particles, ε is the Lennard-Jones potential well depth, σ is the distance where the two particles’ intermolecular potential is equal to zero and α is the Lennard-Jones Softcore potential energy function curvature control parameter, which has previously been recommended to be α=0.5.33
| (3) |
In the six ΔGbound and ΔGfree steps, each step is divided into 21 windows with different coupling parameters (λ), where λ=0 is the initial state and λ=1 is the final state. The energy in each step (ΔG) can be calculated using Equation 4 where the bracket represents the average of the potential energy of the studied system (σ(λ)) for the given λ value, which is calculated using the trapezoid integration method. The λ values in this study are listed in Table 1. Since each of the six steps has 21 windows, there are in total 126 windows in each ligand-pair three-step TI calculation, and each window includes 2 ns of molecular dynamics (MD) simulation production runs, making TI a computationally intensive method (252 ns MD simulation for each ligand-pair three-step TI calculation). The following “Simulation Details” list the MD simulation settings for each window.
Table 1.
The Intermediate States (λ) in This Study
| Window Number | sander | pmemd |
|---|---|---|
| 6 | 0.01, 0.2, 0.4, 0.6, 0.8, 0.99 | 0.01 becomes 0.00 and 0.99 becomes 1.00 |
| 11 | 0.01, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 0.99 | 0.01 becomes 0.00 and 0.99 becomes 1.00 |
| 15 | 0.01, 0.1, 0.2, 0.3, 0.35, 0.4, 0.45, 0.5, 0.55, 0.6, 0.65, 0.7, 0.75, 0.8, 0.9, 0.99 | 0.01 becomes 0.00 and 0.99 becomes 1.00 |
| 17 | 0.01, 0.1, 0.2, 0.3, 0.35, 0.4, 0.45, 0.5, 0.55, 0.6, 0.65, 0.7, 0.75, 0.8, 0.9, 0.99 | 0.01 becomes 0.00 and 0.99 becomes 1.00 |
| 21 | 0.01, 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45, 0.5, 0.55, 0.6, 0.65, 0.7, 0.75, 0.8, 0.85, 0.9, 0.95, 0.99 | 0.01 becomes 0.00 and 0.99 becomes 1.00 |
| (4) |
Free Energy Calculation in the One Step Thermodynamic Integration
In the one step TI, the vdW and electrostatic transformation of the unique regions between ligands A and B both occur in the same step. Therefore, each ΔGbound and ΔGfree contains only one step, and each step contains 21 windows, resulting in a total of 42 windows in each ligand-pair one-step TI calculation (82 ns MD simulation production runs). This could greatly reduce computational costs. However, the simultaneous electrostatic and vdW transformation events in the one step TI could result in high system instabilities, which is overcome in this study by activating both Lennard-Jones (Equation 3) and Coulomb Softcore potentials (Equation 5) simultaneously,33 where qa and qb are electrostatic charges of transformed atoms in two ligands and β is the Coulomb Softcore potential energy function curvature control parameter, which has previously been recommended to be β =12 Å2.33
| (5) |
Simulation Details
The minimization was composed of the following four steps, following a protocol suggested in a recent TI study.17 The four step minimizations all shared the same settings in 2,500 steps of steepest descent minimization, followed by 2,500 steps of conjugate gradient minimization with 10 Å non-bonded cutoffs. The only differences in each minimization step were the restraint force constant. Firstly, solutes (protein-ligand complexes or ligands) were restrained with a 500 kcal mol−1 Å−2 force constant while no restraints were put on solvents. In the second, third and fourth minimization steps, the restraint force constant on solutes was reduced from 10 to 2 to 0 kcal mol−1 Å−2, while solvents were allowed to move freely. Next, a 100 ps density equilibration (NVT ensemble) was performed. The system was heated from 0 to 300K, and the Langevin temperature control was activated with a 2 ps−1 collision frequency, using 10 Å non-bonded cutoffs, the SHAKE algorithm and 2 fs time steps. The solutes were restrained with a 10 kcal mol−1 Å−2 force constant while solvents were allowed to relax. This was followed by a 100 ps constant pressure (1 bar) and temperature (300K) equilibration and a 2 ns production run (NPT ensemble) without any restraints on the system.
Accuracy and Error Measurement
The uncertainty in this study was estimated from the root-mean-square deviation (RMSD) in Equation 6, where ΔΔGexperimental is described in the “Experimental Enzymatic Setting” part and Equation 1, and n is the number of ligand-pair transformations. The coefficient of determination (R2) is used to describe the relationship between the experimental and predicted binding free energy differences of ligand-pairs. The standard error of the mean (SEM) is used for error estimate, as shown in Equation 7, where σ is the population standard deviation and n is the number of the samples in the population.
| (6) |
| (7) |
Results and Discussion
Comparison of Amber Algorithms and Transformation Steps in Accuracy
The TI calculation results using both old and new Amber MD engines, sander and pmemd, are listed in Table 2. Only four ligand-pair transformations (Figure 1, part 1) were extensively tested in the first part of this study due to the high computational costs of TI using sander, particularly with a large number of windows in a solvated protein-ligand complex system (about 32,000 atoms).
Table 2.
Calculated and Experimental ΔΔGbind (kcal/mol) of Benzimidazole Inhibitors in FtFabI using 2 ns MD simulations
| Amber Algorithm | Transformation Step | No. of Windows | 91→177 ΔΔG(expt)a = −1.27 | 91→135 ΔΔG(expt) = −1.04 | 135→138 ΔΔG(expt) = −1.53 | 135→166 ΔΔG(expt) = 0.76 | RMSDall (kcal/mol) | RMSD (kcal/mol)b |
|---|---|---|---|---|---|---|---|---|
| Sander | 3 | 6 | −0.47±0.06c | −0.91±0.07 | −1.99±0.12 | 0.28±0.08 | 0.52±0.09 | 0.47±0.07 |
| Sander | 3 | 11 | −0.41±0.06 | −0.39±0.07 | −1.98±0.13 | 1.46±0.08 | 0.68±0.09 | 0.64±0.07 |
| Sander | 3 | 15 | −0.27±0.06 | −0.50±0.07 | −1.56±0.13 | 1.83±0.08 | 0.78±0.09 | 0.78±0.07 |
| Sander | 3 | 17 | −0.30±0.05 | −0.55±0.07 | −1.47±0.13 | 1.72±0.08 | 0.73±0.09 | 0.73±0.07 |
| Sander | 3 | 21 | −0.22±0.06 | −0.51±0.07 | −1.19±0.18 | 1.83±0.08 | 0.81±0.11 | 0.80±0.07 |
|
| ||||||||
| Sander | 1 | 6 | −0.68±0.06 | −0.70±0.13 | −0.76±0.20 | 2.13±0.17 | 0.86±0.15 | 0.76±0.13 |
| Sander | 1 | 11 | −0.39±0.06 | −0.73±0.13 | −0.95±0.20 | 2.05±0.17 | 0.85±0.15 | 0.80±0.13 |
| Sander | 1 | 15 | −0.41±0.06 | −0.69±0.13 | −0.74±0.20 | 2.11±0.17 | 0.91±0.15 | 0.82±0.13 |
| Sander | 1 | 17 | −0.38±0.06 | −0.65±0.13 | −0.77±0.20 | 2.06±0.17 | 0.90±0.15 | 0.81±0.13 |
| Sander | 1 | 21 | −0.33±0.06 | −0.60±0.13 | −0.76±0.20 | 2.11±0.18 | 0.93±0.15 | 0.85±0.13 |
|
| ||||||||
| pmemd | 3 | 6 | −0.81±0.04 | −0.77±0.07 | −2.76±0.08 | 1.10±0.11 | 0.69±0.08 | 0.32±0.08 |
| pmemd | 3 | 11 | −0.47±0.04 | −0.55±0.05 | −1.97±0.09 | 1.15±0.08 | 0.55±0.07 | 0.51±0.06 |
| pmemd | 3 | 15 | −0.41±0.04 | −0.67±0.05 | −1.71±0.09 | 1.43±0.08 | 0.58±0.07 | 0.58±0.06 |
| pmemd | 3 | 17 | −0.34±0.04 | −0.69±0.05 | −1.69±0.09 | 1.50±0.08 | 0.62±0.07 | 0.62±0.06 |
| pmemd | 3 | 21 | −0.35±0.04 | −0.89±0.05 | −1.46±0.09 | 1.45±0.08 | 0.58±0.07 | 0.58±0.06 |
|
| ||||||||
| pmemd | 1 | 6 | −0.01±0.07 | −0.87±0.08 | −2.14±0.14 | 3.01±0.13 | 1.33±0.11 | 1.29±0.10 |
| pmemd | 1 | 11 | 0.06±0.07 | −0.97±0.08 | −2.11±0.14 | 2.41±0.13 | 1.10±0.11 | 1.06±0.10 |
| pmemd | 1 | 15 | 0.05±0.06 | −0.87±0.08 | −1.66±0.14 | 1.99±0.13 | 0.91±0.11 | 0.91±0.09 |
| pmemd | 1 | 17 | −0.01±0.06 | −0.89±0.08 | −1.72±0.14 | 2.05±0.13 | 0.91±0.11 | 0.91±0.09 |
| pmemd | 1 | 21 | −0.06±0.06 | −0.96±0.08 | −1.63±0.13 | 2.06±0.12 | 0.89±0.10 | 0.89±0.09 |
Experimental ΔΔ G values.
