Abstract
Class I myosins are widely expressed with roles in endocytosis and cell migration in a variety of cell types. Dictyostelium express multiple myosin Is, including three short-tailed (Myo1A, Myo1E, Myo1F) and three long-tailed (Myo1B, Myo1C, Myo1D). Here we report the molecular basis of the specific localizations of short-tailed Myo1A, Myo1E and Myo1F compared to our previously determined localization of long-tailed Myo1B. Myo1A and Myo1B have common and unique localizations consistent with the various features of their tail region; specifically the BH sites in their tails are required for their association with the plasma membrane and heads are sufficient for relocalization to the front of polarized cells. Myo1A does not localize to actin waves and macropinocytic protrusions, in agreement with the absence of a tail region which is required for these localizations of Myo1B. However, in spite of the overall similarity of their domain structures, the cellular distributions of Myo1E and Myo1F are quite different from Myo1A. Myo1E and Myo1F, but not Myo1A, are associated with macropinocytic cups and actin waves. The localizations of Myo1E and Myo1F in macropinocytic structures and actin waves differ from the localization of Myo1B. Myo1B colocalizes with F-actin in the actin waves and at the tips of mature macropinocytic cups whereas Myo1E and Myo1F are in the interior of actin waves and along the entire surface of macropinocytic cups. Our results point to different mechanisms of targeting of short- and long-tailed myosin Is, and are consistent with these myosins having both shared and divergent cellular functions.
Keywords: unconventional myosins, BH site, membrane binding, macropinocytosis, actin waves
Introduction
Class I myosins are among the most conserved and wide spread members of the myosin superfamily [Berg et al., 2001; Odronitz and Kollmar, 2006, 2007; Sebe-Pedros et al., 2014]. They are found in almost all eukaryotic species where they are involved in diverse cellular processes including, among others, cell motility, endocytosis, vesicle shedding, channel gating and cell signaling [Bond et al., 2013; Kim and Flavell, 2008; McConnell and Tyska, 2010]. However, studying the detailed functions of these myosins is often complicated by the fact that most organisms have multiple myosin I isoforms, e.g. eight in humans and seven in Dictyostelium [Berg et al., 2001; Kollmar, 2006], with at least partially redundant functions [Falk et al., 2003; Jung et al., 1996; Novak et al., 1995]. An understanding of the biological roles of the myosins would be facilitated by a detailed characterization and comparison of their different localizations in cells, the structural basis of those localizations and how these may correlate with unique and shared functions of the myosins. Dictyostelium amoebae are extremely motile and undergo dramatic changes in cell morphology accompanied by relocation of numerous proteins [Bagorda et al., 2006], including myosin Is (for examples see [Brzeska et al., 2012; Brzeska et al., 2014], and Fig. S1). This dynamic morphology makes Dictyostelium an excellent model for studying the molecular basis for the targeting of individual myosin I family members to different compartments in a single cell.
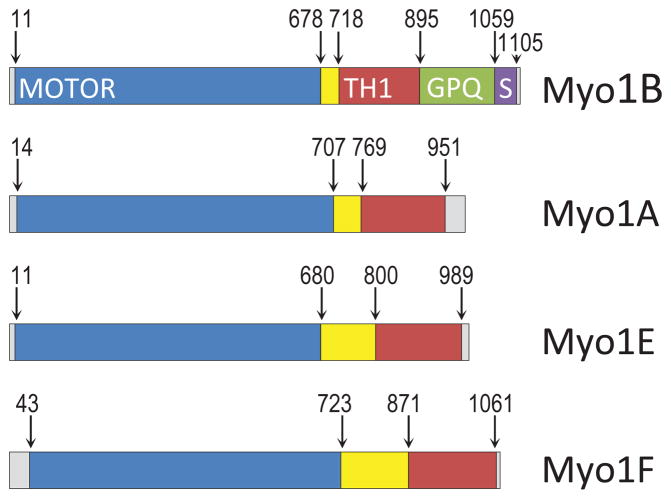
Almost all class I myosins have a single heavy chain consisting of a motor, neck and tail [Berg et al., 2001; Odronitz and Kollmar, 2007]. The motor contains actin-dependent motor activity with an ATP-sensitive actin-binding site and an actin-activated ATPase site; the neck region binds light chains [Greenberg and Ostap, 2013; McConnell and Tyska, 2010]; the tail provides sites of interactions with other cellular components. All of the myosin I tails have an N-terminal basic region, also called a tail homology region 1 (TH1), that binds acidic phospholipids [McConnell and Tyska, 2010; Odronitz and Kollmar, 2007] (Fig. 1). Binding of acidic lipids through the basic region of the tail is a unique property of myosin Is and it is essential for most of their functions [McConnell and Tyska, 2010]. Lipid binding can be by PIP2-, or PIP3-specific PH domains [Hokanson et al., 2006; Komaba and Coluccio, 2010; Lu et al., 2015] or by less specific interactions proportional to the net negative charge of phospholipids [Brzeska et al., 2012; Brzeska et al., 2010; Brzeska et al., 2008; Feeser et al., 2010; Mazerik and Tyska, 2012]. The seven Dictyostelium myosin Is include three long-tailed myosins (Myo1B, Myo1C, Myo1D), three short-tailed myosins (Myo1A, Myo1E, Myo1F), and one myosin without a tail (Myo1K) that has an additional actin-binding site within its motor domain and binds membranes through a C-terminal farnesylated site [Dieckmann et al., 2010; Kollmar, 2006; Schwarz et al., 2000]. In some older papers these myosins were named MIA, MIB, MIC, MID, MIE, MIF and MIK.
Fig. 1. Myosin Is used in this study.
The boundaries of the motor (blue), TH1 (red), GPQ (green) and SH3 (purple) regions are marked according to Cymobase [Kollmar, 2006; Odronitz and Kollmar, 2006, 2007] and based on Pfam v.28. The region between the motor domain and TH1 domain contains the neck that contains light chain binding site(s). See text for exact boundaries of Myo1A mutants.
We have shown previously that non-specific binding to acidic lipids of Dictyostelium Myo1B requires a short basic-hydrophobic region, the BH site, located within the TH-1 domain of tails [Brzeska et al., 2012; Brzeska et al., 2010; Brzeska et al., 2008]. In long-tailed myosins (Myo1B in Fig. 1) the basic region is followed by a Gly, Pro, Gln (GPQ)-rich region that is an ATP-insensitive actin-binding site [Rosenfeld and Rener, 1994] and an SH3 domain that interacts with Acan125/CARMIL [Jung et al., 2001; Xu et al., 1995] and perhaps other proteins [Cheng et al., 2012; Krendel et al., 2007; Maxeiner et al., 2015]. The short-tailed myosins (Myo1A, Myo1E and Myo1F in Fig. 1) have neither the GPQ region nor the SH3 domain; their tails contain only the TH1 domain.
The cellular localization of Dictyostelium Myo1B is highly dynamic, paralleling the rapidly changing cell morphology [Brzeska et al., 2012; Brzeska et al., 2014]. Myo1B localizes uniformly to the plasma membrane of non-motile or slowly moving cells. In actively moving cells, Myo1B relocates to other structures such as pseudopods, actin waves (dynamic structures associated with the basal plasma membrane), endocytic protrusions, cell-cell contacts, and to the front of polarized, chemotaxing cells. The BH site [Brzeska et al., 2010; Brzeska et al., 2008] is required for Myo1B localization to the plasma membrane [Brzeska et al., 2012] whereas both the BH site and the GPQ region are required for Myo1B localization to actin waves and macropinocytic protrusions [Brzeska et al., 2014]. The head is required and sufficient for localization at the front of polarized cells [Brzeska et al., 2012]. Thus, the combination of the specific properties of the individual tail domains and the head determine the localization of Myo1B.
Long-tailed Myo1B and short-tailed Myo1A have overlapping functions. Mutants lacking either of these myosin Is are defective in migration and pseudopod formation [Jung and Hammer, 1990; Titus et al., 1993], and they work cooperatively during macropinocytic uptake of fluids [Jung et al., 1996; Novak et al., 1995]. These observations suggested that these two myosins would have some similar localizations. On the other hand, the similar overall domain organization of the short-tailed Dictyostelium Myo1A, Myo1E and Myo1F suggested that the localizations of the short-tailed myosins would be similar to each other, but different from several of the localizations of long-tailed Myo1B, because the short-tailed myosins lack the GPQ and SH3 regions (Fig. 1). We found that the dynamic localizations of Myo1A were as predicted from the structural differences between Myo1A and Myo1B. Unexpectedly, however, the localizations of Myo1E and Myo1F differed from the localizations of Myo1A; most strikingly Myo1E and Myo1F, but not Myo1A, localized to the center of the actin waves and to macropinocytic cups but in a manner different than Myo1B.
