Summary
Pseudomonas aeruginosa is an important bacterial opportunistic pathogen, presenting a significant threat towards individuals with underlying diseases such as cystic fibrosis. The transcription factor AmrZ regulates expression of multiple P. aeruginosa virulence factors. AmrZ belongs to the ribbon-helix-helix protein superfamily, in which many members function as dimers, yet others form higher-order oligomers. In this study, four independent approaches were undertaken and demonstrated that the primary AmrZ form in solution is tetrameric. Deletion of the AmrZ C-terminal domain leads to loss of tetramerization and reduced DNA binding to both activated and repressed target promoters. Additionally, the C-terminal domain is essential for efficient AmrZ-mediated activation and repression of its targets.
Keywords: bacteria, biofilm biology, gene expression/regulation, microbial genetics, microbial communities
Introduction
The Gram-negative bacterium Pseudomonas aeruginosa is ubiquitous. It can be widely found in fresh water and soil, and as an opportunistic pathogen, also imposes a threat to immunocompromised humans. P. aeruginosa versatility is achieved by rapidly adapting to new environments, in which multiple levels of regulation are involved. A large pool of transcription factors participate in this regulation, and one such factor AmrZ (Alginate and Motility Regulator Z) functions as a global activator and repressor. Well-studied AmrZ targets include those involved in exopolysaccharide production (alginate, Psl and Pel), flagella, twitching motility, and metabolism of the second messenger bis-(3’, 5’) cyclic di-guanylate (c-di-GMP) (Baynham and Wozniak, 1996; Ramsey et al., 2005; Baynham et al., 2006; Tart et al., 2006; Jones et al., 2013, 2014; Martínez-Granero et al., 2014). For instance, AmrZ is necessary for transcription of the alginate biosynthesis operon, the promoter of which is designated as PalgD since algD is the first gene of this operon. Alginate is responsible for the mucoid phenotype and contributes to increased resistance to antimicrobials and host immunity (Gacesa, 1998; Baynham et al., 1999; Ramsey and Wozniak, 2005). AmrZ also represses transcription of gcbA (also referred to as adcA or PA4843), which encodes a diguanylate cyclase enzyme that synthesizes c-di-GMP (Jones et al., 2014; Petrova et al., 2014). Deletion of gcbA results in increased motility and reduced biofilm production (Jones et al., 2014; Petrova et al., 2014). The ΔamrZ mutant displays increased levels of Psl and c-di-GMP, and therefore forms enhanced biofilms compared to the wild type strain (Jones et al., 2013, 2014).
The 12.34 kDa protein AmrZ is composed of three segments: a flexible N terminus (residues 1911), a ribbon-helix-helix domain (residues 12-66), and a C-terminal domain (CTD, residues 67-108) (Figure 1A). The ribbon-helix-helix domain mediates DNA binding and its structure is highly similar to other members, such as Arc, Mnt and MetJ, in the Arc superfamily (Vershons et al., 1985; Knight et al., 1989; Schreiter and Drennan, 2007; Waligora et al., 2010). All Arc superfamily proteins are phage or bacterial transcription factors, which bind to DNA via N-terminal ribbon-helix-helix domains. C-terminal domains of different Arc proteins however, mediate diverse functions including ligand binding and oligomerization (Schreiter and Drennan, 2007). Arc proteins form dimers or higher order oligomers, which are often necessary for their functions (Waldburger and Sauer, 1995; Schreiter and Drennan, 2007). In our previous studies, glutaraldehyde cross-linking experiments suggested that cross-linked AmrZ forms oligomers independent of its N terminus (Waligora et al., 2010). However, it remains unknown which AmrZ domain(s) mediates oligomerization, or what roles this may play during AmrZ-mediated activation or repression.