RMSD excluding the non-converged 135→138 transformation.
The error estimate using the standard error of the mean in Equation 7.
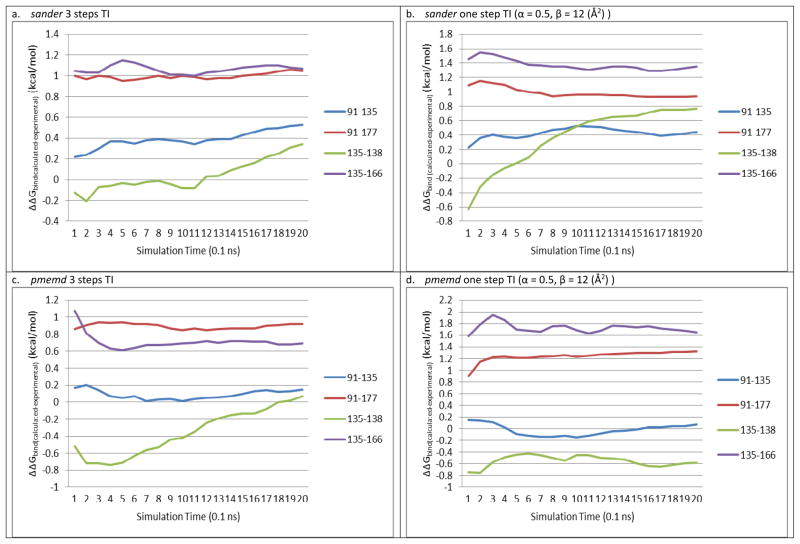
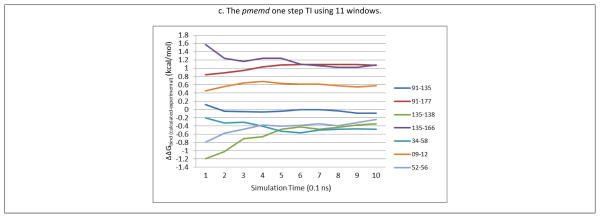
Recent studies have shown that some protein-ligand systems may require long TI simulations (≥ 3.5 ns production run per window) to reach convergence,29,32 while some other systems converge in a relatively shorter time frame (after 0.5~2 ns of production runs per window).7,53 Therefore, we first perform a 3.5 ns, one-step pmemd TI with 11 and 21 windows for four of the transformations, and plot the ΔΔGbind (calculated- experimental) and cumulative MD simulation length to check convergence, a simple convergence test recommended by the AMBER community and other studies7,53, as shown in Supporting Figure 1. It can readily be seen that the predicted binding free energy difference exhibits no significant change after 1 ns of simulation length (within an uncertainty of 0.2 kcal/mol), in both 11 and 21 windows cases. However, it is obviously still possible that true convergence (global minimum) might only be reached after a few hundred ns of TI, which cannot be achieved within a reasonable time frame with current computational power. More efficient TI protocols in the future, such as TI running on GPUs, might enable longer TI simulations to answer this question. In the current study, due to the goal of surveying multiple ligands with limited computational resources, we only perform 2ns of TI in different environment settings in this study. The plots of calculated ΔΔGbind (calculated- experimental) and cumulative MD simulation lengths suggest that all TI calculations using sander/pmemd and the one-step/three-step transformations in this study converged after 1 ns MD production runs (within an uncertainty of 0.2 kcal/mol), with the exception of the 135→138 transformation (Figure 2). This result agrees with another recent study of the Mycobacterium tuberculosis InhA (MtFabI): triclosan analogues system, where TI results converged after 600 ps of production runs.53
Figure 2.
Plots of calculated ΔΔGbind(calculated- experimental) (kcal/mol) vs. cumulative MD simulation lengths using 11 windows
It is known that TI converges poorly if large numbers of atoms or groups are involved in a transformation. Since the 135→138 transforms two hydrogen atoms into two methyl groups, it is not surprising that the 135→138 transformation does not converge well except in the one-step TI using pmemd. In Table 2, RMSDs including and excluding the 135→138 transformation are both listed. In the following discussion, RMSDall represents all four transformations included RMSD, while RMSDexcluded 135→138 means the 135→138 transformation is excluded.
In Table 2, both sander and pmemd in the three-step TI offer satisfactory prediction compared to experimental binding free energy (RMSDall ≤ 0.81 kcal/mol), while pmemd performs slightly better than sander in the three-step TI using 11 or more intermediate states (ΔRMSD ≤ 0.2~0.3 kcal/mol). This may be a consequence of pmemd allowing the initial and final states (λ values) to be 0 and 1 respectively, while sander only allows values very close to 0 and 1 (Table 2).40 Therefore, numerical integration in pmemd results requires no extrapolation and cancels its associated errors.
Similarly, the difference between sander and pmemd in the one-step TI using 11 or more intermediate states is trivial (ΔRMSD ≤ 0.26 kcal/mol). However, the one-step TI using either sander or pmemd (RMSDexcluded 135→138 ranging from 0.76~1.29 kcal/mol) delivers less agreement with experimental values than the three-step TI (RMSD excluded 135→138 ranging from 0.32~0.80 kcal/mol). This may be a consequence of the one-step TI allowing simultaneous vdW and electrostatic transformations, and relying on both Lennard-Jones (Equation 3) and Coulomb Softcore potentials (Equation 5) together to enhance system stability. However, the two Softcore potentials in the one-step TI have only been tested in predicting solvation energy of small molecules33. Therefore, the default setting of Lennard-Jones (Equation 3) and Coulomb Softcore potentials (Equation 5) in the one-step TI may need more optimization for this system. More details are discussed below in the “Optimization of the One-Step TI” section.
Accuracy Dependence on Simulation Length
Since computational expense is one of the major hurdles for thermodynamic integration and other alchemical free energy calculation methods (FEP, BAR, etc.), shortening the simulation lengths and increasing the computational efficiency will be of high interest. Because the ΔGbind(experimental-calculated) converge after 1 ns of simulations (Figure 2) (within an uncertainty of 0.2 kcal/mol, and excepting the 135→138 transformation), it is of interest to see if the TI accuracy is maintained using 1 ns MD simulations; the results are listed in Table 3. In Table 2 & 3, it is clearly shown that TI using 1 ns MD simulations has slightly lower RMSD values than 2 ns MD simulations in most circumstances (excluding the non-converged 135→138 transformation), with ΔRMSD(1ns–2ns) ranging from −0.14 to 0 kcal/mol, (Table 3, the ΔRMSD(1ns–2ns) column). This result suggests that TI using 1 ns MD simulations offers sufficient sampling and accuracy. On the other hand, if only one trajectory is used for each window, the extra 1 ns simulation of TI using 2 ns MD simulation may not sample more useful conformations, but may amplify inaccuracies in the force field and slightly degrade the prediction results, as suggested in prior studies.2,7
Table 3.
