Abstract
Merkel cell carcinoma is a rare but highly aggressive cutaneous neuroendocrine carcinoma. Cytokeratin-20 (CK20) is expressed in approximately 95% of Merkel cell carcinomas and is useful for distinction from morphologically similar entities including metastatic small cell lung carcinoma. Lack of CK20 expression may make diagnosis of Merkel cell carcinoma more challenging, and has unknown biological significance. Approximately 80% of CK20-positive Merkel cell carcinomas are associated with the oncogenic Merkel cell polyomavirus. Merkel cell carcinomas lacking Merkel cell polyomavirus display distinct genetic changes from Merkel cell polyomavirus-positive Merkel cell carcinoma, including RB1 inactivating mutations. Unlike CK20-positive Merkel cell carcinoma, the majority of CK20-negative Merkel cell carcinomas are Merkel cell polyomavirus-negative, suggesting CK20-negative Merkel cell carcinomas predominantly arise through virus-independent pathway(s) and may harbor additional genetic differences from conventional Merkel cell carcinoma. Hence, we analyzed 15 CK20-negative Merkel cell carcinoma tumors (ten Merkel cell polyomavirus-negative, four Merkel cell polyomavirus-positive, and one undetermined) using the Ion Ampliseq Comprehensive Cancer Panel, which assesses copy number alterations and mutations in 409 cancer-relevant genes. Twelve tumors displayed prioritized high-level chromosomal gains or losses (average 1.9 per tumor). Non-synonymous high confidence somatic mutations were detected in 14 tumors (average 11.9 per tumor). Assessing all somatic coding mutations, an ultraviolet-signature mutational profile was present, and more prevalent in Merkel cell polyomavirus-negative tumors. Recurrent deleterious tumor suppressor mutations affected TP53 (9/15, 60%), RB1 (3/15, 20%), and BAP1 (2/15, 13%). Oncogenic activating mutations included PIK3CA (3/15, 20%), AKT1 (1/15, 7%)) and EZH2 (1/15, 7%). In conclusion, CK20-negative Merkel cell carcinoma display overlapping genetic changes with CK20-positive Merkel cell carcinoma, including RB1 mutations restricted to Merkel cell polyomavirus-negative tumors. However, some CK20-negative Merkel cell carcinomas harbor mutations not previously described in Merkel cell carcinoma. Hence, CK20-negative Merkel cell carcinomas harbor diverse oncogenic drivers which may represent therapeutic targets in individual tumors.
Keywords: Merkel cell carcinoma, cytokeratin 20, retinoblastoma, Merkel cell polyomavirus, ultraviolet mutations
Introduction
Merkel cell carcinoma, a primary cutaneous neuroendocrine carcinoma, is among the most deadly of skin cancers with a mortality rate of 30-75% (1). Although rare, the incidence has nearly tripled over the past several decades, which may be due in part to increased diagnostic recognition (1). In 2008, Feng and colleagues identified a novel tumorigenic virus, Merkel cell polyomavirus, that is clonally integrated in 80% of Merkel cell carcinomas (2). Evidence suggests that Merkel cell polyomavirus promotes tumorigenesis via oncogenic action of viral large T antigen and small T antigen (3-6). In polyomavirus-negative Merkel cell carcinoma, recent evidence suggests that major players in the pathogenesis include ultraviolet light and loss of the tumor suppressor RB1 (7, 8).
Cytokeratin 20 (CK20) labels 95% of Merkel cell carcinomas, typically in a paranuclear dot pattern (9-11). Accurate diagnosis becomes more challenging in the approximately 5% of Merkel cell carcinomas that lack CK20 staining (9, 10). In such cases, additional immunohistochemistry and careful clinical workup is necessary to exclude the possibility of metastasis from a non-cutaneous small cell carcinoma.
We have previously shown that CK20-negative Merkel cell carcinoma often lacks detectable Merkel cell polyomavirus sequence (12). In the parotid, the presence or absence of CK20 expression may be used to divide parotid small cell carcinomas into two subtypes: CK20-positive “Merkel cell-type”, and the more aggressive CK20-negative “pulmonary type” (13). These observations suggest that the absence of CK20 expression in cutaneous Merkel cell carcinoma may have molecular and clinical significance. To better understand the molecular changes in CK20-negative Merkel cell carcinomas, we analyzed a previously assembled cohort of 15 CK20-negative Merkel cell carcinomas by next generation sequencing to characterize mutational and copy number changes in these tumors.
Methods
Case selection
Studies were conducted according to previously approved Institutional Review Board Protocols. Selection and histopathologic characterization of CK20-negative Merkel cell carcinomas used in this study has been previously described (12), with the exception of one additional case (MC15) which was a high-grade neuroendocrine carcinoma presenting in the soft tissue of the neck that stained positively for cytokeratins, synaptophysin, and neuron specific enolase, and was negative for CK7, CK20, thyroid transcription factor-1, and melanocytic markers. Tumors with adequate material were also evaluated by immunohistochemistry for LTAg as previously described (14), which was concordant with PCR results in all but one case (MC11). Clinicopathologic features of tumors in this study are summarized in Supplemental Table S1.