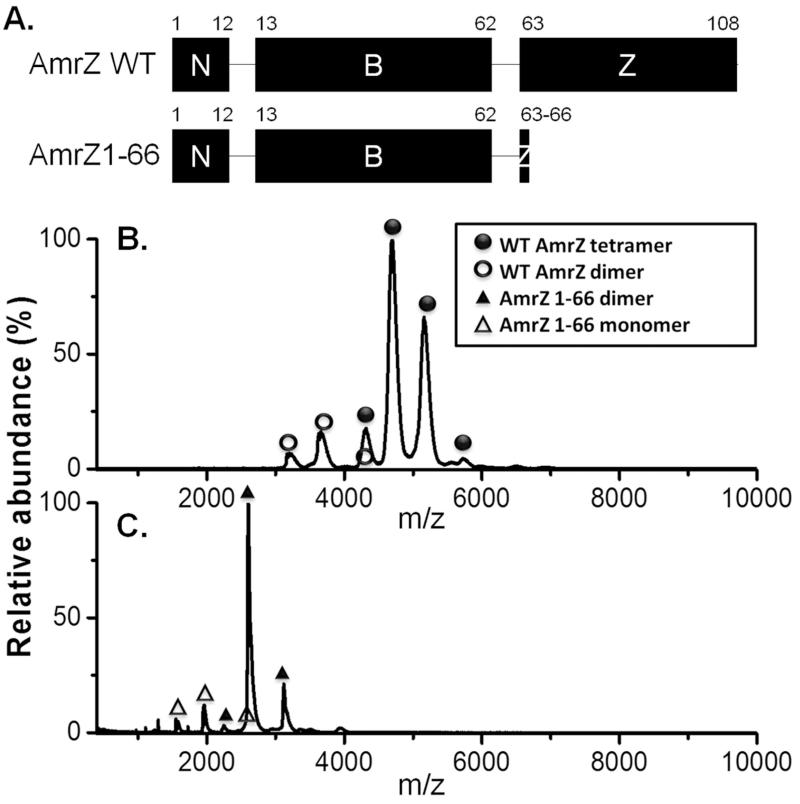
Figure 1. The C-terminal domain of AmrZ mediates tetramerization.
(A.) Proposed domains of full length AmrZ and AmrZ1-66. N: N terminus; B: DNA binding domain; Z: oligomerization domain. Numbers above indicate residues flanking each domain. (B.) & (C.) Nano-ESI MS measurements of purified AmrZ (B.) and AmrZ1-66 (C.). Purified proteins were buffered exchanged into 500 mM ammonium acetate and analyzed by nano-ESI MS. The X-axis represents mass/charge (m/z) ratios, while the Y-axis was calculated by normalizing to the highest signal intensity. A certain protein/complex may have different charges, resulting in a series of peaks in MS. This information was used to calculate molecular masses of different AmrZ oligomeric species. Electrospray ionization produces charged droplets, and desorption and/or desolvation helps formation of gas phase ions. Proteins have multiple protonation sites and will be protonated with different charges as solvent evaporates. Therefore, in ESI MS, protein analytes usually have a distribution of charge states. Molecular weights (MW) of proteins are calculated based on the equation: m/z = (MW + nH+)/n, and the average of all charge states is considered the MW of a specific protein.
In this report, results from multiple biochemical and genetic approaches reveal that AmrZ exists primarily as tetramers in solution. Oligomerization requires an intact AmrZ CTD, the loss of which leads to reduced DNA binding as well as a diminished ability of AmrZ to mediate activation and repression.
Results and Discussion
AmrZ forms tetramers in solution via its C-terminal domain
In the Arc superfamily, the basic functional unit is dimeric; however, proteins such as Mnt and TrwA form tetramers and even higher-order oligomers when bound to DNA (Schreiter and Drennan, 2007). In this study nano-electrospray ionization mass spectrometry (nano-ESI MS) was used to determine the AmrZ oligomeric state in solution. Mass spectrometry precisely measures the size of a molecule and nano-ESI MS allows the detection of proteins or protein complexes when they are still folded (Karas et al., 2000; Ma et al., 2014). As shown in Figure 1B, most peaks correspond to an AmrZ complex with the size of approximately 51.0 kDa. Due to tag cleavage following purification, recombinant AmrZ contains three extra residues, (Gly-Pro-His), resulting in its monomeric molecular weight as 12.63 kDa (Pryor et al., 2012). The AmrZ complex seen in MS therefore reflects an AmrZ tetramer. AmrZ tetramerization was confirmed by size exclusion chromatography (SEC) (Figure S1), a dominant negative in vivo genetic test (Figure S2), and a glutaraldehyde crosslinking experiment (Figure S4).
