Abstract
During tumor growth and angiogenesis there is a dynamic remodelling of tissue architecture often accompanied by the release of extracellular matrix constituents full of biological activity. One of the key constituents of the tumor microenvironment is the large heparan sulfate proteoglycan perlecan. This proteoglycan, strategically located at cell surfaces and within basement membranes, is a well-defined pro-angiogenic molecule when intact. However, when partially processed by proteases released during cancer remodelling and invasion, the C-terminal fragment of perlecan, known as endorepellin, has opposite effects than its parent molecule. Endorepellin is a potent inhibitor of angiogenesis by exerting a dual receptor antagonism by simultaneously engaging VEGFR2 and α2β1 integrin. Signaling through the α2β1 integrin leads to actin disassembly and block of endothelial cell migration, necessary for capillary morphogenesis. Signaling through the VEGFR2 induces dephosphorylation of the receptor via activation of SHP-1 and suppression of downstream proangiogenic effectors, especially attenuating VEGFA expression. A novel and emerging role of endorepellin is its ability to evoke autophagy by activating Peg3 and various canonical autophagic markers. This effect is specific for endothelial cells as these are the primary cells expressing both VEGFR2 and α2β1 integrin. Thus, an endogenous fragment of a ubiquitous proteoglycan can regulate both angiogenesis and autophagy through a dual receptor antagonism. The biological properties of this natural endogenous protein place endorepellin as a potential therapeutic agent against cancer or diseases where angiogenesis is prominent.
Keywords: Perlecan, LG domains, angiogenesis, autophagy, Peg3
Introduction
The extracellular matrix (ECM) is a biological structure that is found in all multi-cellular organisms. It provides physical scaffolding through adhesive interactions between adjacent cells, and also biochemical support by mediating a number of signalling processes through the direct interaction with various growth factor receptors located on ECM-associated cells (1-3). The ECM is the primary feature that dictates organismal architecture and many of its biological roles are maintained within various animal lineages. These roles are predominantly cell adhesion and cell-to-cell communication. However, due to an independent evolution rate between multi-cellular lineages, ECM constituents often differ among organisms and animals of differing species (4) and have the ability to utilize a wide variety of biological roles, which they use to fit the biological task they require (5). A major class of molecules found within the ECM is proteoglycans, an expanding family of gene products that now encompass 43 distinct genes belonging to four families (6). Deregulated expression of many proteoglycans has been closely linked to the onset of a number of pathologies including wound healing, tumor growth, fibrosis and abnormal angiogenesis (7). This emphasizes the importance of a balanced and controlled expression of these biologically active proteoglycans. This minireview will focus on the roles of the proteoglycan perlecan and its C-terminal fragment, endorepellin, in cancer, angiogenesis and autophagy.
Perlecan genetics
Heparan sulfate proteoglycans (HSPGs) are a special class of proteoglycans, which govern crucial events in embryonic development, inflammation, wound repair and cancer. Perlecan, also known as HSPG2, is one of the largest proteoglycans found in the body (8,9). It was originally isolated from mouse basement membrane (10) and given its eponym from its appearance of a “beads on a string” on rotary shadow electron microscopy. It was later discovered that the cell-surface HSPG of human colon carcinoma cells was essentially the same as perlecan (11-13). It has a protein core of ~470 kDa and contains three HS chains located at the N-terminus (Figure 1A), each weighing approximately 30 kDa. Perlecan is encoded by the HSPG2 gene located on the short arm (p) of chromosome 1, specifically 1p36 (13). This large gene covers >120 kb of continuous DNA and contains 97 protein-encoding exons (14). The HSPG2 gene has a complex organization at the promoter level (15,16), and is an early response gene that is transcriptionally inhibited by interferon γ (17) and induced by TGFβ (18) and phorbol ester (19). Its promoter complexity is further enhanced by the potential generation of alternatively-spliced mRNA variants reported to occur in mast cells (20,21).
Figure 1.
Schematic representation of the multimodular perlecan and its C-terminal endorepellin. (A) Perlecan is large multidomain proteoglycan consisting of 5 domains. It contains 3 HS chains towards the N-terminus. Domain I contains a SEA module. Domain II is made up of four LDL repeats and the first IgG. Following this domain, is Domain III which is made up of 3 LG repeats, each being separated by 3 laminin-like EGF repeats. Domain IV solely contains a further 20 IgG repeats. The C-terminal Domain V, termed endorepellin, has a similar structure to Domain III as it contains 3 laminin repeats; however, each of these is separated by 2 laminin-like EGF repeats. (B) Refined crystal structure of LG3 domain of endorepellin. Image was generated using PDB:3SH4 and presented using PyMol software. LG3 is constructed as a 15-stranded anti-parallel β sandwich forming a jellyroll fold characteristic of LG domains. The right image represents a 90° rotation along the y axis. This region is known to interact with α2β1 integrin located on endothelial cells. The two α helixes are illustrated in red, the 12 β sheets are yellow and the loops are green. Of particular note, Asp4197 is highlighted in magenta (circled) and is found at the N-terminus of the protein. This amino acid is suggested to be important in the processing of endorepellin. A mutation in this amino acid inhibits endorepellin activity. Thus, this site is hypothesised to be a potential binding site for the α2β1 integrin.
The expression profile of perlecan during development follows a non-random and defined pattern. Within early phases, perlecan can be found within the endothelial cells of the cardiac tissue such as the heart and blood vessels, followed later by being expressed in the liver, kidney and spleen (22). Lack of perlecan in developing Hspg2−/− mouse embryo leads to severe brain, heart, and skeletal dysplasia responsible for death between embryonic stage E10.5 and birth, primarily because of blood leakage into the pericardial cavity (23,24). Importantly, this is around the stage of development when perlecan expression is induced. Moreover, Hspg2−/− mice develop severe skeletal dysplasia characterized by shortened bones and craniofacial abnormalities, and die shortly after birth of respiratory failure due to the cartilage defects of the rib cage (23,24).
Mutations in the human perlecan gene can cause at least two autosomal recessive skeletal disorders, the dyssegmental dysplasia Silverman-Handmaker type (DDSH) (25,26), and the Schwartz-Jampel syndrome, with myotonic myopathy and chondrodysplasia (SJS) (27-29). DDSH new-borns display massive skeletal abnormalities and die shortly after birth. SJS is represented by a range of mutations with most patients reaching adulthood exhibiting a spectrum of disorders related to the degree of haplo-insufficiency, notably severe muscle stiffness (29). In addition to the disorganization of the growth plate and chondro-osseous junction of bones caused by the absence of perlecan, there are other distortions in the growth of joints including the temporo-mandibular joint, myotonia and electrical disturbances of muscle and modifications to the structures of facial bones (30). Moreover, perlecan deficiency in hypomorphic mutant mice influences bone formation by enhancing osteogenesis and concurrently affects mineralization, leading to increased brittleness at older ages (31).
The absence of perlecan affects FGF signalling in the growth plate (23). Hspg2 mutant mice with a knock-in mutation Cys1532Tyr, found in human SJS, have a phenotype similar to SJS (32). These findings provide robust genetic evidence demonstrating the critical need for perlecan expression and also the importance of the protein core in cardiovascular development. Recent studies designed to unravel the cause of the leakage into the pericardial cavity have shown that basement membranes lacking perlecan deteriorate in the heart with accompanying loss of cell–cell attachment in the ventricle and outflow tract (33).
Perlecan protein core and its functions
The protein core of perlecan is composed of five distinct modules which are schematically illustrated in Figure 1A. Perlecan regulates several biological processes by interacting with growth factor receptors and soluble growth factors through its HS chains or protein core (34-36). Perlecan is widely conserved across animals and is one of the few gene products that are found in both vascular and avascular tissues (22,37-40). Due to its large size and complex structure, perlecan has many roles. Among these are: cell adhesion and invasion (41-45), inhibition of smooth muscle cell proliferation (46-48), cardiovascular development (24), lipid metabolism (49-51), corneal epithelial structure (52), epidermal and osteophyte formation (53,54), cartilage homeostasis (55), endochondral ossification (56), apoptosis (57), lens capsule homeostasis (58), and synaptogenesis (59,60). However the most notable role is its ability to promote vessel formation (40,61-65) and angiogenesis (9,66-71). This occurs through the binding of several pro-angiogenic factors to the HS chains or the protein core (61,64,72-75). Moreover, perlecan binds to several growth factors (76-78), including progranulin (79), a protein inducing angiogenesis and cancer growth (80,81). Perlecan can be processed by MMP7 at the invasive tumor microenvironment, thereby acting as a molecular switch to alter prostate carcinoma and favoring cell invasion (45).
One of the most intriguing functions of perlecan is its involvement in blood vessel formation (1,82). The mRNA levels of perlecan are high in endothelial cells of the developing mouse embryo (22), and further increase after recruitment of pericytes to the endothelial tubes. Perlecan also supports the maintenance of brain and skin subendothelial basement membrane and promotes vascular formation and angiogenesis by modulating FGF2 activity (83). Hspg2Δ3/Δ3 mice harboring a 45-bp in-frame deletion of Hspg2 exon 3, which removes the attachment sites for HS chains in Domain I, are viable but have small eyes and their lenses degenerate within 3 weeks of birth (58). Hspg2Δ3/Δ3 mice also possess impaired angiogenesis, delayed wound healing and retarded tumor growth (84). Thus, perlecan HS chains play an important role in the regulation of wound healing and tumor growth. Perlecan knockdown in zebrafish reveals abnormal phenotypes with a pronounced curvature of the tail and trunk caused by severe defects in the musculoskeletal system and inhibition of angiogenesis (40). Recently, it has been shown that perlecan deficiency promotes endothelial cell dysfunction by reducing the expression of endothelial-specific nitric oxide synthase (85), thereby controlling vascular relaxation.