Targeted next-generation sequencing
For the majority of cases, previously purified tumor DNA was adequate for sequencing (12). For cases requiring additional DNA extraction, an H&E stained section was used as a guide for dissection from a minimum of 4 formalin-fixed, paraffin-embedded 10-micron sections by a board-certified dermatopathologist (P.W.H.) to obtain a minimal tumor purity of 60%. DNA extraction, Ion Torrent-based next generation sequencing, and data analysis were performed as previously described (15), using the Ion Ampliseq Comprehensive Cancer Panel that enables copy number and mutational analysis through targeting the complete coding sequence of 409 cancer relevant genes. Briefly, DNA was extracted using the Qiagen FFPE AllPrep DNA/RNA kit. Barcoded libraries were generated from 40ng of DNA per sample using the Ion Ampliseq library kit 2.0 with Ion Ampliseq Comprehensive Cancer Panel primers. Templates were prepared using the Ion PI Template OT2 200 Kit v3 (Life Technologies, Foster City, CA) on the Ion One Touch 2. Sequencing of multiplexed templates was performed using the Ion Torrent Proton on Ion PIv2 chips using the Ion PI Sequencing 200 Kit v3 (Life Technologies, Foster City, CA) according to the manufacturer's instructions. Data analysis was performed using in-house developed, previously validated pipelines using Torrent Suite 4.0.2, with alignment by TMAP and variant calling using the Torrent Variant Caller plugin. Annotated variants were filtered to remove synonymous or non-coding variants, poorly supported calls, and panel-specific/sequencing artifacts. Variants present in the ESP6500 or phase I 1000G dataset, or in the Exome Aggregation Consortium at >0.1% were filtered out as likely germ line alterations (16-18). Potential driving somatic alterations were prioritized using the COSMIC Database. For ultraviolet signature analyses, synonymous as well as non-synonymous somatic variants were considered. Quality control metrics are listed in Supplemental Table S2. Statistical analyses were performed on Graphpad Prism 6.0 software using Student's t-test or Mann-Whitney U nonparametric test.
Validation of single nucleotide variants by Sanger sequencing
Selected single nucleotide variations affecting AKT1, PIK3CA, RB1, TP53, EZH2, PTCH1, TSC1, and BAP1 were validated by PCR amplification using new or previously reported primer pairs (Supplemental Table S3)(19-21), followed by Sanger sequencing at the University of Michigan Sequencing Core. Chromatograms were visualized using Sequence Scanner 2 software.
Results
Clinicopathologic Features of CK20-Negative Merkel Cell Carcinoma
Clinicopathologic characteristics of the sequencing cohort are described in Supplemental Table S1. Twelve were primary tumors, and three were metastatic tumors. Two primary-metastasis pairs were tested. All fifteen tumors were CK20 negative. We have previously described the morphologic and clinical features of the majority of these tumors (12). As previously reported, there was no definitive morphologic difference between our cohort and CK20 positive Merkel cell carcinomas. By immunohistochemistry and/or previously reported quantitative PCR, ten tumors (from nine patients) were Merkel cell polyomavirus-negative, four tumors (from three patients) were Merkel cell polyomavirus-positive, and one lacked sufficient remaining material for Merkel cell polyomavirus detection (Supplemental Table S1)(12).
Copy Number Alterations
First, we analyzed global changes including chromosomal gains/losses and non-synonymous mutations. Ten tumors displayed prioritized high-level chromosomal gains or losses involving tumor suppressors or oncogenes (Supplemental Tables S1 and S4, Supplemental Figure S1). Recurrent copy number changes included gains of chromosome 8 and loss of chromosomes 5 and 21. The most frequent alteration involved gain of AKT1 on chromosome 14q32.33 in 5/15 (33.3%) of tumors. Gain of MYCL1, previously reported in Merkel cell carcinoma (22), was detected in one tumor (1/15, 6.7%) and a gain of MYC was identified in two tumors (2/15, 13.3%).
