Abstract
Early research on the cyanobacterial clock focused on characterizing the genes that are needed to keep, entrain, and convey time within the cell. As the scope of assays used in molecular genetics expanded to capture systems-level properties (i.e. RNA-seq, ChIP-seq, metabolomics, high-throughput screening of genetic variants), so did our understanding of how the clock fits within and influences a broader cellular context. Here we review the work that has established a global perspective of the clock with a focus on: (1) an emerging network-centric view of clock architecture, (2) mechanistic insights on how temporal and environmental cues are transmitted and integrated within this network, (3) the systematic alteration of gene expression and cellular metabolism by the clock, and (4) insights on the evolution of temporal control in cyanobacteria.
Keywords: Circadian Rhythms, kai Oscillator, RpaA, Global Regulation
1. Introduction
1.1. The Simplest Circadian Clock
Circadian control over biological processes is pervasive among eukaryotes and cyanobacteria, suggesting a strong evolutionary advantage for anticipating regular fluctuations in the environment (63, 61, 10). This advantage has been experimentally demonstrated in cyanobacteria, where cultures that have the same circadian period as external light sources out-compete cultures with periods that differ (61, 19). In eukaryotes, several mechanisms for timekeeping have been elucidated (63, 67). While the components of these clocks vary between species, all of the eukaryotic systems are thought to fundamentally keep time through interlocking transcription / translation feedback loops, suggesting a common evolved network motif (63). The cyanobacterial clock is unique in its architecture, in that transcriptional feedback is not essential for the maintenance of time (85, 59). Nonetheless, the cyanobacterial timing system still exhibits the fundamental properties or persistence, entrainment, and temperature compensation established for eukaryotic circadian clocks (13), making it a valuable and simpler system to study circadian rhythms.
The majority of work on the cyanobacterial clock has been in Synechococcus elongatus PCC 7942 (S. elongatus), which has several key properties including: a simple cellular structure and genome size (2.7Mb, about 2700 genes) which reduces the complexity of genetic interactions; natural competence enabling systematic genetic manipulation (72, 26); and a powerful luciferase-based reporter system that can be utilized to quantify circadian rhythms from hundreds to thousands of genomic variants concurrently (44, 42, 52, 74). These properties and the standardization of high-throughput molecular assays that capture systems-level attributes (i.e. RNA-seq, ChIP-seq, metabolomics) make it possible to not only study the mechanism of the clock, but determine how it fits within and influences a broader cellular context.
1.2. Components of the Cyanobacterial Clock
Several reviews on the molecular mechanism of the S. elongatus clock have been published (53, 35, 15). We will not go into the level of detail previously covered, but will briefly summarize key components of the clock for reference. As previously mentioned, circadian rhythms in S. elongatus can be quantified using a luciferase-based reporter assay, where either the bacterial or firefly luciferase gene is placed under control of a circadian regulated promoter, and its expression is measured over time as changes in bioluminescence (44). To identify the genes that compose and influence the clock, random chemical and transposon mutagenesis screens were performed on reporter strains and mutants with altered circadian rhythms were selected and characterized (45, 26, 38, 71, 37). After extensive screening, only 19 genes have been shown to reliably alter circadian rhythms when knocked-out or over-expressed (Table 1), suggesting that the circadian phenotype is determined by a relatively small number of genes, and is fairly robust to genomic perturbations.
Table 1. Genetic effectors of circadian rhythms.
| Gene | How it was Discovered | Knockout Phenotype | Interaction Partners |
|---|---|---|---|
| kaiB | EMS (45) and Comp Screen (29) | Arrhythmic (45, 29) | KaiA(86), KaiC(8), CikA(7) |
| kaiC | EMS (45) and Comp Screen (29) | Arrhythmic (45, 29) | KaiA(65), KaiB(8), SasA(33), CikA(32), ATP / ADP (68) |
| kaiA | EMS (45) and Comp Screen (29) | Arrhythmic (45, 29) | KaiB(86), KaiC(65), LdpA(31), quinone (40) |
| ldpA | TN Mut Screen (37) | Per -1h, Light insensitive | KaiA(31), CikA(31), SasA(31), FeS Cluster(37) |
| pex | KaiC Mut Sup Screen (47) | Per -1h, Abnormal Resetting (47, 78) |
DNA: kaiA Promoter (1) |
| prkE | Y2H Screen with CikA (51) | Abnormal Resetting (51) | CikA(51) |
| cikA | TN Mut Screen (71) | Per -2.5h, No Phase Reset, Low Amp (71) |
KaiC(32), KaiB(7), RpaA(21), LdpA(31), quinone(32) |
| sasA | Y2H Screen with KaiC (33) | Per -3h, Low Amp (33) | KaiC (33), RpaA(21), LdpA(31), RpaB(39) |
| rpaA | Response Reg KO Screen (79) | Arrhythmic (55) | CikA(21), SasA(21), DNA: kaiBC Promoter(55) |
| rpaB | Similar to RpaA (22) | Nonviable (22) | SasA (39), DNA: kaiBC Promoter(22) |
| labA | KaiC Mut Sup Screen (81) | Low Amp, Per - WT (81) | |
| cpmA | TN Mut Screen (38) | Shifts Phase of some reporters 10h (38) |
|
| crm | TN Mut Screen (6) | Arrhythmic (6) | |
| clpX | TN Mut Screen (26) | Period Increase (26) | |
| clpPII | TN Mut Screen (26) | Period Increase (26) | |
| nht1 | Y2H Screen with CikA (51) | Per +1h when over exp. (51) | CikA(51) |
| ircA | Y2H Screen with CikA (51) | Phase +8h when over exp. (51) | CikA(51) |
| cdpA | Y2H Screen with CikA (51) | Phase +6h when over exp. (51) | CikA(51) |
| lalA | Similar to LabA (82) | Dec Amp when over exp. (82) |
Abbreviations: EMS - EMS Mutagenesis Screen. Comp Screen - Complement Screen. TN Mut Screen - Transposon Mutagenesis Screen. Mut Sup Screen - Mutation Suppressor Screen. Y2H Screen - Yeast Two Hybrid Screen. Response Reg KO Screen - Response Regulator Knockout Screen. Per - Period. Amp - Amplitude. Over Exp - Over Expressed.