We next determined which part of AmrZ mediates oligomerization. Previous work showed that removal of N-terminal residues 1-11 does not prevent AmrZ oligomerization, and the ribbon-helix-helix domain (residues 12-66) is responsible for DNA binding and dimerization (Figure 1A) (Waligora et al., 2010; Pryor et al., 2012). Specifically, AmrZ1-66 was used in determining the crystal structure of DNA-bound AmrZ, and this truncation variant interacts with DNA as a dimer of dimers (Pryor et al., 2012). We therefore hypothesized that the AmrZ CTD mediates tetramerization. To test this, we determined the oligomeric state of purified AmrZ in the absence of the CTD (AmrZ1-66). Nano-ESI MS revealed that AmrZ1-66 exists as dimers and monomers, but no tetramer was observed (Figure 1C). This is consistent with data obtained from protein cross-linking experiments (Figure S4). These results collectively showed that the AmrZ CTD mediates tetramerization.
To unveil how the AmrZ CTD contributes to tetramerization, we used multiple tools to predict the secondary structure and properties of this domain. The NCBI conserved domain database failed to identify any conserved domain homologous to this CTD (Marchler-Bauer et al., 2014). The online prediction tools PSIPRED (Figure S5A) and Jpred (B. Xu, unpublished) predicted that most residues in the AmrZ CTD form two α-helices (Jones, 1999; Drozdetskiy et al., 2015). The α-helix, which mediates DNA binding and is essential for transmembrane domain formation, is the most common secondary structure of proteins (Breitwieser, 2004). Many proteins, such as G-protein coupled receptors, form dimers or oligomers via helix-helix packing (Breitwieser, 2004). Therefore, these two α-helices are likely to be responsible for AmrZ oligomerization. Typically a small number of residues, called hot spot residues, are important for successful helix-helix interactions (Bullock et al., 2011). Although hydrophobic and aromatic amino acids constitute the majority of hot spot residues, polar and charged residues such as arginine remain significant players at monomer-monomer interfaces (Bogan and Thorn, 1998; Bullock et al., 2011; Matthews et al., 2012). The ExPASy ProtScale tool (Gasteiger et al., 2005) predicted most residues within the AmrZ CTD domain are polar or hydrophilic (Figure S5B). In addition, overall this domain is hydrophilic, with the grand average of hydropathicity at −0.512 (minus values suggest hydrophilic) (ProtParam; (Kyte and Doolittle, 1982; Gasteiger et al., 2005)). The AmrZ CTD contains five arginine residues, which, together with other polar residues, may contribute to oligomerization through hydrogen bonding and salt bridges (Ali and Imperiali, 2005; Matthews et al., 2012).
Taken together, we used four independent approaches (glutaraldehyde cross-linking, SEC, nano-ESI MS, and genetics) to demonstrate the oligomeric state of native AmrZ as tetrameric, and identified that the CTD mediates tetramerization.