Results
Localization of Myo1A
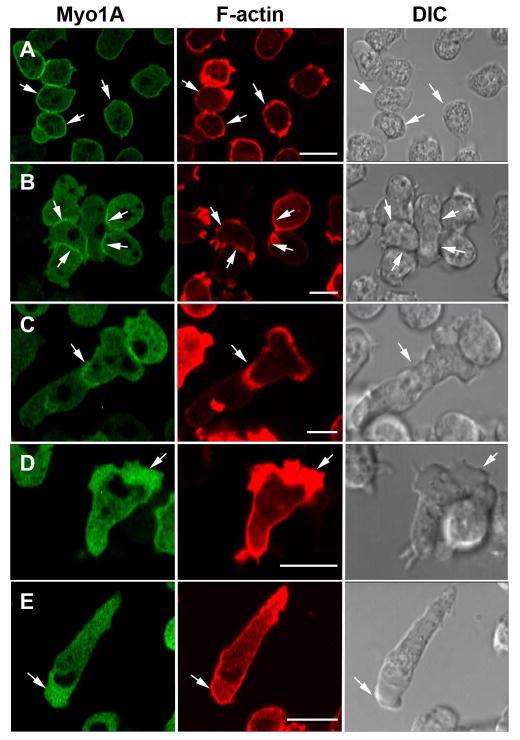
In the experiments described below cells were cotransfected with two separate plasmids, each carrying different antibiotic resistance and each encoding a different protein with GFP or RFP fused to its N-terminus. Expressed GFP-Myo1A localized uniformly to the plasma membrane of non-motile and slowly moving Myo1A-null cells (Fig. 2A), at positions of cell-cell contacts in randomly moving cells (Fig. 2B), in the regions of close cell-cell contacts of chemotaxing cells (Fig. 2C), diffusely in pseudopods (Fig. 2D) and the front of elongated chemotaxing cells (Fig. 2E). Myo1B has also been found to localize to these regions of the cell [Brzeska et al., 2012; Brzeska et al., 2014]. However, in contrast to Myo1B, Myo1A did not localize to actin waves (Fig. 3A) or macropinocytic protrusions (Fig. 4A), which is consistent with Myo1A lacking a GPQ domain (Fig. 1) that is required for these localizations of Myo1B [Brzeska et al., 2012; Brzeska et al., 2014]. In fact, the Myo1A localization is strikingly similar to the localization of truncated Myo1B that has only the motor, neck and TH1 domains [Brzeska et al., 2012; Brzeska et al., 2014].
Fig. 2. Localizations of Myo1A that are similar to the localization of Myo1B.
GFP-Myo1A and the F-actin marker, RFP-lifeact, were coexpressed in Myo1A-null cells. Myo1A is present at: (A) the plasma membrane of non-motile cells, (B) the sites of cell-cell contacts, (C) regions of cell-cell contact between chemotaxing cells, (D) in the cortex of pseudopods of randomly moving cells, and (E) at the front of chemotaxing cells. All the above localizations are similar to that for Myo1B. Panel A shows fixed cells, all other panels show live cells. Arrows point to the structures of interest. Bars are 10 μm.
Fig. 3. Myo1A is absent from actin waves.
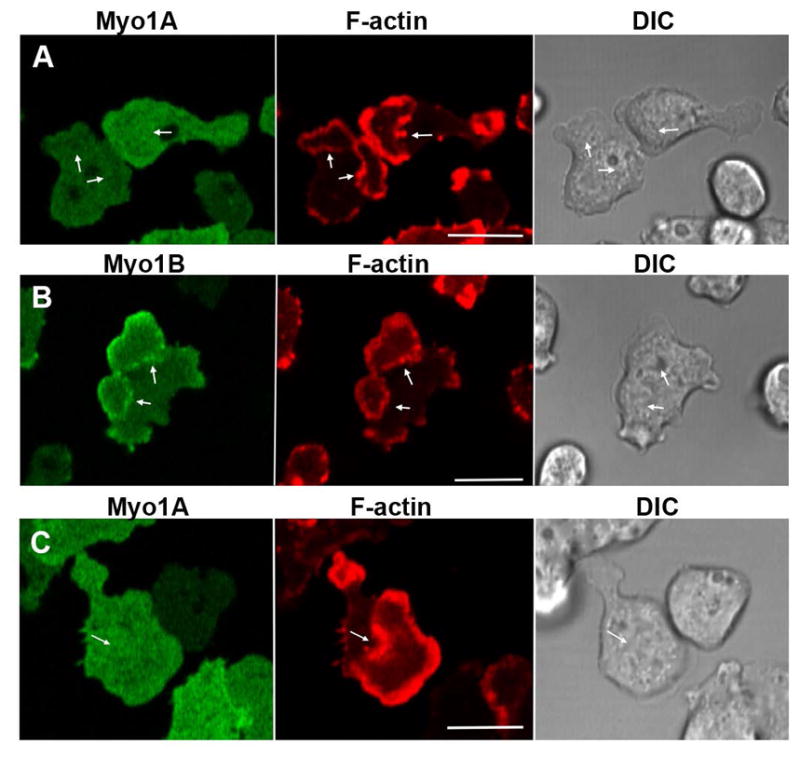
GFP-Myo1A or GFP-Myo1B was coexpressed with RFP-lifeact in Myo1A-null or Myo1B-null cells. Actin waves were induced by 1 μM latrunculin. (A) Myo1A does not localize to actin waves in Myo1A-null cells. (B) Myo1B does localize to actin waves in Myo1A-null cells. (C) Myo1A does not localize to actin waves in Myo1B-null cells. Images of live cells are shown. Arrows indicate positions of actin waves identified by RFP-lifeact fluorescence. Bars are 10 μm.
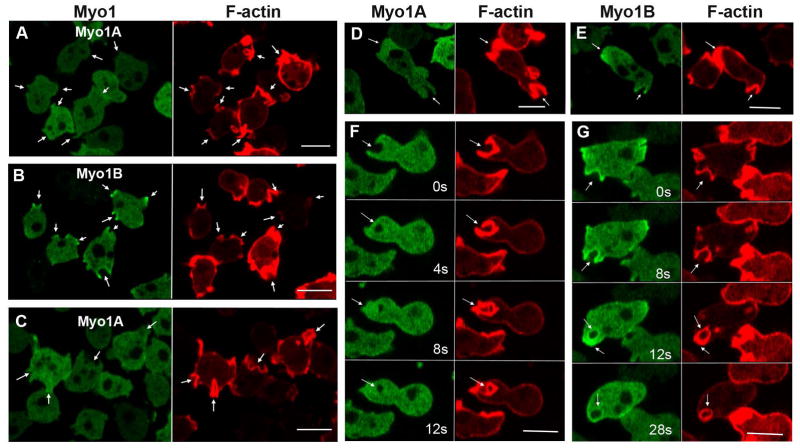
Fig. 4. Myo1A is absent from macropinocytic protrusions.
GFP-Myo1A or GFP-Myo1B was co-expressed with RFP-lifeact in Myo1A-null and Myo1B-null cells, and their distributions observed in live cells undergoing macropinocytosis. (A) Myo1A does not localize to macropinocytic protrusions in Myo1A-null cells. (B) Myo1B does localize to macropinocytic protrusions in Myo1A-cells. (C) Myo1A does not localize to macropinocytic protrusions in Myo1B-null cells. (D) Myo1A does localize to a pseudopods but not to macropinocytic cups in Myo1A-null cells. (E) Myo1B localizes to pseudopod and to a macropinocytic cup in a Myo1A-null cell. (F) Formation of a macropinocytic cup in a Myo1B-null cell coexpressing Myo1A and lifeact. Images were taken at times indicated in the figure. Myo1A does not localize to the cup. (G) Formation of a macropinocytic cup in a Myo1B-null cell coexpressing Myo1B and lifeact. Myo1B does localize to the cup. Images were taken at times indicated in the figure. Arrows point to macropinocytic protrusions. Bars are 10 μm.
The differences in Myo1A and Myo1B localization are well illustrated in Figs. 4D and E which show single cells simultaneously forming pseudopods and macropinocytic cups. Both Myo1A (Fig. 4D) and Myo1B (Fig. 4E) are present in the pseudopod but only Myo1B is present in the macropinocytic cup. The formation of macropinocytic protrusions and cups, and the accompanying relocalization of Myo1B to these protrusions, is highly dynamic (Fig. 4G and Fig. S1). Myo1B localized uniformly in newly forming cups (Fig. 4G, 0 s) and relocated to cup edges during cup closure (Fig. 4G, 8 s). Myo1B was enriched at the site of vesicle internalization (Fig. 4G 12 s) and, for a short time, remained associated with the ingested vesicle (Fig. 4G, 28 s). Myo1A was absent from macropinocytic cups during the entire process (Fig. 4F) with only occasional very slight enrichment at the site of vesicle closure (Fig. 4F, 8 s).
The potential for redistribution of Myo1A in the absence of Myo1B, and vice versa, was tested by expressing GFP-Myo1B in Myo1A-null cells and GFP-Myo1A in Myo1B-null cells. Myo1A did not localize either to actin waves or to macropinocytic protrusions when expressed in Myo1B-null cells (Figs. 3C and 4C), and Myo1B localized to actin waves (Fig. 3B) and macropinocytic protrusions (Fig. 4B), and underwent the same dynamic changes in Myo1A-null cells as it did when expressed in Myo1B-null cells (Fig. S1). Also, Myo1A clearly localized to the same compartments in wild type AX2 cells as in Myo1A-null cells, and Myo1B localized to macropinocytic protrusions and actin waves in AX2 cells in a manner similar to that in Myo1B-null cells (Fig. S2). These results show that the localizations of expressed Myo1A and Myo1B are not dramatically affected by the presence or absence of their equivalent endogenous isoforms and point to the targeting specificity of individual myosin Is.
Localization of Myo1A Mutants
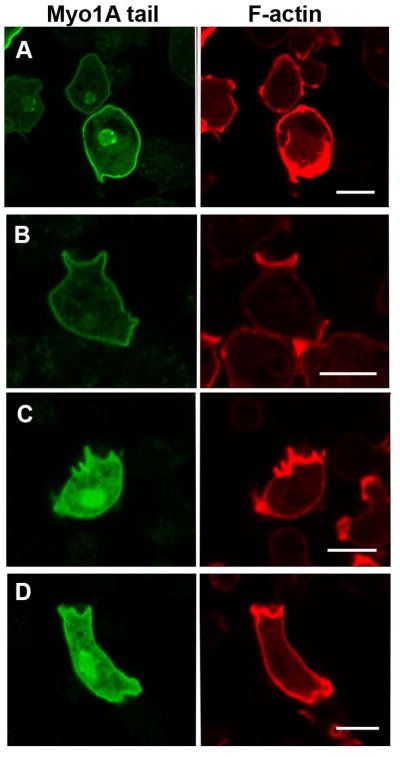
The role of the tail in Myo1A localization was examined by expressing GFP-Myo1 tail alone, residues 772–994 (Fig. 1), in Myo1A null cells. The Myo1A tail localized uniformly to the plasma membrane of non-motile Myo1A-null cells (Fig. 5A) and Myo1B-null cells (Fig. 5B), and remained associated all along the plasma membrane in randomly moving and chemotaxing cells (Figs. 5C and 5D). Again, this is quite similar to what has been observed for the localization of the Myo1B tail region, consistent with the presence of a basic TH1 region in both myosins (Fig. 1) that was shown to be required for Myo1B localization to the plasma membrane [Brzeska et al., 2012]. However, the localization of Myo1A tail to the plasma membrane appeared to be less striking than for Myo1B tail [Brzeska et al., 2012], especially in live starved cells; the background fluorescence of Myo1A tail in the cytosol was higher than that of Myo1B tail. We also observed nuclear localization of Myo1A tail (Figs. 5A, C, D) that was presumably artificial since full-length Myo1A did not localize to the nucleus.