Mutational Profiles
A total of 179 high-confidence non-synonymous somatic mutations were detected in 14 of the 15 tumors, with an average of 11.9 non-synonymous mutations/tumor (Supplemental Table S1). When assessing all coding mutations (non-synonymous and synonymous) to evaluate mutational signatures, Merkel cell polyomavirus-negative Merkel cell carcinoma displayed a higher mutation burden (Figure 1A-B, Supplemental Table S1). For these mutation events, we characterized the UV- signature mutational profile, specifically the fraction of mutations that involve C >T transitions at dipyrimidine sites (23). We found that C > T transitions were the most prevalent single base substitution (Supplemental Table S1). C > T transitions were a significantly higher proportion of total single nucleotide variants in Merkel cell polyomavirus-negative tumors relative to Merkel cell polyomavirus-positive tumors (p = 0.004) (Supplemental Table S1). On average, C > T transitions at dipyrimidine sites were elevated above the level predicted by random chance in Merkel cell polyomavirus-negative Merkel cell carcinomas (p=0.001, df=9, t=4.767), but not Merkel cell polyomavirus-positive Merkel cell carcinomas (p=0.724, df=4, t=-0.379) by one-sample t-test (Figure 1C-D, Supplemental Table S1). CC > TT tandem substitutions, another form of UV-signature mutation, were detected in 9/10 (90%) Merkel cell polyomavirus-negative Merkel cell carcinomas and only 1/4 (25%) Merkel cell polyomavirus-positive Merkel cell carcinomas (Supplemental Table S1). Unlike small cell lung carcinoma, C>A (tobacco signature) transitions were rare in CK20-negative Merkel cell carcinoma (Supplemental Figure S2) (24, 25). Hence, in most cases, CK20-negative Merkel cell carcinomas are similar to CK20-positive tumors in harboring either high UV-signature mutation burden or presence of Merkel cell polyomavirus (Supplemental Figure S2).
Figure 1. Mutational profiles of CK20-negative Merkel cell carcinoma.
(A) Mutation burden in CK20-negative Merkel cell carcinoma tumors, with UV mutations represented by C>T at (C/TpC/T) sites. High-confidence synonynmous and non-synonymous mutations are included. (B) Total number of mutations in CK20-negative Merkel cell carcinoma, with significantly higer mutation burden in Merkel cell polyomavirus-negative tumors (Mann-Whitney U test). Means are indicated by solid lines. (C) Relative proportion of mutation types in CK20-negative Merkel cell carcinoma, with UV mutations represented by C>T at (C/TpC/T) sites. (D) C>T mutations occuring at dipyrimidine sites (UV-signature mutations) as a fraction of total C>T mutations in CK20-negative Merkel cell carcinoma. Merkel cell polyomavirus-negative tumors display significantly higher fraction of UV mutations than Merkel cell polyomavirus-positive tumors (Student's t-test). Dashed line indicates the fraction predicted by random chance (75%). Means are indicated by solid lines. Pair: primary-metastasis pairs. Age: age in years at diagnosis. *Asterisk indicates p <0.05. MCPyV: Merkel cell polyomavirus. SNV: single nucleotide variant.
One Merkel cell polyomavirus-positive tumor (MC11) displayed a UV-signature mutational profile and TP53 inactivating mutations, features common to Merkel cell polyomavirus-negative tumors in this cohort and our previous study (8). This tumor was positive for Merkel cell polyomavirus DNA by two of three PCR primer pairs, confirmed by Sanger sequencing (12). By immunohistochemistry, LTAg protein expression was not detected in this tumor (Supplemental Figure S3A). Furthermore, this tumor displayed foci of squamous differentiation, a feature reported as being restricted to Merkel cell polyomavirus-negative tumors (Supplemental Figure S3B-C) (26). Therefore, although this tumor harbors Merkel cell polyomavirus DNA, many other features are more consistent with a Merkel cell polyomavirus-negative tumor, in keeping with the absence of detectable levels of LTAg protein.
Tumor Suppressor Mutations
For tumor suppressors, multiple recurrent mutations were identified which were more prevalent in Merkel cell polyomavirus-negative tumors (p=0.03) (Figure 2). These included mutations in tumor suppressor genes not previously implicated in Merkel cell carcinoma including APC, TET2, and BAP1 as well as those known to be inactivated in Merkel cell carcinoma including TP53 and RB1 (Figure 2, Supplemental Table S3). APC mutations were detected in 2 tumors (2/15, 13.3%). TET2, which has been associated with melanoma susceptibility (27), was mutated in 2/15 (13.3%) cases. The tumor suppressor BAP1 displayed an inactivating mutation in both tumors of a primary-metastasis pair. TP53 mutations (9/15, 60%) were located in inactivation hotspots (Supplemental Figure S4) and were consistent with UV-signature mutations in the majority of cases. TP53 mutations were almost exclusively detected in Merkel cell polyomavirus-negative tumors, similar to previous reports (8, 28). RB1 mutations (3/15, 20%) with predicted functional significance were restricted to Merkel cell polyomavirus-negative tumors, also similar to previous reports (7, 8) (Figure 2, Supplemental Figure S4).
Figure 2. Mutational landscape of CK20-negative Merkel cell carcinomas.
Copy number alterations and mutations were filtered for predicted functional significance in cancer. MCPyV: Merkel cell polyomavirus. FP indel: frame-preserving insertion/deletion. FS indel: frameshift insertion/deletion. Pair: primary-metastasis pairs. Age: age in years at diagnosis.