Impact of the AmrZ C-terminal domain on DNA binding, activation and repression
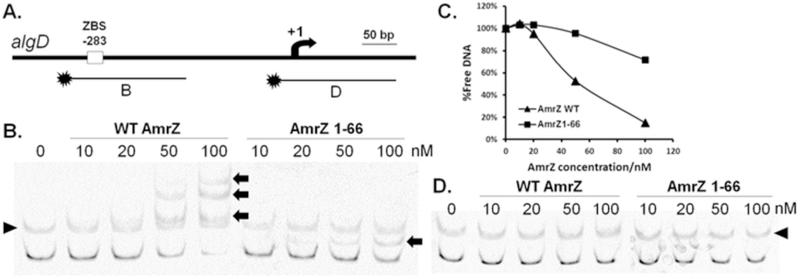
We next sought to determine effects of removing the AmrZ CTD on DNA binding. Purified AmrZ and AmrZ1-66 were tested for DNA binding to promoters of AmrZ targets. A [6FAM]-labeled DNA fragment containing the AmrZ binding site (ZBS) was amplified from its activated target PalgD (Baynham and Wozniak, 1996), and AmrZ binding determined by EMSA (Electrophoretic Mobility Shift Assay) (Figure 2A). Results showed that, compared to full length AmrZ, AmrZ1-66 has significantly reduced binding to PalgD (Figure 2B&C). However, the CTD is not absolutely necessary for DNA binding, since DNA binding activities were seen when higher AmrZ1-66 concentrations were used (Figure 2B&C). Both AmrZ and AmrZ1-66 recognize the DNA target specifically, as no binding was seen when a non-specific DNA was tested in this assay (Figure 2D). Similar results were observed when the repressed target PgcbA was used (Figure S6), indicating this reduced DNA binding of AmrZ1-66 is likely to be conserved for different AmrZ targets.
Figure 2. Truncation of the C-terminal domain results in reduced binding affinity to PalgD.
[6FAM]-labeled DNA fragments (10 nM in each reaction) with (B) or without (D) the ZBS in PalgD were incubated with various concentrations of AmrZ or AmrZ1-66 at room temperature for 20 min and resolved by 4% non-denaturing PAGE on ice (B. & D.). Relative amounts of free DNA in (B.) were quantified via densitometry of non-shift bands using ImageJ (v1.46r) and plotted against AmrZ concentrations (C.). AmrZ monomeric concentrations ranged between 10 nM and 100 nM. The arrow in (A.) represents the PalgD transcription start site. Arrows in (B.) indicate DNA mobility shifts induced by AmrZ or AmrZ1-66 binding. Triangles in (B. & D.) point to non-specific bands, which are also present in no protein lanes.
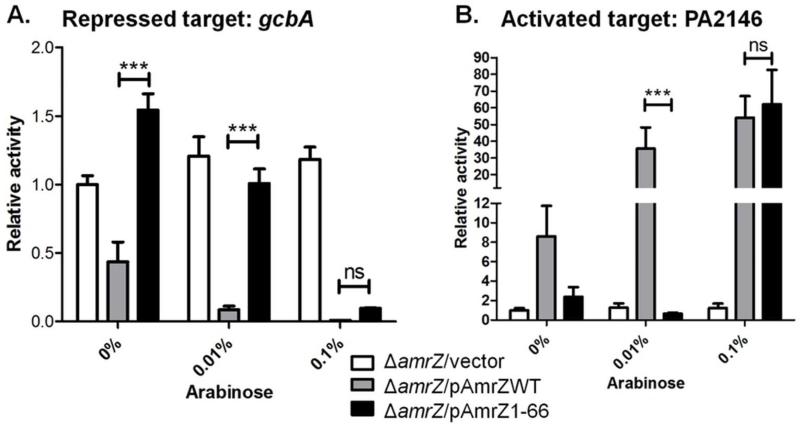
Finally, we sought to investigate the significance of the AmrZ CTD in vivo in the type strain PAO1. In a previous study, we showed that AmrZ activates PA2146 and represses gcbA transcription (Jones et al., 2014). The hypothetical gene PA2146 is 92% similar to Escherichia coli yciG, which is associated with bacterial responses to glucose (Sunya et al., 2012). Moreover, PA2146 expression is repressed when P. aeruginosa is treated with inhibitory compounds such as protoanemonin and azithromycin (Kai et al., 2009; Bobadilla Fazzini et al., 2013). We compared the ability of AmrZ1-66 and AmrZ to regulate these two targets. In PAO1 ΔamrZ with plasmids containing AmrZ or AmrZ1-66, mRNA levels of PA2146 and gcbA were quantified at various arabinose concentrations. Consistent with findings in RNA-Seq (Jones et al., 2014), gcbA transcription remained de-repressed in the PAO1 ΔamrZ strain when the empty vector pHERD20T was introduced. However, even at 0% arabinose, pHERD20T-WT AmrZ (pAmrZWT) was sufficient to restore AmrZ-mediated repression of gcbA, while regulation by AmrZ1-66 was not observed until 0.1% arabinose was used (Figure 3A). Similar results were observed with the activated target PA2146, as it required a much higher amount of AmrZ1-66 for transcription compared to full length AmrZ (Figure 3B). Reduced efficiency by AmrZ1-66 was also observed during AmrZ-mediated activation of PalgD (Figure S8) as well as twitching motility (Table S2).