Calculated and Experimental ΔΔGbind (kcal/mol) of Benzimidazole Inhibitors in FtFabI using 1 ns MD simulations
| Amber Algorithm | Transformation Step | No. of Windows | 91→177 ΔΔG(expt)a = −1.27 | 91→135 ΔΔG(expt) = −1.04 | 135→138 ΔΔG(expt) = −1.53 | 135→166 ΔΔG(expt) = 0.76 | RMSD (kcal/mol) | RMSDall (kcal/mol)b | ΔRMSD (1ns–2ns) (kcal/mol)c |
|---|---|---|---|---|---|---|---|---|---|
| Sander | 3 | 6 | −0.45±0.08d | −1.37±0.10 | −2.26±0.17 | 0.74±0.11 | 0.57±0.12 | 0.44±0.10 | −0.03 |
| Sander | 3 | 11 | −0.47±0.08 | −0.88±0.10 | −2.34±0.18 | 1.33±0.11 | 0.64±0.12 | 0.50±0.10 | −0.14 |
| Sander | 3 | 15 | −0.33±0.08 | −0.80±0.10 | −2.01±0.18 | 1.69±0.11 | 0.71±0.12 | 0.67±0.10 | −0.11 |
| Sander | 3 | 17 | −0.36±0.08 | −0.72±0.10 | −1.89±0.18 | 1.71±0.11 | 0.70±0.12 | 0.68±0.10 | −0.05 |
| Sander | 3 | 21 | −0.27±0.08 | −0.67±0.10 | −1.61±0.13 | 1.77±0.12 | 0.74±0.11 | 0.73±0.10 | −0.07 |
|
| |||||||||
| Sander | 1 | 6 | −0.78±0.09 | −0.62±0.19 | −1.14±0.27 | 1.86±0.24 | 0.67±0.21 | 0.64±0.18 | −0.12 |
| Sander | 1 | 11 | −0.43±0.08 | −0.65±0.19 | −1.12±0.27 | 2.05±0.24 | 0.82±0.21 | 0.79±0.18 | −0.01 |
| Sander | 1 | 15 | −0.44±0.08 | −0.61±0.19 | −0.95±0.27 | 2.03±0.24 | 0.84±0.21 | 0.79±0.18 | −0.03 |
| Sander | 1 | 17 | −0.41±0.08 | −0.55±0.19 | −1.00±0.27 | 2.00±0.24 | 0.84±0.21 | 0.79±0.18 | −0.02 |
| Sander | 1 | 21 | −0.31±0.08 | −0.51±0.18 | −1.01±0.28 | 2.09±0.25 | 0.90±0.21 | 0.86±0.18 | 0.01 |
|
| |||||||||
| pmemd | 3 | 6 | −0.87±0.05 | −0.82±0.07 | −3.10±0.12 | 1.06±0.11 | 0.83±0.09 | 0.27±0.08 | −0.05 |
| pmemd | 3 | 11 | −0.60±0.05 | −0.73±0.07 | −2.46±0.12 | 1.20±0.11 | 0.63±0.09 | 0.43±0.08 | −0.08 |
| pmemd | 3 | 15 | −0.52±0.05 | −0.83±0.07 | −2.16±0.12 | 1.48±0.12 | 0.62±0.10 | 0.53±0.09 | −0.05 |
| pmemd | 3 | 17 | −0.41±0.05 | −0.80±0.07 | −2.15±0.12 | 1.48±0.12 | 0.65±0.10 | 0.57±0.09 | −0.05 |
| pmemd | 3 | 21 | −0.42±0.05 | −1.03±0.07 | −1.95±0.12 | 1.45±0.11 | 0.59±0.09 | 0.55±0.08 | −0.03 |
|
| |||||||||
| pmemd | 1 | 6 | −0.18±0.10 | −1.05±0.11 | −1.75±0.19 | 2.80±0.18 | 1.16±0.15 | 1.16±0.13 | −0.13 |
| pmemd | 1 | 11 | −0.04±0.10 | −1.19±0.11 | −1.98±0.19 | 2.45±0.18 | 1.07±0.15 | 1.05±0.13 | −0.01 |
| pmemd | 1 | 15 | −0.01±0.09 | −1.11±0.11 | −1.67±0.19 | 2.11±0.18 | 0.93±0.15 | 0.92±0.13 | 0.01 |
| pmemd | 1 | 17 | −0.05±0.09 | −1.12±0.11 | −1.79±0.19 | 2.14±0.18 | 0.93±0.15 | 0.92±0.13 | 0.01 |
| pmemd | 1 | 21 | −0.1±0.09 | −1.06±0.11 | −1.68±0.19 | 2.09±0.17 | 0.89±0.15 | 0.89±0.13 | 0 |
Experimental ΔΔG values
RMSD excluding the 135→ 138.
RMSD (excluding the 135→ 138) in 1 ns MD simulation trajectories minus RMSD (excluding the non-converged 135→ 138) in 2 ns MD simulation trajectories.
The error estimate using the standard error of the mean in Equation 7.
Effect of the Number of Intermediate States on Accuracy
TI calculations with six intermediate states provide the best agreement with experimental results (lowest RMSDexcluded 135→138 values) for both the sander three-step/one-step TI and the pmemd three-step TI using either 1 or 2 ns MD simulations (Tables 2 & 3). Using pmemd for the one-step TI is the only circumstance where a larger number of intermediate states offers a better performance (lower RMSD values). However, 21 intermediate states (including all four transformations) only provides 0.18 kcal/mol lower RMSD values than 11 intermediate states in the pmemd one-step TI, but with almost twice the computational cost. This does not justify the usage of large numbers of intermediate states. On the other hand, 6 intermediate states in the pmemd one-step TI yields an unsatisfactory RMSDexcluded 135→138 of 1.29 kcal/mol (with a cutoff of ~1 kcal/mol), while 11 and 15 intermediate states provides a broader line accuracy (RMSDall =1.07 and 0.93 kcal/mol correspondingly). Therefore, 11 or 15 intermediate states seem to offer the best balance between predictive accuracy and efficiency in the pmemd one-step TI.
Effect of Amber Algorithms, Transformation Steps, Number of Intermediate States and Simulation Length on Efficiency
If one averages the speed of pmemd and sander over all intermediate states and calculates their relative efficiency using Equation 840, pmemd provides a calculation 3.1 times faster than sander, similar to the previous reports by the pmemd TI developers (previously reported: pmemd about 2.9 times faster than sander).40 Moreover, pmemd has several additional advantages: (1) sander scaling performance is optimal only if the number of cores is a power of two, while pmemd has no such limitation. This will allow TI calculations to be more efficient in cluster machines where the number of cores per node is not necessarily a power of 2;40 (2) sander only allows minimization of a vdW transformation step on two cores (in both the one-step and three-step TI) while pmemd does not have such a limitation.40 Since TI equilibrium and production runs immediately after a minimization require more than two cores to finish a TI job in a reasonable time, users can request the same number of cores in TI minimization, equilibrium and production runs in pmemd for convenience;40 and (3) pmemd is more scalable to larger numbers of cores than sander.40 Putting all these factors (queue times, etc.) together, TI calculations in pmemd are about 4–5 times faster than in sander, using the University of Illinois at Chicago (UIC) Extreme Computing machine, with 16 Intel Xeon E5-2670 2.6 GHz CPUs per node and QDR infiniband communication between nodes.
| (8) |
TI efficiency decreases linearly with larger numbers of transformation steps, intermediate states and longer simulation length: the one-step TI is three times faster than the three-step TI (assuming both cases converge); 11 intermediate states is about twice as efficient as 21 intermediate states, and TI using 1 ns MD simulations is twice as fast as 2 ns. In this case, the one-step TI using pmemd, 11 intermediate states and 1 ns simulations would be the most efficient method, considering RMSDexcluded 135→138 values closer to 1 kcal/mol (Table 3). Using 96 Intel Xeon E5-2670 2.6 GHz CPU cores with QDR infiniband between nodes for the 11 windows one-step TI correspondingly, one pair of ligand TI calculations can be finished in 10 hours. However, the one-step TI is not as accurate as the three-step TI as mentioned above (Table 3). If the pmemd three-step TI calculation using 6 intermediate states and 1 ns MD simulations is implemented using 96 Intel Xeon E5-2670 2.6 GHz CPUs on the UIC Extreme machine, it requires 13.8 hours excluding any queue time. Since using a large number of cores might prolong the queue time on a cluster machine, and might not be easily available, there is strong interest to optimize the efficient one-step TI protocol to improve its accuracy.