Oncogenic Mutations
For oncogenic driver mutations, CK20-negative Merkel cell carcinomas were similar to CK20-positive Merkel cell carcinomas and displayed heterogeneous oncogenic driver mutations (Figure 2). The PI3K pathway displayed activating mutations in a subset of tumors. PIK3CA mutations were found in 3/15 (20%), including hotspot activating mutations E542K and H1047L. One tumor displayed the AKT1 (E17K) activating mutation. Of interest, one tumor harbored an EZH2 (Y641F) activating mutation not previously described in Merkel cell carcinoma. EZH2 is an oncogenic histone methyltransferase that promotes tumorigenesis by epigenetic repression of tumor suppressor genes, and is under investigation as a therapeutic target (29).
Comparison of Matched Primary and Metastatic Tumors
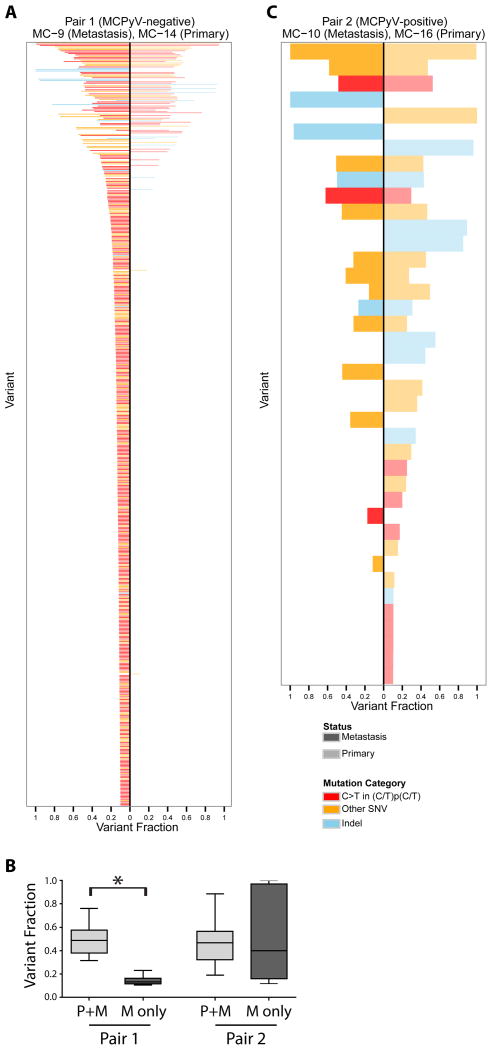
As predicted, paired primary-metastatic cases in our cohort displayed largely overlapping mutational profiles (Figures 2, 3). One primary-metastasis pair (MC9/MC14) was Merkel cell polyomavirus-negative. The metastasis in this case was a satellite metastasis that was detected and excised three months after excision of the primary tumor, without intervening radiotherapy or chemotherapy. This metastasis displayed a 7-fold higher mutational burden relative to the primary, consistent with accumulation of mutations during or after metastasis (Figure 3A, B). The other pair (MC10/MC16) was Merkel cell polyomavirus-positive. The metastasis in this case was an in-transit metastasis that developed four years after excision of the primary tumor. There was no radiotherapy or chemotherapy between excisions of the primary and metastasis. In this case, in contrast, the metastatic tumor did not accumulate a high mutational burden (Figure 3 B, C). For the Merkel cell polyomavirus-negative primary-metastasis pair, mutations detected in both the primary and the metastasis demonstrated an average variant fraction of 50.0% (standard deviation 16.4%) in both the primary and metastatic tumors, as expected for heterozygous mutations present in all tumor cells given the high tumor content (although inclusion of rare germ line alterations cannot be entirely excluded). Notably, mutations detected in the metastasis but not the primary tumor occurred at an average variant fraction of 16.3% (standard deviation 11.1%), suggesting heterozygous mutations present in approximately one-third of tumor cells (Figure 3B). This finding suggests the emergence of a more highly mutated tumor subclone within the metastasis of this Merkel cell polyomavirus-negative case.
Figure 3. Comparison of matched primary-metastatic CK20-negative Merkel cell carcinomas demonstrates clonal evolution in a Merkel cell polyomavirus-negative Merkel cell carcinoma.
(A) High-confidence mutations detected in a primary-metastasis Merkel cell carcinoma pair negative for Merkel cell polyomavirus, ranked by summed variant fraction. Each bar represents a single mutation. (B) Variant fractions of mutations detected in metastatic tumors, comparing mutations specific to the metastasis (M only) relative to mutations shared with the primary tumor (P+M), in both pairs. (C) High-confidence single nucleotide alterations detected in a primary-metastasis Merkel cell carcinoma pair positive for Merkel cell polyomavirus, ranked by summed variant fraction. P+M: mutations shared between primary and metastasis. M only: mutations detected only in the metastasis and not the primary. MCPyV: Merkel cell polyomavirus. SNV: single nucleotide variant. *Asterisk indicates p <0.05.