The pro-angiogenic activity of perlecan is thought to be achieved primarily through interacting with FGF receptors using the three HS chains present in the N-terminal Domain I (61). This interaction actively increases cell proliferation, motility, and adhesion (44,61). Often, the HS chains of perlecan can be liberated by either partial proteolysis of the protein core (86) or by the activity of heparanase (87). The latter enzyme is known to cleave HS generating relatively large fragments (88) that are full of HS-binding mitogens and pro-angiogenic factors (89). Perlecan HS chains are key factors for angiogenesis in an experimental animal model of hind-limb ischemia (71). In this animal model, the HS side chains in perlecan are important mediators of the angiogenic response to ischemia through a mechanism that involves induction of VEGF expression (71). On the other hand, the HS chains of perlecan, which are processed by heparanase released by tumor and inflammatory cells (90-94), have a profound inhibitory role on smooth muscle cell proliferation in vivo (46,47). Thus, there is a fine balance of activator and inhibitor effects at the N-terminus of perlecan, further stressing the biological complexity of this macromolecule.
Deregulated expression of perlecan is reported to be instrumental in cancer progression (84,95,96). For instance, in some cancers of the ovaries perlecan expression is lost from the basement membrane (97). In others, such as melanomas, oral squamous carcinomas and hepatocellular carcinomas, perlecan is markedly upregulated (95,98,99). In the latter case, knockdown of perlecan in carcinoma cells isolated from various tumor sites reduces cell migration and adhesion (98). Moreover, knockdown of perlecan in metastatic prostate cancer cells reduces in vivo tumor growth (100). Perlecan is overexpressed in the desmoplastic prostate cancer stroma and this is mediated by TNFα-mediated transcriptional induction of perlecan (101). Thus, perlecan transcription could be a part of an innate response of the tumor stroma to cancer invasion.
Collectively, these findings posit perlecan and its HS chains as key regulators of angiogenesis during tumor progression. Because perlecan acts as a pro- and anti-angiogenic HSPG, aberrant perlecan levels and/or abnormal processing during cancer invasion could have profound effects on tumor progression and metastasis.
Endorepellin, the C-terminal angiostatic fragment of perlecan
The most notable region of this HSPG is its C-terminal Domain V (Figure 1A), also known as endorepellin to signify its anti-endothelial properties (102). Endorepellin is an 85-kDa glycoprotein consisting of three laminin-like globular (LG) repeats separated by epidermal growth factor (EGF)-like module doublets. The crystal structure of the LG3 domain has been recently solved (103). This region is made up of about 200 amino acids and is constructed as a 15-stranded anti-parallel β sandwich forming a jellyroll fold, characteristic of LG domains (Figure 1B). Notably, Asp4197 located near the N-terminus of LG3, might be a potential binding site for the α2β1 integrin, as mutation of this residue LG3 activity (103).
Endorepellin is not produced alone but must be proteolitically cleaved from secreted perlecan, and as such endorepellin has its own repertoire of actions. Generation of bioactive endorepellin is mediated by the activity of Cathepsin L (104), the same enzyme that is also involved in the generation of endostatin. One can envisage a scenario where, during remodelling of basement membranes, Cathepsin L can dually affect angiogenesis. In contrast, LG3 is highly sensitive to BMP1-Tolloid-like family of MMPs (105). Once cleaved, endorepellin is found in similar regions as perlecan, primarily within the basement membrane of epithelial-lining organs and along the cardiovascular system (106). Notably increased levels of endorepellin have been found in sarcopenia, the age-related loss of skeletal muscle mass and function (107). Endorepellin works by binding with high affinity to receptors located on endothelial cells and, therefore, influences the cells lining of the vessels. The difference in the receptors targeted by LG modules and the actions they provoke are critically assessed in the next sections.
Endorepellin signaling
One of the key features of liberated soluble endorepellin is its ability to signal through various receptors including integrins and VEGFRs (108). This signaling ability of perlecan/endorepellin is also extended to its perlecan ortholog in Drosophila named Trol, for terribly reduced optical lobes. Trol regulates multiple developmental stages (109) as well as FGF-signaling output during mesoderm development (110). Signaling through perlecan regulates prostate cancer progression through Sonic Hedgehog (111,112). In mammalians, both perlecan and endorepellin can bind VEGFR1 and VEGFR2, at sites independent of the VEGFA binding site (113). VEGFA is able to bind to either receptor at picomolar concentrations; however VEGFA has approximately 5-fold higher binding capacity for VEGFR1 (Kd~100 pM) than VEGFR2 (Kd~480 pM) (114). Notably, endorepellin binds with a similar affinity to both receptors as VEGFA (114) and this binding capacity is greater than the whole glycanated parental proteoglycan. In competition binding assays, highly-purified, human recombinant endorepellin cannot be displaced from either VEGFR1 or VEGFR2 by molar excesses of recombinant VEGFA (114). This demonstrates that endorepellin binds to a discrete region of VEGFR2 ectodomain that doesn’t overlap with the VEGF ligand binding site.
Initially, it was demonstrated that endorepellin binds to the α2β1 integrin on endothelial cells (115-119) and platelets (117) where it enhances collagen-platelet response. In addition, endorepellin affects the global proteomic profile of endothelial cells (120). It is well established that the α2β1 integrin is an important receptor regulating various aspects of angiogenesis (121-123), and is required for endorepellin’s biological activity, both in vitro and in tumor xenografts (124). However, it is now known that endorepellin binds to both VEGFR2 and α2β1 on endothelial cells (114,125). This causes internalization and downregulation of both receptors within 10 minutes of binding; a concept referred to as “dual receptor antagonism” (114). Importantly, only the LG1/2 domain of endorepellin can interact with VEGFR2, whereas, only the LG3 domain is capable to interact with the α2β1 integrin (125,126). Indeed, cells expressing α2β1 integrin, but lacking VEGFR2, are incapable of responding to endorepellin (114). LG1/2 and the LG3 domains are able to elicit different responses independently of each other (126). This suggests that cells that lack α2β1 but do express VEGFR2, may actually be able to respond to endorepellin but specifically only to the LG1/2 domain.
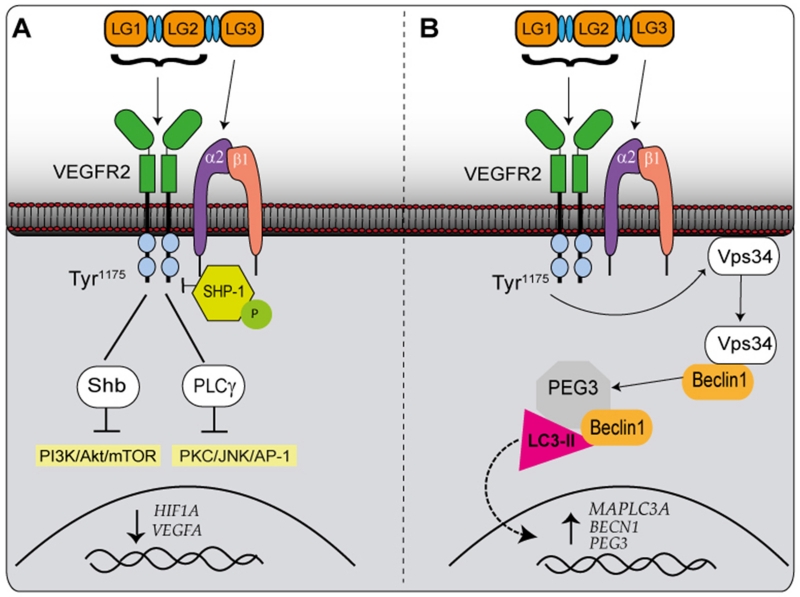
The downstream cascades from the dual signalling by endorepellin have been investigated using an antibody array capable of probing the phosphorylation status of numerous receptor tyrosine kinases (RTKs) (119). True to its angiostatic activity, endorepellin reduces the phosphorylation of key receptors involved with angiogenesis including VEGFR1/2, EGFR and ROR2 (119). These findings were further validated when the effects of endorepellin were hampered following functional blockage of the key endorepellin receptor, α2β1. Following up to 30 minutes of endorepellin treatment, the α2 subunit was immune-precipitated and revealed that Src homology protein phosphatase-1 (SHP-1) was functionally linked to the α2 subunit (119). Moreover, following endorepellin treatment, SHP-1 phosphorylation also increases, suggesting that SHP-1 is a key downstream target of α2β1 following VEGFR2 engagement by soluble endorepellin. Endorepellin also causes SHP-1 to actively translocate from the plasma membrane to the nucleus where it is known to influence the transcription of early response genes (119). These findings were validated in vivo, as endothelial cells isolated from α2β1−/− mice following endorepellin treatment have significantly reduced VEGFR2 phosphorylation (119).
Endorepellin signalling is able to influence the activity of two important signalling branches, the PI3K/Akt/mTOR and PKC/JNK/AP-1 pathways (Figure 2A), both of which are thought to be primarily governed by SHP-1 bioactivity (113). Under normal physiological conditions, PLCγ and the adaptor protein Shb are bound to VEGFR2 at Tyr1175. When these proteins become phosphorylated they activate the PI3K/Akt/mTOR and PKC/JNK/AP-1 pathways, respectively (Figure 2A). As well as being known to translocate to the nucleus to regulate genes, SHP-1 is able to dephosphorylate VEGFR2 at Tyr1175 , which in turn causes the physical dissociation of Shb and PLCγ from VEGFR2, resulting in decreased signalling of PI3K/Akt/mTOR and PKC/JNK/AP-1 pathways (113). Normal signalling of VEGFA is able to promote the recruitment of PLCγ to VEGFR2. However, treatment with endorepellin can inhibit the effects of VEGFA on PLCγ (113). As endorepellin and VEGFA do not compete for the same binding region on VEGFR2, this would suggest that endorepellin carries a greater signalling potential than VEGFA, likely by allosterically inhibiting the receptor or preventing its dimerization. More studies need to be performed to decipher the complexity of downstream signalling and its components.
Figure 2.
