SUMMARY
Transcription factors (TFs) are thought to function with partners to achieve specificity and precise quantitative outputs. In the developing heart, heterotypic TF interactions, such as between the T-box TF TBX5 and the homeodomain TF NKX2-5, have been proposed as a mechanism for human congenital heart defects. We report extensive and complex interdependent genomic occupancy of TBX5, NKX2-5, and the zinc finger TF GATA4, coordinately controlling cardiac gene expression, differentiation, and morphogenesis. Interdependent binding serves not only to co-regulate gene expression, but also to prevent TFs from distributing to ectopic loci and activate lineage-inappropriate genes. We define preferential motif arrangements for TBX5 and NKX2-5 cooperative binding sites, supported at the atomic level by their co-crystal structure bound to DNA, revealing direct interaction between the two factors, and induced DNA bending. Complex interdependent binding mechanisms reveal tightly regulated TF genomic distribution and define a combinatorial logic for heterotypic TF regulation of differentiation.
INTRODUCTION
Transcriptional regulation during differentiation and organogenesis relies on the fine quantitative regulation of thousands of genes. Within a particular cell type, defined sets of DNA-binding transcription factors (TFs) functioning on defined genomic elements are involved in determining cellular identity (Davidson, 2010; Levo and Segal, 2014). Combinatorial interactions of small numbers of TFs have been proposed to underlie tissue-specific gene expression in Drosophila embryonic mesoderm development and in mammalian organogenesis (De Val et al., 2008; He et al., 2011; Junion et al., 2012; Siersbaek et al., 2014; Stefflova et al., 2013; Tijssen et al., 2011; Tsankov et al., 2015; Wilson et al., 2010; Zaret and Grompe, 2008; Zinzen et al., 2009). Despite the suggestion of combined function from occupancy or gain of function biochemical studies (eg. Jolma et al., 2015; Spitz and Furlong, 2012), the functional output of interactions between developmentally important TFs and their genomic and structural basis have not been explored on a broad scale or in sufficient detail. The nature and importance of heterotypic TF interactions are therefore incompletely understood.
The T-box TF TBX5 and the homeodomain TF NKX2-5 regulate several aspects of heart development (Bruneau et al., 2001; Lyons et al., 1995; Pashmforoush et al., 2004; Tanaka et al., 1999). TBX5 and NKX2-5 can physically interact, and their potential for co-activation of target genes is apparent in their synergistic action in vitro on reporter constructs (Bruneau et al., 2001; Hiroi et al., 2001). This interaction has been proposed to be the basis for the similar congenital heart defects caused by mutations in either TBX5 or NKX2-5 in humans (Basson et al., 1997; Li et al., 1997; Schott et al., 1998). Similarly, mutations in the zinc finger TF gene GATA4 result in congenital heart defects, and GATA4 has been proposed to be a functional partner of TBX5 and NKX2-5 (Durocher et al., 1997; Garg et al., 2003). Indeed, mice doubly heterozygous for null alleles of Tbx5 and Nkx2-5 or Tbx5 and Gata4 have defects in heart formation that are more severe than those caused by each individual mutation (Maitra et al., 2008; Moskowitz et al., 2007). Relative doses of TBX5, NKX2-5, and GATA4 are clearly important, but it is not known what the overlapping roles are for these important cardiac TFs, or how this interaction is coordinated at the genomic or molecular level. TBX5 and GATA4 can, together with additional factors, induce cardiac gene expression programs de novo (Ieda et al., 2010; Qian et al., 2012; Song et al., 2012; Takeuchi and Bruneau, 2009); thus they are clearly positioned in a robust gene regulatory network. Drosophila orthologs of TBX5, NKX2-5, and GATA4 function together as a collective unit with shared binding that does not rely on motif grammar (Junion et al., 2012). Whether this mechanism is retained in mammals is not known. A mechanistic understanding of the collaborative function of TBX5, NKX2-5, and GATA4 will have important implications for understanding congenital heart disease and for developing strategies for cardiac regeneration, and more broadly to understand fundamental principles of heterotypic TF interactions.
Here we examined the overlapping function of TBX5 and NKX2-5 in mouse cardiac differentiation and morphogenesis and their relationship with GATA4. We find that they function cooperatively across the genome, participating in a complex and dynamic TF network regulating programs of gene expression required for expansion of cardiac progenitors and for cardiac differentiation. This cooperative interaction relies on the interdependent binding of the three TFs. The crystal structure of the ternary complex of TBX5 and NKX2-5 and a bona fide DNA target provides an atomic structural basis for this interdependent interaction. We also find that, surprisingly, in the absence of TBX5 or NKX2-5 (or both), the remaining TFs redistribute to other genomic sites associated with genes with increased expression in the TF null cells. Thus, in addition to cooperative binding for gene activation, the interdependent binding of heterotypic factors serves to prevent TF distribution to lineage-inappropriate sites. This redistribution relies on the establishment of new DNA/protein interactions able to induce ectopic gene activation, and unveils a tightly regulated genomic distribution that may underlie the dysregulation of gene expression in congenital heart defects. The genomic interactions between TBX5 and NKX2-5, dictated by a preferential motif arrangement, define a molecular and genomic logic for the regulation of mammalian differentiation by heterotypic TFs.
RESULTS
Impaired Cardiac Differentiation in Tbx5/Nkx2-5 Null Embryos
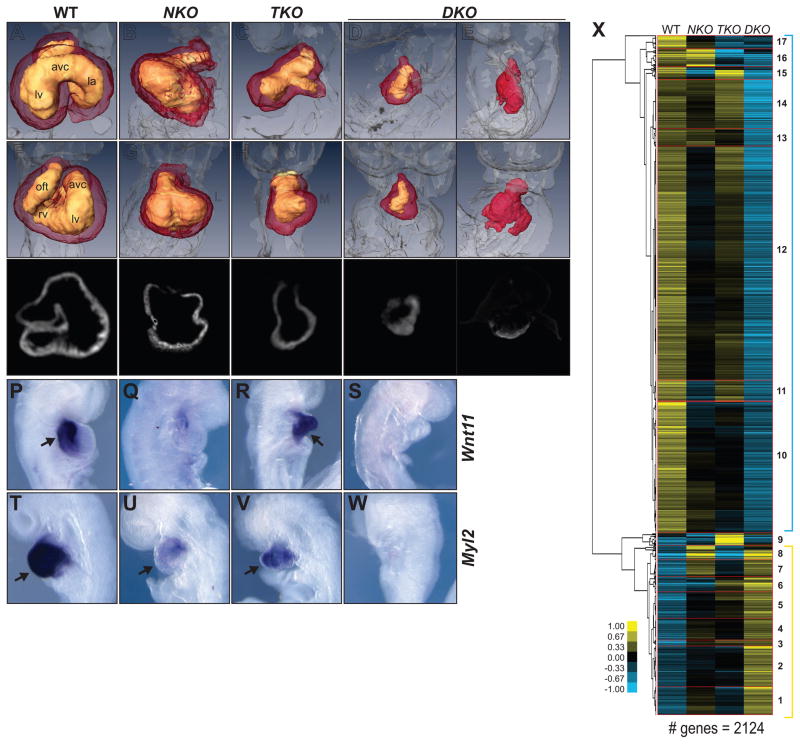
Perinatal death of mice doubly heterozygous for Tbx5 and Nkx2-5 null alleles (Moskowitz et al., 2007) precludes a breeding scheme to generate embryos lacking both factors. Using mice carrying a null allele of Nkx2-5 (Tanaka et al., 1999) and a conditional allele of Tbx5 (Mori et al., 2006), we derived wildtype (WT), Nkx2-5 null (NKO), Tbx5 null (TKO) and Tbx5;Nkx2-5 double null (DKO) embryonic stem (ES) cells (Figure S1A). These ES cells were used to generate entirely ES cell–derived WT, single and double KO embryos (Figure S1B). TKO and NKO embryonic day (E) 9.5 embryos recapitulated previously reported phenotypes (Bruneau et al., 2001; Tanaka et al., 1999) (Figure 1A–O). DKO ES cells yielded embryos that died by E9.5 and had, in the most extreme cases, only a small patch of quiescent or very slowly beating heart tissue (Figure 1D,E,I,J,N,O). Marker analysis confirmed the significant increase in phenotypic severity in the DKO embryos (Figure 1P–W). Thus, combined loss of Tbx5 and Nkx2-5 results in a dramatic restriction in the potential for the embryonic heart to form beyond its initial differentiation.