Discussion
Recent studies have advanced our understanding of genetic alterations in Merkel cell carcinoma. The majority harbor genomic integration of Merkel cell polyomavirus sequences, which undergo characteristic tumor-specific truncating mutations (2, 30). In contrast, Merkel cell carcinomas lacking detectable Merkel cell polyomavirus have a high mutation burden characterized by a predominance of UV-signature mutations, with recurrent inactivation of tumor suppressors including TP53, RB1, and NOTCH (7, 8). Merkel cell polyomavirus-negative tumors are heterogeneous with regard to oncogenic drivers, with potential roles for oncogenes including PIK3CA, HRAS, and KNSTRN (8, 31). Based on these observations, it has been proposed that Merkel cell carcinoma may arise via viral-mediated or UV-associated pathways (8).
CK20 expression is a useful diagnostic finding in the majority of Merkel cellcarcinomas. However, a small minority of tumors lacks CK20 expression (9-11). The mechanism for loss or absence of CK20 expression in Merkel cell carcinoma is poorly understood. Loss of CK20 expression with tumor progression has been reported (32), however the majority of CK20-negative tumors in our study were primary tumors. Previously published findings in Merkel cell carcinoma cell lines derived from CK20-negative tumors in this study reveals that loss of CK20 expression occurs at the mRNA level in one cell line, but at the protein level, to variable degrees, in others (Supplemental Table S5)(33). Furthermore, cell lines with low/absent CK20 frequently displayed decreased CK8 and chromogranin A expression, but not other lineage markers such as ATOH1, synaptophysin, or neuron-specific enolase (Supplemental Table S5)(33). These observations suggest that CK20 loss, in different tumors, may reflect changes in expression/stability of either CK20 mRNA or protein. Although the sample size is small, these observations also suggest that loss of CK20 expression may be associated with expression changes in a subset of other lineage markers. In addition, we recently showed that CK20-negative cutaneous Merkel cell carcinoma was infrequently associated with Merkel cell polyomavirus, suggesting these tumors arise via a virus-independent pathway (12). Together, these observations suggest that Merkel cell carcinomas lacking CK20 expression may harbor additional molecular differences relative to CK20-positive Merkel cell carcinomas.
Herein, through next generation sequencing of 409 cancer-related genes, we found that CK20-negative Merkel cell carcinomas share important molecular similarities with CK20-positive Merkel cell carcinoma. Copy number alterations similar to CK20-positive Merkel cell carcinoma were observed, such as gain of chromosome 1 including MYCL1 (22). Gains of AKT1 were the most frequent high-level oncogene gain, and were relatively more common in our cohort (seen in 33% of samples) than previously reported in Merkel cell carcinoma (4% of samples) (22). The majority of tumors in our cohort displayed a predominance of C to T transition mutations with a UV signature mutational profile in agreement with a previous study of CK20-positive Merkel cell carcinoma (34). TP53 and RB1 mutations were highly recurrent in Merkel cell polyomavirus-negative tumors, in agreement with previous reports (7, 8, 28). In agreement with a previous sequencing study of CK20-positive Merkel cell carcinoma, TP53 was the most frequently mutated tumor suppressor (8).
In addition, we identified mutations in tumor suppressors TET2, APC, and BAP1. TET2 is a tumor suppressor gene that epigenetically regulates gene expression by catalyzing conversion of methylcytosine to 5-hydroxymethylcytosine (27, 35). BAP1 tumor suppressor is a deubiquitinating enzyme that undergoes somatic mutation in many cancers and has been linked to a hereditary cancer syndrome associated with mesothelioma and uveal melanoma, among other tumors (36-38). EZH2, a histone methyltransferase in the Polycomb Repressor Complex, undergoes oncogenic activating mutations affecting Y641 (also reported as Y646) in lymphoma and melanoma (21, 29, 39). Therapies targeting EZH2 in malignancy are under active investigation (40).
Clonal evolution, the continued accumulation of genetic changes within a tumor during progression, results in intratumoral heterogeneity, and has been shown to have important implications for tumor biology, prognosis, and therapeutic response (15, 41). Two primary-metastasis pairs (MC9/MC14 and MC10/MC16) were included in our cohort and were evaluated for the possibility of clonal evolution. One of these was a Merkel cell polyomavirus-positive tumor, which demonstrated no significant increase in mutational burden in the metastasis. The other was a Merkel cell polyomavirus-negative tumor, which demonstrated a much higher mutational burden in the metastasis than the primary tumor. Most mutations restricted to the metastasis occurred at a variant fraction of approximately 16%, reflecting a more highly mutated subclone of tumor cells comprising approximately 32% of the metastasis. Mutations unique to the metastasis included truncating mutations of tumor suppressors DCC, ATR, and KMT2A. It is unknown whether these genes play roles in Merkel cell carcinoma metastasis and clonal evolution. Although limited to a single tumor, our findings indicate that clonal evolution and intratumoral heterogeneity occur in Merkel cell carcinoma, similar to other tumors (41).