Schematic representation of the signalling pathways involved in endorepellin-evoked intracellular signalling network. (A) The anti-angiogenic effects of endorepellin. Endorepellin signals through VEGFR2 via its LG1/2 domain and α2β1 via LG3. This causes phosphorylation of SHP-1, which relocates from the plasma membrane to the α2 subunit of the integrin. This subsequently causes dephosphorylation of VEGFR2 at Tyr1175, which prevents the Shb and PLCγ from binding to VEGFR2 and therefore prevents the activation of PI3K/AKt/mTOR and PKC/JNK/AP-1 pathways. These pathways prevent the transcription of HIF1A and VEGFA. (B) The pro-autophagic effects of endorepellin. Endorepellin signals through VEGFR2 and α2β1in the same manner as previously described. Although the involvement of SHP-1 is not yet elucidated, Tyr1175 is again dephosphorylated causing Vps34 to re-locate from the plasma membrane and interact with Beclin1 in autophagosomes. Beclin 1 then co-localizes with Peg3 and LC3-II. The signalling of endorepellin also results in increased mRNA expression of MAPLC3A, BECN1 and Peg3.
Anti-angiogenic properties of endorepellin
Due to the high level expression in cardiovascular tissues and vessels, and the ability of endorepellin to signal through VEGFR2, angiogenesis is the most widely reported process endorepellin is able to influence (113,114,119). Despite being a key region within the pro-angiogenic parent molecule perlecan, endorepellin is a potent mediator in repressing angiogenesis both in vitro (113,114) and in vivo (119).
HIF-1α is a key transcription factor that is commonly upregulated when cells detect reduced oxygen content. This, in turn, promotes angiogenesis to replenish hypoxic regions. Endorepellin suppresses HIF-1α levels in porcine aortic endothelial cells (PAE) stably overexpressing VEGFR2 (113). Importantly, this downregulation is transcriptionally regulated as cells transfected with luciferase driven by the human HIF1A promoter have a dose- and time-dependent suppression of the promoter activity following endorepellin treatment (113). In support of these findings, blocking VEGFR2, via either small molecule inhibitor or siRNA, abrogates the effects of endorepellin, demonstrating that this bioactivity is mediated through this receptor. Importantly, soluble endorepellin suppresses HIF1A and VEGFA mRNA and inhibits the secretion of VEGFA under hypoxic conditions (113).
There is robust in vivo evidence that supports a role for endorepellin as anti-cancer and anti-tumor angiogenic factor. When mice bearing either squamous cell or Lewis lung carcinomas, are systemically treated with human recombinant endorepellin at clinical dosages (5 mg/kg), the exogenously administered endorepellin specifically localizes to the tumour vascular system and peri-vascular regions where it remains for several days (116). This leads to regression of tumour angiogenesis and also induces a more hypoxic tumour microenvironment, whilst decreasing the tumour metabolism and the mitotic index (116). These findings put forward bold evidence that endorepellin is able to inhibit tumour growth through a number of angiogenic-mediated processes, but also that systemic administration of endorepellin is able to specifically target the tumor vascular system, thereby providing a proof for targeted tumour specificity. In support of these studies in mammals, exogenous administration of endorepellin to zebrafish with morpholino-mediated suppression of perlecan expression, can rescue the severe vascular phenotype (40). Although this again provides evidence to suggest the effects of endorepellin on angiogenesis, it also suggests that endorepellin may have potential therapeutic effects on cardiovascular diseases as well as cancer.
Pro-autophagic role of endorepellin
We recently discovered that endorepellin acts as a potent inducer of an intracellular catabolic process known as autophagy (126). These effects are also regulated through interaction with VEGFR2 and α2β1 (Figure 2B) suggesting that these receptors also hold the downstream signaling machinery to regulate several pathways outside of angiogenesis (126).
Autophagy is a catabolic lysosomal process that is conserved across all eukaryotes (127). It is a highly-controlled process, which can often be described as pro-survival, where a cell uses self-eating as an alternate energy source by degrading and recycling cytosolic components (128). Autophagy is stimulated during various pathological and physiological states, such as starvation. In cells starved for nutrients or growth factors, autophagy provides crucial nutrients and energy that enhance cell survival through the breakdown of cytosolic components. Other cellular stresses, including hypoxia, growth factor withdrawal, and ER stress, can also induce autophagy, often to enhance cell survival (129-131). The autophagic pathway, essential for embryonic development and organogenesis (132,133), can be deregulated in several disorders, including metabolic and infectious diseases, neurodegenerative disorders, angiogenesis and cancer (134). According to an emerging hypothesis, autophagy provides an anti-carcinogenic function in primary cells by safeguarding against metabolic stress through the homeostatic turnover of mitochondria and the clearance of protein aggregates. In established tumors, autophagy may confer a survival advantage on tumor cells that are under metabolic stress, as a result of a high proliferation rate and exposure to hypoxia from insufficient vascularization (135). However, there is also strong evidence that sustained angiogenesis evoked by angiostatic proteins negatively affect tumor angiogenesis and cancer growth (136).
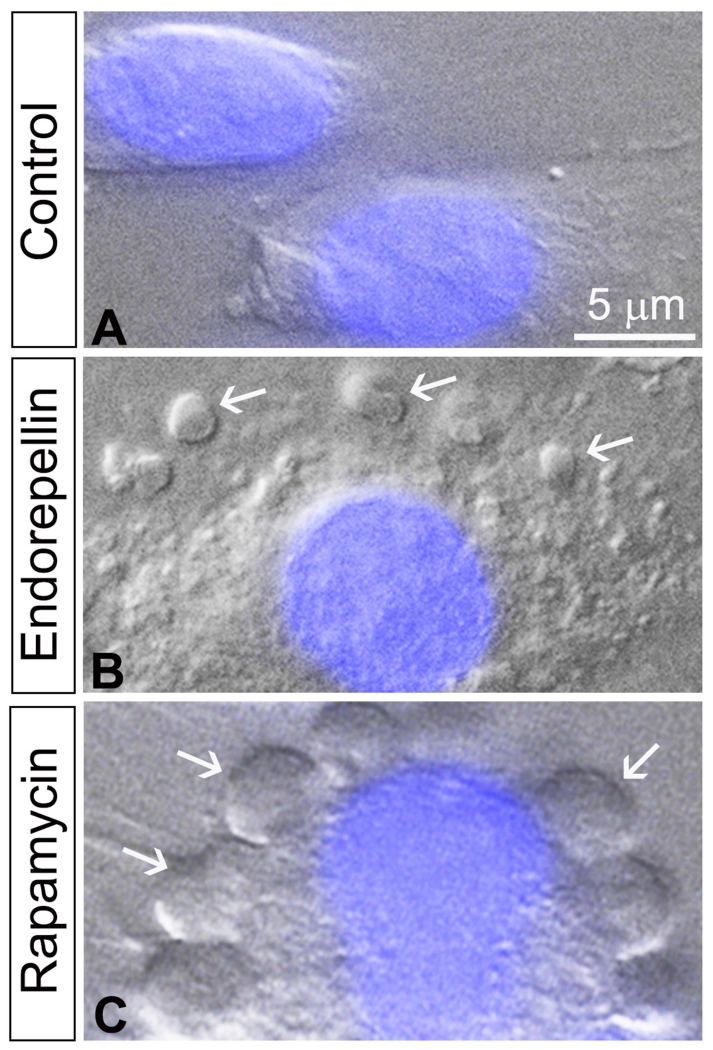
Notably, other secreted ECM products, including decorin, endostatin and kringle 5, have been shown to trigger autophagy (137-139). Endorepellin causes the formation of well-developed, large autophagosomes in HUVEC that are visible using differential interference contrast (DIC) microscopy (Figure 3B), that are similar to those generated by canonical autophagy-inducing factors such as rapamycin (Figure 3C), and nutrient deprivation (126). Importantly, the number of these structures increases in a dose-dependent manner following endorepellin treatment (126). Two key markers of autophagy are Beclin1 and LC3 (140). Beclin1 is an important binding partner of class III PI3K-Vps34. Under normal conditions Vps34 is found located at the plasma membrane; however, when autophagy is initiated, Vps34 is mobilized to the phagophores, the initial formation stage of autophagosomes (141). LC3 exists in two forms, LC3-I and LC3-II. The latter is the cytosolic form which is conjugated to phosphatidylethanolamine and is recruited to the autophagosomal membranes (140). The stimulatory effects of endorepellin on these canonical catabolic markers provide the strongest evidence of endorepellin-mediated autophagy.
Figure 3.
Endorepellin induces autophagy in endothelial cells through an mTOR-dependent pathway. Representative images of HUVECs following 6 h treatment with vehicle (A) 200 nM endorepellin (B) or 100 nM rapamycin (C) in nutrient-enriched conditions. The images were captured with differential interference contrast microscopy and fluorescence to detect the nucleus stained in blue by DAPI. Notice the formation of multiple autophagosomes (white arrows) in the cytoplasm of the cells in both endorepellin- and rapamycin-treated endothelial cells vis-à-vis vehicle-treated controls.
Following treatment with endorepellin, there is a change in the distribution of Vps34 from the plasma membrane to the cytoplasm (126). Interestingly, once redistributed, endorepellin evokes the movement of Vps34 into large intracytoplasmic vacuoles, which are Beclin 1 and LC3-II positive (126). Although the levels of Vps34 do not significantly change, the levels of Beclin 1, LC3 and Peg3 are all increased (126). We previously reported (142) that Peg3 is required for decorin-mediated induction of autophagy and proposed that Peg3 should be regarded as a marker of autophagy. Interestingly, when Peg3 is depleted, the ability of endorepellin to increase Beclin 1 and LC3 is totally abrogated, consistent with a primary role for Peg3 in the autophagic process (126). Notably, endorepellin causes co-localization of Peg3 with both autophagic markers LC3-II and Beclin 1 (126). These data suggest that not only may Peg3 be a novel autophagic protein, but it may also be one of the most reliable markers as it is responsible for the induction of Beclin1 and LC3.