The core circadian oscillator in S. elongatus is encoded by three neighboring genes, kaiA, kaiB, and kaiC, two of which are expressed on a single message as kaiBC (29). Their products were identified as parts of the clock early on, as inactivation of any of them abolishes rhythms. KaiC is an autokinase, autophosphatase, and ATPase; in complex with KaiA and KaiB, KaiC displays a daily rhythm of phosphorylation at residues Ser431 and Thr432 (60, 95, 69). KaiA stimulates KaiC autophosphorylation and KaiB opposes KaiA’s stimulatory activity leading to KaiC dephosphorylation (90). The 24-h KaiC phosphorylation pattern can be reconstituted in vitro by merely combining the three Kai proteins and ATP, suggesting that the phosphorylation cycle is the fundamental timekeeping mechanism in cyanobacteria (59). However, the low ATPase activity of KaiC, about 10-30 ATP hydrolyzed per monomer per day, also oscillates in a circadian manner, is intrinsically temperature-compensated, and determines circadian period length, suggestive of a timekeeping role that may be separable from the cycle of KaiC’s phosphorylation state (84).
Synchronization of the oscillator to the external environment is mediated in large part by CikA (circadian input kinase A) and KaiA, both of which have been shown to indirectly sense light through the redox state of the pool of plastoquinone: a co-factor whose status varies with photosynthetic activity (32, 94, 40). The binding of oxidized quinones by KaiA causes it to aggregate, leading to a decrease in KaiC’s autophosphorylation efficiency (94). How the binding of quinone by CikA impacts entrainment has not been as thoroughly investigated, but CikA has been shown to modify the phosphorylation state of KaiC, and is less stable when bound to quinone (32). The clearest evidence for CikA’s involvement in entrainment is the failure of cikA-null strains to reset the clock after a 5-h dark pulse, a potent resetting signal in WT cells (71). Alterations to the ATP / ADP ratio (which, like cellular redox state, varies with photosynthetic activity) also affect entrainment. Phase resetting in the in vitro oscillator can be achieved simply by altering this ratio, presumably by directly affecting the autophosphorylation efficiency of KaiC (68).
Entrainment and light sensitivity defects have been observed in ldpA-, pex-, and prkE- null strains, but the roles of these genes in the clock are not well established. LdpA (light dependent period A) copurifies with CikA and KaiA, and like those proteins also binds a redox-active cofactor, suggesting that it is involved in entrainment by a photosynthesis-sensing mechanism (31). This idea is supported by the circadian phenotype observed in ldpA-null strains, in which the period is 1 hour shorter and does not vary with light intensity, as does circadian period in WT (37). Knockouts of either pex (period extender) or prkE (phase resetting kinase E) result in an altered phase response of the clock after a dark pulse (78, 51). The Pex protein contains a DNA-binding domain, and binds the kaiA promoter in vitro (1). The resetting defect in a pex-null background may be explained by a misregulation of kaiA gene expression. It is unclear through what mechanism PrkE functions, but it may be mediated through CikA, as they were shown to interact in a yeast two-hybrid assay (51).
Temporal information from the cyanobacterial oscillator is transmitted to downstream genes via the histidine protein kinase SasA (Synechococcus adaptive sensor A), whose autophosphorylation is stimulated by interaction with KaiC (33, 79). Phosphorylated SasA in turn transfers a phosphoryl group to RpaA (regulator of phycobilisome association A) (21, 2), a transcription factor that directly regulates the expression of approximately 100 genes, as determined by ChIP-seq (55). Moreover, RpaA indirectly regulates the expression of nearly all genes in the genome as shown by the complete loss of rhythms of S. elongatus transcripts in an rpaA-null strain as determined by RNA-seq (55). Disruption of sasA also results in severely damped gene expression rhythms (33, 79). Surprisingly, the phosphorylation state of RpaA, and subsequently its activity, have been shown to be dependent on CikA, which was primarily thought to be involved in entrainment (21). Furthermore, the transcription factor RpaB appears to influence the RpaA phosphorylation state (16), and binds to DNA binding sites that overlap those of RpaA, perhaps occluding its binding to promoters including that of kaiBC (22, 55). As RpaA is emerging as the integration point of circadian and environmental input into the regulation of gene expression (16), much of this review will focus on its mechanism of phosphorylation, and global impact on transcription and metabolism.
Additional genes influence the circadian control of gene expression, but their mechanisms are not well characterized (Table 1). LabA (low amplitude and bright) and the product of a paralogous gene, LalA (labA-like A), have both been shown to affect the amplitude of rhythms (81, 82), presumably through altering the redox state of the cell via FMNH metabolism (82); however, no direct association between these proteins and the known clock components has been observed. CpmA (circadian phase modifier A) was identified as affecting phasic expression of the kaiA, psbAI and psbAII promoters (38). Deletions of cpmA are characterized by a severe growth phenotype, suggesting that the lack of the cpmA gene product may disrupt general cellular metabolism.
2. A Network-Centric View of the Clock
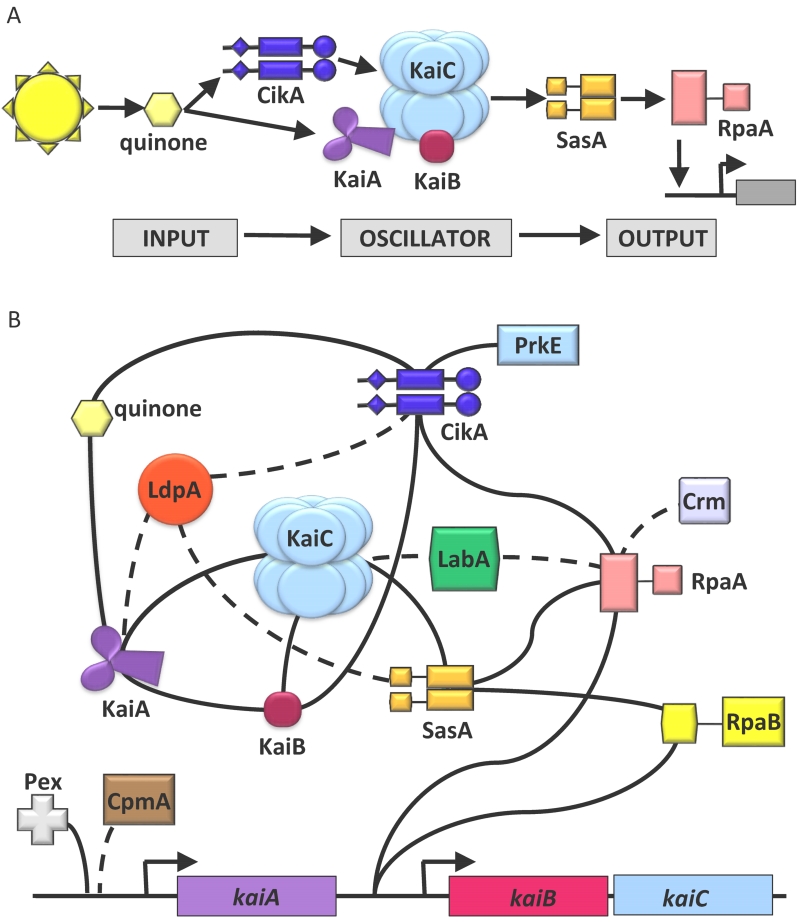
The dominant representation of the clock has been as a linear architecture that consists of (1) an input pathway that reacts to cellular and environmental cues and entrains (2) the core oscillator, which subsequently controls cellular transcription through (3) an output pathway (53) (Figure 1A). Recent findings have begun to dissolve this construct, and suggest that the clock is better represented as a highly connected network of proteins where input and output are dependent upon overlapping interactions (73). To illustrate the high degree of connectivity between components, we list the binding partners for all clock genes in Table 1 and schematically show the known network of clock gene products in Figure 1B.