Fig. 5. The Myo1A tail localizes to the plasma membrane.
GFP-Myo1A tail and RFP-lifeact were coexpressed in Myo1A-null and Myo1B-null cells. (A) Myo1A tail in non-motile Myo1A-null cells. (B) Myo1A tail in a non-motile Myo1B-null cell. (C) Myo1A tail in a randomly moving Myo1B-null cell. (D) Myo1A tail in a chemotaxing Myo1B-null cell. (A) and (B) show fixed cells. (C) and (D) show live cells. Bars are 10 μm.
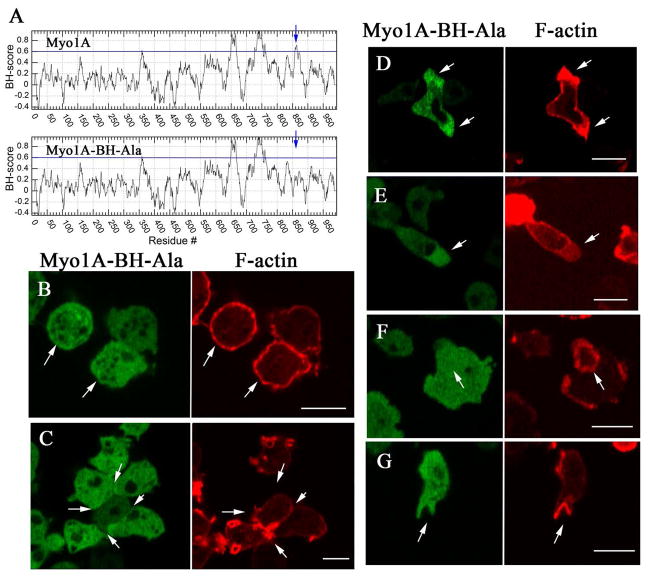
Analysis of the full length Myo1A sequence by BH search [Brzeska et al., 2010] reveals the presence of a BH peak (856PKKDGSWFAIKRKVP870) at a position within the TH1 domain similar to the BH site of Myo1B (Fig. 6A). To determine if the localization of Myo1A to the plasma membrane depends on the BH site, the four Lys residues of the BH site of full-length Myo1A were mutated to Ala (856PAADGSWFAIARAVP870). These mutations eliminated the BH peak (Myo1A-BH-Ala mutant in Fig. 6A) and drastically reduced Myo1A association with the plasma membrane (Fig. 6B) and sites of cell-cell contacts (Fig. 6C). Thus, similarly to Myo1B, the BH site is required for plasma membrane association of Myo1A. The Myo1A-BH-Ala mutant, like wild-type Myo1A, did localize to pseudopods (Fig. 6D) and at the front of elongated cells (Fig. 6E), for which only the motor domain of Myo1B was required [Brzeska et al., 2012]. As expected, the Myo1A-BH-Ala mutant, like wild-type Myo1A, did not associate with actin waves (Fig. 6F) or with macropinocytic cups (Fig. 6G).
Fig. 6. Plasma membrane localization of Myo1A requires the BH site.
GFP-Myo1A-BH-Ala and RFP-lifeact were coexpressed in Myo1A-null cells. (A) BH plots of Myo1A and Myo1A-BH-Ala mutant run with window 19. The position of the BH peak is marked with an arrow. The regions with BH values above the horizontal line at 0.6 are considered to be positive BH peaks and the mutated BH site in the tail is marked with an arrow. (B) Myo1A-BH-Ala does not localize to the plasma membrane of non-motile cells. (C) Myo1A-BH-Ala does not localize to the sites of cell-cell contacts of non-motile cells. (D) Myo1A-BH-Ala localizes to pseudopods. (E) Myo1A-BH-Ala localizes to the front of chemotaxing cells. (F) Myo1A-BH-Ala does not localize to actin waves. (G) Myo1A-BH-Ala does not localize to macropinocytic cups. Arrows point to the structures of interest. (B) shows fixed cells, and (C)–(G) show live cells. Bars are 10 μm.
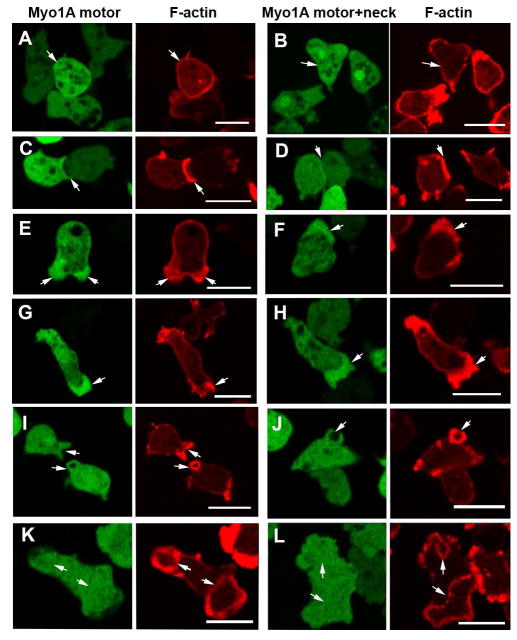
The neck regions of some class I myosins have been shown to bind lipids in vitro in addition to binding calmodulin [Benesh et al., 2010; Brzeska et al., 2010; Chen et al., 2012; Swanljung-Collins and Collins, 1992; Tang et al., 2002] and the neck region of at least one myosin I has been implicated in its binding to intracellular receptors [Cyr et al., 2002]. The Myo1A neck is longer than the neck of Myo1B, with two IQ motifs that bind calmodulins, whereas the neck of Myo1B has a single IQ motif that binds a smaller Myo1B-specific light chain [Crawley et al., 2011]. For these reasons, the possible role of the Myo1A neck, which also has a candidate BH site (Fig. 2A), in membrane binding was tested by expressing the Myo1A motor domain with and without the neck region (residues 1–771 and 1–727, respectively) in Myo1A-null cells. Neither construct associated with the plasma membrane of non-motile cells (Figs. 7A and B) or with the membrane at sites of cell-cell contacts (Figs. 7C and D). Thus, the neck region of Myo1A is insufficient for binding Myo1A to the plasma membrane. On the other hand, the motor domain, both with and without the neck region, localized to pseudopods (Figs. 7E and F), and to the front of elongated cells (Figs. 7G and H) in agreement with the previous finding for Myo1B that the head is sufficient for these localizations. As expected, from the localization of Myo1A and the lack of a BH site and a GPQ-region, neither construct localized to macropinocytic cups (Figs. 7I, J) or to actin waves (Figs. 7K and L). Hence, the localizations of both the Myo1A motor domain and Myo1A motor+neck were the same as the previously determined localization of the Myo1B motor+neck [Brzeska et al., 2012; Brzeska et al., 2014].
Fig. 7. Myo1A motor domain localizes to pseudopods and the cell front.
Myo1A GFP-Motor or GFP-Motor+Neck was coexpressed with RFP-lifeact in GFP-A-null cells. Both Myo1A truncation mutants are absent from the plasma membrane of non-motile cells (A, B) and sites of cell-cell contacts (C, D). Both Myo1A truncation mutants localize to pseudopods (E, F) and to the front of chemotaxing cells (G, H). Like full length Myo1A, neither Myo1A mutant localizes to either macropinocytic cups (I, J) or to actin waves (K, L). (A) and (B) show non-motile fixed cells, the other panels show live cells. Arrows point to sites of interest. Bars are 10 μm.
Summarizing, short-tailed Myo1A localizes to the plasma membrane and the actin-rich front of polarized, migrating cells. Membrane targeting requires the BH site in the tail, and localization to the pseudopod is dictated by the motor region. These findings are similar to what we had found for the roles of the head and BH site in the localization of long-tailed Myo1B [Brzeska et al., 2012; Brzeska et al., 2014], and are consistent with the common roles of Myo1A and Myo1B in cell migration [Jung and Hammer, 1990; Titus et al., 1993]. Myo1A does not localize to the regions where localization of long-tailed Myo1B requires the GPQ region, which short-tailed Myo1A does not have.
Localization of Myo1E and Myo1F
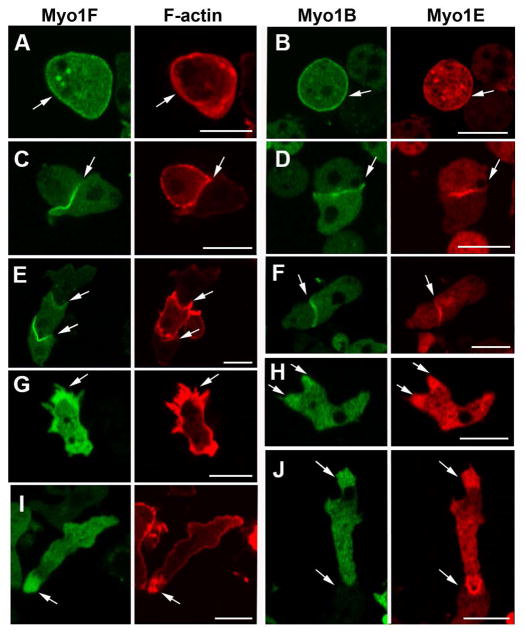
The overall similarity in sequence and domain organization of Myo1E and Myo1F to each other and to Myo1A (Fig. 1) suggested that the three Dictyostelium short-tailed myosin Is would have the same intracellular localizations. While some aspects of Myo1E localization have previously been reported [Chen et al., 2012; Durrwang et al., 2006] the localization of Myo1F has not been studied and the dynamics of the localization of these two myosins have not been described. Therefore, we coexpressed GFP-Myo1F with RFP-lifeact and coexpressed RFP-Myo1E with GFP-Myo1B. Myo1E and Myo1F both localized sharply to the plasma membrane of non-motile and slowly moving cells (Figs. 8A and B). Myo1F and Myo1E were also found at the membrane of cell-cell contacts of randomly moving (Figs. 8C and D) and chemotaxing cells (Figs. 8E, F and J). Both Myo1F and Myo1E localized diffusely to pseudopods (Figs. 8G and H), and to the front of chemotaxing cells (Figs. 8I and J). Thus, as anticipated, Myo1E and Myo1F were found in the same locations as Myo1A.