Since immunohistochemical expression of thyroid transcription factor-1 is not completely sensitive or specific for distinguishing small cell lung carcinoma from Merkel cell carcinoma in many cases (9, 10, 32, 42, 43), our findings may be useful in discriminating CK20-negative Merkel cell carcinoma from a cutaneous metastasis of small cell lung carcinoma. Most small cell lung carcinomas are predicted to be Merkel cell polyomavirus-negative and harbor non-UV mutations of TP53 (26, 44-47). In contrast, most cutaneous Merkel cell carcinomas are predicted to harbor Merkel cell polyomavirus, UV-signature TP53 mutations, or both. However, mutational analysis of any single gene may not be fully sensitive or specific to distinguish Merkel cell carcinoma from small cell lung carcinoma. Broader mutational profiling achieved by expanded next-generation sequencing approaches may be ideal in such cases.
Recently, Pulitzer et al. performed mutational analysis of five Merkel cell carcinomas with squamous differentiation (combined tumors) and found UV-associated mutations as well as TP53 and RB1 mutations (48). These combined tumors were all Merkel cell polyomavirus-negative. The results from Pulitzer et al. are in line with this study and our previous study (8), with Merkel cell polyomavirus-negative tumors displaying UV-signature mutation spectra. Interestingly, in our cohort, one tumor with squamous differentiation harbored detectable Merkel cell polyomavirus DNA, but lacked LTAg expression by immunohistochemistry, and displayed a UV-signature mutational profile similar to Merkel cell polyomavirus-negative Merkel cell carcinoma. The detection of Merkel cell polyomavirus in a combined Merkel cell carcinoma is unexpected based on previous reports (26, 46). However, Merkel cell polyomavirus-positive Merkel cell carcinoma tumors with low Merkel cell polyomavirus viral load and/or lack of LTAg protein expression have been reported to be more similar to Merkel cell polyomavirus-negative tumors with regard to RB1 expression, TP53 expression, and background solar damage (49, 50).
A potential limitation of our study is possible inclusion of germline mutations in our data set. However, high priority mutations in our data set were compared against germ line sequencing databases (1000 Genomes, Exome Sequencing Project and ExAC datasets)(2, 9, 17, 18) with stringent cutoffs, suggesting somatic events. Furthermore, no patient was known to harbor a hereditary cancer syndrome. A further limitation is the potential for detection of UV-associated mutations in background epidermis (51). Although we cannot absolutely exclude this possibility, tumor purity was sufficiently high in our samples that mutations in the background epidermis would be predicted to be detected only at very low levels. Furthermore, UV-signature mutational profiles were detected regardless of whether samples were collected from skin, soft tissue, or lymph nodes.
In summary, we find that CK20-negative Merkel cell carcinomas harbor similarities to CK20-positive Merkel cell carcinoma, including RB1 mutations restricted to Merkel cell polyomavirus-negative tumors, MYCL1 gains, and PI3K pathway activation. However, a subset of CK20-negative Merkel cell carcinomas harbor genetic changes not previously described in Merkel cell carcinoma, which may be prognostically or therapeutically relevant. Furthermore, CK20-negative Merkel cell carcinomas frequently harbor UV-signature mutations which might allow for molecular distinction from extracutaneous small cell carcinomas.
Supplementary Material
Acknowledgments
This project was supported in part by the Anatomic Pathology Project Fund of the University of Michigan, and by the Pathology Department of the Cleveland Clinic. P.W.H. is supported by the Dermatopathology Research Career Development Award of the Dermatology Foundation. S.A.T. is supported by the A. Alfred Taubman Medical Institute. A.A.D. and M.E.V. are supported by NIH grants CA183084 and CA189352, with additional support from the University of Michigan Cancer Center Support Grant CA046592 (A.A.D.) and a Research Scholar Award (American Skin Association) (M.E.V.).
Footnotes
Disclosure/Conflicts of Interest: None.