Figure 1. The clock is a highly integrated network of proteins.
(A) The clock has traditionally been thought of as separable, sequential processes: input, oscillator, output. (B) Recent findings have shown these processes share many proteins and are highly dependent on each other. A better clock representation is as a highly integrated network of proteins. Solid lines represent verified, physical interactions between network components. Dashed lines represent suspected interactions. This figure is a schematic representation of the data in Table 1.
2.1. Dissolving the construct of separable input and output pathways
The first evidence of a highly integrated clock network came when CikA, traditionally thought of as an input protein, was found to be important for clock output, showing that the same protein could be involved in both processes. The prevailing understanding of clock output and RpaA-mediated transcriptional control operated under the linear paradigm that KaiC stimulates phosphorylation of SasA, which phosphorylates and activates RpaA, which regulates the expression of the majority of genes in the genome. This concept changed when Gutu and O’Shea characterized the circadian phosphorylation profile of RpaA in vitro and in vivo (21). Here, they recapitulated findings that phosphorylation of RpaA by SasA in vitro is enhanced in the presence of KaiC (33). More significantly, they demonstrated that CikA acts as a phosphatase on RpaA in vitro, and this activity is highly stimulated in the presence of KaiBC. Through examination of phosphomimetic alleles that “lock” KaiC into a specific phosphorylation state, the authors concluded that the pSpT-phosphostate of KaiC stimulates SasA phosphorylation, whereas the pST-KaiC-KaiB complex stimulates dephosphorylation of RpaA by CikA, supporting a simple model of circadian RpaA activation wherein SasA and CikA exert opposing effects to produce an oscillation of RpaA phosphorylation. This study alone illustrates the dependence of output on multiple, coordinated interactions: CikA with RpaA, SasA with RpaA, CikA with KaiB and KaiC, SasA with KaiC.
While several genes have been identified that influence entrainment, the details of clock input are not as well understood. To better characterize what genetic interactions are important to both input and output, and how sensitive the network is to alterations in these interactions, we performed a second-site suppressor mutagenesis screen on a cikA-null strain (73). As a cikA deletion mutant exhibits defects in both clock output and entrainment (2.5-h decrease in period, low amplitude oscillations, lack of resetting ability after a dark pulse (71)), we sought to identify mutations that could rescue either (73). We recovered two independent, single-nucleotide mutations in sasA (traditionally only attributed to clock output) that restored normal rhythms and, surprisingly, also the ability to reset circadian phase after a 5-h dark pulse in a cikA-null background. These properties were maintained even when the kaiBC promoter was removed from SasA-RpaA control, suggesting that suppression of cikA-null phenotypes was not simply explained by an attenuation of the transcription-translation feedback loop. This result showed that clock entrainment is sensitive to alterations in the biochemical properties of the clock output protein SasA, and conceptually, further connected input and output.
The strongest evidence that input and output are linked comes from KaiA/ KaiB/ KaiC / SasA cooperative binding studies (86, 7). KaiB and SasA compete for binding on a region of KaiC termed the B-loop (86). As previously mentioned, KaiB binding to KaiC stimulates KaiC dephosphorylation, and SasA binding leads to SasA autophosphorylation and subsequently the activation of RpaA. Tseng et al. showed that KaiB - KaiC binding is stimulated by KaiA, which leads to a subsequent decrease in SasA - KaiC binding through competitive inhibition (86). This result shows how an altered interaction between an input sensing protein (KaiA) and other proteins in the network could indirectly alter output dynamics. We expect that this effect is reciprocated in the sasA mutants identified in the second-site suppressor screen mentioned above. The phase resetting input may have been rescued in a cikA-null background by altering the dynamics of the KaiB and SasA competition with KaiC, and subsequently the interactions of KaiA with the oscillator.
Chang et al. elucidated the nature of this competition with the extraordinary discovery that KaiB is a metamorphic protein that switches between two structural conformations (7). Although the sequence similarity between KaiB and the N-terminal domain of SasA has long been recognized (33), crystal structures showed that KaiB adopts a unique fold (the ground state, gs-KaiB) distinct from the thioredoxin-like fold of SasA (25, 17, 34). Chang and colleagues determined via NMR spectroscopy that KaiB also has a structural conformation identical to the N-terminal fold of SasA, termed the fold-switch state (fs-KaiB), which can be stabilized with a single point mutation. When KaiB is locked into the fs-KaiB state, it has a competitive binding advantage for KaiC over SasA, which results in a decrease in SasA kinase activity and an increase in CikA phosphatase activity as measured by RpaA phosphorylation levels (7). The slow transition of gs-KaiB to the fs-KaiB state introduces a time delay that allows KaiA-activated KaiC phosphorylation to mature before KaiB binds and induces KaiC dephosphorylation by sequestering KaiA. These findings reveal KaiB as a key link between the oscillator and the CikA/SasA/RpaA output system, and implicate KaiB as a potential post-translational point of feedback from the output apparatus on the clock.