Fig. 8. Localizations of Myo1F and Myo1E that are similar to the localization of Myo1A.
GFP-Myo1F and RFP-Myo1E were coexpressed with RFP-lifeact or RFP-Myo1E, respectively, in Myo1B-null cells. Myo1F and Myo1E, as well as Myo1B, localize to: (A, B) the plasma membrane, (C, D) sites of cell-cell contacts, (E, F) sites of cell-cell contact between chemotaxing cells, (G, H) pseudopods, and (I, J) the front of chemotaxing cells. (A and B) show fixed non-motile cells, and (C–J) show live cells. Arrows point to the sites of interest. Bars are 10 μm.
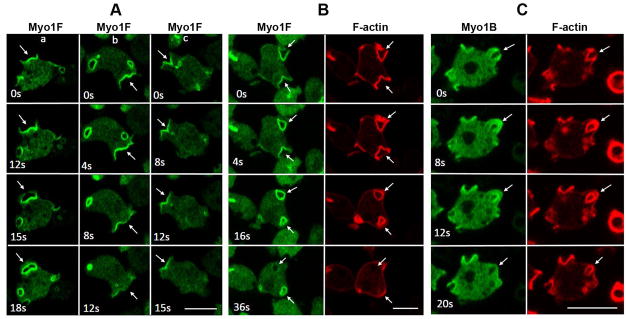
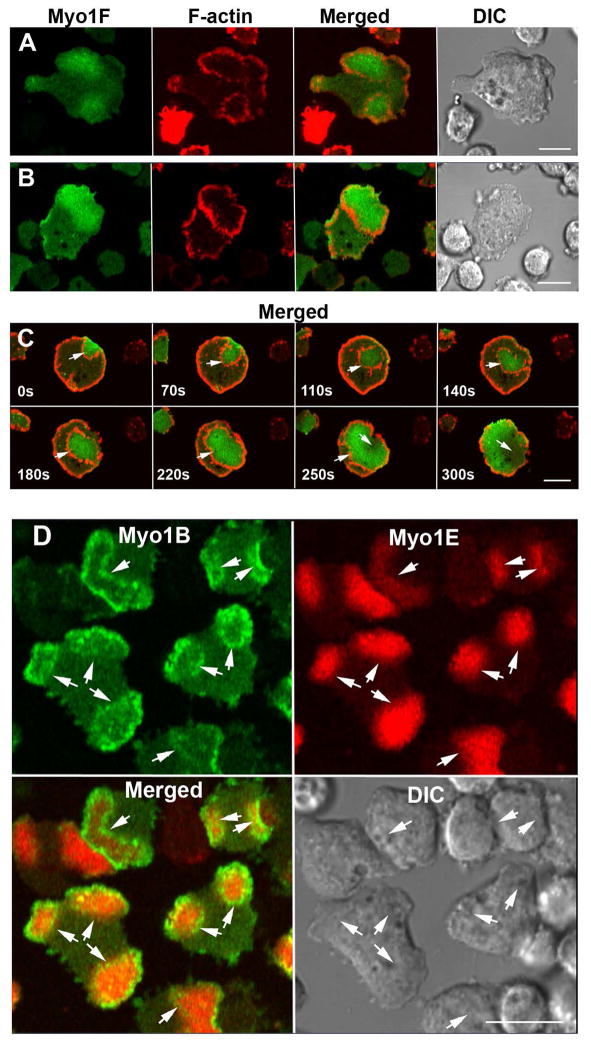
Unexpectedly, however, both Myo1F and Myo1E were also found to localize to macropinocytic structures (Figs. 9 and 10) from which Myo1A was almost completely absent (Figs. 4A and F). The cortex of Dictyostelium amoebae is highly dynamic and cells routinely form curved macropinocytic cups and flat protrusions/ruffles. The flat protrusions may turn into macropinocytic cups that can become internalized media-filled vesicles (Fig. 9A, column a) or disappear without forming cups (Fig. 9A, column b), and partially formed cups may convert into flat protrusions (Fig. 9A, column c; Fig. S3).
Fig. 9. Localization of Myo1F in macropinocytic structures.
GFP-Myo1F or GFP-Myo1B were coexpressed with RFP-lifeact in Myo1B-null cells. Images of live cells were taken at the times indicated in the panels. (A) Localization of Myo1F during formation of various kinds of extensions. Column (a) flat protrusion turning into a macropinocytic cup. Column (b) flat protrusion disappearing without turning into a macropinocytic cup. Column (c) macropinocytic cup turning into a flat protrusion. (B) Localization of Myo1F and F-actin during internalization of a pinocytic vesicle. (C) Localization of Myo1B and F-actin during internalization of a pinocytic vesicle. Arrows point to the locations of interest (see text). Bars are 10 μm.
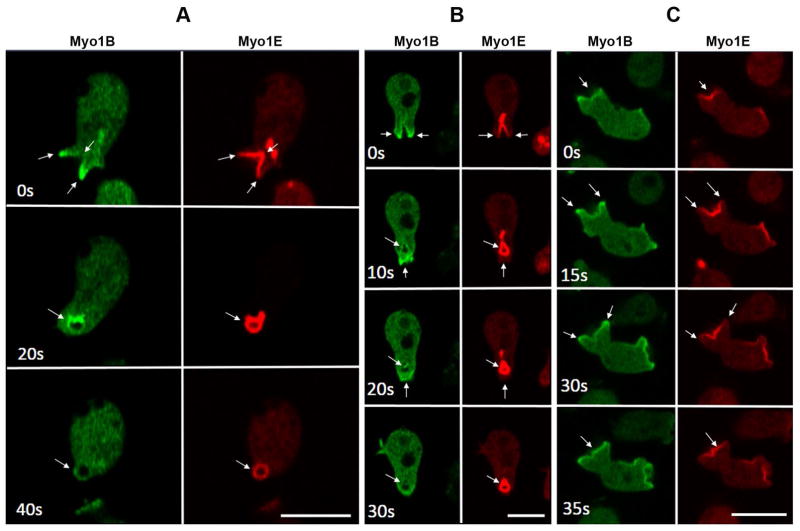
Fig. 10. Different Localizations of Myo1E and Myo1B during macropinocytosis.
RFP-Myo1E and GFP-Myo1B were coexpressed in Myo1B-null cells. Images of live cells were taken at times indicated in the panels. Different characterictics of macropinocytic structures are shown in panels (A), (B) and (C), see text for description. Arrows point to the structures of interest. Bars are 10 μm.
Although both Myo1F and Myo1B localized to pinocytic structures their specific localizations differed. At the final stages of cup formation, Myo1F was present along the entire cup surface but fading toward the leading edges (Figs. 9A column c, 0 s and 9B, 0 s) whereas Myo1B was strongly enriched at the edges of the cups (Fig. 9C, 0 s). Upon cup closure, Myo1B was enriched at the site of closure (Fig 9C, 8 s) whereas Myo1F remained uniformly distributed on the membrane of the internalized vesicle (Figs. 9B, 4 s and 16 s). Myo1F associated with internalized vesicles more strongly than Myo1B (Fig. 9B 16 s, and Fig. 9C, 12 s), often remaining associated with internalized vesicles longer than F-actin (Fig. 9B, 36 s) whereas Myo1B usually disappeared from the internalized vesicles earlier than F-actin (Fig. 9C, 20 s).
As illustrated in cells coexpressing Myo1E and Myo1B (Fig. 10), the localization of Myo1E in pinocytic structures was the same as observed for Myo1F and different from that of Myo1B. Myo1E, like Myo1F, was observed along most of the cup surface, but more weakly or absent at the leading edges where Myo1B was greatly enriched (Fig. 10A, 0 s; Fig. 10B, 0 s; Fig. 10C, 15 s). After vesicle internalization, Myo1E was absent from the site of vesicle closure where Myo1B was enriched (Fig. 10B 10 s and 20 s), and Myo1E remained associated with internalized vesicles longer than Myo1B (Fig. 10A, 40 s; Fig. 10B, 20 s and 30 s). In situations in which the pinocytic cup failed to close and reverted into a flat protrusion (Fig 10C), Myo1E and Myo1B colocalized in flat protrusions/ruffles before the cup started to form (Fig. 10C, 0s) and after the cup reverted to a flat protrusion (Fig. 10C, 35 s). It was only when the cup started to close (Fig. 10C, 15 s) that the locations of Myo1E and Myo1B differed significantly with Myo1B at the cup edges surrounding Myo1F in the cup interior. Additional examples are shown in Fig. S4.