References
- 1.Pulitzer MP, Amin BD, Busam KJ. Merkel cell carcinoma: review. Adv Anat Pathol. 2009;16:135–44. doi: 10.1097/PAP.0b013e3181a12f5a. [DOI] [PubMed] [Google Scholar]
- 2.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang Y, Moore PS. Merkel cell carcinoma: a virus-induced human cancer. Annu Rev Pathol. 2012;7:123–44. doi: 10.1146/annurev-pathol-011110-130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verhaegen ME, Mangelberger D, Harms PW, et al. Merkel Cell Polyomavirus Small T Antigen Is Oncogenic in Transgenic Mice. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spurgeon ME, Cheng J, Bronson RT, Lambert PF, DeCaprio JA. Tumorigenic activity of merkel cell polyomavirus T antigens expressed in the stratified epithelium of mice. Cancer Res. 2015;75:1068–79. doi: 10.1158/0008-5472.CAN-14-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wendzicki JA, Moore PS, Chang Y. Large T and small T antigens of Merkel cell polyomavirus. Curr Opin Virol. 2015;11:38–43. doi: 10.1016/j.coviro.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimino PJ, Robirds DH, Tripp SR, et al. Retinoblastoma gene mutations detected by whole exome sequencing of Merkel cell carcinoma. Mod Pathol. 2014 doi: 10.1038/modpathol.2013.235. [DOI] [PubMed] [Google Scholar]
- 8.Harms PW, Vats P, Verhaegen ME, et al. The Distinctive Mutational Spectra of Polyomavirus-Negative Merkel Cell Carcinoma. Cancer Research. 2015 doi: 10.1158/0008-5472.CAN-15-0702. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bobos M, Hytiroglou P, Kostopoulos I, Karkavelas G, Papadimitriou CS. Immunohistochemical distinction between merkel cell carcinoma and small cell carcinoma of the lung. Am J Dermatopathol. 2006;28:99–104. doi: 10.1097/01.dad.0000183701.67366.c7. [DOI] [PubMed] [Google Scholar]
- 10.Hanly AJ, Elgart GW, Jorda M, Smith J, Nadji M. Analysis of thyroid transcription factor-1 and cytokeratin 20 separates merkel cell carcinoma from small cell carcinoma of lung. J Cutan Pathol. 2000;27:118–20. doi: 10.1034/j.1600-0560.2000.027003118.x. [DOI] [PubMed] [Google Scholar]
- 11.Moll R, Lowe A, Laufer J, Franke WW. Cytokeratin 20 in human carcinomas. A new histodiagnostic marker detected by monoclonal antibodies. Am J Pathol. 1992;140:427–47. [PMC free article] [PubMed] [Google Scholar]
- 12.Miner AG, Patel RM, Wilson DA, et al. Cytokeratin 20-negative Merkel cell carcinoma is infrequently associated with the Merkel cell polyomavirus. Mod Pathol. 2014 doi: 10.1038/modpathol.2014.148. [DOI] [PubMed] [Google Scholar]
- 13.Nagao T, Gaffey TA, Olsen KD, Serizawa H, Lewis JE. Small cell carcinoma of the major salivary glands: clinicopathologic study with emphasis on cytokeratin 20 immunoreactivity and clinical outcome. Am J Surg Pathol. 2004;28:762–70. doi: 10.1097/01.pas.0000126776.65815.48. [DOI] [PubMed] [Google Scholar]
- 14.Fisher CA, Harms PW, McHugh JB, et al. Small cell carcinoma in the parotid harboring Merkel cell polyomavirus. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;118:703–12. doi: 10.1016/j.oooo.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Warrick JI, Hovelson DH, Amin A, et al. Tumor evolution and progression in multifocal and paired non-invasive/invasiveurothelial carcinoma. Virchows Arch. 2015;466:297–311. doi: 10.1007/s00428-014-1699-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu W, O'Connor TD, Jun G, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-codingvariants. Nature. 2013;493:216–20. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genomes Project C, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Exome Aggregation Consortium. Lek M, Karczewski K, et al. Analysis of protein-coding genetic variation in 60,706 humans. 2015 doi: 10.1038/nature19057. [cited 2015 11/12/2015] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan RJ, Nitta M, Borger D, et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS One. 2011;6:e28585. doi: 10.1371/journal.pone.0028585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harms PW, Fullen DR, Patel RM, et al. Cutaneous basal cell carcinosarcomas: evidence of clonality and recurrent chromosomal losses. Hum Pathol. 2015;46:690–7. doi: 10.1016/j.humpath.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Harms PW, Hristov AC, Kim D, et al. Activating mutations of the oncogene EZH2 in cutaneous melanoma revealed by next generation sequencing. Hum Pathol Case Reports. 2014;1:21–28. [Google Scholar]