Observations that entrainment and resetting ability, traditionally defined as a function of the input pathway, are influenced by alterations in input, output, and the Kai oscillator itself demonstrate the high degree of interconnection between clock components and the temporal modulation of circadian phase, and point to intermolecular interaction kinetics as a defining feature of circadian clock activity. However, transcriptional feedback of clock output on the expression levels of the kai genes also affects phase setting and clock robustness. As previously mentioned, time is kept in eukaryotic systems through interlocking transcription/ translation feedback loops (TTFL) (63), but this mechanism was expected to be less important in cyanobacteria because a solely post-translational oscillator (PTO) is sufficient to keep time in vitro (59). Indeed, evidence suggests that the TTFL in S. elongatus is not essential for timekeeping by the Kai PTO, but negative feedback from the Kai oscillator maintains Kai proteins at levels necessary for robust oscillations and phase stability (5). Hosokawa et al. proposed that the TTFL modulates the KaiC:KaiA ratio such that the PTO is characterized by lower amplitude and shorter period at subjective dawn (28). This attenuated state of the PTO, when the ratio of KaiC to KaiA is at trough level, is more sensitive to dark-induced phase resetting than the state present at subjective dusk, when the relative level of KaiC peaks. The TTFL is also required for maintaining phase synchrony in a dividing population; circadian oscillations in a growing culture of cells harboring only a PTO rapidly lose synchrony of oscillations (83). Thus, although transcriptional feedback regulation is not an essential determinant of clock network function, it appears to play an important supportive role in preserving clock properties in the cellular context.
2.2. Temporal Output through an RpaA hub
The discovery that SasA and CikA have antagonistic roles in RpaA phosphorylation presented a significantly clarified model of circadian output; however, other data paint a more complex picture. RpaA phosphorylation and transcriptional rhythms persist in the absence of CikA, implying that RpaA dephosphorylation can occur through additional mechanisms, possibly via autophosphatase activity or that of SasA. Furthermore, a substantial portion of cellular RpaA is continuously phosphorylated in the absence of KaiC in vivo, suggesting that the dephosphorylation of RpaA, and subsequent decrease in its transcriptional activity, is more dependent on signals from the Kai oscillator than is RpaA activation (6). This view is in contrast to the dominant output model that the oscillator induces transcriptional activation rather than repression. Intriguingly, cells that express a KaiC variant unable to undergo phosphorylation cycling, poised at the stage where we now realize KaiB and SasA compete for KaiC binding, still exhibited transcriptional rhythms of gene expression, albeit with a 48-h rather than 24-h period (87). Nakahira et al. showed that there is a global down-regulation of gene expression when kaiC is over-expressed (58), supporting this paradigm shift. However, Ito et al. showed that only genes whose expression peaks at dusk are repressed by kaiC over-expression, while dawn-peaking genes are up-regulated (30). Xu et al. tested the effects of kaiA over-expression, which leads to KaiC hyper-phosphorylation, and found the reciprocal global expression changes to those seen when kaiC levels are increased: dusk genes are up-regulated, dawn genes are down-regulated (96). Given these new results, they proposed a “yin-yang” regulation model of gene expression, wherein the opposing states of transcriptional regulation are dependent on two different states of the oscillator. Mechanistically, however, it was still not clear what constitutes the “active” state of the oscillator that transduces temporal information to downstream elements, and if this signal is activating or repressive.
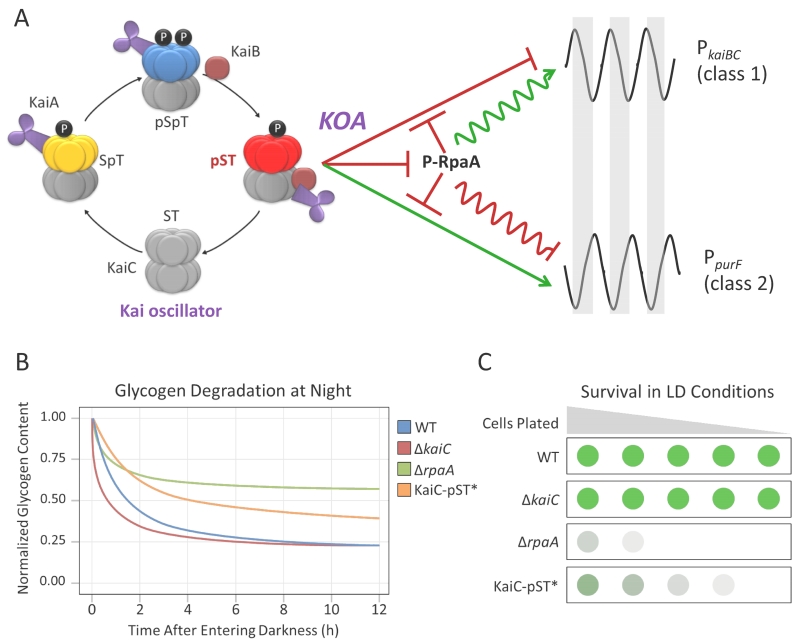
Paddock et al. sought to define the specific signaling state(s) of the oscillator by measuring reporter gene expression in strains that carry KaiC mimics of each of the four KaiC phosphorylation states (ST, SpT, pSpT, and pST), and comparing these values to those in strains lacking KaiC. They defined this metric as Kai-complex output activity (KOA) (62). Surprisingly, only one phosphomimetic of KaiC shows significantly altered gene expression levels relative to a kaiC-null mutant (i.e. has a high KOA). In strains lacking the oscillator, expression from the canonical class 1 (dusk-peaking) promoter kaiBC is locked at the peak level of WT oscillations, whereas expression from the class 2 promoter purF (dawn-peaking) is locked at the trough. Expression of phophomimetics of the ST, SpT, and pSpT states all preserved this pattern typical of no KaiC. However, the KaiC-ET variant (mimicking the pST state) locked class 1 promoter activity low and class 2 activity high, a reversal of the clockless pattern. In WT, high KOA coincides with early subjective dawn, at the trough of class 1 promoter expression and peak of class 2 promoter expression. At this time the pST state is the most prevalent phosphoform of KaiC. Maximal KOA was observed only in the presence of KaiB, further supportive of the idea that the output activity of KaiC arises from interactions with a broader network of players. These results were recapitulated by a simple mathematical model wherein P-RpaA represses KOA at dusk, with peak KOA coinciding with the peak of class 2 promoter activity at dawn. A simplified model of Kai output activity is shown in Figure 2A. It should be noted that all phosphoforms of KaiC are detectable during circadian cycles in WT in vivo, suggesting that the increases and decreases in the pST state account for the sine waves of expression from class 1 and class 2 genes.