The localizations of Myo1F (Fig. 11 A–C) and Myo1E (Fig. 11D) in actin waves were also examined. Again in contrast to what was observed for Myo1A, both Myo1E and Myo1F were associated with waves. Their localizations were similar to each other but strikingly different from the localization of Myo1B (Fig. 11D). Whereas, Myo1B colocalized with F-actin (Bretschneider et al., 2009; Brzeska et al., 2014) in a sharp, partially patched pattern (Fig. 11D), Myo1F (Figs. 11 A–C) and Myo1E (Fig. 11D) localized diffusely within the region enclosed by Myo1B and actin (see also supplemental movies M1–M5). The difference between Myo1B and Myo1E localization in actin waves is illustrated in cells coexpressing these two myosins where Myo1B can clearly be seen to be encircling the more expansive wave-associated Myo1E (Fig. 11D)). This bull’s-eye distribution of Myo1F and Myo1B in actin waves is best illustrated in the merged image in Fig 11D.
Fig. 11. Myo1F and Myo1E localize to the interior of actin waves.
Cells were cotransfected with GFP-Myo1F and RFP-lifeact (A–C) or with RFP-Myo1E and GFP-Myo1B (D). Actin waves were induced with 1 μM latrunculin. (A) Myo1F-null cells expressing Myo1F and lifeact. (B) Myo1B-null cells expressing Myo1F and lifeact. (C) AX2 cells expressing Myo1F and lifeact, snapshots of wave propagation taken at times indicated in the panel. (D) Myo1B-null cells expressing Myo1E and Myo1B. Images of live cells are shown. Arrows point to actin waves. Bars are 10 μm.
Myo1F localized only to the region enclosed by actin waves in Myo1F-null, Myo1B-null, and wild type AX2 cells (Figs. 11A, 11B and 11C, respectively) and, in contrast to Myo1B, did not associate with the actin; i.e. Myo1F does not locate to the region normally occupied by Myo1B even when Myo1B is absent and the localization of expressed Myo1F is not affected by the presence of endogenous Myo1F.
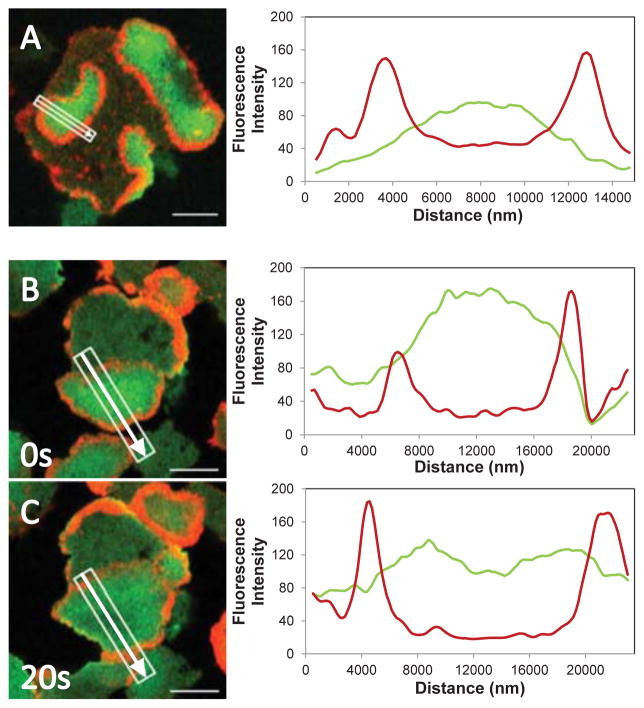
Myo1F remained in the region encircled by the F-actin wave during the entire period of wave expansion but the fluorescence eventually decayed beginning at the region where Myo1F first appeared (Figs. 11 C, 250 s and 300 s). Line scans of Myo1F and F-actin fluorescence in circular actin waves clearly show a peak of Myo1F fluorescence in between two sharp peaks of actin fluorescence (Fig. 12A, B). The Myo1F fluorescence diminished as the wave expanded, the decay starting at the center of the region encircled by the wave (compare Figs. 12B 0 s and 12C 20 s).
Fig. 12. Linescans of Myo1F and F-actin fluorescence in actin waves.
GFP-Myo1F and RFP-lifeact were coexpressed in Myo1B-null cells and actin waves were induced with 1 μM latrunculin. Merged cell images are shown at the left and line scans of GFP-Myo1F and RFP-lifeact fluorescence are shown at the right. (A) Cell with multiple waves, the Myo1F fluorescence peak is located in the middle of the region encircled by a wave. This image corresponds to supplemental movie M2. (B) and (C) actin wave in the same cell at 0 s and 20 s. During wave expansion Myo1F fluorescence decayed in the middle of the wave. The white rectangle shows the region scanned, the arrow indicates the direction of scanning. Images of live cells are shown. Bars are 10 μm.
Discussion
Dictyostelium amoebae actively extend pseudopodia during random and directed motility, and the axenic strains are highly endocytic, robustly producing macropinocytic crowns. The seven myosin Is of Dictyostelium have roles in both migration and endocytosis, contributing to each process to a different extent, and some of their functions often overlap [Falk et al., 2003; Jung et al., 1996; Novak et al., 1995]. Consistent with their cellular roles, the individual myosin Is localize to the leading edge and to endocytic membranes: for Myo1B [Bretschneider et al., 2009; Brzeska et al., 2012; Brzeska et al., 2014; Clarke et al., 2010; Fukui et al., 1989; Morita et al., 1996; Neuhaus and Soldati, 2000; Novak et al., 1995], for Myo1C [Jung et al., 2001; Rump et al., 2011], for Myo1D [Morita et al., 1996], for Myo1E [Chen et al., 2012; Durrwang et al., 2006], for Myo1K [Dieckmann et al., 2010; Schwarz et al., 2000] and this manuscript for Myo1A, Myo1E and Myo1F. However the localization of family members with respect to each other within a single cell had not been investigated. In the current work, we explore the localizations of the three Dictyostelium short-tailed myosin Is, Myo1A, Myo1E and Myo1F, relative to each other and in comparison to the well-studied localization of long-tailed Myo1B. Additionally, the molecular basis of targeting of Myo1A has been determined and compared to that of Myo1B to gain a fuller understanding of the molecular determinants of intracellular targeting of this family of motors.
The similarities and differences of the localization of Myo1A compared to Myo1B are consistent with the various features of their tail regions. Myo1A localizes sharply to the plasma membrane of freshly plated cells, at cell-cell contacts and at the sites of close contacts between chemotaxing cells, as does Myo1B. The BH site in the TH1 domain of the tail is required for these localizations of both myosins. Both Myo1A and Myo1B also localize diffusely at pseudopods and at the front of chemotaxing cells. The head of both myosins is required and sufficient for these localizations, and for their dissociation from the plasma membrane and subsequent relocation. The tail alone remains associated with the plasma membrane during chemotaxis. The principal difference between Myo1A and Myo1B localizations is that Myo1B, but not Myo1A, localizes to actin waves and macropinocytic structures. The latter localizations of long-tailed Myo1B require both the BH site and the GPQ region; short-tailed Myo1A does not have a GPQ region. The common localizations of Myo1A and Myo1B to the pseudopod is consistent with each of these myosin Is playing a role in controlling pseudopod formation during migration [Titus et al., 1993; Wessels et al., 1991; Wessels et al., 1996].
We had anticipated that the localizations of short-tailed Myo1E and Myo1F would be very similar to the localizations of Myo1A as each of their tails contains only a TH1 domain. This is indeed the case in migrating cells where all three short-tailed myosins (as well as long-tailed Myo1B) localize to the plasma membrane, to sites of cell-cell contacts, to pseudopods of randomly moving cells, and to the front of chemotaxing cells. However, a major difference between the localizations of Myo1A, Myo1E, and Myo1F is that only the latter two are found on the membrane surface of macropinocytic cups and within the region bordered by actin-rich waves that form on the bottom of the cell in the presence of low concentrations of latrunculin. Myo1A is clearly absent from these regions of the cell. It is important to note that the localizations of Myo1E and Myo1F in macropinocytic structures and in relation to actin waves differ from that of Myo1B. Myo1B colocalizes with F-actin in the actin waves and at the tips of mature macropinocytic cups whereas Myo1E and Myo1F are in the interior of actin waves, and along the entire surface of macropinocytic cups except for the leading edges where Myo1B is most concentrated. Thus, in spite of the overall similarity of their domain structure, the cellular distributions of the three short-tailed myosin Is, are quite different, consistent with their having both shared and divergent cellular functions [Chen et al., 2012; Durrwang et al., 2006; Falk et al., 2003; Titus et al., 1993].
The localizations of Myo1E and Myo1F in macropinocytic cups and inside actin waves coincide with the previously described localizations of PI(3,4,5)P3 in these structures [Clarke et al., 2010; Dormann et al., 2004; Gerhardt et al., 2014; Gerisch et al., 2009; Gerisch et al., 2011; Hoeller et al., 2013; Swanson, 2008; Veltman et al., 2014]. The decay of Myo1F fluorescence in the center of expanding waves is also similar to that reported for the PI(3,4,5)P3 (compare Fig. 12A in this paper to Figs. 4A and 4B in [Gerhardt et al., 2014] and Fig. 12C in this paper to Fig 5C in [Gerhardt et al., 2014]). Together with the demonstrated affinity of Myo1E and Myo1F, but not Myo1A and Myo1B, for PI(3,4,5)P3 in vitro [Chen et al., 2012; Zhang et al., 2010] it strongly suggests that binding to PI(3,4,5)P3, presumably through a PI(3,4,5)P3-specific PH domain, is the main determinant of Myo1E and Myo1F targeting to these two structures in vivo.
In contrast to Myo1E and Myo1F the localization of Myo1B in macropinocytic structures and in actin waves depends on both its BH site, that binds acidic lipids nonspecifically, and its GPQ-domain, that binds F-actin. Actin waves, edges of micropinocytic cups and sites of internalization of pinocytic vesicles, where Myo1B localizes, are strongly enriched in F-actin [Bretschneider et al., 2004; Clarke et al., 2010] and not enriched in PIP3 or PIP2 [Clarke et al., 2010; Gerisch et al., 2011; Hoeller et al., 2013]. Thus, Myo1B is targeted to actin waves and macropinocytic structures by different mechanisms than Myo1E and Myo1F.