Figure 1. Tbx5/Nkx2-5 Deletion Impairs Cardiac Development.
(A–E) Lateral and (F–J) frontal view of OPT reconstructions of WT (A,F), NKO (B,G), TKO (C,H) and DKO (D–E,I–J) E8.75 hearts. Right ventricle (rv), left ventricle (lv), atrio-ventricluar canal (avc), out flow tract (oft).
(K–O) cTNT immunostaining- optical transversal sections of WT (K), NKO (L), TKO (M) and DKO (N,O) E8.75 hearts.
(P–S) Transcription of Wnt11, and (T–W) Myl2 in WT (P,T), NKO (Q,U), TKO (R,V) and DKO (S,W) E8.75 hearts. Arrows indicate the signal.
(X) Hierarchical clustering of microarray analysis performed in WT, NKO, TKO and DKO E8.75 hearts. Clusters are indicated by numbers.
Genetic Program Regulated by TBX5 and NKX2-5
We examined gene expression by microarray analysis of heart tissues from E8.75 ES cell–derived embryos (Figure 1X and Table S1,S2) and confirmed microarray results by qRT-PCR of representative genes (Figure S1C). Concordant with the role of TBX5 and NKX2-5 as transcriptional activators (Bruneau et al., 2001; Durocher et al., 1996) a majority of transcripts were downregulated in DKO hearts, spread among subgroups with different patterns of response to each genetic perturbation (clusters 10–17 - blue bracket)(Bruneau et al., 2001; Hiroi et al., 2001). A second major group with upregulated gene expression in DKO hearts was identified, highlighting the existence of TBX5- and/or NKX2-5-dependent repressed genes (clusters 1–8 - yellow bracket). Although major groups showed regulatory contexts depended on only one or the other TF (clusters 1,6–8,11,15,16), complex patterns were also identified (eg. cluster 9), reflecting complexity in the genetic interaction between Tbx5 and Nkx2-5.
Dynamic Gene Expression Regulated by TBX5 and NKX2-5 During CM Differentiation
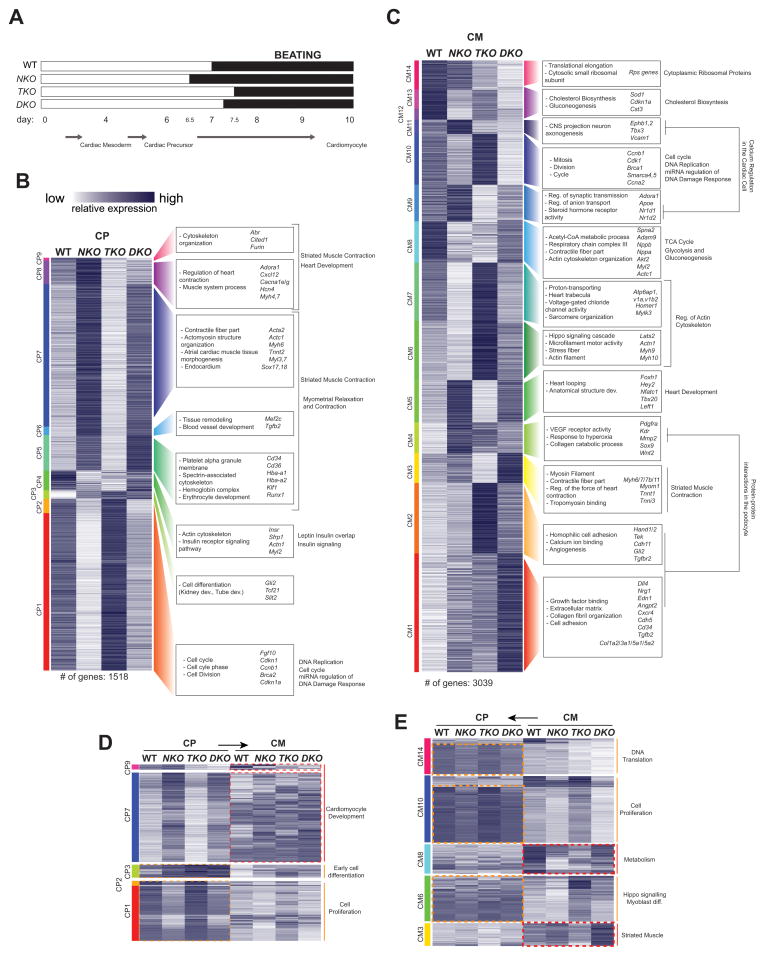
For a detailed understanding of the genetic programs regulated by these factors during cardiomyocyte differentiation and to overcome early lethality of the mutant embryos, we used a directed differentiation system (Kattman et al., 2011; Wamstad et al., 2012) to generate cardiac precursors (CP) and cardiomyocytes (CM) from WT, TKO, NKO, and DKO ES cell lines (Figure S1A). The directed differentiation protocol yielded cultures enriched (80–90%) in cardiac Troponin T (cTnT) (+) beating CMs for all genotypes.
Dynamics of differentiation were different across the four genotypes. NKO cells begin spontaneous contractions 12 hours earlier than WT, and TKO do so 12 later. DKO cells begin beating around 6 hours later than WT, suggesting a major role for TBX5 in the induction of CM differentiation and for NKX2-5 as a repressor of premature cardiac differentiation (Figure 2A, (Prall et al., 2007)). We examined gene expression by RNA-seq at CP and CM stages, and clustered transcripts by expression pattern at each specific stage (Figure 2B–C, Table S1,2). Although no correlation was found between TKO cells and embryonic gene expression analysis (likely due to the TKO embryonic phenotype, which is largely a hypoplasia of Tbx5-expressing tissue - Bruneau et al., 2001 and this report), for the NKO and DKO cells, expression changes in genes detected in the in vivo microarray data and across each stage correlated well (Figure S2C,D).
Figure 2. Transcriptional Analysis of Tbx5/Nkx2-5 Null Cells during Cardiomyocyte Differentiation.
(A) Dynamics of CM differentiation in WT and KO cells.
(B–C) Clustering of differentially expressed coding and non-coding RNAs in WT, NKO, TKO and DKO cells at CP (B) and CM (C) stages of CM differentiation. Enriched GO terms and representative genes are shown to the right.
(D–E) Re-clustering of specific RNA-seq expression clusters at CP (D) or CM (E) looking at both differentiation stages at the same time.
The analysis of changes in Gene Ontology (GO) terms between clusters/stages allowed us to understand temporal dynamics of Tbx5/Nkx2-5-dependent regulation of gene expression (Figure 2B–C, Table S2). Concordant with early NKO differentiation, two major groups of Nkx2-5-dependent genes were identified at CP: repressed genes (CP1,2) associated with cell division and cycle progression, and upregulated genes (CP5,6,7,8,9) associated with cardiac differentiation and function. Two clusters in this second group (CP8,9) presented a different behaviour between NKO and DKO cells, suggesting that genes in these two clusters could be responsible for the different beating initiation between NKO and DKO cells. The dynamics of expression of these clusters showed predominant early expression of downregulated cell proliferation genes (CP1,2), in contrast to later predominant expression of upregulated CM differentiation specific genes (CP7,9) (Figures 2D, S2A), finding a similar temporal pattern in genes associated to these functions identified at CM (Figures 2E, S2B). This stage specificity is more obvious in cluster CP9, whose genes are highly expressed in WT cells at CM, activated by both NKX2-5 and TBX5, yet are repressed by NKX2-5 at CP. These results confirm the role of NKX2-5 as a repressor of premature cardiac differentiation (Prall et al., 2007). At the CM stage, although NKX2-5 seems to still promote cell proliferation (CM10) and repress differentiation (CM5,9,11), TBX5 functions as a repressor of proliferative genes (Cluster CM10), and is primarily required for CM differentiation (CM3,5,9,11). We also found clusters with increased or ectopic gene expression in single and double KO cells. For example, cluster CM1 comprised genes with an additive repressor effect of TBX5 and NKX2-5, and was highly enriched for endocardial genes (e.g., Dll4, Edn1, Cdh5, Nrg1, Angpt2, Cd34), and collagen and extracellular matrix genes, suggesting a role for TBX5 and NKX2-5 in repressing alternate cell lineages during cardiac differentiation.