Figure 2. Transcriptional and metabolic rhythms are regulated by the KaiC-pST phosphostate.
(A) Peak Kai complex oscillator activity (KOA) coincides with the pST state of KaiC and occurs circa subjective dawn, repressing class 1 and activating class 2 promoter output. In addition, the pST state of KaiC exerts a repressive effect on RpaA phosphorylation, further amplifying the effects of KOA on the promoter outputs. Gray bars represent subjective night. Panel A is adapted from Paddock et al. (62). (B) Model of glycogen degradation during a 12-h dark period in WT, ΔkaiC, ΔrpaA, and KaiC-ET strains. KaiC-ET is a phosphomimetic of the pST state, and is labeled as “KaiC-pST*” in panels B and C. The WT and ΔkaiC strains exhibit normal glycogen degradation and end the night period with a similar terminal glycogen content. The ΔrpaA and KaiC-pST* strains exhibit a similar phenotype to each other in that glycogen degradation slows earlier in the evening and both strains degrade less of their overall glycogen content relative to WT cells. (c) Model of a dilution series to test strain viability under LD growth conditions. Darker circles indicate more robust growth. WT and ΔkaiC do not show any growth phenotype when grown under LD conditions. The ΔrpaA and KaiC-pST* strains show significantly attenuated growth under the same conditions. Panels B and C were adapted from Diamond et al. (9). Metabolic and physiological experiments indicate that KaiC-pST* phenocopies ΔrpaA, thus locking the clock at peak KOA has similar effects to a removal of RpaA activity.
This elegant study demonstrated that the output signal from the oscillator is indeed negative for the majority of genes, occurs at subjective dawn (coinciding with the pST state of KaiC), and represses the default high expression of class 1 genes while simultaneously activating default low-expressing class 2 genes. Furthermore, this work hints at the role of other players in the output pathway. An increase in KOA was detected from the purF promoter even when RpaA was absent, and although P-RpaA tracks reliably in antiphase with peak KOA levels in WT, the phosphorylation state of RpaA did not correlate with the changes in gene expression that were observed using KaiC phosphomimetics. These results are indicative of the presence of a KaiC-dependent, RpaA-independent mechanism that affects gene expression. Nonetheless, locking the clock at peak KOA correlates with rpaA-null phenotypes of glycogen degradation and growth impairment during light:dark (LD) cycles (9), illustrating that a high Kai output value has cellular effects similar to the removal of RpaA activity (Figure 2B,C).
2.3. The Balance of Power: Integrating the Clock and Environmental Cues in Cellular Processes
Although there is a clear advantage for the cell to be able to anticipate regular external fluctuations (such as diel changes in light and temperature) (61), it also needs to be able to respond to unpredictable environmental changes. How the cell integrates information from both the clock and the environment to control the expression levels of genes has been an important, outstanding question in the field. Increasing evidence suggests that this integration occurs through the complex dynamics of RpaA and the related transcription factor RpaB.
Two-component signaling pathways, comprising a histidine kinase and a cognate response regulator protein, are key to cellular signaling in bacteria (75). Phosphotransfer from the environmental-sensing histidine kinase to an Asp residue on the receiver domain of the response regulator (RR) activates or represses binding of the RR to regulatory nucleotide sequences on the chromosome. In cyanobacteria, the NblS-RpaB two-component system, which is essential for viability, is the most widely conserved and most prominent player in the global transcriptional regulation of stress responses. In addition to its roles in heat, cold, salt, and osmotic stress (56), RpaB has also been implicated in control of regulatory targets affecting cell size and morphology (57), suggesting that, like RpaA, RpaB controls a wide range of genes. Furthermore, the initial identification of RpaA and RpaB as regulators of energy transfer from the light-harvesting phycobiliproteins (2) adds to the breadth of roles that these RRs play in cellular physiology.
RpaB has been shown to specifically recognize the HLR1 (high-light response) DNA-binding site (23). Using chromatin immunoprecipitation (ChIP), Hanaoka et al. showed that RpaB binding to HLR1 is dependent on light intensity, such that it binds and represses transcription under normal conditions (low light), and dissociates and allows transcription under high light (23). Exploring the dynamics of RpaB further, Moronta-Barrios and colleagues revealed that exposure to high light stress induces dephosphorylation of RpaB (56). RpaB phosphorylation cycles only in LD conditions, indicating that the RpaB cycle is driven by environmental cues rather than the circadian clock. However, evidence for intersection with the clock system arose in a study of regulatory interactions, which demonstrated phosphotransfer in vitro from SasA to both RpaA and RpaB, as well as another response regulator, SrrA, related to the NblS/RpaB pathway (39). This finding raised the possibility of cross-talk between the SasA-RpaA clock output system and RpaB, which was subsequently solidified by Espinosa et al. (16). They showed that overexpression of the N-terminal receiver domain of RpaB alone substantially decreases the RpaA phosphorylation ratio, and alters the periods of both class 1 and class 2 reporters, showing a clear dependency between these two systems. Moreover, the pattern of LD-driven P-RpaB is altered when KaiC is absent and there is no clock.
The intersection of RpaB with the SasA/RpaA clock output system is not restricted to modulations of RpaA phosphorylation levels. RpaB has also been shown to bind regulatory elements that overlap RpaA binding sites in several promoters including kaiBC and purF (16). The competition between RpaA and RpaB for binding at these promoters elucidates the transcriptional mechanism of negative output regulation at the kaiBC locus and is likely responsible for the RpaA-independent repression of purF observed in Paddock et al. (62). In this manner, RpaB integrates environmental information into clock output while not directly perturbing the oscillator itself. Whole-genome ChIP analysis of RpaB should uncover the extent to which RpaA and RpaB share binding targets and lend additional insight into the interconnections between these pathways. It is likely that other connections will be uncovered that link environmental signaling pathways to the core circadian regulon as the broader cellular network is studied further.