We found also that the localization of a myosin I to a particular compartment could be highly specific. For example, Myo1A did not relocalize to actin waves nor to macropinocytic structures in Myo1B-null cells, and Myo1F remained localized inside actin waves in Myo1B-null cells. So, at least in these structures, the short-tailed myosin Is are unlikely to compensate for the absence of Myo1B. This is in contrast to what has been observed for the two mammalian short-tailed myosin Is where, in the absence of Myo1a, Myo1d relocalizes to sites in intestinal brush border microvilli that are specific for Myo1a in the wild type animal [Benesh et al., 2010].
Our data on the localizations of Myo1B, Myo1A, Myo1D, and Myo1F are in general agreement with their postulated functions. Analyses of myosin I null cell lines missing single or multiple myosin Is and of wild type cells overexpressing myosin Is or their mutants [Chen et al., 2012; Dai et al., 1999; Durrwang et al., 2006; Jung and Hammer, 1990; Jung et al., 1996; Novak et al., 1995; Novak and Titus, 1997, 1998; Rivero, 2008; Schwarz et al., 2000; Soldati, 2003; Titus et al., 1995] have revealed numerous cellular activities of Dictyostelium myosin Is, including cell motility, endocytosis, and cortical tension. Although some of the results varied between different laboratories, the data clearly show that more than one myosin I is involved in each of the above processes (for reviews see [Ostap and Pollard, 1996; Rivero, 2008; Soldati, 2003; Titus, 2000]). It seems likely that myosin Is play critical roles in linking the actin network to the plasma membrane, either anchoring the actin filaments or providing a contractile force. Alternatively or additionally some myosin Is may recruit regulators of actin dynamics [Kim and Flavell, 2008; McConnell and Tyska, 2010].
Different myosin I isoforms may have redundant functions or may play different roles at different stages of the same process. As one example, Myo1A, Myo1B and Myo1F colocalize at the front of chemotaxing cells and in pseudopods and Myo1A-null, Myo1B-null and Myo1F-null cells all exhibit motility defects [Falk et al., 2003; Titus et al., 1993; Wessels et al., 1991; Wessels et al., 1996]; no information is currently available for the motility of Myo1E-null cells. The motility impairments are most likely due to defects in pseudopod formation. Quantitative analysis reveals that Myo1A-, Myo1B- and Myo1F-null cells extend more pseudopodia and turn more frequently than control cells [Falk et al., 2003; Titus et al., 1993; Wessels et al., 1991]. Simultaneous deletion of two myosin Is causes progressively decreased rates of motility that likely are the result of increasingly aberrant control over pseudopod formation, as has been observed for the Myo1A-/Myo1B- and Myo1A-/Myo1F-mutants [Falk et al., 2003]. Thus, multiple myosin Is contribute to pseudopod formation and migration working in concert with each other to produce the correct number, size and placement of these critical protrusions [Falk et al., 2003; Titus et al., 1993; Wessels et al., 1991; Wessels et al., 1996].
A number of Dictyostelium myosin Is had been shown to be present in endocytic structures. These include Myo1B [Bretschneider et al., 2009; Brzeska et al., 2012; Clarke et al., 2010; Fukui et al., 1989; Neuhaus and Soldati, 2000], Myo1C [Jung et al., 2001; Rump et al., 2011], Myo1E [Chen et al., 2012; Durrwang et al., 2006] and Myo1K [Dieckmann et al., 2010; Schwarz et al., 2000], and we have now shown that Myo1F also localizes to macropinocytic structures while Myo1A does not. It is unlikely that the functions of all of these myosin Is are redundant because they include both, short- and long-tailed members, their specific localizations differ, and null mutations have different effects. Myo1A-null, Myo1B-null, Myo1C-null, and Myo1D-null cells show only moderate inhibition of pinocytosis (0–20%) but a much stronger inhibition (about 60%) was observed for Myo1A/Myo1B-null, Myo1B/Myo1C-null and Myo1B/Myo1C/Myo1D-null cells [Jung et al., 1996; Novak et al., 1995; Ostap and Pollard, 1996], i.e. the double mutant phenotypes are not simply a predictable combination of the single mutant phenotypes. Our finding that the location of Myo1B within macropinocytic structures differs from the localization of Myo1E and Myo1F is consistent with non-redundancy of their functions. However, since we did not observe Myo1A in macropinocytic structures, it is not clear why the absence of Myo1A impairs pinocytosis. It could be that a Myo1A function affects pinocytosis indirectly; a good candidate being the maintenance of cortical tension [Dai et al., 1999].
Myo1A-/Myo1B- and Myo1B-/Myo1C-double mutants exhibit notable decreases in cortical tension that are not observed for Myo1A-, Myo1B- or Myo1C-single mutants, and an excess of either Myo1B or Myo1C results in increased cortical tension [Dai et al., 1999]. These results suggest that there must be a precise balance of myosin I activities to maintain the necessary cortical dynamics. The observation that endocytic defects correlate with changes in cortical tension indicates that control of the cortical actin cytoskeleton tension by Myo1A, Myo1B and Myo1C (and likely other myosin Is) has an important role in determining rates of macropinocytic uptake. The localization of both Myo1B and Myo1A on the plasma membrane agrees with such interpretation. However, it is possible that the two phenotypes are only indirectly related [Dai et al., 1999; Novak and Titus, 1997] and that the myosin Is have different roles in regulating macropinocytic cup dynamics or structure.
Analyses of mutant cell lines revealed that the short-tailed myosin Is that bind to PIP3, Myo1E and Myo1F, cooperate with each other, and with long-tailed Myo1D which also binds to PIP3, in phagocytosis [Chen et al., 2012; Durrwang et al., 2006]. Defects in phagocytosis are often paralleled by defects in pinocytosis [Ostap and Pollard, 1996], but not always. For example, overexpression of Myo1B caused 80% inhibition of pinocytosis and 70% inhibition of phagocytosis [Novak and Titus, 1997], but overexpression of Myo1E caused 30% reduction of pinocytosis and 40% increase of phagocytosis [Durrwang et al., 2006]. Also, a Myo1B null mutant did not have a fluid uptake defect [Jung et al., 1996; Novak et al., 1995] yet exhibited decreased particle uptake and slow growth on bacteria [Jung and Hammer, 1990] whereas wild type, non-axenic strains of Dictyostelium do not internalize fluids, although they are highly phagocytic [Bloomfield et al., 2015].
Myosin Is have been implicated in endocytosis in several different organisms including yeast [Giblin et al., 2011] and Entamoeba [Voigt et al., 1999]. The yeast myosin Is have long tails and they target to the plasma membrane where they work in concert with each other to drive membrane invagination and vesicle scission in an actin-dependent manner [Giblin et al., 2011]. Vertebrate long-tailed Myo1e [Cheng et al., 2012; Krendel et al., 2007; Maxeiner et al., 2015] and short-tailed Myo1g [Dart et al., 2012] each play a crucial role in endocytosis, including Fc-γ receptor-driven phagocytosis [Dart et al., 2012; Maxeiner et al., 2015]. Myosin I was also found to support cortical tension of mammalian cells [Nambiar et al., 2009] consistent with myosin I functions being conserved throughout evolution.
Summarizing our results, Myo1A, Myo1B, Myo1E and Myo1F share a common sharp localization at the plasma membrane of freshly plated cells, at cell-cell contacts and in the front of streaming cells, and a common diffused localization in pseudopods and at the front of chemotaxing cells. Therefore, it is likely that all four myosins contribute to the processes occurring in these cell compartments, perhaps with overlapping functions. On the other hand, Myo1A is absent from actin waves with which Myo1B, Myo1E and Myo1F associate (but Myo1E and Myo1F differently than Myo1B). Therefore, Myo1A is unlikely to contribute to the formation or propagation of actin waves, and the functions of Myo1E and Myo1F in actin waves may be similar to each other but different than the role of Myo1B. Finally, Myo1A is most likely not directly involved in the formation or function of macropinocytic structures, and the roles of Myo1E and Myo1F in macropinocytosis may be similar to each other, but different than the role of Myo1B, especially during cup closure. The different localizations of myosin Is in micropinosomes seems to be of special importance, especially in light of the recent discovery that many Rac-activated cancers use micropinocytosis for feeding [Commisso et al. 2013; Kamphorst et al., 2015] and because the involvement of myosin Is in endocytosis seems to be conserved during evolution.
Materials and methods
DNA Constructs
Standard PCR-based cloning and mutagenesis methods were used to generate all of the expression plasmids used in this study (see Supplemental Table 1). TA cloning of PCR products was carried out using the StrataClone system (Agilent) and PCR mutagenesis was performed using the Q5 site-directed mutagenesis kit (New England BioLabs). The full sequences for each clone were verified (BioMedical Genomics Center). Except where noted, all of the expression plasmids were generated using an expression plasmid that contains a low copy number origin of replication, the Ddp1-based pLittle and pTX-GFP plasmids [Levi et al., 2000; Patterson and Spudich, 1995], a G418 resistance cassette and a gfp gene for generating an N-terminal GFP-fusion protein. The wild type GFP-M Myo1 and GFP-Myo1B Ddp1-based extrachromosomal expression plasmids, pDTa48R and pDTb60, have been described previously [Brzeska et al., 2012; Senda et al., 2001]. An RFP-Myo1E expression plasmid was generated by cloning the full-length Myo1E gene into pDM449, a Ddp1-based expression plasmid that carries a hygromycin resistance cassette [Veltman et al., 2009]. Lifeact [Riedl et al., 2008] fused to RFP was cloned between XhoI and Hind III sites [Veltman et al., 2009; Brzeska et al 2014] of the pDM358 plasmid, a Ddp1-based expression plasmid that carries a hygromycin resistance cassette [Veltman et al., 2009].