Figure 3. The AmrZ C-terminal domain is required for efficient repression (A.) and activation (B.) of its targets.
Arabinose-inducible plasmids encoding AmrZ or AmrZ1-66 were transferred into PAO1 ΔamrZ. After induction by various arabinose concentrations, cells were harvested, and mRNA levels quantified by qRT-PCR. Gene expression was normalized to the reference gene rpoD and relative gene expression was compared to the ΔamrZ strain with the empty vector pHERD20T in the absence of arabinose. Unpaired two-tailed student t-tests were used for statistical analyses of three independent experiments. ***: P<0.001; ns: not significant.
Oligomerization not only provides an economic way to form large complexes, but also enables proteins, such as transcription factors, to flexibly modulate their DNA binding specificity and affinity. Differential sequence specificity can be achieved through oligomerization of different combinations of transcription factors (Matthews et al., 2012). For some transcription factors such as the λ repressor, oligomerization via its CTD significantly enhances binding affinity to its operator sites within the phage genome (Bell et al., 2000). In the present study, we provide evidence that after removing the CTD, AmrZ loses the ability to form tetramers and exhibits reduced DNA binding to both activated and repressed targets, suggesting the significance of the CTD during AmrZ-mediated regulation.
Members in the Arc protein family represent diverse DNA binding proteins in various phage and bacterial species, and some also contain CTDs. CTDs in proteins such as Mnt and TrwA mediate tetramerization (Waldburger and Sauer, 1995; Moncalián and De La Cruz, 2004; Madl et al., 2006), while at least two members (NikR and MetJ) exhibit significantly higher DNA binding activities when their CTDs are bound by ligands (nickel and S-adenosylmethionine, respectively) (Rafferty et al., 1989; Chivers and Sauer, 2002). Sequence alignment using PROMALS3D was performed between AmrZ and other Arc family members including Mnt, TrwA, NikR and MetJ (Pei et al., 2008). However, significant similarity was observed only in their ribbon-helix-helix domains (Figure S9). While our current data support the requirement of the AmrZ CTD for tetramerization, with effects on DNA binding and subsequent gene expression, we cannot exclude a role of the CTD in other functions, such as ligand binding, which requires further investigations.
Conclusions
Overall, we have investigated the significance of the AmrZ CTD for oligomerization, DNA binding affinity, as well as AmrZ-mediated activation and repression. Tetramers composed of four DNA binding-proficient monomers are critical for AmrZ-mediated regulation. Further work is necessary to understand which residues are at the interface between monomers to mediate inter-monomer interactions. Since this CTD does not appear to be present in other proteins, more understanding of this domain may provide insights into a novel oligomerization mechanism and potentially other unknown functions of this domain.
Supplementary Material
Acknowledgement
Public Health Service awards AI097511 and NR013898 (DJW), an NSF instrument development grant DBI0923551 (VHW) and funds provided from the Public Health Preparedness for Infectious Diseases (PHPID) program (phpid.osu.edu) supported this work. BX was partially supported by the student fellowship grant from the Cystic Fibrosis Foundation (XU13H0).