Overall these results highlight an unappreciated complexity of the interaction between Tbx5 and Nkx2-5 during the progression of cardiac differentiation.
Genomic Occupancy of TBX5, NKX2-5 and GATA4 during CM Differentiation
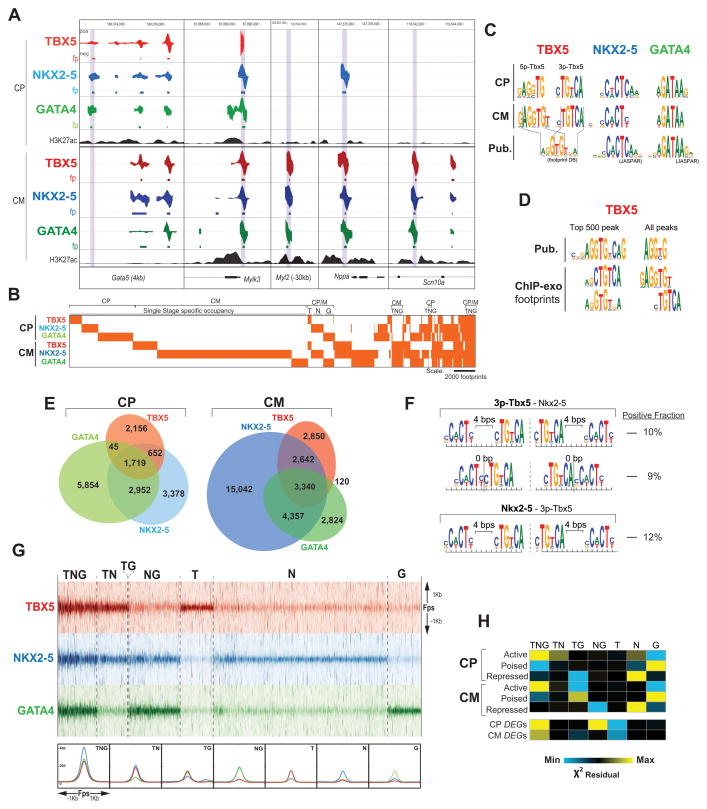
To understand mechanisms underlying the dynamic gene regulation described above, we examined the genomic localization of TBX5, NKX2-5, and GATA4 at CP and CM stages by ChIP-exo (Rhee and Pugh, 2011) (Figure 3A). As negative controls for NKX2-5 and TBX5, we used NKO, TKO, and DKO cells; any footprint (fp) remaining in the KO of the respective factor was discarded. We identified 4,985, 8,718, and 11,000 fps at CP and 8,952, 25,381, and 10,641 fps at CM for TBX5, NKX2-5 and GATA4, respectively. Concordant with dynamics of Tbx5 and Nkx2-5 expression, stage-specific occupancy analysis revealed varied temporal patterns, including single-stage occupancy events and sites occupied in CP and CM (Figure 3B). Taking advantage of the high resolution of ChIP-exo, we characterized TBX5, NKX2-5 and GATA4 binding at each stage performing de novo motif discovery on the fps for each TF. In both stages, the most significantly enriched motifs in NKX2-5 and GATA4 fps matched previously characterized motifs (Figure 3C). For TBX5, the most enriched motif in both stages has a 6-bp consensus sequence CTGTCA, corresponding to the complementary strand of the MEIS1 motif core (TGACAG), which does not match in vitro-derived 8-bp TBX5 consensus motifs (AGGTGTGA; (Ghosh et al., 2001; Jolma et al., 2013)), but is highly similar to the 3′ end of alternate motifs identified for TBX3, TBX5, and TBX20 (He et al., 2011; Sakabe et al., 2012; Shen et al., 2011; van den Boogaard et al., 2012) (Figure 3C). We also found a second significant TBX5 motif (GAGGTG) that shares 5 bp with the reported 8-bp consensus motifs. We refer this secondary motif as 5p-TBX5 and the primary motif as 3p-TBX5. While 5p-TBX5 and 3p-TBX5 appear in the same fps, they are most often not located directly next to each other. Instead they occur without any consistent pattern of spacing except in the strongest TBX5 fps (Top 500 bound loci) where they appear together as a longer combined motif (He et al., 2011) (Figure 3D) previously considered as a single motif. This new TBX5 motif composition suggests the existence of more complex mechanisms regulating TBX5 occupancy during CM differentiation.
Figure 3. Dynamic DNA Occupancy of TBX5, NKX2-5 and GATA4 during Cardiomyocyte Differentiation.
(A) Visualization of TBX5, NKX2-5 and GATA4 ChIP-exo data at CP and CM in representative loci (positive for H3K27ac at CP and/or CM, Wamstad et al., 2012).
(B) Clustering of TBX5, NKX2-5 and GATA4 footprints at CP and CM stages.
(C) Most enriched binding motifs for TBX5, NKX2-5 and GATA4 at CP and CM compared with published motifs.
(D) Most enriched motifs for TBX5 in the top 500 and the entire set of peaks identified for TBX5 in this report and (He et al., 2011).
(E) Stage specific overlapping of TBX5, NKX2-5 and GATA4 footprints.
(F) Motif spacing/orientation pattern between the instances of the co-occurring motifs. Left: Most represented spacing/orientation when the primary motif corresponds to TBX5 (top) or NKX2-5 (bottom). Right: fractions of motif co-occurrences in either orientation.
(G) Top: Tag heatmap for TBX5, NKX2-5 and GATA4 at ±1 kb the midpoint of each footprint in each specific occupancy group at CM. Bottom: Median signal density of each factor in each specific occupancy group.
(H) Overlap of each occupancy group at CP and CM with (top) cardiac enhancers and (bottom) ± 20kb DEGs. Color represents X2 residuals (yellow indicates significant overlap).
See also Figure S3.
TBX5, NKX2-5 and GATA4 Co-Occupy Genomic Loci during CM Differentiation
Our occupancy analysis revealed dynamic and extensive co-binding of TBX5, NKX2-5 and GATA4 at the CP and CM stages (Figure 3B,E), and motif analysis revealed a statistically significant enrichment of motifs for the other two TFs within fps for TBX5, NKX2-5, and GATA4 (Jolma et al., 2013). Examining heterotypic motif spacing and/or orientation pattern between the instances of the co-occurring motifs, we found consistent spacing of NKX2-5 and TBX5 motifs in fps for both TFs more often than expected by chance. Specifically, 3p-TBX5 – NKX2-5 (3p-TBX5 the primary motif) appears frequently with 4-bp (10% of the cases) or 0-bp (i.e., immediately adjacent, 9%) spacing, and NKX2-5 – 3p-TBX5 (NKX2-5 the primary motif) often occurs with 4-bp spacing (12% of the cases) (Figure 3F- E-values < 10−5) (Jolma et al., 2013). In both cases, we found that the motifs tended to co-occur on the same strand with a head-to-head orientation, but we did not find any bias in ordering. No other TF pairs (GATA4 – TBX5 and GATA4 – NKX2-5) have conserved spacing across fps, supporting the notion of cooperative binding of NKX2-5 and TBX5, and indicating a preferential use of the 3p-TBX5 motif in these instances.
Sites where TBX5, NKX2-5 and GATA4 co-occur (TNG sites, Figure 3G) corresponded with the highest signal. These sites have also the highest degree of overlap with active enhancers (Wamstad et al., 2012) at both stages (Figure 3H), and with proximal regions (± 20 kb) to genes with differential expression (DEGs) upon loss of either factor (Figure 3H). Therefore, shared binding of multiple heterotypic TFs is associated with the most active cardiac enhancers and the most active regulation of target genes. An important question about co-occupancy is whether multiple TFs indeed bind the same piece of DNA. We addressed this question by performing Re-ChIP-exo for TBX5 and NKX2-5 at CM in WT cells (Figure S3A - see experimental procedures). TBX5(+) – Nkx2-5(+) single ChIP-exo fps (TN and TNG sites) overlapping Re-ChIP(+) sites (11.1% of the TBX5 – NKX2-5 sites − 6% of TN and 15.2% of TNG sites-) correspond to the highest ChIP-exo signal intensity for each TF (Figure S3B), and TNG Re-ChIP-exo positive sites have the highest association with DEGs (Figure S3C).