Optimization of the pmemd One-Step TI
In a one-step TI calculation, the α and β parameters in Equation 3 & 5 are the Lennard-Jones and Softcore potential energy function curvature control parameters, respectively. These two parameters have thus far only been explored in the solvation energies of small molecules.33 Previous solvation energy studies suggest that decreasing α from 0.5 to 0.2 might improve the accuracy.33,52 Therefore, we initially tested α values from 0.6 to 0.2, decrementing by 0.1, while keeping β constant at 12 Å2 (Table 4).
Table 4.
Calculated and Experimental ΔΔGbind (kcal/mol) of Benzimidazole Inhibitors in FtFabI using the pmemd one-step TI and 1 ns MD simulations
| α in Equation 3 | β in Equation 5 (Å2) | No. of Windows | 91→177 ΔΔG(expt)a = −1.27 | 91→135 ΔΔG(expt) = −1.04 | 135→138 ΔΔG(expt) = −1.53 | 135→166 ΔΔG(expt) = 0.76 | RMSDall (kcal/mol) | RMSD (kcal/mol)b |
|---|---|---|---|---|---|---|---|---|
| 0.6 | 12 | 6 | 0.09±0.11c | −0.63±0.11 | −1.96±0.20 | 2.02±0.18 | 0.97±0.16 | 0.61±0.14 |
| 0.6 | 12 | 11 | −0.16±0.11 | −0.75±0.12 | −1.71±0.20 | 2.26±0.18 | 0.95±0.16 | 0.67±0.14 |
|
| ||||||||
| 0.5 | 12 | 6 | −0.18±0.10 | −1.05±0.11 | −1.75±0.19 | 2.80±0.18 | 1.16±0.15 | 0.64±0.13 |
| 0.5 | 12 | 11 | −0.04±0.10 | −1.19±0.11 | −1.98±0.19 | 2.45±0.18 | 1.07±0.15 | 0.76±0.13 |
|
| ||||||||
| 0.4 | 12 | 6 | −0.21±0.07 | −0.29±0.12 | −2.23±0.18 | 4.28±0.17 | 1.90±0.14 | 0.84±0.13 |
| 0.4 | 12 | 11 | −0.35±0.07 | −0.17±0.12 | −1.92±0.18 | 3.11±0.17 | 1.34±0.14 | 0.74±0.12 |
|
| ||||||||
| 0.3 | 12 | 6 | −0.22±0.06 | −0.53±0.11 | −1.21±0.17 | 2.41±0.17 | 1.02±0.14 | 0.69±0.12 |
| 0.3 | 12 | 11 | −0.24±0.06 | −0.41±0.11 | −1.46±0.17 | 2.17±0.17 | 0.93±0.14 | 0.70±0.12 |
|
| ||||||||
| 0.3 | 15 | 6 | −0.20±0.06 | −1.13±0.11 | −2.16±0.17 | 1.90±0.16 | 0.84±0.13 | 0.72±0.12 |
| 0.3 | 15 | 11 | −0.27±0.06 | −0.84±0.11 | −1.88±0.17 | 2.08±0.17 | 0.85±0.14 | 0.62±0.12 |
|
| ||||||||
| 0.3 | 18 | 6 | −0.42±0.06 | −1.14±0.11 | −2.00±0.18 | 2.23±0.16 | 0.88±0.14 | 0.56±0.12 |
| 0.3 | 18 | 11 | −0.41±0.06 | −0.41±0.12 | −1.88±0.18 | 1.84±0.17 | 0.79±0.14 | 0.65±0.13 |
|
| ||||||||
| 0.3 | 21 | 6 | −0.03±0.07 | −0.88±0.11 | −1.56±0.19 | 1.74±0.17 | 0.79±0.14 | 0.72±0.12 |
| 0.3 | 21 | 11 | 0.02±0.07 | −0.78±0.11 | −1.76±0.18 | 1.90±0.17 | 0.88±0.14 | 0.77±0.12 |
|
| ||||||||
| 0.2 | 12 | 6 | −0.09±0.06 | −0.33±0.12 | −2.47±0.18 | 2.58±0.18 | 1.23±0.14 | 0.96±0.13 |
| 0.2 | 12 | 11 | −0.15±0.06 | −0.57±0.11 | −2.25±0.18 | 2.55±0.18 | 1.14±0.14 | 0.71±0.13 |
Experimental ΔΔG values
RMSD excluding the 135→166 transformation
The error estimate using the standard error of the mean in Equation 7.
In Table 4, the RMSDall values are almost all larger than 1 kcal/mol across various α values, ranging from 0.93 to 1.90 kcal/mol, since the predicted ΔΔGbind values of the 135→166 transformation all significantly overestimated the experimental ΔΔGbind values (by 1.26~2.26 kcal/mol). If the 135→166 transformation is excluded from the RMSD calculation, as shown in the rightest column of Table 4, all RMSDexcluded 135→166 values are below 1 kcal/mol across various α values.
The one-step TI results converged using α = 0.5, 0.4, and 0.3, together with fixed β = 12 (Å2) (if 0.2 kcal/mol error is allowed), as shown in Supporting Figure 2. These three settings also give comparably low RMSD values (particularly if the 135→166 transformation is excluded), while α = 0.3 gives the lowest all-transformations-included RMSD (0.93 kcal/mol in 11 windows). α = 0.2 doesn’t provide converged results (Supporting Figure 2). Using α = 0.6 was not recommended in the initial study,33 and our data also suggests that the one-step TI with α = 0.6 doesn’t converge in either 6 or 11 windows using 1 ns MD simulations (Supporting Figure 2). Therefore, α = 0.3 seems to be the best value for the cases studied here. However, the predicted ΔΔGbind value of the 135→166 transformation still deviates excessively from the experimental value using α = 0.3 and β = 12 (Å2). Since the β value controls the Coulomb Softcore potentials (Equation 5), it was of interest to see if fine-tuning the β value with α=0.3 could offer better performance, particularly for the 135→166 transformation.
The results of incrementing the β value by 3 Å2, from 12 to 21 Å2, while keeping α constant at 0.3, are listed in Table 4 and Supporting Figure 3. Among the various β values, only β = 12 and 18 Å2 provide converged ΔΔGbind of all transformations. Particularly, β = 18 Å2 and α = 0.3 using 11 windows offers the lowest all-transformation-included RMSD (0.79 kcal/mol in 11 windows), while the ΔΔGbind of all transformations converged (Supporting Figure 3). The setting of β = 18 Å2, α = 0.3 and 11 windows also offers the best accuracy for the 135→166 transformation. Therefore, the one-step pmemd TI with α = 0.3 and β = 18 (Å2) using 11 windows seem to be the best one-step TI Softcore potential settings for our FabI-benzimidazole system.
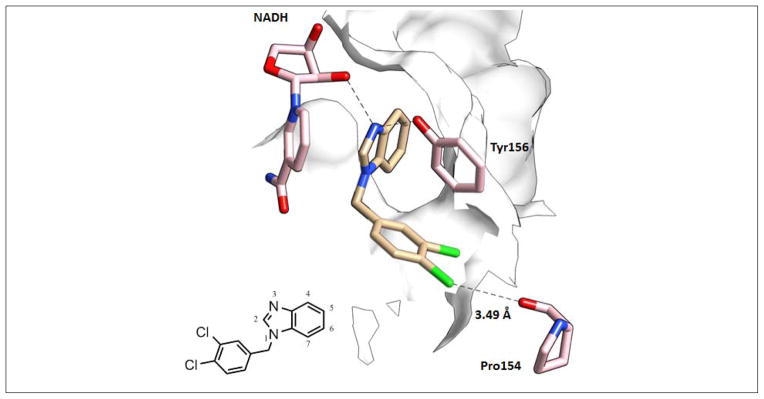
The one-step TI, regardless of various settings, encounters difficulties in the 135→166 transformation. Even though the β = 18 Å2 and α = 0.3 combination in the one-step TI provides one of the lowest RMSDall and best converged prediction results for the 135→166 transformation (1.84 kcal/mol), it is still 1.08 kcal/mol over-estimated from the experimental value. The 135→166 transformation exchanges a hydrogen atom for a methyl group at the 2 position of the benzimidazole ring (Figure 1). Therefore, it would be expected that the vdW interaction is the primary contribution in this transformation. However, the nitrogen atom at the 3 position of the benzimidazole ring forms a key hydrogen bond interaction with the FabI Tyr156 and the hydroxyl group of the NADH nicotinamine ribose (Figure 3). Since the methyl group could attract electrons from the nitrogen atom, the 135→166 transformation is believed to affect not only the vdW interaction, but also the electrostatic distribution of the nitrogen atom, and weaken the key hydrogen bonds. Since vdW radius and charge transformations occur simultaneously in the one-step TI, the one-step TI might not offer an accurate estimate on the 135→166 transformation as the three-step TI: the pmemd three-step TI using 6 windows can offer a significantly better and converged prediction for the 135→166 transformation (1.06 kcal/mol), only 0.3 kcal/mol over-estimated (Table 3). Hence, if a highly accurate prediction is required for the modification of the 2 position on the benzimidazole ring, the pmemd three-step TI using 6 windows would be recommended. If only a quick estimate is required, the pmemd one-step TI using 11 windows, α = 0.3 and β = 18 Å2, can still capture the behaviour of the 2 position modification of the benzimidazole ring with borderline accuracy.