3. How the Clock Fits within a Broader Cellular Context
While significant advances have been made in our understanding of the maintenance and transference of temporal information in S. elongatus, the physiological and metabolic relevance of a functional clock has been largely unexplored. It is clear that control of the clock over cellular processes is extensive, but which processes and to what degree are still open questions. Initial insights can be gleaned from temporal transcriptomics data, a global characterization of RpaA binding by ChIP-seq, and new work that examines circadian rhythms in metabolism. Here, we discuss these experiments and their implications.
3.1. Global Transcriptional Control
Initial microarray and luciferase reporter studies revealed that the majority of S. elongatus genes exhibit circadian fluctuations in their expression levels (48, 30), and that these changes are highly correlated to rhythmic changes in the super-helical density of the chromosome (93, 88). While these findings point to a possible global regulatory mechanism of the clock through the systematic alteration of chromosome topology, they do not reveal the underlying process by which the clock achieves it. Work by Takai et al. showed that, as expected, global transcriptional control is RpaA-mediated, as the inactivation of RpaA eliminates circadian oscillations from a wide range of promoter-driven bioluminescence reporters, and total bioluminescence decreases significantly at promoters controlling dusk-peaking circadian genes (79). However, it was not clear from this work whether RpaA directly regulates the expression of all genes, or acts indirectly through the regulation of other transcription factors or genome compaction rhythms.
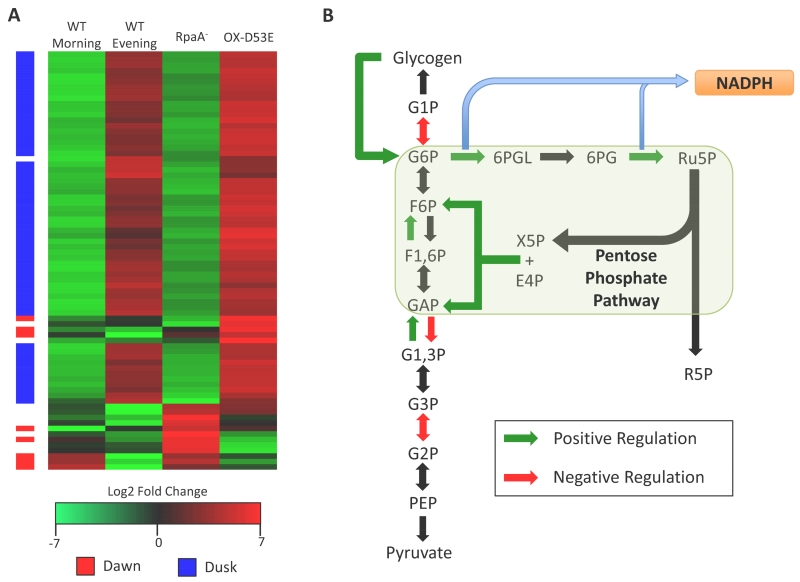
Significant insight was gained when Markson et al. characterized the global gene expression profile of an rpaA-null strain as well as the genomic binding sites of RpaA as measured by ChIP-seq (55). Their data showed that RpaA binds to only about 110 loci in the genome, and the expression levels of only 84 genes are modulated by 2-fold or more in an rpaA-null mutant. Genes in RpaA’s direct regulon include four sigma factors and two transcription factors, which presumably in turn control the rhythmic expression of the majority of cycling genes in the genome. Interestingly, they observed that the deletion of rpaA arrests the global transcription profile in a dawn-like expression state (Figure 3A). That is, genes down-regulated in the mutant are highly enriched in transcripts that peak at dusk, while up-regulated genes are highly enriched in transcripts that peak at dawn. These data support the previously described claim that RpaA-mediated regulation can be either positive or negative (62). Given that 69 of the 84 genes that change more than 2-fold were down-regulated, RpaA appears to be important to drive the expression of many dusk-peaking transcripts. This conclusion is supported by the finding that expressing a constitutively active form of RpaA (RpaA D53E) in an rpaA-null background leads to a rapid shift from a dawn-like to a dusk-like expression state (Figure 3A) (55).
Figure 3. Direct targets of RpaA are dusk-peaking and affect glycogen metabolism.
(A) Heatmap showing the expression of 76 genes that change more than 2-fold in ΔrpaA strains. The columns indicate the expression of these genes at morning and evening circadian time points as well as in a ΔrpaA and ΔrpaA::rpaA(D53E) strain, which has a mutation that simulates constitutively active RpaA. The column on the left of the heatmap indicates if a gene has a normal circadian expression peak at dawn (red) or dusk (blue) based on data from Vijayan et al. (88). The expression data indicate that genes controlled by RpaA are normally activated in the evening and repressed in the morning. Overexpression of the constitutively active form of RpaA shifts the genes to an evening-like expression state. Correlation with the circadian timing of genes indicates that RpaA acts most strongly on dusk-peaking transcripts, and primarily activates dusk transcripts while repressing dawn transcripts. (B) Metabolic pathway diagram summarizing reactions in Glycolysis, Glycogen Metabolism, and the Pentose Phosphate Pathway. Reactions driven by genes positively regulated by RpaA are indicated in green, whereas reactions driven by genes repressed by RpaA are indicated in red. Mapping RpaA transcriptional regulation to these pathways shows the positive effect of RpaA on the oxidative phase of the Pentose Phosphate Pathway. Expression data and RpaA target genes in panels A and B were adapted from Markson et al. (55).
Global gene expression profiling has been done for other cyanobacterial species. Like in S. elongatus, the majority of genes in Prochlorococcus exhibit circadian fluctuations in expression levels when cells are grown in an LD cycle (99). However, less than 10% of Synechocystis PCC6803 transcripts cycle in LD as determined by microarrays, and like RpaA’s direct targets in S. elongatus, these cycling transcripts are mostly dusk-peaking (46). Perhaps the primary role of the clock in cyanobacteria is to temporally modulate the expression of genes that function in the dark through repression and de-repression of RpaA activity. The expansion of clock control over additional cellular processes in species like S. elongatus and Prochlorococcus may have emerged through the absorption of global transcriptional regulators into the RpaA regulon.