These results reveal the high relevance of sites with the highest probability of co-binding by TBX5, NKX2-5, and GATA4 in the control of gene expression during cardiac differentiation.
Interdependent Genomic Occupancy of TBX5, NKX2-5 and GATA4
The complex gene expression patterns in single and double KOs suggested the existence of complex interrelationships between TBX5, NKX2-5 and GATA4. To understand how the occupancy of one factor functionally relates to the binding of others, we performed ChIP-exo for TBX5, NKX2-5 and GATA4 in NKO, TKO and DKO cells (Figure S4A). We found various behaviors for TBX5 and NKX2-5: independent binding, one TF depending on the other’s presence, and interdependent binding (both require the other’s presence). Similarly, GATA4 binding often was dependent on the presence of TBX5, NKX2-5, or both. Unexpectedly, we observed significant ectopic binding of the remaining TFs when the other was ablated (e.g., TBX5 ectopic binding at the Corin and Tbx3 loci in NKO cells; Figure S4A).
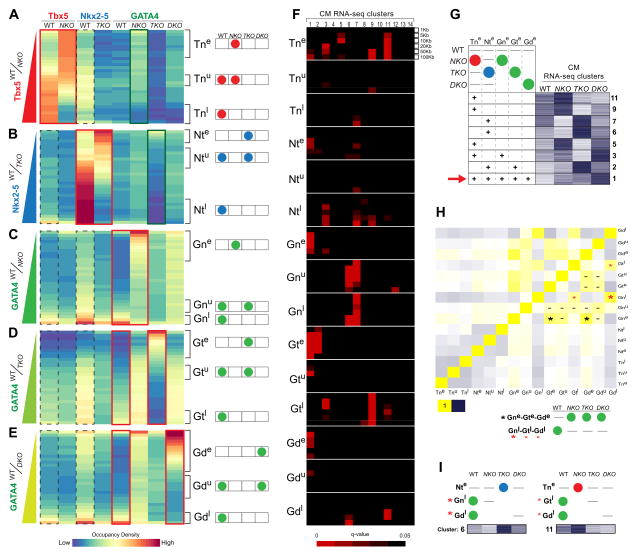
To better classify these behaviors, and interrogate TF binding relationships, TF fps were clustered according to the WT/KO signal ratio (Figure 4A, TBX5 WT/NKO; B, NKX2-5 WT/TKO; C, GATA4 WT/NKO; D, GATA4 WT/TKO; E, GATA4 WT/DKO; red frames), plotting over them the occupancy density for each partner factor for each genotype (Figure 4A–E). Highest partner WT co-occupancy densities (NKX2-5 for TBX5, Figure 4A; TBX5 for NKX2-5, Figure 4B; TBX5 and NKX2-5 for GATA4, Figure 4C–E, dotted black frames) were found around WT fps, independently of their behavior in KO cells, and the lowest was around the ectopic KO occupancy sites. A similar result was found by plotting the TBX5 and NKX2-5 ChIP-exo signal over fps only found in WT cells, in WT and KO cells, and only in KO cells (NKX2-5 signal over TBX5 fps, Figure S4B; TBX5 signal over NKX2-5 fps, Figure S4C), where TBX5-NKX2-5 overlap was found mostly in WT and WT/KO occupancy sites (Figure S4B,C - dotted lines), but most of the ectopically occupied regions were unoccupied in WT cells by the partner factor.
Figure 4. Interdependent Transcription Factor DNA Occupancy Regulates Gene Expression during Cardiomyocyte Differentiation.
(A–E) TFs occupancy density over TBX5 (A), NKX2-5 (B) and GATA4 (C–E) footprints aligned along the ChIP-exo signal gradient between WT and NKO (A, C), TKO, (B, D) or DKO (E) CMs. Ectopic (e), unaffected (u) or lost (l) occupancy of TBX5 in NKO (Tn), NKX2-5 in TKO (Nt), GATA4 in NKO (Gn), GATA4 in TKO (Gt) or GATA4 in DKO (Gd) CMs was indicated with brackets. Right: Scheme of each TF occupancy pattern.
(F) Overlap of each occupancy pattern within 1, 5, 20, 50 and 100 kb proximal to DEGs of each RNA-seq cluster at CM. Red intensity represents q-value enrichment.
(G) Representation of each ectopic occupancy pattern (Tne, Nte, Gne, Gte, Gde) overlap around RNA-seq Heatmap clusters identified in (F). Red arrow indicates the significant overlap of the 5 ectopic occupancy patterns around cluster CM1.
(H) Occupancy patterns direct correlation. Yellow, positive correlation. Blue, inverse correlation. Best correlation was between Gne-Gte-Gde groups (black asterisks) and between Gnl-Gtl-Gdl groups (red asterisks). Gata4 unaffected occupancy groups significantly overlap with most of the Gata4 occupancy groups (lines).
(I) Most represented occupancy patterns in two representative gene expression clusters (CM6, 11).
See also Figure S4.
Of note, TBX5/NKX2-5 NKO/TKO ectopic occupancy was frequently accompanied by GATA4 (Figure 4A–B-green frames), and in WT cells, highest WT/KO GATA4 signal ratio was associated with highest partner occupancy (Figure 4C–E), highlighting the close partnership existing between GATA4 and TBX5/NKX2-5.
Taken together, these occupancy patterns revealed that 1) lost or preserved partner occupancy in null cells cannot be explained only by the joint/single occupancy of both factors, and 2) ectopic occupancy cannot be explained by direct TBX5 – NKX2-5 – GATA4 competition, leaving the mechanisms underlying these behaviors still unclear.
Interdependent Transcription Factors Genomic Occupancy Is Essential For CM Differentiation
To understand how the different behaviors in TF occupancy affect gene expression, we defined 15 groups of occupied regions: five groups defined by the changes in TBX5 occupancy in NKO cells (Tn), NKX2-5 in TKO cells (Nt), GATA4 in NKO cells (Gn), GATA4 in TKO cells (Gt) and GATA4 in DKO cells (Gd). Each of these groups was subdivided into ectopic occupancy in KO cells (e), unaffected occupancy (u) and loss of occupancy (l) (Figure 4A–E, scheme at right). We analyzed the statistical enrichment of each group in proximal regions (1, 5, 10, 20, 50 and 100 kb) to DEGs identified by RNA-seq (Figures 4F, S4D).
Groups corresponding to ectopic occupancy of TBX5 (Tne), NKX2-5 (Nte) or GATA4 (Gne,Gte and Gde) were significantly represented near DEGs corresponding to clusters CM1,2,3,5,6,7,9,11 (Figure 4F–G, S4D), which are characterized by upregulated gene expression in those genotypes where the ectopic TF occupancy was significantly found (Figure 4G). In particular, the expression cluster characterized for the presence of upregulated genes in all the KO backgrounds (CM1) was enriched for all groups of sites exhibiting ectopic TF binding (Tne, Nte, Gne, Gte, Gde) (Figure 4G-red arrow). These observations strongly suggest that abnormally increased gene expression in KO cardiomyocytes can be explained in multiple loci by ectopic TF occupancy and therefore, highlight the importance of understanding the mechanisms preventing this ectopic occupancy.
Interestingly, the three different groups for GATA4 ectopic occupancy (Gne, Gte and Gde) showed the highest direct correlation (GATA4 ectopic occupancy in the same location in different genotypes; Figure 4H-black asterisks). This implies the existence of multiple sites where GATA4 ectopic occupancy proximal to DEGs is suppressed in the presence of NKX2-5 and TBX5. The complexity of this functional interdependency is confirmed by the significant correlation between the groups where GATA4 binding was lost in single or double KO cells (Figure 4H-red asterisks) and found significantly enriched around DEGs. These results pointed to complex interdependence between TFs. Clear examples for this complexity are clusters 6 and 11 (Figure 4I), where opposite behaviors in different TFs occupancy in different KO backgrounds result in opposite changes in gene expression in each genotype.
Among CM RNA-seq clusters, only CM4, 10, 12, 13 and 14 showed no enrichment in any group of occupancy sites (Figure S4D). GO analysis of these clusters did not show enrichment in specific processes of cardiomyocyte differentiation (Figure 2C and Table S2) indicating that we are able to distinguish between those genes (and processes) where TBX5, NKX2-5 and GATA4 act directly or indirectly.