As described above, endorepellin has three main LG domains exhibiting profound differences in the receptors they target and effects they provoke. LG1/2 binds with high affinity to VEGFR2, whereas, the LG3 domain binds α2β1 (114). The importance of these three domains is profound in directing whether endorepellin induces autophagy or inhibits angiogenesis. LG1/2, independently of LG3, induces the expression of PEG3, BECN1 and MAPLC3A genes. In contrast, signalling by LG3 alone actually reduces the levels of all the aforementioned genes (126). This demonstrates that only endorepellin domains LG1/2 are able to promote autophagy, which is solely dependent on VEGFR2 (Figure 2B). In contrast, the LG3 domain signals through α2β1 and works to repress autophagy. Therefore, the ability of endorepellin to induce autophagy is based on VEGFR2, while its anti-angiogenic property is based upon dual receptor antagonism, involving VEGFR2 and α2β1 integrin.
Since full length endorepellin is able to induce autophagy (126), it would suggest that signalling of LG1/2 through VEGFR2 carries a greater signalling potential and can override the effects of LG3 and the α2β1 to reduce autophagy. Further evidence of this signalling being solely dependent on VEGFR2 is provided when α2β1 is blocked using anti-α2β1 blocking antibodies. In this setting, there are no changes in the endorepellin-induced increases in PEG3, BECN1 or MAPLC3A (126). However, when VEGFR2 is silenced, the ability of endorepellin to increase PEG3, BECN1 and MAPLC3A is markedly suppressed (126). Thus, endorepellin evokes a catabolic program in endothelial cells and this aberrant autophagy could be detrimental to the formation of cancer-associated blood vessels.
Conclusions
Angiogenesis is an imperative aspect of normal human development. However, this process is often aberrant in many forms of cancer where the tumors induce the development of a florid blood supply in order to support their progressive increase in growth. The proteoglycan perlecan and its C-terminal fragment endorepellin best describe the fundamental balance required to maintain the angiogenic system. Perlecan proteoglycan is, overall, a pro-angiogenic molecule that is expressed in the cardiovascular system during development and in the adult life. In contrast, endorepellin which is found in similar regions as perlecan, is strongly anti-angiogenic. Perlecan is a large multidomain proteoglycan that has major effects in the development of the cardiovascular system from an embryonic stage of development, and into adulthood where perlecan is necessary for proper blood vessel maintenance. Perlecan-derived endorepellin signals through a dual receptor antagonism by modulating the activity of both VEGFR2 and α2β1 integrin, a process that ultimately leads to a suppression of powerful pro-angiogenic cues, including HIF-1α and VEGFA. The effects of endorepellin have been demonstrated in a number of in vitro and in vivo models placing it in the category of a molecule with true therapeutic potential. More importantly, exogenous endorepellin specifically targets the tumour vasculature thereby retarding cancer growth, metabolism and angiogenesis. Moreover, as endorepellin targets the α2β1 integrin, a powerful pro-angiogenic receptor that is overexpressed in actively-proliferating endothelial cells, it exerts a profound effect on tumor angiogenesis vis-à-vis normal blood vessels which usually express only low levels of this integrin. As more protein-based therapies to treat various pathologies start to be utilized in the clinics, a natural endogenous inhibitor of angiogenesis and pro-autophagic factor such as endorepellin could be of potential advantage. The use of endorepellin would carry very low risk to patients, as it is non-toxic and does not target normal vasculature (116).
Already the potential diagnostic and prognostic values of LG3 domain of endorepellin have been utilized, as LG3 has been reported to be a potential clinical biomarker in patients with premature rupture of fetal membranes (143), renal failure (144,145), and in vascular allograft rejection (146). Endorepellin and LG3 have been detected in the urine of children with sleep apnea (147), in the media conditioned by apoptotic endothelial cells (57,148,149), and in the secretome of pancreatic and colon carcinoma cells (105,150-152). Notably, circulating LG3 levels are reduced in breast cancer patients, suggesting that LG3 might be a useful biomarker for cancer progression and invasion (153). Although the discovery of endorepellin is still in its early stages, increasing evidence on the potential anti-cancer implications this protein can have is progressing. Granted that before endorepellin can be considered in the clinics, much more field work is required. The novel approach of targeted therapy using an endogenous protein that bares little toxicity, and aims to use the body’s own defence to directly curtail tumors nutrients, certainly represents an exciting avenue to explore in cancer treatment.
Acknowledgments
We like to thank Michaela Agapiou for critical reading of the manuscript. The original research was supported in part by National Institutes of Health Grants RO1 CA39481, RO1 CA47282 and RO1 CA164462.
Nomenclature
- ECM
extracellular matrix
- GAG
glycosaminoglycan
- HS
heparan sulfate
- HSPG
HS proteoglycan
- GAG
glycosaminoglycan
- VEGF
vascular endothelial cell growth factor
- VEGFR
VEGF receptor
- MMP
matrix metalloproteinase
- EGF
epidermal growth factor
- LG
laminin-like globular domain
- SHP1
Src homology protein phosphatase-1
- PLCγ
phospholipase Cγ
- Shb
SH2 domain-containing adaptor protein B
- PI3K
phosphatidyl inositide 3-kinases
- mTOR
mammalian target of rapamycin
- JNK
c-Jun N-terminal kinase
- PKC
protein kinase C
- AP1
activating protein 1
- HIF-1α
hypoxia-inducible factor 1α
- DIC
differential interference contrast microscopy
- HUVEC
human umbilical vein endothelial cell
- LC3
microtubule-associated protein 1A/1B-light chain 3
- VPS34
class III phosphoinositide kinase
- Peg3
paternally expressed gene 3
- PAE
porcine aortic endothelial cells
- DDSH
dyssegmental dysplasia Silverman-Handmaker type
- SJS
Schwartz-Jampel syndrome
- RTK
receptor tyrosine kinase
Footnotes
Declaration of interest
The authors declare that they have no competing interests. All the authors were involved in drafting the article, and approved the final version to be published.
References
- 1.Iozzo RV. Basement membrane proteoglycans: from cellar to ceiling. Nat Rev Mol Cell Biol. 2005;6:646–656. doi: 10.1038/nrm1702. [DOI] [PubMed] [Google Scholar]
- 2.Farach-Carson MC, Carson DD. Perlecan - a multifunctional extracellular proteoglycan scaffold. Glycobiology. 2007;17:897–905. doi: 10.1093/glycob/cwm043. [DOI] [PubMed] [Google Scholar]
- 3.Wilusz RE, Sanchez-Adams J, Guilak F. The structure and function of the pericellular matrix of articular cartilage. Matrix Biol. 2014;39:25–32. doi: 10.1016/j.matbio.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abedin M, King N. Diverse evolutionary paths to cell adhesion. Trends Cell Biol. 2010;20:734–742. doi: 10.1016/j.tcb.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iozzo RV, Murdoch AD. Proteoglycans of the extracellular environment: clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB J. 1996;10:598–614. [PubMed] [Google Scholar]
- 6.Iozzo RV, Schaefer L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015;42 doi: 10.1016/j.matbio.2015.02.003. In press. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iozzo RV, Schaefer L. Proteoglycans in health and disease: Novel regulatory signaling mechanisms evoked by the small leucine-rich proteoglycans. FEBS J. 2010;277:3864–3875. doi: 10.1111/j.1742-4658.2010.07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 9.Iozzo RV, Cohen IR, Grässel S, Murdoch AD. The biology of perlecan: the multifaceted heparan sulphate proteoglycan of basement membranes and pericellular matrices. Biochem J. 1994;302:625–639. doi: 10.1042/bj3020625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassell JR, Robey PG, Barrach HJ, Wilczek J, Rennard SI, Martin GR. Isolation of a heparan sulfate-containing proteoglycan from basement membrane. Proc Natl Acad Sci USA. 1980;77:4494–4498. doi: 10.1073/pnas.77.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murdoch AD, Dodge GR, Cohen I, Tuan RS, Iozzo RV. Primary structure of the human heparan sulfate proteoglycan from basement membrane (HSPG2/perlecan). A chimeric molecule with multiple domains homologous to the low density lipoprotein receptor, laminin, neural cell adhesion molecules, and epidermal growth factor. J Biol Chem. 1992;267:8544–8557. [PubMed] [Google Scholar]
- 12.Iozzo RV, Hassell JR. Identification of the precursor protein for the heparan sulfate proteoglycan of human colon carcinoma cells and its post-translational modifications. Arch Biochem Biophys. 1989;269:239–249. doi: 10.1016/0003-9861(89)90105-7. [DOI] [PubMed] [Google Scholar]