Figure 3.
The binding mode of the compound 135 from the in-house crystal structure (pdb code: 3UIC). The nitrogen atom at the 3 position of the benzimidazole ring forms key hydrogen bonds with the NADH nicotinamide ribose (truncated for clarity) and Tyr156. The 5 and 6 positions of the benzimidazole ring fall into a lipophilic cavity, formed by the Ile91, Ala92, Phe93, Ala94, Pro95, and Ala197 side chains. The para-chloride might form a weak halogen bond with the carbonyl backbone of Pro154, as discussed in the “Further Validation and Medicinal Chemistry Perspective” section. The meta-chloride is in a hydrophobic pocket formed by the Leu99 and Ile200 side chains.
Further Validation and Medicinal Chemistry Perspective
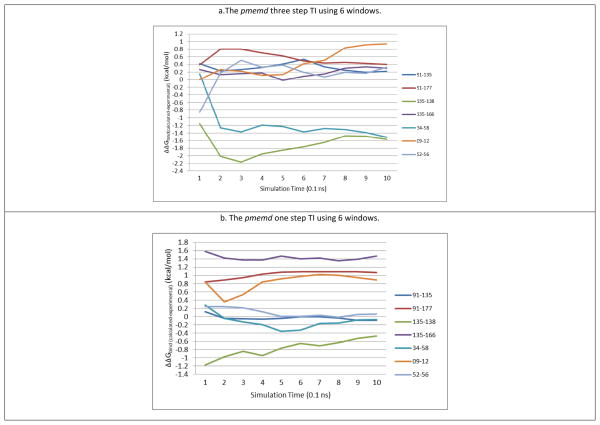
The additional three ligand-pair transformations shown in Part 2 of Figure 147 were tested to further validate the best performing and the most efficient one-step pmemd TI (α = 0.3, β = 18 Å2, 11 windows) and the three-step pmemd TI (6 windows), both using 1 ns MD simulations. The results are shown in Table 5 and Figure 4.
Table 5.
Calculated and Experimental ΔΔG (kcal/mol) of Benzimidazole Inhibitors in FtFabI using QM-MM/GBSA and TI
| Transformation | ΔΔGexpt (kcal/mol)a | ΔΔGTI(kcal/mol) pmemd 3 steps 6 windows | ΔΔGTI(kcal/mol) pmemd 1 steps 6 windows b | ΔΔGTI(kcal/mol) pmemd 1 steps 11 windows | ΔΔGQM-MM/GBSA (kcal/mol)c | ΔΔHQM-MM/GBSA (kcal/mol)d |
|---|---|---|---|---|---|---|
| 91→177 | −1.27 | −0.87±0.05e | −0.42±0.06 | −0.41±0.06 | −5.99±0.92 | −5.05±0.08 |
| 91→135 | −1.04 | −0.82±0.07 | −1.14±0.11 | −0.41±0.12 | −3.91±0.91 | −3.84±0.07 |
| 135→138 | −1.53 | −3.10±0.12 | −2.00±0.18 | −1.88±0.18 | −1.39±0.73 | −4.63±0.07 |
| 135→166 | 0.76 | 1.06±0.11 | 2.23±0.16 | 1.84±0.17 | 1.38±0.84 | 2.02±0.07 |
| 34→58 | 1.48 | −0.08±0.13 | 1.41±0.22 | 1.00±0.17 | 6.18±2.01 | 2.43±0.07 |
| 09–12F→11–12F | −0.12 | 0.82±0.13 | 0.77±0.22 | 0.46±0.22 | −3.13±1.78 | −1.61±0.08 |
| 52→56 | −0.53 | −0.20±0.08 | −0.47±0.14 | −0.77±0.14 | 0.53±1.34 | −0.56±0.08 |
|
| ||||||
| RMSDall (kcal/mol) | N/A | 0.94g/ 0.49h | 0.75g/ 0.52i | 0.66g/ 0.53i | 3.00 | 2.28 |
| R2 f | N/A | 0.48g/ 0.83h | 0.80g/ 0.84i | 0.73g/ 0.77i | 0.68 | 0.91 |
Experimental ΔΔ G values.
α in Equation 3 = 0.3, β in Equation 5 = 18 (Å2).
The QM/MM-GBSA results use 2,400 frames evenly extracted from a 6 ns molecular dynamic simulations trajectories, the GBneck2 method, the PM3 Hamiltonian, and mbondi2 effective radii setting from our previous study. ΔΔ G(A→B)=(ΔH B -TΔS B)- (ΔH A -TΔS A).
The QM/MM-GBSA results use 2,400 frames evenly extracted from a 6 ns molecular dynamic simulations trajectories, the GBneck2 method, the PM3 Hamiltonian, and mbondi2 effective radii setting from our previous study. ΔΔ H(A→B)=ΔH B- ΔH A.
The error estimate using the standard error of the mean in Equation 7.
R2 is the coefficient of determination between ΔΔ Gexp and ΔΔ Gcalculated.
Calculated including all transformations.
Calculated including the 135→138 and 34→58 transformations results from the one-step pmemd TI using 11 windows.
Calculated including the 135→166 transformation result from the three-step pmemd TI using 6 windows.
Figure 4.
Plots of calculated ΔΔGbind(experimental-calculated) (kcal/mol) vs. cumulative MD simulation lengths using the pmemd three and one step TI (α = 0.3, β = 18 (Å2) ).
In the three-step pmemd TI with 6 windows calculations, the 34→58 transformations do not converge, as shown in Figure 4a. Therefore, in Table 5, the RMSD including all transformations is on the high end (0.94 kcal/mol) and the coefficient of determination is low (R2 = 0.48) due to the 135→138 and non-converged 34→58 outlier transformations. When the RMSDall was calculated including the 135→138 and 34→58 transformations results from the one-step pmemd TI in 11 windows (seven transformations), the result improved significantly (RMSDall =0.48 kcal/mol and R2 = 0.83). On the other hand, the one-step pmemd TI using 11 windows and 1 ns MD simulations converged for all seven transformations, while the 135→138 transformation didn’t converge in the one-step pmemd TI using 6 windows. The one-step pmemd TI using both 6 and 11 windows offered quite satisfactory results (RMSD = 0.75 and 0.66 kcal/mol). Moreover, the 135→166 transformation has been shown to be difficult for the one-step pmemd TI, as discussed in the previous section. If the one-step pmemd TI result is re-calculated using the 135→166 transformation result from the three-step pmemd TI in 6 windows, the RMSD of the new one-step pmemd TI can be as low as 0.52 and 0.53 kcal/mol for 6 and 11 windows, respectively.