Altogether these data suggest complex interplay between TBX5, NKX2-5, and GATA4, where phenotypes associated with absence of TBX5/NKX2-5 are not only due the lack of the mutated TF, or the concomitant loss of cooperative binding of partner TFs, but also to the redistribution of other TFs across the genome.
TBX5–NKX2-5 Interdependent Occupancy
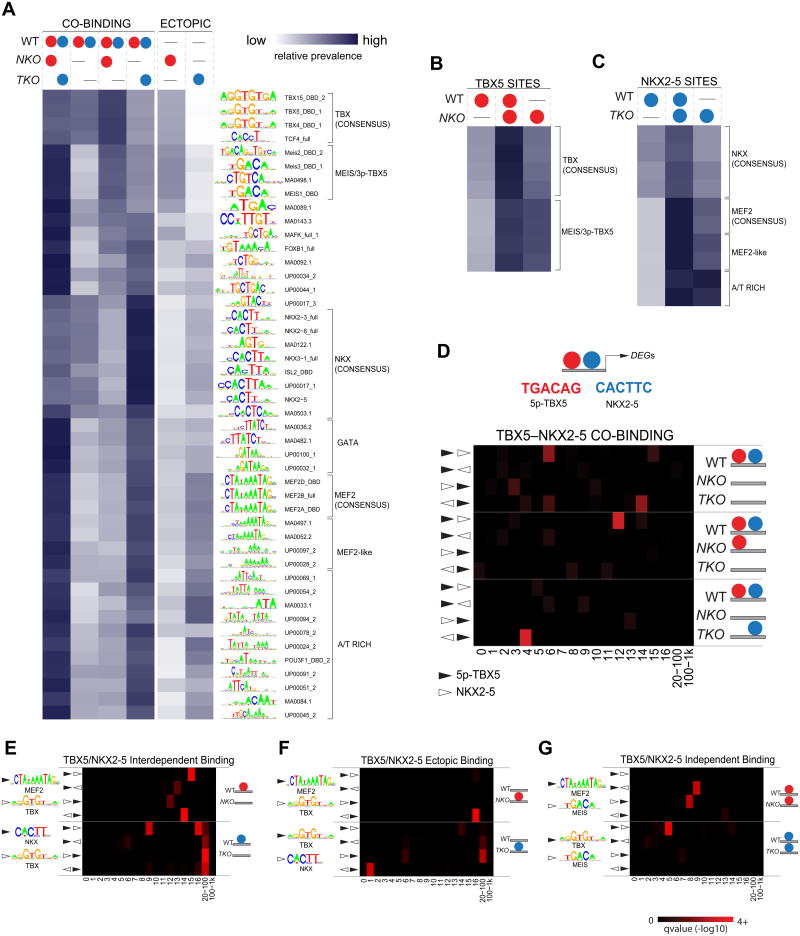
To understand the mechanisms regulating TFs co-binding, interdependence and ectopic TF occupancy, we attempted to uncover a potential specific binding motif arrangement responsible for the various TF interrelationships, by analyzing the relative prevalence of binding motifs. For co-occurring TBX5 and NKX2-5 sites, we analyzed independently the different behaviors associated with single KO contexts (Figure 5A, 1st – 4th columns). Independent TBX5–NKX2-5 sites had a broad range of represented motifs, although with less prevalence of consensus NKX (CACTT core) or TBX (GGTGTGA) motifs, and high occurrence of GATA, MEF2 or MEIS (5p-TBX5) recognition sequences (Figure 5A, 1st column). This suggests that although TBX5 and NKX2-5 consensus motifs are present at some of these sites, TBX5–NKX2-5 independent co-occupancy might depend on interactions with third partners. The opposite pattern was found in interdependent co-binding sites, with each factor’s consensus being enriched, suggesting that even in presence of these strong binding motifs, the binding of one factor relies on the presence of the other (Figure 5A, 2nd column). This observation was supported by the different relative motif composition characterized single-dependency sites: consensus TBX- or NKX motifs were highly overrepresented in sites where NKX2-5 is lost in TKO, but TBX5 was retained in NKO cells (Figure 5A, 3nd column), or in sites where NKX2-5 is retained in TKO but TBX5 is lost in NKO cells (Figure 5A, 4th column), respectively. These results clearly indicate that varied TF co-occupancy behaviors can be explained by potential anchor factors determined by a specific binding motif composition.
Figure 5. Specific Binding Motif Composition Regulates TBX5-NKX2-5 Genome- Wide Distribution.
(A) Heatmap showing the relative prevalence of selected motifs in TBX5 – NKX2-5 co-binding sites where occupancy is unaffected in KO cells (1st column), lost in both (2nd column), lost only in TKO (3rd column) or NKO (4th column) CMs; and sites with ectopic occupancy of TBX5 in NKO (5th column) or NKX2-5 in TKO (6th column) CMs.
(B,C) Relative prevalence of selected motifs in TBX5(B) or NKX2-5(C) sites where occupancy is lost (1st column), unaffected (2nd column) or ectopic (3rd column) in NKO (B) or TKO (C)CMs.
(D) Most represented motifs for co-occurring TBX5 – NKX2-5 and overrepresented distance/orientation for this motif pair in different TBX5 – NKX2-5 co-binding patterns.
(E–G) Overrepresented distance/orientation in specific motif pairs in TBX5/NKX2-5 sites where occupancy is lost (E), ectopic (F) or unaffected (G) in NKO/TKO CMs. Red intensity represents q-value in D–H
Ectopic occupancy sites were characterized by a similar motif composition as single-dependency co-occupancy sites but with opposite relative prevalence (Figure 5A, 3rd and 5th column for TBX5, 4rd and 6th column for NKX2-5). Thus, secondary motifs represented in single-dependency co-occupancy sites, MEIS/5p-TBX5 and MEF2-A/T rich motifs for TBX5 and NKX2-5, respectively (Figure 5A, 3rd and 4th columns), were the most prevalent in TBX5 and NKX2-5 ectopic occupancy (Figure 5A, 5th and 6th columns). This pattern was even more obvious analyzing each factor independently. For TBX5 and NKX2-5 general occupancy (Figure 5B,C), while consensus motifs are highly represented in TF independent sites (Figure 5B,C, middle columns) and equally low prevalent in dependent and ectopic sites (Figure 5B,C, left and right columns), secondary motifs were highly represented in both independent and ectopic sites (Figure 5B,C, middle and right columns). Similar results were obtained comparing GATA4 TBX5–NKX2-5- dependent/independent occupancy (Figure S5A). These observations suggest that secondary motifs are more dependent on the genomic context than consensus motifs. This context dependency becomes more obvious analyzing differences in motif prevalence for ectopic GATA4 occupancy, where we detect different motif enrichments depending on the lack of Tbx5, Nkx2-5 or both (Figure S5B).
Altogether, these results suggest that ectopic TF redistribution relies on the establishment of new motif/partner interactions, interactions that directly depend on the complex behavior of different partners in each TBX5/NKX2-5 KO context. This implies a tightly co-regulated TFs co-binding, where in many sites, the interaction between different TF prevents them, direct or indirectly, from redistributing to less favored genomic interactions and inducing ectopic gene activation. This mechanism is likely to be relevant to many heterotypic TF interactions.
Binding motif conformation regulates Heterotypic TFs Occupancy
Complex TF interactions seem to rely on a specific binding motif composition. To identify specific motif arrangements that might regulate the establishment of TF interactions, we performed a spacing/orientation/order analysis of the most represented pair of motifs for each factor (3p-TBX5 and NKX2-5 consensus motif) in TBX5–NKX2-5 co-occurrence groups (Figure 5D). In contrast to interdependent co-binding sites, where varied spacing/orientations were allowed, single-dependency TBX5–NKX2-5 co-binding sites showed, respectively, a very specific enriched configuration of this motif-pair. Thus, a 12bp gap /5’-5’ orientation was found in TBX5-dependent NKX2-5-independent sites, and a 4bp gap /3’–5’ in NKX2-5-dependent TBX5-independent sites (Figure 5D). These findings strongly suggest the existence of a preferential motif distribution facilitating heterotypic TF interactions for specific contexts and interdependent behaviors.