We thank Dr. Thomas Hollis for kindly providing AmrZ and AmrZ1-66, Dr. Joe J. Harrison for providing the E. coli S17λpir strain, and Dr. Christopher J. Jones for pCJ4. We are grateful to Sheri Dellos-Nolan and Dr. Meenu Mishra for critical reading of this manuscript. We also acknowledge Dr. Nrusingh Mohapatra for assistance with β-galactosidase assays.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Ali MH, Imperiali B. Protein oligomerization: How and why. Bioorganic Med. Chem. 2005;13:5013–5020. doi: 10.1016/j.bmc.2005.05.037. [DOI] [PubMed] [Google Scholar]
- Baynham P, Brown AL, Hall LL, Wozniak DJ. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 1999;33:1069–1080. doi: 10.1046/j.1365-2958.1999.01550.x. [DOI] [PubMed] [Google Scholar]
- Baynham PJ, Ramsey DM, Gvozdyev BV, Cordonnier EM, Wozniak DJ. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 2006;188:132–140. doi: 10.1128/JB.188.1.132-140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynham PJ, Wozniak DJ. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 1996;22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- Bell CE, Frescura P, Hochschild A, Lewis M. Crystal structure of the λ repressor C-terminal domain provides a model for cooperative operator binding. Cell. 2000;101:801–811. doi: 10.1016/s0092-8674(00)80891-0. [DOI] [PubMed] [Google Scholar]
- Bobadilla Fazzini RA, Skindersoe ME, Bielecki P, Puchalka J, Givskov M, Martins dos Santos VAP. Protoanemonin: a natural quorum sensing inhibitor that selectively activates iron starvation response. Environ. Microbiol. 2013;15:111–120. doi: 10.1111/j.1462-2920.2012.02792.x. [DOI] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Breitwieser GE. G protein-coupled receptor oligomerization: implications for G protein activation and cell signaling. Circ. Res. 2004;94:17–27. doi: 10.1161/01.RES.0000110420.68526.19. [DOI] [PubMed] [Google Scholar]
- Bullock BN, Jochim AL, Arora PS. Assessing helical protein interfaces for inhibitor design. J. Am. Chem. Soc. 2011;133:14220–14223. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers PT, Sauer RT. NikR repressor: High-affinity nickel binding to the C-terminal domain regulates binding to operator DNA. Chem. Biol. 2002;9:1141–1148. doi: 10.1016/s1074-5521(02)00241-7. [DOI] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015:1–6. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacesa P. Bacterial alginate biosynthesis--recent progress and future prospects. Microbiology. 1998;144(Pt 5):1133–43. doi: 10.1099/00221287-144-5-1133. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins M, Appel R, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker J, editor. The Proteomics Protocols Handbook SE & 52. Humana Press; 2005. pp. 571–607. [Google Scholar]
- Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 2014;10:e1003984. doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Ryder CR, Mann EE, Wozniak DJ. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J. Bacteriol. 2013;195:1637–1644. doi: 10.1128/JB.02190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kai T, Tateda K, Kimura S, Ishii Y, Ito H, Yoshida H, et al. A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI or rhlI, in Pseudomonas aeruginosa. Pulm. Pharmacol. Ther. 2009;22:483–6. doi: 10.1016/j.pupt.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Karas M, Bahr U, Dülcks T. Nano-electrospray ionization mass spectrometry: addressing analytical problems beyond routine. Fresenius. J. Anal. Chem. 2000;366:669–676. doi: 10.1007/s002160051561. [DOI] [PubMed] [Google Scholar]