The TI results here are also consistent with our structure-activity relationship (SAR) (Tables 5 & 6). The in-house crystal structure (pdb code: 3UIC) suggests that the distance between the para-chloride in compound 91 and the backbone carbonyl group of Pro154 is 3.49 Å,46 slightly outside the halogen bond distance range (carbonyl to chloride: 3.00 – 3.40 Å), which is calculated in the previous perspective studies (Figure 3).54,55 However, the halogen bond distance range in carbonyl to bromide is slightly larger (3.00 – 3.50 Å), and the center of para-bromide in compound 177 and the backbone carbonyl group of Pro154 is still 3.49 Å. Therefore, the in-house structural study suggests that the para-chloride and para- bromide in compound 91 and 177 might form a mixture of halogen bond and vdW interactions in this hydrophobic pocket.46 In both of our in-house SAR and TI calculation results, the para-chloride to bromide substitution in the 91→177 transformation improves the binding affinity (Table 6, ΔΔGexpt = −1.27 (kcal/mol), ΔΔGTI = −0.87±0.05 (kcal/mol)). The TI result also suggests that the binding free energy difference in the 91→177 transformation is mostly from vdW interactions (83%) with minor electrostatic interactions (17%), as shown in Table 6. Because a halogen bond consists primarily of electrostatic interactions,54 the 17% of electrostatic interactions here might support the concept of a halogen bond and vdW interactions mixture, where vdW interactions are still the major ones. It has been reported that the traditional biomolecular modeling force fields might result in up to 1–2 kcal/mol error in halogen bonding calculations.56 This might explain why the ΔΔGTI of 91→177 transformation in this study deviates −0.4~−0.8 kcal/mol from the corresponding ΔΔGexpt. The recently customized force fields for halogen boding, such as the positive extra-point approach in the AMBER general force field (GAFF)57,58 and the OPLS-AA56 force field, and the polarizable ellipsoidal approach59 compatible with the AMBER GAFF, or even the computationally expensive density function theory approach60 might provide more accurate insights of the potential halogen bonds between FabI benzimidazole inhibitors and FabI, a well-known halogen bond favoring enzyme,54 but comparative studies of halogen bonding potentials were beyond the scope of this study, and thus we simply note the potential uncertainty.
Table 6.
The Breakdown of the Calculated ΔΔGbind (kcal/mol) of Benzimidazole Inhibitors in FtFabI Using the Three-Step pmemd TI, 1 ns MD Simulations TI, and 6 Windows
| Transformation | ΔΔGStep1 charge removal (kcal/mol) | ΔΔGStep2 vdW radius change (kcal/mol) | ΔΔGStep3 charge adding on (kcal/mol) | ΔΔGTI(kcal/mol) pmemd 3 steps 6 windows | ΔΔGexpt (kcal/mol)a |
|---|---|---|---|---|---|
| 91→177 | −0.21±0.04b | −0.74±0.09 | 0.06±0.03 | −0.87±0.05e | −1.27 |
| 91→135 | −0.15±0.05 | −0.25±0.13 | −0.42±0.03 | −0.82±0.07 | −1.04 |
| 135→138 | 0.33±0.12 | −3.75±0.22 | 0.32±0.02 | −3.10±0.12 | −1.53 |
| 135→166 | −0.54±0.05 | 1.85±0.17 | −0.25±0.11 | 1.06±0.11 | 0.76 |
| 34→58 | 0.12±0.01 | 1.70±0.17 | −1.90±0.20 | −0.08±0.13 | 1.48 |
| 09-12F→11-12F | 0.63±0.06 | 0.49±0.23 | −0.30±0.09 | 0.82±0.13 | −0.12 |
| 52→56 | −0.01±0.06 | −0.34±0.16 | 0.15±0.01 | −0.20±0.08 | −0.53 |
Experimental ΔΔGbind values.
The error estimate using the standard error of the mean in Equation 7.
ΔΔGStep1+ΔΔGStep2+ΔΔGStep3=ΔΔGTI. Any slightly unmatched values are due to rounding error.
Moreover, the TI data agree with the experimental results that the meta-chloride (91→135) and meta-methyl (52→56) substitutions are also binding favorably because the meta-chloride and methyl provides hydrophobic interactions to the FabI hydrophobic region comprised of the Ile200 and Leu99 side chains. Comparing to the meta-chloride and meta-methyl, the TI result in the 09–12F→11–12F transformation shows a positive ΔΔGstep2 (0.49 kcal/mol in Table 6), and indicates that the methoxy group induces unfavorable binding free energy from a steric effect. The summation of the ΔΔGstep1 and ΔΔGstep3 in the 09–12F→11–12F transformation is positive as well (0.33 kcal/mol), which is a binding-unfavorable ΔΔGelectrostatic, possibly from introducing a more polar methoxy group in a lipophilic region. These observations are consistent with our previous SAR and structural biology analysis (pdb code: 4J3F and 4J4T).47
On the other hand, the 5 and 6 positions of the benzimidazole ring are within an empty hydrophobic pocket (Figure 3). Here the ΔΔGbind of the 135→138 transformation is highly negative, −3.10 kcal/mol (Table 6), suggesting that introducing any hydrophobic group, two methyl groups here as an example, favors the binding. Moreover, the ΔΔGstep2 of the 135→138 transformation contributes the most to the ΔΔGbind, indicating that the hydrophobic vdW interactions here play a key role (Table 6). In contrast, a more polar substitution in this hydrophobic pocket, such as the 34 → 58 transformation, reduces the binding affinity (Table 6).
The positive ΔΔGstep2 of the 135→166 transformation (1.85 kcal/mol in Table 6) contributes the most to the ΔΔGbind in the 135→166 transformation (1.06 kcal/mol in Table 6), indicating that a steric clash of the methyl group on the 2 position of the benzimidazole ring lowers the binding affinity, as previously noted (pdb code: 3UIC).46 Moreover, our previous structure metabolic stability relationship (SMR) analysis also determined that a methyl group or a hydrogen bond acceptor substitution in the 2 position of the benzimidazole ring compromises the metabolic stability, limiting substitution options at this position.45
Interestingly, our prior analysis has shown that the QM-MM/GBSA method using 2,400 frames evenly extracted from 6 ns molecular dynamic simulations trajectories, the GBneck2 method, the PM3 Hamiltonian, and the mbondi2 effective radii setting, can rank the benzimidazole binding affinity accurately (R2 between predicted and experimental binding free energy is equal to 0.88).2 However, QM-MM/GBSA fails to quantify the binding free energy differences between two similar benzimidazole ligands, since the RMSD including all seven transformations using QM-MM/GBSA is as high as 3.00 kcal/mol (Table 5). If the inaccurate entropy calculation results from our former study 2 are excluded and only the enthalpy differences between two similar benzimidazole ligands are included into the RMSD calculation, the RMSD including all seven transformations using QM-MM/GBSA is still not satisfactory (RMSDall = 2.28 kcal/mol, Table 5). Former studies also suggested that implicit solvent methods, MM/PBSA and (QM−)MM/GBSA, take even more computational resources than explicit solvent methods (TI, FEP, BAR, etc.) in order to reach a converged and satisfactory quantification of binding free energy differences.7 Therefore, implicit solvent methods are more suitable for binding affinity ranking rather than binding free energy difference quantification.
Conclusions
In this study, we have shown TI to be affordable for a real world, large scale prediction. The one-step pmemd TI using 1 ns MD simulations and 6 windows can be as efficient as such implicit solvent methods as QM-MM/GBSA. The one-step pmemd TI using 1 ns MD simulations and 6 windows can finish in 5 hours for one protein-ligand binding pair transformation using 96 Intel Xeon E5-2670 2.6 GHz CPUs on the UIC Extreme Computing machine. In comparison, our former study suggests that QM-MM/GBSA, using the GBneck2 method, the PM3 Hamiltonian, the mbondi2 radii setting, and 2,400 frames extracted evenly from 6 ns MD simulations trajectories, requires 4.5 hours.2 Besides the improvement in TI efficiency, TI offers higher accuracy in prediction of binding free energy differences between two similar ligands than does QM-MM/GBSA (RMSD 0.75 kcal/mol versus 3.07 kcal/mol). This result is highly encouraging as alchemical free energy calculation methods, such as TI, have been notorious for their slow predictive speed. It usually ended up requiring longer real world time than a synthetic chemist making a small modification on a certain scaffold. However, as the advance of the methodology and protocols highlighted in the former and current studies, universities and pharmaceutical companies equipped with small or medium size computer clusters can utilize TI to predict large scale putative ligand transformations and provide useful insights for a drug discovery team.