We extended our spacing/orientation analysis to the general occupancy patterns for each factor. Amongst others, specific arrangements for consensus TBX, NKX, MEF2 and MEIS pairs were identified associated with different TBX5/NKX2-5 dependent or independent occupancy patterns, but also with ectopic TBX5/NKX2-5 sites (Figure 5E–G, S6).
Altogether, these results support the existence of specific permissive binding motif pair distributions allowing heterotypic TF interactions. The same analysis was performed distinguishing between sites proximal (20kb) to DEGs, distal to DEGs or the entire set of occupancy sites, obtaining the same statistical enrichments for the same motif pairs. This result showed the strength of this TF-occupancy regulatory mechanism.
Crystal Structure of NKX2-5/TBX5 Fusion Protein Bound to the Nppa Promoter Region
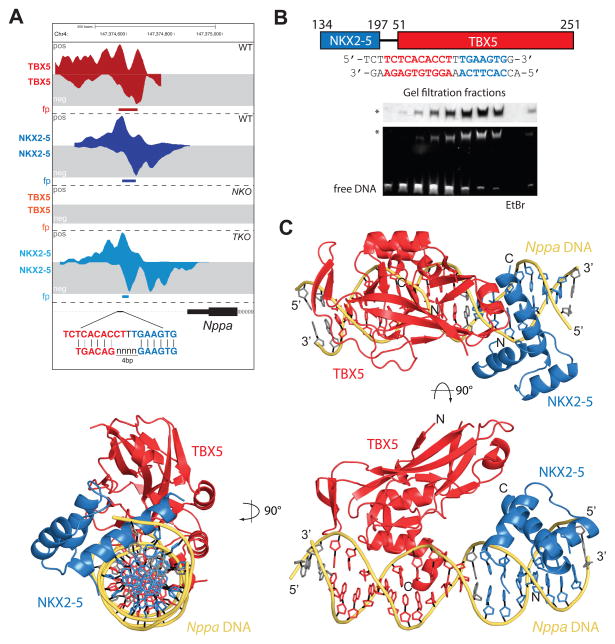
A good example of TF interactions allowed by specific binding motif arrangement is the proximal promoter of Nppa, that consistent with previous gain-of-function studies that demonstrated co-activation of this regulatory element (Bruneau et al., 2001; Hiroi et al., 2001), is co-occupied by TBX5 and NKX2-5 (Figure 3A, 6A). TBX5 binding at this site depends on NKX2-5, showing cooperative binding, and has the most preferential motif arrangement defined for loci with this interdependent behavior (Figure 6A, S3). We structurally characterized the ternary complex of TBX5 and NKX2-5 on the Nppa promoter by X-ray crystallography. After unsuccessful initial trials to crystallize the TBX5 T-box domain (TBD; amino acids (aa) 51–251) and the NKX2-5 homeodomain (HD; aa 134–197) bound to the Nppa promoter region (positions −252 to −234), we engineered a fusion protein between NKX2-5HD and TBX5TBD with a poly-serine linker (Figure 6B). The NKX2-5(aa134-197)15SerTBX5(aa51-251) construct (NKX-TBXlinked) expressed well in E. coli, was purified to homogeneity and stably associated with DNA (Figure 6B). We obtained crystals of the TBX5-NKX2-5-DNA complex fractions by vapor diffusion that diffracted to 3.0Å resolution. The crystal structure was solved by molecular replacement with the known individual crystal structures and refined to Rwork/Rfree values of 19.0/24.1% (Table S3).
Figure 6. Crystal Structure of NKX2-5 and TBX5 Bound to the Nppa Promoter.
(A) Visualization of TBX5 and NKX2-5 occupancy at the Nppa promoter in WT, NKO and TKO cells at CM stage. Comparison between most represented motifs for TBX5–NKX2-5 co-binding sites and motifs identified for TBX5 and NKX2-5 at this region
(B) Schematic overview of NKX-TBXlinked indicating residue numbers of the individual domains and the linker region (top). Sequence of the DNA fragment used for co-crystallization indicating the respective recognition motifs of NKX2-5 (blue) and TBX5 (red). Native gel analyses of protein (Coomassie) and DNA (EtBr) in the eluted size exclusion chromatography fractions. TBX5-NKX2-5-DNA complex is indicated by an asterisk
(C) Structure of NKX-TBXlinked bound to the Nppa promoter region in cartoon representation in three orthogonal views. TBX5TBD is shown in red, NKX2-5HD is shown in blue and DNA is shown in yellow/grey. Orientation and location of respective N-/C-termini and 5′/3′ ends are indicated.
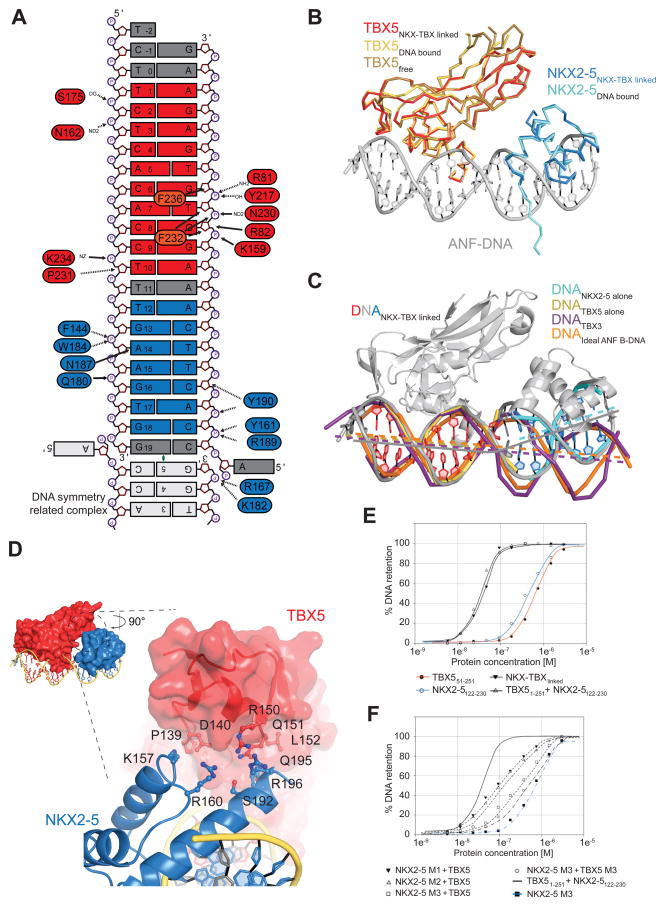
The structure shows that TBX5 and NKX2-5 specifically bind to their respective recognition sequences in the Nppa promoter, which harbors both sites in succession (Figure 6C). The linker is not visible in our electron density: it is flexible and does not influence the positioning of the individual proteins. We superimposed individual DNA binding regions of TBX5 and NKX2-5 with known crystal structures of the individual proteins bound to DNA (Pradhan et al., 2012; Stirnimann et al., 2010) and could not observe changes in the orientation of the residues involved in DNA binding (Figure S7A–B). In detail, α-helix 3 of TBX5TBD (aa 232–237) is structured and specific residues make direct contacts with the DNA bases (F236, F232). For NKX2-5, we also found almost identical interactions with residues F144, Q180, K182, W184, Q186, N187, R189, Y190 contacting the DNA (Figure 7A). Some of the contacts in the high-resolution structures of the binary TBX5-DNA and NKX2-5-DNA complexes (Pradhan et al., 2012; Stirnimann et al., 2010) are mediated by water molecules that cannot be reliably positioned at the resolution obtained for the TBX5-NKX2-5-DNA complex. Nonetheless, the orientations of the individual side chains of the residues involved in these water-mediated contacts are almost identical to their orientations in high-resolution structures. We therefore conclude that in the ternary complex the protein-nucleic acid interactions of TBX5 and NKX2-5, with their respective DNA recognition sequences, are identical to the previously determined binary complex structures (Pradhan et al., 2012; Stirnimann et al., 2010). Although we do not observe any global structural rearrangements of the TBX5 and NKX2-5 proteins in comparison to their individual DNA bound species or unbound TBX5 (Figure 7B), the overall conformation of the Nppa promoter DNA significantly deviates from idealized B-DNA or DNA bound to a Tbx3 dimer (Figure 7C). In the ternary complex, the Nppa promoter DNA shows strong bending that might be induced by a small (~150 Å2) and previously unknown protein interaction surface between the TBX5 and NKX2-5 DNA binding domains of the two TFs (Figure 7D). These findings strongly support our observation of specific binding motif distribution allowing stable TF interactions.