Cell Lines, Cell Culturing and Treatment
All cells were grown in HL5 media (Formedium, HLG0101) [Sussman, 1987] supplemented with appropriate antibiotics. The previously described Myo1A-null cells and Myo1F-null cells in thymidine auxothrop background JH10 [Hadwiger and Firtel, 1992; Peterson et al., 1995; Titus et al., 1995] were grown without antibiotic selection while the Myo1B-null strain in the wild type AX2 background [Brzeska et al., 2014] was maintained in 7 μg/ml blasticidin S HCl (Invitrogen). Expression plasmids were transformed into wild type or null mutants by electroporation [Gaudet et al., 2007]. Cells expressing wild type or mutant GFP-Myo1A, GFP-Myo1B and GFP-Myo1F were selected and maintained in media containing 12 μg/ml G418 sulfate (Mediatech). Cells expressing RFP-lifeact or RFP-Myo1E were selected and maintained in media containing 50 μg/ml hygromycin (Invitrogen). Cells coexpressing two proteins were cotransfected with two plasmids, each carrying a single protein and different antibiotic resistance. Cell selection was carried out in the presence of antibiotics dictated by the transfected plasmids and cell types (for example Myo1B-null cells expressing Myo1B and Myo1D were selected and grown in the presence of blasticidin, G418 and hygromycin).
Cells were grown, transfected and prepared for microscopy as described earlier [Brzeska et al., 2014]. Briefly, cells were grown in 10-cm Petri dishes and transferred to chambered cover glass (Nalge Nunc International, 1555383) for microscopy. Starvation was induced by exchanging HL5 media for starvation solution that contained 20 mM phosphate buffer pH 6.2, 0.2 mM CaCl2 and 2 mM MgCl2. Localization of proteins in non-motile or slowly moving cells was determined by recording images of fixed freshly plated cells, localization of actively moving cells during random cell movement was observed in live cells that were starved between 0.5 h and 4 h, and localization in chemotaxing cells was observed in live cells after overnight starvation at 4 °C. Actin waves were induced by treating cells that were starved for 30 min with 1 μM latrunculin (latrunculin A, Sigma) as described [Brzeska et al., 2014], and waves were recorded in live cells.
Image Recording and Processing
Cells were viewed with a Zeiss 780 confocal microscope with a 63x plan Apochromat 1.4 objective. Image recording and profile scanning were done using Zeiss Zen software, and final illustrations were prepared using Zeiss Zen software and Photoshop.
Supplementary Material
Acknowledgments
M. A. Titus was supported by the NSF (MCB-1244235). All other authors were supported by the intramural program of the National Heart, Lung and Blood Institute.
References
- Bagorda A, Mihaylov VA, Parent CA. Chemotaxis: moving forward and holding on to the past. Thromb Haemost. 2006;95:12–21. [PubMed] [Google Scholar]
- Benesh AE, Nambiar R, McConnell RE, Mao S, Tabb DL, Tyska MJ. Differential localization and dynamics of class I myosins in the enterocyte microvillus. Mol Biol Cell. 2010;21:970–978. doi: 10.1091/mbc.E09-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield G, Traynor D, Sander SP, Veltman DM, Pachebat JA, Kay RR. Neurofibromin controls macropinocytosis and phagocytosis in Dictyostelium. eLife. 2015;4 doi: 10.7554/eLife.04940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond LM, Brandstaetter H, Kendrick-Jones J, Buss F. Functional roles for myosin 1c in cellular signaling pathways. Cell Signal. 2013;25:229–235. doi: 10.1016/j.cellsig.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider T, Anderson K, Ecke M, Muller-Taubenberger A, Schroth-Diez B, Ishikawa-Ankerhold HC, Gerisch G. The three-dimensional dynamics of actin waves, a model of cytoskeletal self-organization. Biophys J. 2009;96:2888–2900. doi: 10.1016/j.bpj.2008.12.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider T, Diez S, Anderson K, Heuser J, Clarke M, Muller-Taubenberger A, Kohler J, Gerisch G. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Brzeska H, Guag J, Preston GM, Titus MA, Korn ED. Molecular basis of dynamic relocalization of Dictyostelium myosin IB. J Biol Chem. 2012;287:14923–14936. doi: 10.1074/jbc.M111.318667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H, Guag J, Remmert K, Chacko S, Korn ED. An experimentally based computer search identifies unstructured membrane-binding sites in proteins: application to class I myosins, PAKS, and CARMIL. J Biol Chem. 2010;285:5738–5747. doi: 10.1074/jbc.M109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H, Hwang KJ, Korn ED. Acanthamoeba myosin IC colocalizes with phosphatidylinositol 4,5-bisphosphate at the plasma membrane due to the high concentration of negative charge. J Biol Chem. 2008;283:32014–32023. doi: 10.1074/jbc.M804828200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H, Pridham K, Chery G, Titus MA, Korn ED. The association of myosin IB with actin waves in Dictyostelium requires both the plasma membrane-binding site and actin-binding region in the myosin tail. PLoS One. 2014;9:e94306. doi: 10.1371/journal.pone.0094306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Wang Y, Sesaki H, Iijima M. Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis. Sci Signal. 2012;5:ra10. doi: 10.1126/scisignal.2002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Grassart A, Drubin DG. Myosin 1E coordinates actin assembly and cargo trafficking during clathrin-mediated endocytosis. Mol Biol Cell. 2012;23:2891–2904. doi: 10.1091/mbc.E11-04-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Engel U, Giorgione J, Muller-Taubenberger A, Prassler J, Veltman D, Gerisch G. Curvature recognition and force generation in phagocytosis. BMC Biol. 2010;8:154. doi: 10.1186/1741-7007-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG, Bar-Sagi D. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley SW, Liburd J, Shaw K, Jung Y, Smith SP, Cote GP. Identification of calmodulin and MlcC as light chains for Dictyostelium myosin-I isozymes. Biochemistry. 2011;50:6579–6588. doi: 10.1021/bi2007178. [DOI] [PubMed] [Google Scholar]
- Cyr JL, Dumont RA, Gillespie PG. Myosin-1c interacts with hair-cell receptors through its calmodulin-binding IQ domains. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2002;22:2487–2495. doi: 10.1523/JNEUROSCI.22-07-02487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Ting-Beall HP, Hochmuth RM, Sheetz MP, Titus MA. Myosin I contributes to the generation of resting cortical tension. Biophys J. 1999;77:1168–1176. doi: 10.1016/s0006-3495(99)76968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart AE, Tollis S, Bright MD, Frankel G, Endres RG. The motor protein myosin 1G functions in FcgammaR-mediated phagocytosis. J Cell Sci. 2012;125:6020–6029. doi: 10.1242/jcs.109561. [DOI] [PubMed] [Google Scholar]
- Dieckmann R, von Heyden Y, Kistler C, Gopaldass N, Hausherr S, Crawley SW, Schwarz EC, Diensthuber RP, Cote GP, Tsiavaliaris G, Soldati T. A myosin IK-Abp1-PakB circuit acts as a switch to regulate phagocytosis efficiency. Mol Biol Cell. 2010;21:1505–1518. doi: 10.1091/mbc.E09-06-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Weijer G, Dowler S, Weijer CJ. In vivo analysis of 3-phosphoinositide dynamics during Dictyostelium phagocytosis and chemotaxis. J Cell Sci. 2004;117:6497–6509. doi: 10.1242/jcs.01579. [DOI] [PubMed] [Google Scholar]
- Durrwang U, Fujita-Becker S, Erent M, Kull FJ, Tsiavaliaris G, Geeves MA, Manstein DJ. Dictyostelium myosin-IE is a fast molecular motor involved in phagocytosis. J Cell Sci. 2006;119:550–558. doi: 10.1242/jcs.02774. [DOI] [PubMed] [Google Scholar]
- Falk DL, Wessels D, Jenkins L, Pham T, Kuhl S, Titus MA, Soll DR. Shared, unique and redundant functions of three members of the class I myosins (MyoA, MyoB and MyoF) in motility and chemotaxis in Dictyostelium. J Cell Sci. 2003;116:3985–3999. doi: 10.1242/jcs.00696. [DOI] [PubMed] [Google Scholar]
- Feeser EA, Ignacio CM, Krendel M, Ostap EM. Myo1e binds anionic phospholipids with high affinity. Biochemistry. 2010;49:9353–9360. doi: 10.1021/bi1012657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Lynch TJ, Brzeska H, Korn ED. Myosin I is located at the leading edges of locomoting Dictyostelium amoebae. Nature. 1989;341:328–331. doi: 10.1038/341328a0. [DOI] [PubMed] [Google Scholar]
- Gaudet P, Pilcher KE, Fey P, Chisholm RL. Transformation of Dictyostelium discoideum with plasmid DNA. Nat Protocols. 2007;2:1317–1324. doi: 10.1038/nprot.2007.179. [DOI] [PubMed] [Google Scholar]
- Gerhardt M, Ecke M, Walz M, Stengl A, Beta C, Gerisch G. Actin and PIP3 waves in giant cells reveal the inherent length scale of an excited state. J Cell Sci. 2014;127:4507–4517. doi: 10.1242/jcs.156000. [DOI] [PubMed] [Google Scholar]
- Gerisch G, Ecke M, Schroth-Diez B, Gerwig S, Engel U, Maddera L, Clarke M. Self-organizing actin waves as planar phagocytic cup structures. Cell Adhes Migr. 2009;3:373–382. doi: 10.4161/cam.3.4.9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch G, Ecke M, Wischnewski D, Schroth-Diez B. Different modes of state transitions determine pattern in the Phosphatidylinositide-Actin system. BMC Cell Biol. 2011;12:42. doi: 10.1186/1471-2121-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giblin J, Fernandez-Golbano IM, Idrissi FZ, Geli MI. Function and regulation of Saccharomyces cerevisiae myosins-I in endocytic budding. Biochem Soc Trans. 2011;39:1185–1190. doi: 10.1042/BST0391185. [DOI] [PubMed] [Google Scholar]