One of the major TI speed improvement comes from the TI implementation in a newer and faster Amber MD engine, pmemd, in April 2014.40 This study has shown in a larger scale than the former study that the pmemd TI can be significantly more efficient than TI implemented on the older and slower Amber MD engine, sander, while the pmemd TI still maintains similar accuracy (Tables 2 & 3). The pmemd TI is shown to be 3.1 times faster than sander TI using the conditions of this study. Moreover, pmemd scales better than sander TI, consistent with the former study. Sander also requires the number of cores to be a power of two to reach its optimal speed, while pmemd has no such limitation. This allows pmemd to fully maximize its performance using a machine where the number of cores in a node is not a power of two. Lastly, the sander TI only allows minimization to be run on two cores when activating Softcore potentials, while the following equilibration and production runs still require a larger number of cores to finish computation jobs in a timely manner. This complicates job submission and might prolong the job queue time. Briefly, our experience suggests that the pmemd TI might be 4–5 times faster than the sander TI, including the above factors and the associated queue time, etc. Other TI efficiency improvements included reducing the MD simulation lengths from 2 to 1 ns (Table 2–3) and the number of windows from 21 to 6 (Table 2–5) without compromising the convergence and accuracy of results, consistent with the former pioneering studies. 7,33–37
Moreover, we have tested various vdW and Coulomb Softcore potential controlling parameters (α and β values) in the one-step TI in a protein-ligand system for the first time (Tables 4 and 5). The results suggest that the one-step pmemd TI using 6 windows can offer convergence and accuracy comparable (RMSD = 0.75 kcal/mol) to the three-step pmemd TI (RMSD = 0.83 kcal/mol) when α is equal to 0.3 and β is equal to 18 Å2. Moreover, implementation of the one-step TI can offer three times faster speed than the traditional three-step TI. However, the one-step pmemd TI cannot provide as high accuracy prediction in the 135→166 transformation as the three-step pmemd TI. The hydrogen at the 2 position of the benzimidazole ring transforms into a methyl group in the 135→166 transformation. This 2 position methyl group is believed to attract electrons from the 3 position nitrogen atom of the benzimidazole ring, which forms key hydrogen bond interactions with the enzyme and the NADH cofactor (Figure 3). Therefore, the simultaneous vdW radius and charge transformations in the one-step TI might not be as suitable as the three-step TI for this special case. The three-step pmemd TI using 6 windows and 1 ns MD simulations would be encouraged in any future TI prediction on this 2 position modification (Table 5). Moreover, the one-step pmemd TI using 6 windows might not be ideal for a larger modification such as the 135→138 transformation (two hydrogen atoms transformed into two methyl groups) due to the non-converged result (Figure 4). In such a case, implicit solvent methods might be better to quickly rank the binding affinities of more distant molecules. If a more accurate quantification for a larger modification is required, the three-step pmemd TI using 6 windows or the one-step pmemd TI using 11 windows, which offer the converged 135→ 138 transformation results, might be better than the pmemd one-step pmemd TI using 6 windows. Lastly, except for the special cases in the 135→ 138 and 135→ 166 transformations, the one-step pmemd TI using 6 and 11 windows achieved almost identical accuracy (RMSD = 0.52 and 0.53 kcal/mol). As the TI with 11 windows takes almost twice as long as TI using 6 windows, the one-step pmemd TI using 6 windows and 1 ns MD simulations will be the preferred choice for our lead optimization campaign against F. tuleransis FabI.
Lastly, the TI results here also provide benzimidazole structure-activity relationship insights in the FtFabI active site and also suggest that the para-halogen in benzimidazole inhibitors might form a weak halogen bond with FtFabI. Future study using customized force fields for halogen boding 56–59 or even the computationally expensive density function theory approach60 might provide more details of the potential halogen bond between FabI benzimidazole inhibitors and FabI, which is a well-known halogen bond favoring enzyme.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grant U01 AI077949. Pin-Chih Su was financially supported by the American Heart Association (AHA) pre-doctoral fellowship, 13PRE-14800030. Computational resources were kindly provided by the Texas Advanced Computing Center (TACC), Lonestar cluster at the University of Texas at Austin under the XSEDE Teragrid Grant TG-MCB090168 to MEJ, and by the Extreme Computing Machine at the University of Illinois at Chicago. We thank the amber community (http://archive.ambermd.org/) and Drs. Kirk E. Hevener, Paul Gasper, Paulius Mikulskis, Levi Pierce, William Sinko and Jason Swails for helpful discussions.
Abbreviations
- MM-PBSA
Molecular Mechanics - Poisson-Boltzmann Surface Area
- MM-GBSA
Molecular Mechanics - Generalized Born Surface Area
- QM-MM/GBSA
quantum mechanics Molecular Mechanics - Generalized Born Surface Area
- LIE
Linear Interaction Energy
- TI
thermodynamics integration
- FEP
free energy perturbation
- BAR
Bennett’s acceptance ratio method
- FabI
enoyl acyl reductase
- MD
molecular dynamics
- ns
nano seconds
Footnotes
Additional Supporting Information may be found in the online version of this article.
References and Notes
- 1.Hepburn MJ, Simpson AJ. Expert review of anti-infective therapy. 2008;6(2):231–240. doi: 10.1586/14787210.6.2.231. [DOI] [PubMed] [Google Scholar]
- 2.Su P-CT, Cheng-Chieh Mehboob, Shahila Hevener, Kirk E, Johnson Michael E. Journal of Computational Chemistry. 2015 co-submitted article. [Google Scholar]
- 3.Hou T, Wang J, Li Y, Wang W. Journal of chemical information and modeling. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearlman DA. Journal of medicinal chemistry. 2005;48(24):7796–7807. doi: 10.1021/jm050306m. [DOI] [PubMed] [Google Scholar]
- 5.Rastelli G, Del Rio A, Degliesposti G, Sgobba M. Journal of computational chemistry. 2010;31(4):797–810. doi: 10.1002/jcc.21372. [DOI] [PubMed] [Google Scholar]
- 6.Zhu T, Lee H, Lei H, Jones C, Patel K, Johnson ME, Hevener KE. Journal of chemical information and modeling. 2013;53(3):560–572. doi: 10.1021/ci300502h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genheden S, Nilsson I, Ryde U. Journal of chemical information and modeling. 2011;51(4):947–958. doi: 10.1021/ci100458f. [DOI] [PubMed] [Google Scholar]
- 8.Genheden S, Ryde U. Journal of chemical theory and computation. 2011;7(11):3768–3778. doi: 10.1021/ct200163c. [DOI] [PubMed] [Google Scholar]
- 9.Genheden S, Ryde U. Expert Opinion on Drug Discovery. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun H, Li Y, Tian S, Xu L, Hou T. Physical Chemistry Chemical Physics. 2014;16(31):16719–16729. doi: 10.1039/c4cp01388c. [DOI] [PubMed] [Google Scholar]
- 11.Godschalk F, Genheden S, Soderhjelm P, Ryde U. Physical Chemistry Chemical Physics. 2013;15(20):7731–7739. doi: 10.1039/c3cp00116d. [DOI] [PubMed] [Google Scholar]
- 12.Shirts M. In: Computational Drug Discovery and Design. Baron R, editor. Springer; New York: 2012. pp. 425–467. [Google Scholar]
- 13.Kong X, Brooks CL. The Journal of chemical physics. 1996;105(6):2414–2423. [Google Scholar]
- 14.Knight JL, Brooks CL. Journal of computational chemistry. 2009;30(11):1692–1700. doi: 10.1002/jcc.21295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. Journal of computational chemistry. 1992;13(8):1011–1021. [Google Scholar]