3.2. Global Metabolic Control
Extensive research has been performed on the metabolism of cyanobacteria, and both physiological and biochemical observations indicate that diverse metabolic processes display persistent 24-h rhythms in the absence of external stimuli; these processes include peroxiredoxin oxidation/reduction, oxygen evolution, and biosynthesis and degradation of glycogen (14, 98, 64). Unfortunately, how the clock controls metabolic processes is still poorly understood, as few studies have provided a genetic link between metabolic observations and clock output, and most experiments have been conducted in non-physiological conditions (constant light: LL). Glycogen metabolism is one of the few metabolic systems where rhythmic activity has been genetically linked to the clock and studied in both LL and LD contexts. Furthermore, the regulation of glycogen metabolism by the clock is conserved in higher eukaryotes; the mammalian clock has been shown to regulate liver glycogen synthesis (11), and glycogen metabolism is known to cycle with sleep cycles in mammals and flies (66).
It has been known for some time that glycogen in cyanobacteria accumulates during the day and degrades during the night under LD growth (76). Recent work shows that the circadian clock directly influences the orchestration of glycogen and other carbon metabolism when cells are grown under both LL and LD conditions (9, 64). Glycogen content in S. elongatus grown under LL oscillates with 24-h rhythmicity, and this rhythm is lost in a kaiC-null mutant that lacks a functional oscillator (64). However, when glycogen was tracked for 72 h under LD growth conditions, accumulation oscillated in both WT and kaiC-null strains, showing that environmental light cycles can drive metabolic oscillations even in the absence of the clock. Nonetheless, the accumulation of glycogen during the day period in the kaiC-null strain occurred earlier, was significantly more rapid, and resulted in an overall higher glycogen content than in WT (9). Thus, while environmental cycles can drive rhythms, there is a significant input from the circadian clock as to how these rhythms are modulated. Metabolomics measurements revealed that the kaiC-null mutant significantly over-accumulates primary carbon metabolites in the Oxidative Pentose Phosphate Pathway (OPPP) and glycolysis, such as fructose-6-phosphate, in the morning hours of an LD cycle (9). Several genes in the OPPP are direct RpaA targets (Figure 3B), and two of the most strongly down-regulated genes identified in rpaA-null strains are zwf and gnd, which code for enzymes that catalyze the two NADPH-producing steps of the OPPP (55, 41). The enzyme encoded by zwf, glucose-6-phosphate dehydrogenase, is also the rate-limiting step for entry of carbon into the OPPP, and the control of zwf strongly influences flux through the pathway (97). The detection of increased OPPP activity in kaiC-null strains also supports the hypothesis that the KaiABC complex acts as a repressor on SasA/RpaA-mediated output, specifically during the day period (62). Without repressive output from the clock, RpaA is free to be active during the day and activates genes of the OPPP, resulting in an increased concentration of primary carbon metabolites. Unfortunately, the metabolic consequence of OPPP activation is less clear, as a separate set of metabolites were detected as highly depressed in kaiC-null strains, but their identities are unknown (9).
Although kaiC-null strains grow normally under LD conditions, rpaA- and sasA-null strains do not (79) (Figure 2C). Understanding why rpaA and sasA mutants die in LD may elucidate what types of cellular processes are important for survival under diel conditions. Indeed, RpaA’s control over the Oxidative Pentose Phosphate Pathway (OPPP) and glycogen degradation may be linked to the LD sensitivity of rpaA (and to a lesser extent, sasA) mutants. The OPPP shares several enzymes with the Calvin Cycle, and the balance between these two pathways is strictly controlled by the allosteric regulation of these enzymes and a small protein, CP12, which is found in all plants and cyanobacteria (18, 80). When active, CP12 sequesters the key Calvin Cycle enzyme phosphoribulokinase, which slows the activity of this pathway (18). Maintaining a strict balance between the OPPP and the Calvin Cycle in cyanobacteria is important because of their shared enzymatic steps (36). The shift towards the OPPP during evening hours allows it to become the primary source of NADPH from degradation of stored glycogen when photosynthesis is inactive (97). Interestingly, it has also been shown that the deletion of either zwf or gnd genes in cyanobacteria result in a decrease of viability under LD growth conditions (70, 12). Additionally, rpaA mutants have attenuated glycogen degradation at night, and only degrade about half of their stored glycogen reserves over a 12 h period in the dark (9). Glycogen degradation exhibits a similar phenotype in KaiC-pST phosphomimetic strains where the clock is locked at maximum KOA, reinforcing the observation that KaiC-pST represses RpaA activity (Figure 2B). Experiments have also shown that an inability to store or degrade glycogen leads to loss of viability specifically under LD growth (20). However, it is unclear if additional metabolic pathways are controlled by RpaA, and are important for viability at night, and why OPPP activity is important for nighttime viability.
Studies to elucidate the metabolic properties affected by circadian rhythms highlight both the unique metabolic pathways used by photosynthetic organisms, and the lack of good mass spectral compound libraries for cyanobacteria. Efforts should be made to collect mass spectra of compounds that are relevant to this specific class of organisms, as these data will significantly improve metabolic engineering and modeling efforts. While much work on the control of metabolism by the clock remains to be done, a picture is forming that places the circadian oscillator as important for repression of SasA-RpaA output activity during the day, and active RpaA output as important for night-time metabolic processes. Future work should focus on both the biological relevance of temporally separating these classes of metabolic pathways.
4. New Perspectives on Clock Evolution
4.1. Phenotypic Resiliency of a Flexible Network
There is an interesting distinction between clock conservation in eukaryotes and prokaryotes. In eukaryotes, all characterized clocks have the same network architecture of an interlocking transcription and translation feedback loop, but are highly variable in the proteins used to build these loops (63). In cyanobacteria, all clocks pull from the same set of proteins to keep time, but use them in different configurations. The diversity of prokaryotic clock architectures, as inferred from bioinformatic analysis, mirrors the range of genome sizes, environmental niches, and cellular physiologies found in cyanobacteria. While S. elongatus kaiC and kaiB homologs are found in almost all cyanobacterial species for which a genome sequence is available, not all S. elongatus clock genes, including the core oscillator gene kaiA, are present (4). The absence of these genes in a given species does not necessarily signify the absence of a functional clock. Synechococcus WH7803 does not have a cikA homolog, but shows clear circadian rhythms of cell division that persists for four days in constant light (77). Prochlorococcus lacks homologs of two key S. elongatus clock genes, kaiA and cikA, but still shows diel rhythms of expression for 80% of the genes in the genome (99). However, these rhythms do not persist in constant light conditions, suggesting that Prochlorococcus keeps time via an “hourglass” mechanism that is reset each diel cycle (27). This simplified clock may be sufficient in Prochlorococcus species that are very specialized for static environmental niches (3). Conversely, some rhythmic genera, including Synechocystis and Anabaena, harbor multiple copies of the kaiB and kaiC genes, but it is unclear if these paralogs are involved in running a clock (89). This wide range of clock configurations suggests a high degree of flexibility in the gene network that underlies circadian rhythms in cyanobacteria. By flexibility, we mean that interaction parameters within the network can easily evolve to accommodate alterations to the network (loss or gain) and still maintain a selected phenotypic output. This flexibility was experimentally demonstrated in the cikA suppressor mutagenesis screen described above, where the phenotypic consequences of a cikA knockout (2.5-h decrease in period, entrainment defects) could be rescued by a single mutation in sasA (73). We expect that this flexibility is a property of the highly interconnected gene network, as the loss or mutation of one gene can be compensated for by altering the dynamics of overlapping interactions and functions of other genes within the network.