- 13.Dodge GR, Kovalszky I, Chu M-L, Hassell JR, McBride OW, Yi HF, Iozzo RV. Heparan sulfate proteoglycan of human colon: partial molecular cloning, cellular expression, and mapping of the gene (HSPG2) to the short arm of human chromosome 1. Genomics. 1991;10:673–680. doi: 10.1016/0888-7543(91)90451-j. [DOI] [PubMed] [Google Scholar]
- 14.Cohen IR, Grässel S, Murdoch AD, Iozzo RV. Structural characterization of the complete human perlecan gene and its promoter. Proc Natl Acad Sci USA. 1993;90:10404–10408. doi: 10.1073/pnas.90.21.10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iozzo RV, Pillarisetti J, Sharma B, Murdoch AD, Danielson KG, Uitto J, Mauviel A. Structural and functional characterization of the human perlecan gene promoter. Transcriptional activation by transforming factor-β via a nuclear factor 1-binding element. J Biol Chem. 1997;272:5219–5228. doi: 10.1074/jbc.272.8.5219. [DOI] [PubMed] [Google Scholar]
- 16.Iozzo RV, Danielson KG. Transcriptional and post-transcriptional control of proteoglycan gene expression. Progr Nucl Acids Res Mol Biol. 1999;62:19–53. doi: 10.1016/s0079-6603(08)60504-8. [DOI] [PubMed] [Google Scholar]
- 17.Sharma B, Iozzo RV. Transcriptional silencing of perlecan gene expression by interferon-γ. J Biol Chem. 1998;273:4642–4646. doi: 10.1074/jbc.273.8.4642. [DOI] [PubMed] [Google Scholar]
- 18.Dodge GR, Kovalszky I, Hassell JR, Iozzo RV. Transforming growth factor β alters the expression of heparan sulfate proteoglycan in human colon carcinoma cells. J Biol Chem. 1990;265:18023–18029. [PubMed] [Google Scholar]
- 19.Grässel S, Cohen IR, Murdoch AD, Eichstetter I, Iozzo RV. The proteoglycan perlecan is expressed in the erythroleukemia cell line K562 and is upregulated by sodium butyrate and phorbol ester. Mol Cell Biochem. 1995;145:61–68. doi: 10.1007/BF00925714. [DOI] [PubMed] [Google Scholar]
- 20.Jung M, Lord MS, Cheng B, Lyons JG, Alkhouri H, Hughes JM, McCarthy SJ, Iozzo RV, Whitelock JM. Mast cells produce novel shorter forms of perlecan that contain functional endorepellin: A role in angiogenesis and wound healing. J Biol Chem. 2013;288:3289–3304. doi: 10.1074/jbc.M112.387811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lord MS, Jung M, Cheng B, Whitelock JM. Transcriptional complexity of the HSPG2 gene in the human mast cell line, HMC-1. Matrix Biol. 2014;35:123–131. doi: 10.1016/j.matbio.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Handler M, Yurchenco PD, Iozzo RV. Developmental expression of perlecan during murine embryogenesis. Dev Dyn. 1997;210:130–145. doi: 10.1002/(SICI)1097-0177(199710)210:2<130::AID-AJA6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 23.Arikawa-Hirasawa E, Watanabe E, Takami H, Hassell JR, Yamada Y. Perlecan is essential for cartilage and cephalic development. Nat Genet. 1999;23:354–358. doi: 10.1038/15537. [DOI] [PubMed] [Google Scholar]
- 24.Costell M, Gustafsson E, Aszódi A, Mörgelin M, Bloch W, Hunziker E, Addicks K, Timpl R, Fässler R. Perlecan maintains the integrity of cartilage and some basement membranes. J Cell Biol. 1999;147:1109–1122. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arikawa-Hirasawa E, Wilcox WR, Le AH, Silverman N, Govindraj P, Hassell JR, Yamada Y. Dyssegmental dysplasia, Silverman-Handmaker type, is caused by functional null mutations of the perlecan gene. Nat Genet. 2001;27:431–434. doi: 10.1038/86941. [DOI] [PubMed] [Google Scholar]
- 26.Henriquez JP, Casar JC, Fuentealba L, Carey DJ, Brandan E. Extracellular matrix histone H1 binds to perlecan, is present in regenerating skeletal muscle and stimulates myoblast proliferation. J Cell Sci. 2002;115:2041–2051. doi: 10.1242/jcs.115.10.2041. [DOI] [PubMed] [Google Scholar]
- 27.Nicole S, Davoine C-S, Topaloglu H, Cattolico L, Barral D, Beighton P, Hamida CB, Hammouda H, Cruaud C, White PS, Samson D, Urtizberea JA, Lehmann-Horn F, Weissenbach J, Hentati F, Fontaine B. Perlecan, the major proteoglycan of basement membranes, is altered in patients with Schwartz-Jampel syndrome (chondrodystrophic myotonia) Nat Genet. 2000;26:480–483. doi: 10.1038/82638. [DOI] [PubMed] [Google Scholar]
- 28.Arikawa-Hirasawa E, Le AH, Nishino I, Nonaka I, Ho NC, Francomano CA, Govindraj P, Hassell JR, Devaney JM, Spranger J, Stevenson RE, Iannaccone S, Dalakas MC, Yamada Y. Structural and functional mutations of the perlecan gene cause Schwartz-Jampel syndrome, with myotonic myopathy and chondrodysplasia. Am J Hum Genet. 2002;70:1368–1375. doi: 10.1086/340390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warman ML, Cormier-Daire V, Hall C, Krakow D, Lachman R, LeMerrer M, Mortier G, Mundlos S, Nishimura G, Rimoin DL, Robertson S, Savarirayan R, Sillence D, Spranger J, Unger S, Zabel B, Superti-Furga A. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011;155A:943–968. doi: 10.1002/ajmg.a.33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Echaniz-Laguna A, Rene F, Marcel C, Bangratz M, Fontaine B, Loeffler J-P, Nicole S. Electrophysiological studies in a mouse model of Schwartz-Jampel syndrome demonstrate muscle fiber hyperactivity of peripheral nerve origin. Muscle Nerve. 2009;40:55–61. doi: 10.1002/mus.21253. [DOI] [PubMed] [Google Scholar]
- 31.Lowe DA, Lepori-Bui N, Fomin PV, Sloofman LG, Zhou X, Farach-Carson MC, Wang L, Kirn-Safran CB. Deficiency in perlecan/HSPG2 during bone development enhances osteogenesis and decreases quality of adult bone in mice. Calcif Tissue Int. 2014;95:29–38. doi: 10.1007/s00223-014-9859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodgers KD, Sasaki T, Aszodi A, Jacenko O. Reduced perlecan in mice results in chondrodysplasia resembling Schwartz-Jampel syndrome. Hum Mol Gen. 2007;16:515–528. doi: 10.1093/hmg/ddl484. [DOI] [PubMed] [Google Scholar]
- 33.Sasse P, Malan P, Fleischmann M, Roell W, Gustafsson E, Bostani T, Fan Y, Kolbe T, Breitbach M, Addicks K, Welz A, Brem G, Hescheler J, Aszodi A, Costell M, Bloch W, Fleischmann BK. Perlecan is critical for heart stability. Cardiovasc Res. 2008;80:435–444. doi: 10.1093/cvr/cvn225. [DOI] [PubMed] [Google Scholar]
- 34.Iozzo RV, San Antonio JD. Heparan sulfate proteoglycans: heavy hitters in the angiogenesis arena. J Clin Invest. 2001;108:349–355. doi: 10.1172/JCI13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15:1013–1031. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willis CD, Schaefer L, Iozzo RV. The biology of perlecan and its bioactive modules. In: Karamanos NK, editor. Extracellular Matrix: Pathobiology and Signaling. Walter de Gruyter GmbH & Co. KG; Berlin: 2012. pp. 171–184. [Google Scholar]
- 37.Carson DD, Tang J-P, Julian J. Heparan sulfate proteoglycan (perlecan) expression by mouse embryos during acquisition of attachment competence. Dev Biol. 1993;155:97–106. doi: 10.1006/dbio.1993.1010. [DOI] [PubMed] [Google Scholar]
- 38.Iozzo RV. Biosynthesis of heparan sulfate proteoglycan by human colon carcinoma cells and its localization at the cell surface. J Cell Biol. 1984;99:403–417. doi: 10.1083/jcb.99.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murdoch AD, Liu B, Schwarting R, Tuan RS, Iozzo RV. Widespread expression of perlecan proteoglycan in basement membranes and extracellular matrices of human tissues as detected by a novel monoclonal antibody against domain III and by in situ hybridization. J Histochem Cytochem. 1994;42:239–249. doi: 10.1177/42.2.7507142. [DOI] [PubMed] [Google Scholar]
- 40.Zoeller JJ, McQuillan A, Whitelock J, Ho S-Y, Iozzo RV. A central function for perlecan in skeletal muscle and cardiovascular development. J Cell Biol. 2008;181:381–394. doi: 10.1083/jcb.200708022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitelock JM, Graham LD, Melrose J, Murdoch AD, Iozzo RV, Underwood PA. Human perlecan immunopurified from different endothelial cell sources has different adhesive properties for vascular cells. Matrix Biol. 1999;18:163–178. doi: 10.1016/s0945-053x(99)00014-1. [DOI] [PubMed] [Google Scholar]
- 42.Farach-Carson MC, Brown AC, Lynam M, Safran JB, Carson DD. A novel peptide sequence in perlecan domain IV supports cell adhesion, spreading and FAK activation. Matrix Biol. 2008;27:150–160. doi: 10.1016/j.matbio.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farach-Carson MC, Warren CR, Harrington DA, Carson DD. Border patrol:Insights into the unique role of perlecan/heparan sulfate proteoglycan 2 at cell and tissue borders. Matrix Biol. 2014;34:64–79. doi: 10.1016/j.matbio.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lord MS, Chuang CY, Melrose J, Davies MJ, Iozzo RV, Whitelock JM. The role of vascular-derived perlecan in modulating cell adhesion, proliferation and growth factor signaling. Matrix Biol. 2014;35:112–122. doi: 10.1016/j.matbio.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grindel BJ, Martinez JR, Pennington CL, Muldoon M, Stave J, Chung LW, Farach-Carson MC. Matrilysin/matrix metalloproteinase-7(MMP7) cleavage of perlecan/HSPG2 creates a molecular switch to alter prostate cancer cell behavior. Matrix Biol. 2014;36:64–76. doi: 10.1016/j.matbio.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gotha L, Lim SY, Osherov AB, Wolff R, Qiang B, Erlich I, Nili N, Pillarisetti S, Chang YT, Tran PK, Tryggvason K, Hedin U, Tran-Lundmark K, Advani SL, Gilbert RE, Strauss BH. Heparan sulfate side chains have a critical role in the inhibitory effects of perlecan on vascular smooth muscle cell response to arterial injury. Am J Physiol Heart Circ Physiol. 2014;307:H337–H345. doi: 10.1152/ajpheart.00654.2013. [DOI] [PubMed] [Google Scholar]