- 16.Tzoupis H, Leonis G, Mavromoustakos T, Papadopoulos MG. Journal of chemical theory and computation. 2013;9(3):1754–1764. doi: 10.1021/ct301063k. [DOI] [PubMed] [Google Scholar]
- 17.Leonis G, Steinbrecher T, Papadopoulos MG. Journal of chemical information and modeling. 2013;53(8):2141–2153. doi: 10.1021/ci4002102. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen WL, Bollini M, Thakur VV, Domaoal RA, Spasov KA, Anderson KS. Journal of the American Chemical Society. 2011;133(39):15686–15696. doi: 10.1021/ja2058583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy MR, Erion MD. Journal of the American Chemical Society. 2001;123(26):6246–6252. doi: 10.1021/ja0103288. [DOI] [PubMed] [Google Scholar]
- 20.Reddy MR, Singh UC, Erion MD. Journal of computational chemistry. 2007;28(2):491–494. doi: 10.1002/jcc.20510. [DOI] [PubMed] [Google Scholar]
- 21.Reddy MR, Singh UC, Erion MD. Journal of the American Chemical Society. 2011;133(21):8059–8061. doi: 10.1021/ja201637q. [DOI] [PubMed] [Google Scholar]
- 22.Kang S-g, Das P, McGrane SJ, Martin AJ, Huynh T, Royyuru AK, Taylor AJ, Jones PG, Zhou R. The Journal of Physical Chemistry B. 2014;118(24):6393–6404. doi: 10.1021/jp410232j. [DOI] [PubMed] [Google Scholar]
- 23.Steinbrecher T, Case DA, Labahn A. Journal of medicinal chemistry. 2006;49(6):1837–1844. doi: 10.1021/jm0505720. [DOI] [PubMed] [Google Scholar]
- 24.Steiner D, Oostenbrink C, Diederich F, Zürcher M, van Gunsteren WF. Journal of computational chemistry. 2011;32(9):1801–1812. doi: 10.1002/jcc.21761. [DOI] [PubMed] [Google Scholar]
- 25.Chodera JD, Mobley DL, Shirts MR, Dixon RW, Branson K, Pande VS. Curr Opin Struct Biol. Elsevier Ltd; England: 2011. pp. 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinbrecher T, Labahn A. Current Medicinal Chemistry. 2010;17(8):767–785. doi: 10.2174/092986710790514453. [DOI] [PubMed] [Google Scholar]
- 27.Jorge M, Garrido NM, Queimada AnJ, Economou IG, Macedo EnA. Journal of chemical theory and computation. 2010;6(4):1018–1027. doi: 10.1021/ct900214y. [DOI] [PubMed] [Google Scholar]
- 28.Shyu C, Ytreberg FM. Journal of computational chemistry. 2009;30(14):2297–2304. doi: 10.1002/jcc.21231. [DOI] [PubMed] [Google Scholar]
- 29.Lawrenz M, Wereszczynski J, Ortiz-Sánchez J, Nichols S, McCammon JA. Journal of computer-aided molecular design. 2012;26(5):569–576. doi: 10.1007/s10822-012-9542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mobley D. Journal of computer-aided molecular design. 2012;26(1):93–95. doi: 10.1007/s10822-011-9497-y. [DOI] [PubMed] [Google Scholar]
- 31.Mobley DL, Klimovich PV. The Journal of chemical physics. 2012;137(23):230901. doi: 10.1063/1.4769292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monroe J, Shirts M. Journal of computer-aided molecular design. 2014;28(4):401–415. doi: 10.1007/s10822-014-9716-4. [DOI] [PubMed] [Google Scholar]
- 33.Steinbrecher T, Joung I, Case DA. Journal of computational chemistry. 2011;32(15):3253–3263. doi: 10.1002/jcc.21909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pitera JW, van Gunsteren WF. The Journal of Physical Chemistry B. 2001;105(45):11264–11274. [Google Scholar]
- 35.Özal TA, Peter C, Hess B, van der Vegt NFA. Macromolecules. 2008;41(13):5055–5061. [Google Scholar]
- 36.Jorgensen WL, Thomas LL. Journal of chemical theory and computation. 2008;4(6):869–876. doi: 10.1021/ct800011m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruckner S, Boresch S. Journal of computational chemistry. 2011;32(7):1303–1319. doi: 10.1002/jcc.21713. [DOI] [PubMed] [Google Scholar]
- 38.Bruckner S, Boresch S. Journal of computational chemistry. 2011;32(7):1320–1333. doi: 10.1002/jcc.21712. [DOI] [PubMed] [Google Scholar]
- 39.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. Journal of computational chemistry. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaus JW, Pierce LT, Walker RC, McCammon JA. Journal of chemical theory and computation. 2013;9(9):4131–4139. doi: 10.1021/ct400340s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christ CD, Fox T. Journal of chemical information and modeling. 2013;54(1):108–120. doi: 10.1021/ci4004199. [DOI] [PubMed] [Google Scholar]
- 42.Mikulskis P, Genheden S, Ryde U. Journal of chemical information and modeling. 2014;54(10):2794–2806. doi: 10.1021/ci5004027. [DOI] [PubMed] [Google Scholar]
- 43.Homeyer N, Stoll F, Hillisch A, Gohlke H. Journal of chemical theory and computation. 2014;10(8):3331–3344. doi: 10.1021/ct5000296. [DOI] [PubMed] [Google Scholar]
- 44.Hevener KE, Mehboob S, Su PC, Truong K, Boci T, Deng J, Ghassemi M, Cook JL, Johnson ME. Journal of medicinal chemistry. 2012;55(1):268–279. doi: 10.1021/jm201168g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang YY, Liu Y, Mehboob S, Song JH, Boci T, Johnson ME, Ghosh AK, Jeong H. Xenobiotica; the fate of foreign compounds in biological systems. 2014;44(5):404–416. doi: 10.3109/00498254.2013.850553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehboob S, Hevener KE, Truong K, Boci T, Santarsiero BD, Johnson ME. Journal of medicinal chemistry. 2012;55(12):5933–5941. doi: 10.1021/jm300489v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehboob S, Song J, Hevener KE, Su P-C, Boci T, Brubaker L, Truong L, Mistry T, Deng J, Cook JL, Santarsiero BD, Ghosh AK, Johnson ME. Bioorganic & medicinal chemistry letters. 2015;25(6):1292–1296. doi: 10.1016/j.bmcl.2015.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJG. Inc; Wallingford CT: 2009. [Google Scholar]
- 49.Vanquelef E, Simon S, Marquant G, Garcia E, Klimerak G, Delepine JC, Cieplak P, Dupradeau FY. Nucleic acids research. 2011;39(Web Server issue):W511–517. doi: 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dupradeau FY, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N, Lelong D, Rosanski W, Cieplak P. Physical chemistry chemical physics: PCCP. 2010;12(28):7821–7839. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Journal of computational chemistry. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 52.Case DA, VB, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE, III, Darden TA, Duke RE, Gohlke H, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A, Lee TS, LeGrand S, Luchko T, Luo R, Madej B, Merz KM, Paesani F, Roe DR, Roitberg A, Sagui C, Salomon-Ferrer R, Seabra G, Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Kollman PA. 2014 [Google Scholar]
- 53.Lai C-T, Li H-J, Yu W, Shah S, Bommineni GR, Perrone V, Garcia-Diaz M, Tonge PJ, Simmerling C. Biochemistry. 2015;54(30):4683–4691. doi: 10.1021/acs.biochem.5b00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bissantz C, Kuhn B, Stahl M. Journal of medicinal chemistry. 2010;53(14):5061–5084. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hernandes MZ, Cavalcanti SMT, Moreira DRM, de Azevedo WF, Junior, Leite ACL. Current Drug Targets. 2010;11(3):303–314. doi: 10.2174/138945010790711996. [DOI] [PubMed] [Google Scholar]
- 56.Jorgensen WL, Schyman P. Journal of chemical theory and computation. 2012;8(10):3895–3901. doi: 10.1021/ct300180w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ibrahim MAA. Journal of computational chemistry. 2011;32(12):2564–2574. doi: 10.1002/jcc.21836. [DOI] [PubMed] [Google Scholar]
- 58.Ibrahim MAA. The Journal of Physical Chemistry B. 2012;116(11):3659–3669. doi: 10.1021/jp3003905. [DOI] [PubMed] [Google Scholar]
- 59.Du L, Gao J, Bi F, Wang L, Liu C. Journal of computational chemistry. 2013;34(23):2032–2040. doi: 10.1002/jcc.23362. [DOI] [PubMed] [Google Scholar]
- 60.Lu YX, Zou JW, Fan JC, Zhao WN, Jiang YJ, Yu QS. Journal of computational chemistry. 2009;30(5):725–732. doi: 10.1002/jcc.21094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.