Figure 7. Detailed Analysis of the Interactions between NKX2-5 and TBX5 on Nppa DNA.
(A) Schematic summary of protein-DNA interactions. Only direct contacts between protein and DNA molecules are labeled as no water molecules have been built.
(B) Structures of TBX5TBD bound to DNA (yellow, PDB ID 2X6V) and unbound TBX5TBD (brown; PDB ID 2X6U) superimposed on TBX5TBD from NKX-TBXlinked (red) are shown in ribbon representation. The structure of NKX2-5HD bound to DNA (cyan; PDB ID 3RHQ) superimposed on NKX2-5HD from NKX-TBXlinked are shown in ribbon representation. DNA from the NKX-TBXlinked is shown in cartoon representation (grey).
(C) Structural comparison between DNA molecules from NKX-TBXlinked (red/grey/blue) and individually bound TBX5 (yellow), NKX2-5 (cyan), TBX3 (purple; PDB ID 1H6F), and idealized B-DNA of the Nppa promoter (orange). Lines indicate the central tracks of each individual DNA molecule.
(D) Close-up view of the interface between NKX2-5HD (blue, cartoon) and TBX5TBD (red, surface) highlighting the position of contributing and mutated residues.
(E) Graphs of filter-binding assays using radioactively labeled DNA in combination of different purified proteins (indicated below).
(F) Same as (E) using indicated point mutants. NKX2-5 M1 corresponds to NKX2-5122-230 K157A; NKX2-5 M2 to NKX2-5122-230 Q195A/R196A; NKX2-5 M3 to NKX2-5122-230 K157A/Q195A/R196A; TBX5 M3 to TBX51-251 P139A/D140A/R150A/Q151A.
See also Figure S7.
TBX5 and NKX2-5 Interaction Interface
We identified three pairs of amino acid residues involved in a direct TBX5-NKX2-5 interaction (Figure 7D), and investigated the importance of this newly described interaction on their binding to the Nppa promoter. Two adjacent surfaces in TBX5 have high evolutionary sequence conservation: one patch is conserved among TBX2/3/4/5/6 proteins, and the other is only conserved among TBX5 proteins (Figure S7C). The involved residues in TBX5 reside close to helix α2 and the residues in NKX2-5HD are located at its highly conserved C-terminus. We used a mutational approach to assess the contribution of the interaction itself and individual residues to the binding of NKX2-5 and TBX5 to the Nppa promoter site. Using a filter-binding assay we show that TBX5TBD and NKX2-5HD proteins separately bind the Nppa promoter with apparent KD values of ~500 nM, while together TBX5TBD and NKX2-5HD bind the Nppa promoter with one order of magnitude enhanced affinity (apparent KD ~50 nM). The sigmoidal binding curve suggests cooperative binding. The fusion construct NKX-TBXlinked shows the same enhanced binding affinity as the combination of TBX5TBD and NKX2-5HD demonstrating no effect of covalently linking the two factors (Figure 7E, top panel). Mutating individual residues in the TBX5-NKX2-5 interface into alanine (K157, Q195, R196 in NKX2-5 and P139, D140, R150, Q151 in TBX5) reduced the binding of NKX2-5HD and TBX5TBD to the Nppa promoter, while combining mutations of several residues further enhanced this effect (Figures 7E, bottom panel, S7D).
These results suggest that the newly observed TBX5-NKX2-5 interface greatly contributes to the cooperative binding of both factors to the Nppa promoter. We therefore provide a possible mechanism for the specific interplay between TBX5 and NKX2-5 on this region, and likely for other interdependent TBX5 and NKX2-5 binding elements.
DISCUSSION
We defined the genomic and structural basis for dynamic control of gene expression during cardiomyocyte differentiation by the interdependent heterotypic TFs TBX5 and NKX2-5. Although many studies have reported the capacity of these (and other) TFs to cooperate in the control of gene expression, our results showed that this phenomenon occurs in vivo, with significant more complexity and fine-tuning than anticipated. We find that heart development and cardiac gene expression necessitate tight integration between TBX5, NKX2-5, and, among others, GATA4. This integration directly depends on TF binding motif arrangement, which facilitates physical interactions between TFs.
The combined loss of TBX5 and NKX2-5 in vivo shows that their shared regulatory programs are essential for critical aspects of heart development, including expansion of cardiac progenitors and important elements of the cardiac differentiation program. This genetic interaction has been proposed to be at the root of human congenital heart defects (Bruneau, 2008; Bruneau et al., 2001; Hiroi et al., 2001), and our results support this genetic link, also demonstrating the implication of additional partners in this interaction. A complex but defined motif arrangement dictates the interrelationships between these TFs, resulting in a gene regulatory network deployed during the early stages of heart development to ensure robust tissue-specific gene expression.
In accordance with their dynamic regulation of gene expression, genomic occupancy of TBX5, NKX2-5 and GATA4 follows combinatorial rules that neatly predict their regulatory consequences. Occupancy data from He et al. (2011) for TBX5, NKX2-5, and GATA4 do not agree with the results presented here, perhaps due to their use of a highly divergent transformed atrial cell line and ectopic binding events from overexpression. Nonetheless, a common theme is the correlated binding of the three TFs with active cardiac enhancers. Importantly, we identify a strong relationship between differentially expressed genes and altered TF binding, in the absence of TBX5, NKX2-5, or both. Our results establish the existence of broadly relevant cooperative interactions between heterotypic TFs essential in defining the cardiac differentiation program.
We uncovered a motif logic relying on the different interdependent relationships between both factors. Besides the recognition of specific binding motifs, TF occupancy depends in high degree on the interaction with other TFs, giving rise to TFs complexes where strength of interaction is dictated by the combination of TF-TF and TF-DNA affinities. We have also observed permissive binding motif configurations, highlighting the role of DNA as an instructive partner for pairs of heterotypic TFs (Jolma et al., 2015). Our structural studies proved a biophysical basis for these interactions, and additionally suggest important TF-TF protein interactions away from the DNA-binding interface. Studies in Drosophila of TBX5, NKX2-5 and GATA4 orthologous genes suggest that TF-co-binding could be mediated by motifs without specific orientation/order (Junion et al., 2012). The motif logic that we observe, correlating with shared binding behavior and gene regulation, suggests a complex but well-defined TF binding site arrangement that regulates mammalian cardiac gene expression.
Heterotypic TFs complexes also prevent the establishment of less favored interactions based on the stabilization of new complexes by DNA sequence-recognition and/or physical TFs interaction. The loss of one or more factors unbalances optimal co-activation of target loci and results in concomitant redistribution to less favored genomic interactions and ectopic gene expression. Thus, we propose a dual role of co-binding where interactions between anchor TFs and their partners serve not only to induce gene expression, but also to maintain partner TF localization away from undesired genomic locations. Maintenance of this equilibrium appears to be a crucial mechanism for controlling gene expression and cell fate acquisition.
In summary, we showed that TBX5 and NKX2-5 are extraordinarily interdependent for cardiac gene regulation, have unveiled motif grammar-based rules for their genomic interactions, and defined at an atomic level the biophysical basis for cooperative gene regulation by these important heterotypic TFs. We have discovered a new mode of gene regulation, in which cooperative binding of TFs serves not only to active target genes, but because of TF interdependence and specific motif distribution keeps each factor away from binding to ectopic loci, which when unchecked results in inappropriate gene activation. This paradigm of gene regulatory network control is likely to be applicable to many heterotypic TF interactions that result in tissue-specific gene expression.
EXPERIMENTAL PROCEDURES
Transgenic ES cells
Transgenic mouse lines were previously described (Bruneau et al., 2001; Mori et al., 2006; Tanaka et al., 1999). ES cells were derived from mouse intercrosses following standard methodology (Nagy et al., 2003). Tbx5flox/flox;Nkx2-5+/+ and Tbx5flox/flox;Nkx2-5lacZ/lacZ lines were transiently transfected with a Cre-expressing construct to generate sublines: Tbx5flox/flox;Nkx2-5+/+ (WT), Tbx5flox/flox;Nkx2-5lacZ/lacZ (NKO), Tbx5del/del;Nkx2-5+/+ (TKO), and Tbx5del/del;Nkx2-5lacZ/lacZ (DKO) (Figure S1A). Directed differentiation into cardiomyocytes was performed according to published methods (Kattman et al., 2011; Wamstad et al., 2012). Tetraploid or morula aggregation were used to generate ES cell-derived embryos (Nagy et al 2003).