- 47.Tran P-K, Tran-Lundmark K, Soininen R, Tryggvason K, Thyberg J, Hedin U. Increased intimal hyperplasia and smooth muscle cell proliferation in transgenic mice with heparan sulfate-deficient perlecan. Circ Res. 2004;94:550–558. doi: 10.1161/01.RES.0000117772.86853.34. [DOI] [PubMed] [Google Scholar]
- 48.Nugent MA, Nugent HM, Iozzo RV, Sanchack K, Edelman ER. Perlecan is required to inhibit thrombosis after deep vascular injury and contributes to endothelial cell-mediated inhibition of intimal hyperplasia. Proc Natl Acad Sci USA. 2000;97:6722–6727. doi: 10.1073/pnas.97.12.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iozzo RV. Turnover of heparan sulfate proteoglycan in human colon carcinoma cells. A quantitative biochemical and autoradiographic study. J Biol Chem. 1987;262:1888–1900. [PubMed] [Google Scholar]
- 50.Fuki I, Iozzo RV, Williams KJ. Perlecan heparan sulfate proteoglycan. A novel receptor that mediates a distinct pathway for ligand catabolism. J Biol Chem. 2000;275:25742–25750. doi: 10.1074/jbc.M909173199. [DOI] [PubMed] [Google Scholar]
- 51.Xu Y, Ashline D, Liu L, Tassa C, Shaw SY, Ravid K, Layne MD, Reinhold V, Robbins PW. The glycosylation-dependent interaction of perlecan core protein with LDL: implications for atherosclerosis. J Lipid Res. 2015;56:266–276. doi: 10.1194/jlr.M053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inomata T, Ebihara N, Funaki T, Matsuda A, Wantanabe Y, Ning L, Xu Z, Murakami A, Arikawa-Hirasawa E. Perlecan-deficient mutation impairs corneal epithelial structure. Invest Ophtalmol Vis Sci. 2012;53:1277–1284. doi: 10.1167/iovs.11-8742. [DOI] [PubMed] [Google Scholar]
- 53.Sher I, Zisman-Rozen S, Eliahu L, Whitelock JM, Maas-Szabowski N, Yamada Y, Breitkreutz D, Fusenig NE, Arikawa-Hirasawa E, Iozzo RV, Bergman R, Ron D. Targeting perlecan in human keratinocytes reveals novel roles for perlecan in epidermal formation. J Biol Chem. 2006;281:5178–5187. doi: 10.1074/jbc.M509500200. [DOI] [PubMed] [Google Scholar]
- 54.Kaneko H, Ishijima M, Futami I, Tomikawa-Ichikawa N, Kosaki K, Sadatsuki R, Yamada Y, Kurosawa H, Kaneko K, Arikawa-Hirasawa E. Synovial perlecan is required for osteophyte formation in knee osteoarthritis. Matrix Biol. 2013;32:178–187. doi: 10.1016/j.matbio.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.SundarRaj N, Fite D, Ledbetter S, Chakravarti S, Hassell JR. Perlecan is a component of cartilage matrix and promotes chondrocyte attachment. J Cell Sci. 1995;108:2663–2672. doi: 10.1242/jcs.108.7.2663. [DOI] [PubMed] [Google Scholar]
- 56.Jochmann K, Bachvarova V, Vortkamp A. Reprint of: Heparan sulfate as a regulator of endochondral ossification and osteochondroma development. Matrix Biol. 2014;35:239–247. doi: 10.1016/j.matbio.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 57.Laplante P, Raymond M-A, Labelle A, Abe J-I, Iozzo RV, Hebért M-J. Perlecan proteolysis induces α2β1 integrin and src-family kinases dependent anti-apoptotic pathway in fibroblasts in the absence of focal adhesion kinase activation. J Biol Chem. 2006;281:30383–30392. doi: 10.1074/jbc.M606412200. [DOI] [PubMed] [Google Scholar]
- 58.Rossi M, Morita H, Sormunen R, Airenne S, Kreivi M, Wang L, Fukai N, Olsen BR, Tryggvason K, Soininen R. Heparan sulfate chains of perlecan are indispensable in the lens capsule but not in the kidney. EMBO J. 2003;22:236–245. doi: 10.1093/emboj/cdg019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arikawa-Hirasawa E, Rossi SG, Rotundo RL, Yamada Y. Absence of acetylcholinesterase at the neuromuscular junctions of perlecan-null mice. Nat Neurosci. 2002;5:119–123. doi: 10.1038/nn801. [DOI] [PubMed] [Google Scholar]
- 60.Colombelli C, Palmisano M, Eshed-Eisenbach Y, Zambroni D, Pavoni E, Ferri C, Saccucci S, Nicole S, Soininen R, McKee KK, Yurchenco PD, Peles E, Wrabetz L, Feltri ML. Perlecan is recruited by dystroglycan to nodes of Ranvier and binds the clustering molecule gliomedin. J Cell Biol. 2015;208:313–329. doi: 10.1083/jcb.201403111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aviezer D, Hecht D, Safran M, Eisinger M, David G, Yayon A. Perlecan, basal lamina proteoglycan, promotes basic fibroblast growth factor-receptor binding, mitogenesis, and angiogenesis. Cell. 1994;79:1005–1013. doi: 10.1016/0092-8674(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 62.Aviezer D, Iozzo RV, Noonan DM, Yayon A. Suppression of autocrine and paracrine functions of basic fibroblast growth factor by stable expression of perlecan antisense cDNA. Mol Cell Biol. 1997;17:1938–1946. doi: 10.1128/mcb.17.4.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Segev A, Nili N, Strauss BH. The role of perlecan in arterial injury and angiogenesis. Cardiovasc Res. 2004;63:603–610. doi: 10.1016/j.cardiores.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 64.Zoeller JJ, Whitelock J, Iozzo RV. Perlecan regulates developmental angiogenesis by modulating the VEGF-VEGFR2 axis. Matrix Biol. 2009;28:284–291. doi: 10.1016/j.matbio.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitelock JM, Iozzo RV. Heparan sulfate: a complex polymer charged with biological activity. Chem Rev. 2005;105:2745–2764. doi: 10.1021/cr010213m. [DOI] [PubMed] [Google Scholar]
- 66.Mathiak M, Yenisey C, Grant DS, Sharma B, Iozzo RV. A role for perlecan in the suppression of growth and invasion in fibrosarcoma cells. Cancer Res. 1997;57:2130–2136. [PubMed] [Google Scholar]
- 67.Sharma B, Handler M, Eichstetter I, Whitelock J, Nugent MA, Iozzo RV. Antisense targeting of perlecan blocks tumor growth and angiogenesis in vivo. J Clin Invest. 1998;102:1599–1608. doi: 10.1172/JCI3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang X, Couchman JR. Perlecan and tumor angiogenesis. J Histochem Cytochem. 2003;51:1393–1410. doi: 10.1177/002215540305101101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bix G, Iozzo RV. Novel interactions of perlecan: Unraveling perlecan’s role in angiogenesis. Microsc Res. 2008;71:339–348. doi: 10.1002/jemt.20562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang J, Multhaupt H, Chan E, Schaefer L, Schaefer RM, Couchman JR. Essential contribution of tumor-derived perlecan to epidermal tumor growth and angiogenesis. J Histochem Cytochem. 2004;52:1575–1590. doi: 10.1369/jhc.4A6353.2004. [DOI] [PubMed] [Google Scholar]
- 71.Qiang B, Lim SY, Lekas M, Kuliszewski MA, Wolff R, Osherov AB, Rudenko D, Leong-Poi H, Noyan H, Husain M, Tran K, Tryggvason K, Hedin U, Tran-Lundmark K, Strauss BH. Perlecan heparan sulfate proteoglycan is a critical determinant of angiogenesis in response to mouse hind-limb ischemia. Can J Cardiol. 2014;30:1444–1451. doi: 10.1016/j.cjca.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 72.Ghiselli G, Eichstetter I, Iozzo RV. A role for the perlecan protein core in the activation of the keratinocyte growth factor receptor. Biochem J. 2001;359:153–163. doi: 10.1042/0264-6021:3590153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith SML, West LA, Hassell JR. The core protein of growth plate perlecan binds FGF-18 and alters its mitogenic effect on chondrocytes. Arch Biochem Biophys. 2007;468:244–251. doi: 10.1016/j.abb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vincent TL, McLean CJ, Full LE, Peston D, Saklatvala J. FGF-2 is bound to perlecan in the pericellular matrix of articular cartilage, where it acts as a chondrocyte mechanotransducer. Osteoarthritis Cartilage. 2007;15:752–763. doi: 10.1016/j.joca.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 75.Ishijima M, Suzuki N, Hozumi K, Matsunobu T, Kosaki K, Kaneko H, Hassell JR, Arikawa-Hirasawa E, Yamada Y. Perlecan modulates VEGF signaling and is essential for vascularization in endochondral bone formation. Matrix Biol. 2012;31:234–245. doi: 10.1016/j.matbio.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mongiat M, Fu J, Oldershaw R, Greenhalgh R, Gown A, Iozzo RV. Perlecan protein core interacts with extracellular matrix protein 1 (ECM1), a glycoprotein involved in bone formation and angiogenesis. J Biol Chem. 2003;278:17491–17499. doi: 10.1074/jbc.M210529200. [DOI] [PubMed] [Google Scholar]
- 77.Mongiat M, Taylor K, Otto J, Aho S, Uitto J, Whitelock J, Iozzo RV. The protein core of the proteoglycan perlecan binds specifically to fibroblast growth factor-7. J Biol Chem. 2000;275:7095–7100. doi: 10.1074/jbc.275.10.7095. [DOI] [PubMed] [Google Scholar]