- Knight KL, Bowie JU, Vershon AK, Kelley RD, Sauer RT, Vershong AK, Sauer RT. The Arc and Mnt repressors. A new class of sequence-specific DNA-binding protein. J. Biol. Chem. 1989;264:3639–3642. [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Ma X, Lai LB, Lai SM, Tanimoto A, Foster MP, Wysocki VH, Gopalan V. Uncovering the stoichiometry of Pyrococcus furiosus RNase P, a multi-subunit catalytic ribonucleoprotein complex, by surface-induced dissociation and ion mobility mass spectrometry. Angew. Chemie Int. Ed. 2014;53:11483–11487. doi: 10.1002/anie.201405362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl T, Van Melderen L, Mine N, Respondek M, Oberer M, Keller W, et al. Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA. J. Mol. Biol. 2006;364:170–185. doi: 10.1016/j.jmb.2006.08.082. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Granero F, Redondo-Nieto M, Vesga P, Martín M, Rivilla R. AmrZ is a global transcriptional regulator implicated in iron uptake and environmental adaption in P. fluorescens F113. BMC Genomics. 2014;15:237. doi: 10.1186/1471-2164-15-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J, Sunde M, Gell D, Grant R. Protein dimerization and oligomerization in biology. 2012 [Google Scholar]
- Moncalián G, De La Cruz F. DNA binding properties of protein TrwA, a possible structural variant of the Arc repressor superfamily. Biochim. Biophys. Acta & Proteins Proteomics. 2004;1701:15–23. doi: 10.1016/j.bbapap.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Pei J, Tang M, Grishin NV. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:W30–4. doi: 10.1093/nar/gkn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Cherny KE, Sauer K. The Pseudomonas aeruginosa diguanylate cyclase GcbA, a homolog of P. fluorescens GcbA, promotes initial attachment to surfaces, but not biofilm formation, via regulation of motility. J. Bacteriol. 2014;196:2827–2841. doi: 10.1128/JB.01628-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor EE, Waligora EA, Xu B, Dellos-Nolan S, Wozniak DJ, Hollis T. The transcription factor AmrZ utilizes multiple DNA binding modes to recognize activator and repressor sequences of Pseudomonas aeruginosa virulence genes. PLoS Pathog. 2012;8:e1002648. doi: 10.1371/journal.ppat.1002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafferty JB, Somers WS, Saint-Girons I, Phillips SE. Three-dimensional crystal structures of Escherichia coli met repressor with and without corepressor. Nature. 1989;341:705–710. doi: 10.1038/341705a0. [DOI] [PubMed] [Google Scholar]
- Ramsey DM, Baynham PJ, Wozniak DJ. Binding of Pseudomonas aeruginosa AlgZ to sites upstream of the algZ promoter leads to repression of transcription. J. Bacteriol. 2005;187:4430–4443. doi: 10.1128/JB.187.13.4430-4443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Schreiter ER, Drennan CL. Ribbon-helix-helix transcription factors: variations on a theme. Nat Rev Micro. 2007;5:710–720. doi: 10.1038/nrmicro1717. [DOI] [PubMed] [Google Scholar]
- Sunya S, Gorret N, Delvigne F, Uribelarrea J-L, Molina-Jouve C. Real-time monitoring of metabolic shift and transcriptional induction of yciG::luxCDABE E. coli reporter strain to a glucose pulse of different concentrations. J. Biotechnol. 2012;157:379–90. doi: 10.1016/j.jbiotec.2011.12.009. [DOI] [PubMed] [Google Scholar]
- Tart AH, Blanks MJ, Wozniak DJ. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 2006;188:6483–6489. doi: 10.1128/JB.00636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershons AK, Youderian P, Susskindli M, Sauers RT. The bacteriophage P22 Arc and Mnt repressors. J. Biol. Chem. 1985;260:12124–12129. [PubMed] [Google Scholar]
- Waldburger CD, Sauer RT. Domains of Mnt repressor: roles in tetramer formation, protein stability, and operator DNA binding. Biochemistry. 1995;34:13109–13116. doi: 10.1021/bi00040a023. [DOI] [PubMed] [Google Scholar]
- Waligora EA, Ramsey DM, Pryor EE, Lu H, Hollis T, Sloan GP, et al. AmrZ beta-sheet residues are essential for DNA binding and transcriptional control of Pseudomonas aeruginosa virulence genes. J. Bacteriol. 2010;192:5390–5401. doi: 10.1128/JB.00711-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.