Analyses of gene expression
Affymetrix mouse Gene ST 1.0 arrays were hybridized and scanned according to the manufacturer's recommendations. Linear models were fitted for each gene comparisons of paired genotypes to derive the mutant effect using the limma package in R/Bioconductor. For RNA-seq, total RNA was isolated from 3x106 cells using TRizol Reagent. RNA-seq libraries were prepared according to life technologies Ion Total RNA-seq Kitv2 and paired-end 100bp sequenced on an Illumina HiSeq 2500 instrument.
Chromatin Immunoprecipitation
ChIP-exo was performed according to published methods (Boyer et al., 2005; Serandour et al., 2013; Wamstad et al., 2012), using specific antisera to TBX5 (sc-17866 XS, lot # B1213), NKX2-5 (sc-8697 XS lot # B1213), and GATA4 (sc-1237 XS lot # B1213) on chromatin isolated from 107 cells. Re-ChIP-exo was performed combining published methods (Serandour et al., 2013; Shankaranarayanan et al., 2011) from 4X107 cells.
Structure determination
A fusion protein (NKX-TBXlinked) was produced that included NKX2-5 homeodomain (aa134-197), TBX5 T-box domain (aa51-251) and a 15aa long poly-serine linker between the NKX2-5 C-terminus and the TBX5 N-terminus. Crystals were obtained using the vapor diffusion method; diffraction data to 3.0 Å were collected at ID14-1 at ESRF Grenoble. The structure was solved using several iterative molecular replacement and manual rebuilding steps.
Supplementary Material
Acknowledgments
We thank S. Izumo for providing Nkx2-5lacZ/+ mice. We are grateful to P. Swinton (Gladstone Transgenic Core) for morula injections. We also thank L. Ta and R. Chadwick (Gladstone Genomics Core), A. Williams (Gladstone Bioinformatics Core), and C. Miller (Gladstone Histology Core), D. Hawkins (Gladstone Summer Scholars Program) and G. Capitani (Paul Scherrer Institut, Switzerland) for technical support, and G. Howard for editorial assistance. This work was funded by NIH/NHLBI P01 HL089707 to B.G.B.; NHLBI Bench to Bassinet Program U01HL098179/UM1HL098179 to B.G.B. and K.S.P.; the California Institutes for Regenerative Medicine (RN2-00903, to B.G.B.); and the Lawrence J. and Florence A. DeGeorge Charitable Trust/American Heart Association Established Investigator Award (to B.G.B). L.L-Z was supported by a California Institutes for Regenerative Medicine Fellowship. C.U.S. is grateful for the financial support by the Marie Curie framework program 7 and the European Molecular Biology Organization long-term fellowship program. This work was also supported by an NIH/NCRR grant (C06 RR018928) to the J. David Gladstone Institutes and by William H. Younger, Jr. (B.G.B.).
Footnotes
AUTHOR CONTRIBUTIONS
L.L-Z. generated most of the experimental data, performed most of the analyses, and wrote the manuscript. C.U.S obtained crystals and solved the crystal structure. C.U.S. and S.G. refined and analyzed the crystal structure. E.M.S., B.L.K., and M.M. isolated ES cells under supervision of A.N. B.L.K. obtained ES cell derived embryos, did in situ hybridizations and extracted embryonic mRNA for microarray analysis. S.T. did computational analyses. F.B. did filter binding assays. M.A.H.S. participated in motif analysis. D.H. participated in ChIP experiments. K.S.P. supervised computational analyses, with participation of A.K.H. C.W.M. supervised structural and biophysical analyses. B.G.B. imaged embryos, supervised and coordinated the project, and wrote the manuscript. All authors participated in the writing of the manuscript.
ACCESSION NUMBERS
ChIP-exo and RNA-seq data are available in experiments 353R (RNA-seq) and 189R1(ChIP-exo) at https://b2b.hci.utah.edu/gnomex/. The GEO/SRA series entry is GSE72223 for the Microarray data and SRP062699 for ChIP-exo and RNA-seq. The atomic coordinates and structure factors of the NKX2-5-TBX5-Nppa DNA complex have been deposited with the European Protein Data Bank (PDBe) under accession code 5FLV.
Supplemental Information includes Extended Experimental Procedures, seven figures, and three tables that can be found with this article online at -
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al. Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell. 2005 doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner D, Gessler M, Nemer M, Seidman CE, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, et al. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. Embo J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Chen CY, Ardati A, Schwartz RJ, Nemer M. The atrial natriuretic factor promoter is a downstream target for Nkx- 2.5 in the myocardium. Mol Cell Biol. 1996;16:4648–4655. doi: 10.1128/mcb.16.9.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Ghosh TK, Packham EA, Bonser AJ, Robinson TE, Cross SJ, Brook JD. Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Hum Mol Genet. 2001;10:1983–1994. doi: 10.1093/hmg/10.18.1983. [DOI] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015 doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EE. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148:473–486. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-Specific Optimization of Activin/Nodal and BMP Signaling Promotes Cardiac Differentiation of Mouse and Human Pluripotent Stem Cell Lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Levo M, Segal E. In pursuit of design principles of regulatory sequences. Nat Rev Genet. 2014;15:453–468. doi: 10.1038/nrg3684. [DOI] [PubMed] [Google Scholar]
- Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D, Garg V. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori AD, Zhu Y, Vahora I, Koshiba-Takeuchi K, Davidson L, Pizard A, Seidman JG, Seidman CE, Chen XJ, Henkelman RM, et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev Biol. 2006;297:566–586. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, et al. A molecular pathway including id2, tbx5, and nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. A Laboratory Manual. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. Manipulating the Mouse Embryo. [Google Scholar]
- Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, Evans SM, Clark B, Feramisco JR, Giles W, et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- Pradhan L, Genis C, Scone P, Weinberg EO, Kasahara H, Nam HJ. Crystal structure of the human NKX2.5 homeodomain in complex with DNA target. Biochemistry. 2012;51:6312–6319. doi: 10.1021/bi300849c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prall OW, Menon MK, Sollaway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls second heart field progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012 doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe NJ, Aneas I, Shen T, Shokri L, Park SY, Bulyk ML, Evans SM, Nobrega MA. Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Hum Mol Genet. 2012;21:2194–2204. doi: 10.1093/hmg/dds034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron B, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Serandour AA, Brown GD, Cohen JD, Carroll JS. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol. 2013;14:R147. doi: 10.1186/gb-2013-14-12-r147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaranarayanan P, Mendoza-Parra MA, Walia M, Wang L, Li N, Trindade LM, Gronemeyer H. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nature methods. 2011;8:565–567. doi: 10.1038/nmeth.1626. [DOI] [PubMed] [Google Scholar]
- Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, Smemo S, Westlund JM, Cheng H, Dalton N, Gu Y, et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121:4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, Poulsen LL, Rogowska-Wrzesinska A, Jensen ON, Mandrup S. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell reports. 2014;7:1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- Song K, Nam Y, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Stefflova K, Thybert D, Wilson MD, Streeter I, Aleksic J, Karagianni P, Brazma A, Adams DJ, Talianidis I, Marioni JC, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013;154:530–540. doi: 10.1016/j.cell.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnimann CU, Ptchelkine D, Grimm C, Muller CW. Structural basis of TBX5-DNA recognition: the T-box domain in its DNA-bound and -unbound form. Journal of molecular biology. 2010;400:71–81. doi: 10.1016/j.jmb.2010.04.052. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova M, Yamazaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Tijssen MR, Cvejic A, Joshi A, Hannah RL, Ferreira R, Forrai A, Bellissimo DC, Oram SH, Smethurst PA, Wilson NK, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, Gnirke A, Meissner A. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518:344–349. doi: 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boogaard M, Wong LY, Tessadori F, Bakker ML, Dreizehnter LK, Wakker V, Bezzina CR, t Hoen PA, Bakkers J, Barnett P, et al. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest. 2012;122:2519–2530. doi: 10.1172/JCI62613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell stem cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.