- 78.Mongiat M, Otto J, Oldershaw R, Ferrer F, Sato JD, Iozzo RV. Fibroblast growth factor-binding protein is a novel partner for perlecan protein core. J Biol Chem. 2001;276:10263–10271. doi: 10.1074/jbc.M011493200. [DOI] [PubMed] [Google Scholar]
- 79.Gonzalez EM, Mongiat M, Slater SJ, Baffa R, Iozzo RV. A novel interaction between perlecan protein core and progranulin: Potential effects on tumor growth. J Biol Chem. 2003;278:38113–38116. doi: 10.1074/jbc.C300310200. [DOI] [PubMed] [Google Scholar]
- 80.Monami G, Gonzalez EM, Hellman M, Gomella LG, Baffa R, Iozzo RV, Morrione A. Proepithelin promotes migration and invasion of 5637 bladder cancer cells through the activation of ERK 1/2 and the formation of a paxillin/FAK/ERK complex. Cancer Res. 2006;66:7103–7110. doi: 10.1158/0008-5472.CAN-06-0633. [DOI] [PubMed] [Google Scholar]
- 81.Monami G, Emiliozzi V, Bitto A, Lovat F, Xu S-Q, Goldoni S, Fassan M, Serrero G, Gomella LG, Baffa R, Iozzo RV, Morrione A. Proepithelin regulates prostate cancer cell biology by promoting cell growth, migration, and anchorage-independent growth. Am J Pathol. 2009;174:1037–1047. doi: 10.2353/ajpath.2009.080735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iozzo RV, Zoeller JJ, Nyström A. Basement membrane proteoglycans: Modulators par excellence of cancer growth and angiogenesis. Mol Cells. 2009;27:503–513. doi: 10.1007/s10059-009-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gustafsson E, Almonte-Becerril M, Bloch W, Costell M. Perlecan maintains microvessel integrity in vivo and modulates their formation in vitro. PLoS ONE. 2013;8:e53715. doi: 10.1371/journal.pone.0053715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou Z, Wang J, Cao R, Morita H, Soininen R, Chan KM, Liu B, Cao Y, Tryggvason K. Impaired angiogenesis, delayed wound healing and retarded tumor growth in perlecan heparan sulfate-deficient mice. Cancer Res. 2004;64:4699–4702. doi: 10.1158/0008-5472.CAN-04-0810. [DOI] [PubMed] [Google Scholar]
- 85.Nonaka R, Iesaki T, de VS, Daida H, Okada T, Sasaki T, Arikawa-Hirasawa E. Perlecan deficiency causes endothelial dysfunction by reducing the expression of endothelial nitric oxide synthase. Physiol Rep. 2015:3. doi: 10.14814/phy2.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Whitelock JM, Murdoch AD, Iozzo RV, Underwood PA. The degradation of human endothelial cell-derived perlecan and release of bound basic fibroblast growth factor by stromelysin, collagenase, plasmin and heparanases. J Biol Chem. 1996;271:10079–10086. doi: 10.1074/jbc.271.17.10079. [DOI] [PubMed] [Google Scholar]
- 87.Reiland J, Sanderson RD, Waguespack M, Barker SA, Long R, Carson DD, Marchetti D. Heparanase degrades syndecan-1 and perlecan heparan sulfate: functional implications for tumor cell invasion. J Biol Chem. 2004;279:8047–8055. doi: 10.1074/jbc.M304872200. [DOI] [PubMed] [Google Scholar]
- 88.Sanderson RD, Iozzo RV. Targeting heparanase for cancer therapy at the tumor-matrix interface. Matrix Biol. 2012;31:283–284. doi: 10.1016/j.matbio.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 89.Peysselon F, Ricard-Blum S. Heparin-protein interactions: from affinity and kinetics to biological roles. Application to an interaction network regulating angiogenesis. Matrix Biol. 2014;35:73–81. doi: 10.1016/j.matbio.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 90.Vlodavsky I, Iozzo RV, Sanderson RD. Heparanase: multiple functions in inflammation, diabetes and atherosclerosis. Matrix Biol. 2013;32:220–222. doi: 10.1016/j.matbio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 91.Vlodavsky I, Blich M, Li JP, Sanderson RD, Ilan N. Involvement of heparanase in atherosclerosis and other vessel wall pathologies. Matrix Biol. 2013;32:241–251. doi: 10.1016/j.matbio.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peterson SB, Liu J. Multi-faceted substrate specificity of heparanase. Matrix Biol. 2013;32:223–227. doi: 10.1016/j.matbio.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 93.Goldberg R, Meirovitz A, Hirshoren N, Bulvik R, Binder A, Rubinstein AM, Elkin M. Versatile role of heparanase in inflammation. Matrix Biol. 2013;32:234–240. doi: 10.1016/j.matbio.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parish CR, Freeman C, Ziolkowski AF, He YQ, Sutcliffe EL, Zafar A, Rao S, Simeonovic CJ. Unexpected new roles for heparanase in Type 1 diabetes and immune gene regulation. Matrix Biol. 2013;32:228–233. doi: 10.1016/j.matbio.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 95.Cohen IR, Murdoch AD, Naso MF, Marchetti D, Berd D, Iozzo RV. Abnormal expression of perlecan proteoglycan in metastatic melanomas. Cancer Res. 1994;54:5771–5774. [PubMed] [Google Scholar]
- 96.Iozzo RV, Cohen I. Altered proteoglycan gene expression and the tumor stroma. Experientia. 1993;49:447–455. doi: 10.1007/BF01923588. [DOI] [PubMed] [Google Scholar]
- 97.Davies EJ, Backhall FH, Shanks JH, David G, McGown AT, Swindell R, Slade RJ, Martin-Hirsch P, Gallagher JT, Jayson GC. Distribution and clinical significance of heparan sulfate proteoblycans in ovarian cancer. Clin Cancer Res. 2004;10:5178–5186. doi: 10.1158/1078-0432.CCR-03-0103. [DOI] [PubMed] [Google Scholar]
- 98.Kawahara R, Granato DC, Carnielli CM, Cervigne NK, Oliveria CE, Martinez CA, Yokoo S, Fonseca FP, Lopes M, Santos-Silva AR, Graner E, Coletta RD, Paes Leme AF. Agrin and perlecan mediate tumorigenic processes in oral squamous cell carcinoma. PLoS One. 2014;9:e115004. doi: 10.1371/journal.pone.0115004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Duncan MB. Extracellular matrix transcriptome dynamics in hepatocellular carcinoma. Matrix Biol. 2013;32:393–398. doi: 10.1016/j.matbio.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 100.Savoré C, Zhang C, Muir C, Liu R, Wyrwa J, Shu J, Zhau HE, Chung LW, Carson DD, Farach-Carson MC. Perlecan knockdown in metastatic prostate cancer cells reduces heparin-binding growth factor responses in vitro and tumor growth in vivo. Clin Exp Metastasis. 2005;22:377–390. doi: 10.1007/s10585-005-2339-3. [DOI] [PubMed] [Google Scholar]
- 101.Warren CR, Grindel BJ, Francis L, Carson DD, Farach-Carson MC. Transcriptional activation by NFkappaB increases perlecan/HSPG2 expression in the desmoplastic prostate tumor microenvironment. J Cell Biochem. 2014;115:1322–1333. doi: 10.1002/jcb.24788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mongiat M, Sweeney S, San Antonio JD, Fu J, Iozzo RV. Endorepellin, a novel inhibitor of angiogenesis derived from the C terminus of perlecan. J Biol Chem. 2003;278:4238–4249. doi: 10.1074/jbc.M210445200. [DOI] [PubMed] [Google Scholar]
- 103.Le BV, Kim H, Choi J, Kim J-H, Hahn M-J, Lee C, Kim KK, Hwang H-Y. Crystal Structure of the LG3 domain of endorepellin, an angiogenesis inhibitor. J Mol Biol. 2011;414:231–242. doi: 10.1016/j.jmb.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 104.Cailhier J-F, Sirois I, Raymond M-A, Lepage S, Laplante P, Brassard N, Prat A, Iozzo RV, Pshezhetsky AV, Hebért M-J. Caspase-3 activation triggers extracellular release of cathepsin L and endorepellin proteolysis. J Biol Chem. 2008;283:27220–27229. doi: 10.1074/jbc.M801164200. [DOI] [PubMed] [Google Scholar]
- 105.Gonzalez EM, Reed CC, Bix G, Fu J, Zhang Y, Gopalakrishnan B, Greenspan DS, Iozzo RV. BMP-1/Tolloid-like metalloproteases process endorepellin, the angiostatic C-terminal fragment of perlecan. J Biol Chem. 2005;280:7080–7087. doi: 10.1074/jbc.M409841200. [DOI] [PubMed] [Google Scholar]
- 106.O’Riordan E, Orlova TN, Mendelev N, Patschan D, Kemp R, Chander PN, Hu R, Hao G, Gross SS, Iozzo RV, Delaney V, Goligorsky MS. Urinary proteomic analysis of chronic renal allograft nephropathy. Proteomics Clin Appl. 2008;2:1025–1035. doi: 10.1002/prca.200780137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang H, Listrat A, Meunier B, Gueugneau M, Coudy-Gandilhon C, Combaret L, Taillandier D, Polge C, Attaix D, Lethias C, Lee K, Goh KL, Bechet D. Apoptosis in capillary endothelial cells in ageing skeletal muscle. Aging Cell. 2014;13:254–262. doi: 10.1111/acel.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Whitelock JM, Melrose J, Iozzo RV. Diverse cell signaling events modulated by perlecan. Biochemistry. 2008;47:11174–11183. doi: 10.1021/bi8013938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lindner JR, Hillman PR, Barrett AL, Jackson MC, Perry TL, Park Y, Datta S. The Drosophila Perlecan gene trol regulates multiple signaling pathways in different developmental contexts. BMC Dev Biol. 2007;7:121. doi: 10.1186/1471-213X-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Trisnadi N, Stathopoulos A. Ectopic Expression Screen Identifies Genes Affecting Drosophila Mesoderm Development Including the HSPG Trol. G3 (Bethesda ) 2014;5:301–313. doi: 10.1534/g3.114.015891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Datta MW, Hernandez AM, Schlicht MJ, Kahler AJ, DeGueme AM, Dhir R, Shah RB, Farach-Carson C, Barrett A, Datta S. Perlecan, a candidate gene for the CAPB locus, regulates prostate cancer cell growth via the Sonic Hedgehog pathway. Mol Cancer. 