- 22.Paulson KG, Lemos BD, Feng B, et al. Array-CGH reveals recurrent genomic changes in Merkel cell carcinoma including amplification of L-Myc. J Invest Dermatol. 2009;129:1547–55. doi: 10.1038/jid.2008.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brash DE. UV signature mutations. Photochem Photobiol. 2015;91:15–26. doi: 10.1111/php.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeifer GP, Denissenko MF, Olivier M, et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–51. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 25.Poon SL, McPherson JR, Tan P, Teh BT, Rozen SG. Mutation signatures of carcinogen exposure: genome-wide detection and new opportunities for cancer prevention. Genome Med. 2014;6:24. doi: 10.1186/gm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Busam KJ, Jungbluth AA, Rekthman N, et al. Merkel cell polyomavirus expression in merkel cell carcinomas and its absence in combined tumors and pulmonary neuroendocrine carcinomas. Am J Surg Pathol. 2009;33:1378–85. doi: 10.1097/PAS.0b013e3181aa30a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song F, Amos CI, Lee JE, et al. Identification of a melanoma susceptibility locus and somatic mutation in TET2. Carcinogenesis. 2014;35:2097–101. doi: 10.1093/carcin/bgu140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sihto H, Kukko H, Koljonen V, et al. Merkel cell polyomavirus infection, large T antigen, retinoblastoma protein and outcome in Merkel cell carcinoma. Clin Cancer Res. 2011;17:4806–13. doi: 10.1158/1078-0432.CCR-10-3363. [DOI] [PubMed] [Google Scholar]
- 29.Deb G, Singh AK, Gupta S. EZH2: Not EZHY (Easy) to Deal. Mol Cancer Res. 2014;12:639–53. doi: 10.1158/1541-7786.MCR-13-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shuda M, Feng H, Kwun HJ, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105:16272–7. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nardi V, Song Y, Santamaria-Barria JA, et al. Activation of PI3K signaling in Merkel cell carcinoma. Clin Cancer Res. 2012;18:1227–36. doi: 10.1158/1078-0432.CCR-11-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shalin SC, Cifarelli CP, Suen JY, Gao L. Loss of cytokeratin 20 and acquisition of thyroid transcription factor-1 expression in a merkel cell carcinoma metastasis to the brain. Am J Dermatopathol. 2014;36:904–6. doi: 10.1097/DAD.0000000000000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhaegen ME, Mangelberger D, Weick JW, et al. Merkel Cell Carcinoma Dependence on Bcl-2 Family Members for Survival. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harms PW, Vats P, Verhaegen ME, et al. The Distinctive Mutational Spectra of Polyomavirus-Negative Merkel Cell Carcinoma. Cancer Res. 2015;75:3720–7. doi: 10.1158/0008-5472.CAN-15-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W. The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia. 2014;28:485–96. doi: 10.1038/leu.2013.337. [DOI] [PubMed] [Google Scholar]
- 36.Murali R, Wiesner T, Scolyer RA. Tumours associated with BAP1 mutations. Pathology. 2013;45:116–26. doi: 10.1097/PAT.0b013e32835d0efb. [DOI] [PubMed] [Google Scholar]
- 37.Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–5. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–21. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 41.Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational implications of tumor heterogeneity. Clin Cancer Res. 2015;21:1258–66. doi: 10.1158/1078-0432.CCR-14-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koba S, Inoue T, Okawa T, et al. Merkel cell carcinoma with cytokeratin 20-negative and thyroid transcription factor-1-positive immunostaining admixed with squamous cell carcinoma. J Dermatol Sci. 2011;64:77–9. doi: 10.1016/j.jdermsci.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Reddi DM, Puri PK. Expression of focal TTF-1 expression in a case of CK7/CK20-positive Merkel cell carcinoma. J Cutan Pathol. 2013;40:431–3. doi: 10.1111/cup.12079. [DOI] [PubMed] [Google Scholar]
- 44.Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–6. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ly TY, Walsh NM, Pasternak S. The spectrum of Merkel cell polyomavirus expression in Merkel cell carcinoma, in a variety of cutaneous neoplasms, and in neuroendocrine carcinomas from different anatomical sites. Hum Pathol. 2012;43:557–66. doi: 10.1016/j.humpath.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Paik JY, Hall G, Clarkson A, et al. Immunohistochemistry for Merkel cell polyomavirus is highly specific but not sensitive for the diagnosis of Merkel cell carcinoma in the Australian population. Hum Pathol. 2011;42:1385–90. doi: 10.1016/j.humpath.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 48.Pulitzer MP, Brannon AR, Berger MF, et al. Cutaneous squamous and neuroendocrine carcinoma: genetically and immunohistochemically different from Merkel cell carcinoma. Mod Pathol. 2015 doi: 10.1038/modpathol.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leroux-Kozal V, Leveque N, Brodard V, et al. Merkel cell carcinoma: histopathologic and prognostic features according to theimmunohistochemical expression of Merkel cell polyomavirus large T antigen correlated with viral load. Hum Pathol. 2015;46:443–53. doi: 10.1016/j.humpath.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Bhatia K, Goedert JJ, Modali R, Preiss L, Ayers LW. Merkel cell carcinoma subgroups by Merkel cell polyomavirus DNA relative abundance and oncogene expression. Int J Cancer. 2010;126:2240–6. doi: 10.1002/ijc.24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martincorena I, Roshan A, Gerstung M, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–6. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.