- Greenberg MJ, Ostap EM. Regulation and control of myosin-I by the motor and light chain-binding domains. Trends Cell Biol. 2013;23:81–89. doi: 10.1016/j.tcb.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger JA, Firtel RA. Analysis of G alpha 4, a G-protein subunit required for multicellular development in Dictyostelium. Genes Dev. 1992;6:38–49. doi: 10.1101/gad.6.1.38. [DOI] [PubMed] [Google Scholar]
- Hoeller O, Bolourani P, Clark J, Stephens LR, Hawkins PT, Weiner OD, Weeks G, Kay RR. Two distinct functions for PI3-kinases in macropinocytosis. J Cell Sci. 2013;126:4296–4307. doi: 10.1242/jcs.134015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol Biol Cell. 2006;17:4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Hammer JA., 3rd Generation and characterization of Dictyostelium cells deficient in a myosin I heavy chain isoform. J Cell Biol. 1990;110:1955–1964. doi: 10.1083/jcb.110.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Remmert K, Wu X, Volosky JM, Hammer JA., 3rd The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol. 2001;153:1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Wu X, Hammer JA., 3rd Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J Cell Biol. 1996;133:305–323. doi: 10.1083/jcb.133.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, Thompson CB, Rabinowitz JD. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SV, Flavell RA. Myosin I: from yeast to human. Cell Mol Life Sci. 2008;65:2128–2137. doi: 10.1007/s00018-008-7435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar M. Thirteen is enough: the myosins of Dictyostelium discoideum and their light chains. Genomics. 2006;7:183. doi: 10.1186/1471-2164-7-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaba S, Coluccio LM. Localization of myosin 1b to actin protrusions requires phosphoinositide binding. J Biol Chem. 2010;285:27686–27693. doi: 10.1074/jbc.M109.087270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M, Osterweil EK, Mooseker MS. Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis. FEBS Lett. 2007;581:644–650. doi: 10.1016/j.febslet.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi S, Polyakov M, Egelhoff TT. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- Lu Q, Li J, Ye F, Zhang M. Structure of myosin-1c tail bound to calmodulin provides insights into calcium-mediated conformational coupling. Nat Struct Mol Biol. 2015;22:81–88. doi: 10.1038/nsmb.2923. [DOI] [PubMed] [Google Scholar]
- Maxeiner S, Shi N, Schalla C, Aydin G, Hoss M, Vogel S, Zenke M, Sechi AS. Crucial role for the LSP1-myosin1e bimolecular complex in the regulation of Fcgamma receptor-driven phagocytosis. Mol Biol Cell. 2015;26:1652–1664. doi: 10.1091/mbc.E14-05-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazerik JN, Tyska MJ. Myosin-1A targets to microvilli using multiple membrane binding motifs in the tail homology 1 (TH1) domain. J Biol Chem. 2012;287:13104–13115. doi: 10.1074/jbc.M111.336313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell RE, Tyska MJ. Leveraging the membrane - cytoskeleton interface with myosin-1. Trends Cell Biol. 2010;20:418–426. doi: 10.1016/j.tcb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita YS, Jung G, Hammer JA, 3rd, Fukui Y. Localization of Dictyostelium myoB and myoD to filopodia and cell-cell contact sites using isoform-specific antibodies. Eur J Cell Biol. 1996;71:371–379. [PubMed] [Google Scholar]
- Nambiar R, McConnell RE, Tyska MJ. Control of cell membrane tension by myosin-I. Proc Natl Acad Sci U S A. 2009;106:11972–11977. doi: 10.1073/pnas.0901641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus EM, Soldati T. A myosin I is involved in membrane recycling from early endosomes. J Cell Biol. 2000;150:1013–1026. doi: 10.1083/jcb.150.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak KD, Peterson MD, Reedy MC, Titus MA. Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J Cell Biol. 1995;131:1205–1221. doi: 10.1083/jcb.131.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak KD, Titus MA. Myosin I overexpression impairs cell migration. J Cell Biol. 1997;136:633–647. doi: 10.1083/jcb.136.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak KD, Titus MA. The myosin I SH3 domain and TEDS rule phosphorylation site are required for in vivo function. Mol Biol Cell. 1998;9:75–88. doi: 10.1091/mbc.9.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Pfarao: a web application for protein family analysis customized for cytoskeletal and motor proteins (CyMoBase) BMC Genomics. 2006;7:300. doi: 10.1186/1471-2164-7-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2, 269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostap EM, Pollard TD. Overlapping functions of myosin-I isoforms? J Cell Biol. 1996;133:221–224. doi: 10.1083/jcb.133.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B, Spudich JA. A novel positive selection for identifying cold-sensitive myosin II mutants in Dictyostelium. Genetics. 1995;140:505–515. doi: 10.1093/genetics/140.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MD, Novak KD, Reedy MC, Ruman JI, Titus MA. Molecular genetic analysis of myoC, a Dictyostelium myosin I. J Cell Sci. 1995;108(Pt 3):1093–1103. doi: 10.1242/jcs.108.3.1093. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero F. Endocytosis and the actin cytoskeleton in Dictyostelium discoideum. Int Rev cell Mol Biol. 2008;267:343–397. doi: 10.1016/S1937-6448(08)00633-3. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Rener B. The GPQ-rich segment of Dictyostelium myosin IB contains an actin binding site. Biochemistry. 1994;33:2322–2328. doi: 10.1021/bi00174a045. [DOI] [PubMed] [Google Scholar]
- Rump A, Scholz T, Thiel C, Hartmann FK, Uta P, Hinrichs MH, Taft MH, Tsiavaliaris G. Myosin-1C associates with microtubules and stabilizes the mitotic spindle during cell division. J Cell Sci. 2011;124:2521–2528. doi: 10.1242/jcs.084335. [DOI] [PubMed] [Google Scholar]
- Schwarz EC, Neuhaus EM, Kistler C, Henkel AW, Soldati T. Dictyostelium myosin IK is involved in the maintenance of cortical tension and affects motility and phagocytosis. J Cell Sci. 2000;113:621–633. doi: 10.1242/jcs.113.4.621. [DOI] [PubMed] [Google Scholar]
- Sébé-Pedrós A, Grau-Bové X, Richards TA, Ruiz-Trillo I. Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol Evol. 2014;6:290–305. doi: 10.1093/gbe/evu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda S, Lee SF, Cote GP, Titus MA. Recruitment of a specific amoeboid myosin I isoform to the plasma membrane in chemotactic Dictyostelium cells. J Biol Chem. 2001;276:2898–2904. doi: 10.1074/jbc.M008059200. [DOI] [PubMed] [Google Scholar]
- Soldati T. Unconventional myosins, actin dynamics and endocytosis: a menage a trois? Traffic (Copenhagen, Denmark) 2003;4:358–366. doi: 10.1034/j.1600-0854.2003.t01-1-00095.x. [DOI] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Swanljung-Collins H, Collins JH. Phosphorylation of brush border myosin I by protein kinase C is regulated by Ca(2+)-stimulated binding of myosin I to phosphatidylserine concerted with calmodulin dissociation. J Biol Chem. 1992;267:3445–3454. [PubMed] [Google Scholar]
- Swanson JA. Shaping cups into phagosomes and macropinosomes. Nat Rev Mol Cell Biol. 2008;9:639–649. doi: 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Lin T, Ostap EM. Dynamics of myo1c (myosin-ibeta) lipid binding and dissociation. J Biol Chem. 2002;277:42763–42768. doi: 10.1074/jbc.M206388200. [DOI] [PubMed] [Google Scholar]
- Titus MA. The role of unconventional myosins in Dictyostelium endocytosis. J Eukaryot Microbiol. 2000;47:191–196. doi: 10.1111/j.1550-7408.2000.tb00038.x. [DOI] [PubMed] [Google Scholar]
- Titus MA, Novak KD, Hanes GP, Urioste AS. Molecular genetic analysis of myoF, a new Dictyostelium myosin I gene. Biophys J. 1995;68:152S–155S. discussion 156S–157S. [PMC free article] [PubMed] [Google Scholar]
- Titus MA, Wessels D, Spudich JA, Soll D. The unconventional myosin encoded by the myoA gene plays a role in Dictyostelium motility. Mol Biol Cell. 1993;4:233–246. doi: 10.1091/mbc.4.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman DM, Akar G, Bosgraaf L, Van Haastert PJ. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 2009;61:110–118. doi: 10.1016/j.plasmid.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Lemieux MG, Knecht DA, Insall RH. PIP(3)-dependent macropinocytosis is incompatible with chemotaxis. J Cell Biol. 2014;204:497–505. doi: 10.1083/jcb.201309081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt H, Olivo JC, Sansonetti P, Guillen N. Myosin IB from Entamoeba histolytica is involved in phagocytosis of human erythrocytes. J Cell Sci. 1999;112:1191–1201. doi: 10.1242/jcs.112.8.1191. [DOI] [PubMed] [Google Scholar]
- Wessels D, Murray J, Jung G, Hammer JA, 3rd, Soll DR. Myosin IB null mutants of Dictyostelium exhibit abnormalities in motility. Cell Motil Cytoskeleton. 1991;20:301–315. doi: 10.1002/cm.970200406. [DOI] [PubMed] [Google Scholar]
- Wessels D, Titus M, Soll DR. A Dictyostelium myosin I plays a crucial role in regulating the frequency of pseudopods formed on the substratum. Cell Motil Cytoskeleton. 1996;33:64–79. doi: 10.1002/(SICI)1097-0169(1996)33:1<64::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Xu P, Zot AS, Zot HG. Identification of Acan125 as a myosin-I-binding protein present with myosin-I on cellular organelles of Acanthamoeba. J Biol Chem. 1995;270:25316–25319. doi: 10.1074/jbc.270.43.25316. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wang Y, Sesaki H, Iijima M. Proteomic identification of phosphatidylinositol (3,4,5) triphosphate-binding proteins in Dictyostelium discoideum. Proc Natl Acad Sci U S A. 2010;107:11829–11834. doi: 10.1073/pnas.1006153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.