2006;5:9. doi: 10.1186/1476-4598-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Datta S, Pierce M, Datta MW. Perlecan signaling: Helping hedgehog stimulate prostate cancer growth. Int J Biochem Cell Biol. 2006;38:1855–1861. doi: 10.1016/j.biocel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 113.Goyal A, Poluzzi C, Willis AC, Smythies J, Shellard A, Neill T, Iozzo RV. Endorepellin affects angiogenesis by antagonizing diverse VEGFR2- evoked signaling pathways: transcriptional repression of HIF-1α and VEGFA and concurrent inhibition of NFAT1 activation. J Biol Chem. 2012;287:43543–43556. doi: 10.1074/jbc.M112.401786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goyal A, Pal N, Concannon M, Paulk M, Doran M, Poluzzi C, Sekiguchi K, Whitelock JM, Neill T, Iozzo RV. Endorepellin, the angiostatic module of perlecan, interacts with both the α2β1 integrin and vascular endothelial growth factor receptor 2 (VEGFR2) J Biol Chem. 2011;286:25947–25962. doi: 10.1074/jbc.M111.243626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bix G, Fu J, Gonzalez E, Macro L, Barker A, Campbell S, Zutter MM, Santoro SA, Kim JK, Höök M, Reed CC, Iozzo RV. Endorepellin causes endothelial cell disassembly of actin cytoskeleton and focal adhesions through the α2β1 integrin. J Cell Biol. 2004;166:97–109. doi: 10.1083/jcb.200401150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bix G, Castello R, Burrows M, Zoeller JJ, Weech M, Iozzo RA, Cardi C, Thakur MT, Barker CA, Camphausen KC, Iozzo RV. Endorepellin in vivo: targeting the tumor vasculature and retarding cancer growth and metabolism. J Natl Cancer Inst. 2006;98:1634–1646. doi: 10.1093/jnci/djj441. [DOI] [PubMed] [Google Scholar]
- 117.Bix G, Iozzo RA, Woodall B, Burrows M, McQuillan A, Campbell S, Fields GB, Iozzo RV. Endorepellin, the C-terminal angiostatic module of perlecan, enhances collagen-platelet responses via the α2β1 integrin receptor. Blood. 2007;109:3745–3748. doi: 10.1182/blood-2006-08-039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bix G, Iozzo RV. Matrix revolutions: "tails" of basement-membrane components with angiostatic functions. Trends Cell Biol. 2005;15:52–60. doi: 10.1016/j.tcb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 119.Nyström A, Shaik ZP, Gullberg D, Krieg T, Eckes B, Zent R, Pozzi A, Iozzo RV. Role of tyrosine phosphatase SHP-1 in the mechanism of endorepellin angiostatic activity. Blood. 2009;114:4897–4906. doi: 10.1182/blood-2009-02-207134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zoeller JJ, Iozzo RV. Proteomic profiling of endorepellin angiostatic activity on human endothelial cells. Proteome Sci. 2008;6:7. doi: 10.1186/1477-5956-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Senger DR, Claffey KP, Benes JE, Perruzzi CA, Sergiou AP, Detmar M. Angiogenesis promoted by vascular endothelial growth factor: regulation through α1β1 and α2β1 integrins. Proc Natl Acad Sci USA. 1997;94:13612–13617. doi: 10.1073/pnas.94.25.13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sweeney SM, DiLullo G, Slater SJ, Martinez J, Iozzo RV, Lauer-Fields JL, Fields GB, San Antonio JD. Angiogenesis in collagen I requires α2β1 ligation of a GFP*GER sequence and possible p38 MAPK activation and focal adhesion disassembly. J Biol Chem. 2003;278:30516–30524. doi: 10.1074/jbc.M304237200. [DOI] [PubMed] [Google Scholar]
- 123.San Antonio JD, Zoeller JJ, Habursky K, Turner K, Pimtong W, Burrows M, Choi S, Basra S, Bennett JS, DeGrado WF, Iozzo RV. A key role for the integrin α2β1 in experimental and developmental angiogenesis. Am J Pathol. 2009;175:1338–1347. doi: 10.2353/ajpath.2009.090234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Woodall BP, Nyström A, Iozzo RA, Eble JA, Niland S, Krieg T, Eckes B, Pozzi A, Iozzo RV. Integrin α2β1 is the required receptor for endorepellin angiostatic activity. J Biol Chem. 2008;283:2335–2343. doi: 10.1074/jbc.M708364200. [DOI] [PubMed] [Google Scholar]
- 125.Willis CD, Poluzzi C, Mongiat M, Iozzo RV. Endorepellin laminin-like globular repeat 1/2 domains bind Ig3-5 of vascular endothelial growth factor(VEGF) receptor 2 and block pro-angiogenic signaling by VEGFA in endothelial cells. FEBS J. 2013;280:2271–2294. doi: 10.1111/febs.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Poluzzi C, Casulli J, Goyal A, Mercer TJ, Neill T, Iozzo RV. Endorepellin evokes autophagy in endothelial cells. J Biol Chem. 2014;289:16114–16128. doi: 10.1074/jbc.M114.556530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 131.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 132.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 133.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nature Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ramakrishnan S, Nguygen TMB, Subramanian IV, Kelekar A. Autophagy and angiogenesis inhibition. Autophagy. 2007;3:512–515. doi: 10.4161/auto.4734. [DOI] [PubMed] [Google Scholar]
- 137.Nguyen TMB, Subramanian IV, Xiao X, Ghosh G, Nguyen P, Kelekar A, Ramakrishnan S. Endostatin induces autophagy in endothelial cells by modulating Beclin 1 and β-catenin levels. J Cell Mol Med. 2009;13:3687–3698. doi: 10.1111/j.1582-4934.2009.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lock R, Debnath J. Extracellular matrix regulation of autophagy. Curr Opin Cell Biol. 2008;20:583–588. doi: 10.1016/j.ceb.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Neill T, Schaefer L, Iozzo RV. Instructive roles of extracellular matrix on autophagy. Am J Pathol. 2014;184:2146–2153. doi: 10.1016/j.ajpath.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 142.Buraschi S, Neill T, Goyal A, Poluzzi C, Smythies J, Owens RT, Schaefer L, Torres AT, Iozzo RV. Decorin causes autophagy in endothelial cells via Peg3. Proc Natl Acad Sci USA. 2013;110:E2582–E2591. doi: 10.1073/pnas.1305732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Thadikkaran L, Crettaz D, Siegenthaler MA, Gallot D, Sapin V, Iozzo RV, Queloz PA, Schneider P, Tissot JD. The role of proteomics in the assessment of premature rupture of fetal membranes. Clin Chim Acta. 2005;360:27–36. doi: 10.1016/j.cccn.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 144.Parker TJ, Sampson DL, Broszczak D, Chng YL, Carter SL, Leavesley DI, Parker AW, Upton Z. A fragment of the LG3 peptide of endorepellin is present in the urine of physically active mining workers: a potential marker of physical activity. PLoS ONE. 2012;7:e33714. doi: 10.1371/journal.pone.0033714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Surin B, Sachon E, Rougier J-P, Steverlynck C, Garreau C, Lelongt B, Ronco P, Piedagnel R. LG3 fragment of endorepellin is a possible biomarker of severity in lgA nephropathy. Proteomics. 2013 doi: 10.1002/pmic.201200267. In Press. [DOI] [PubMed] [Google Scholar]
- 146.Soulez M, Pilon E-A, Dieudé M, Cardinal H, Brassard N, Qi S, Wu S-J, Durocher Y, Madore F, Perreault C, Hébert M-J. The perlecan fragment LG3 is a novel regulator of obliterative remodeling associated with allograft vascular rejection. Circ Res. 2012;110:94–104. doi: 10.1161/CIRCRESAHA.111.250431. [DOI] [PubMed] [Google Scholar]
- 147.Krishna J, Shah ZA, Merchant M, Klein JB, Gozal D. Urinary protein expression patterns in children with sleep-disordered breathing: preliminary findings. Sleep Med. 2006;7:221–227. doi: 10.1016/j.sleep.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 148.Raymond M-A, Désormeaux A, Laplante P, Vigneault N, Filep JG, Landry K, Pshezhetsky AV, Hébert M-J. Apoptosis of endothelial cells triggers a caspase-dependent anti-apoptotic paracrine loop active on vascular smooth muscle cells. FASEB J. 2004;18:705–707. doi: 10.1096/fj.03-0573fje. [DOI] [PubMed] [Google Scholar]
- 149.Laplante P, Raymond MA, Gagnon G, Vigneault N, Sasseville AM, Langelier Y, Bernard M, Raymond Y, Hebért M-J. Novel fibrogenic pathways are activated in response to endothelial apoptosis: implications in the pathophysiology of systemic sclerosis. J Immunol. 2005;174:5740–5749. doi: 10.4049/jimmunol.174.9.5740. [DOI] [PubMed] [Google Scholar]
- 150.Mauri P, Scarpa A, Nascimbeni AC, Benazzi L, Parmagnani E, Mafficini A, Della Peruta M, Bassi C, Miyazaki K, Sorio C. Identification of proteins released by pancreatic cancer cells by multidimensional protein identification technology: a strategy for identification of novel cancer markers. FASEB J. 2005;19:1125–1127. doi: 10.1096/fj.04-3000fje. [DOI] [PubMed] [Google Scholar]
- 151.Grønborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteom. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 152.Aspinall-O’Dea M, Costello E. The pancreatic cancer proteome-recent advances and future promise. Proteomics Clin Appl. 2007;1:1066–1079. doi: 10.1002/prca.200700144. [DOI] [PubMed] [Google Scholar]
- 153.Chang JW, Kang U-B, Kim DH, Yi JK, Lee JW, Noh D-Y, Lee C, Yu M-H. Identification of circulating endorepellin LG3 fragment: Potential use as a serological biomarker for breast cancer. Proteomics Clin Appl. 2008;2:23–32. doi: 10.1002/prca.200780049. [DOI] [PubMed] [Google Scholar]