Whatever mechanism enables the network to be resilient to the loss of core clock genes may also enable the apparent general robustness of the circadian phenotype to genomic perturbations. This robustness is implied by the fact that only 19 genes have been found to have significant effects on the circadian phenotype after extensive screening of random and targeted mutants (Table 1). It should be noted, however, that these screens were generally done on whole gene knockouts in constant light, and a broader range of genes that influence phenotype may emerge under different screening conditions. If we assume that disruptive genetic mutations correlate to altered cellular and metabolic states, the rarity of such mutations would suggest that the clock is insensitive to many physiological perturbations. This physiological robustness is further supported by the persistence of the clock in dividing cells: S. elongatus doubling time can be as short as 5-6 h (43). That is not to say that the clock is operating in a vacuum within the cell, as it is clearly sensitive to changes in cellular redox state (as detected by the oxidation state of the plastoquinone pool (40)), and energy abundance (as detected by changes in ATP/ADP ratio (68)), as previously described.
4.2. Models for How the Clock Improves Cellular Fitness
The ubiquity of circadian rhythms in eukaryotes and cyanobacteria suggests that there is a clear fitness advantage for anticipating regular fluctuations in the environment. This benefit has been demonstrated in S. elongatus where cells containing normally functioning clocks quickly outcompete arrhythmic mutants when co-cultured in LD cycles (61). Interestingly, this advantage is lost in constant light conditions, and rhythmic cells actually show a decrease in fitness relative to arrhythmic ones. This finding suggests that while the clock provides some value to the cell, its value is “extrinsic”; that is, the clock gains its adaptive worth only under specific environmental conditions, and does not have an inherent benefit to the physiology of the organism otherwise (92). Ouyang et al. showed that this value is maximized when the free running period of the clock matches the period of an external light cycle (61). They demonstrated that circadian clock mutants that had either long or short periods would outcompete WT cells if co-cultured in an LD cycle that matched their respective intrinsic periods. However, the mechanism by which the cell gains an advantage by matching external light cycles is not clear. Three models have been proposed to explain this gain in fitness conferred by the clock: the “diffusible inhibitor model”, the “cell-to-cell communication model”, and “the limiting resource model”. Here we summarize these models.
The diffusible inhibitor model postulates that cyanobacteria secrete a diffusible molecule in a rhythmic fashion that can inhibit the growth of other cells. This model assumes that an inhibitor would be secreted during the day period and subsequently cells would become susceptible at night, or vice versa. If the circadian rhythm is absent or out of phase with the environmental cycle, the times of secretion and sensitivity would not align correctly, resulting in the inhibition of a circadian mutant in co-culture with WT cells. This model has been tested by measuring the growth rates of two strains cultured in chambers separated by a semi-permeable membrane (50). At 0.2 μm, the membrane pores were large enough to allow small molecules and proteins to pass through. It was found that the growth rate was statistically indistinguishable for both WT and an arrhythmic mutant when they were cultured in neighboring chambers separated by a 0.2 μm pore membrane (50). Although it is possible that an inhibitor was not able to pass between the culture vessels, this evidence seems to refute a diffusible inhibitor model. An alternative proposed hypothesis is that the inhibitor is not diffusible, and is instead bound to the cell surface.
The cell-to-cell communication model postulates that through molecule diffusion and some mechanism such as quorum sensing, or a yet undiscovered cell surface interaction, cyanobacteria can coordinate their rhythmic activity within a population (91). It is known that some cyanobacteria have homologues for quorum sensing genes, but research on cell-to-cell communication has been limited and experimental evidence for this model is lacking.
The limiting resource model suggests that the clock provides an advantage in using some limited environmental resource by phasing the metabolism to an optimal part of the daily cycle for resource collection and/or utilization. Even though there are no significant differences in growth rates between WT and clock mutants when grown separately, simulated competition experiments suggest that very small growth differences can account for the observed fitness advantage of clock-containing cells in LD (24). This advantage may be condition dependent, as circadian timing may control some metabolic or transport process, and perturbation of process timing may give no detectable phenotype until cells compete for a specific resource. This situation may be similar to that of a competition between an enzyme and a mutant variant, where substrate binding is weakened and enzymatic activity is identical. If substrate is abundant in this example, then both WT and mutant enzymes will work at their maximum rates. However, if the mutant and WT enzyme are incubated together they will both work at their maximum rate until substrate becomes limiting for one of them due to a lower binding affnity.
Future studies to address any of the given models should begin with a more comprehensive understanding of both transcription and physiology in LD cycles as well as the transcriptional and metabolic processes that are directly regulated by the circadian clock. Although some work has investigated the metabolism and gene expression of cyanobacteria under diurnal growth conditions, there is scant information available for S. elongatus, where the clock is the best studied.
5. Considerations for Future Experiments
Our detailed understanding of the prokaryotic circadian clock stems from 20 years of insightful research from many laboratory groups with complementary expertise. As informative as this early work has been, there is room for improvement with new tools and hindsight, as well as exciting opportunities to gain a holistic understanding of the clock. It is becoming increasingly clear that the clock is built on a highly integrated network of proteins. Further experiments are required to fully probe the nature and breadth of this network. Single gene knockouts are blunt genetic tools that may miss potentially important interactions, especially in S. elongatus where a significant portion of the genome is expected to be essential for viability. Subtler perturbations to genes through random mutagenesis, or through systematically varying gene dosage may elucidate a broader network of interactions. Additional advantages may come through mutational studies in a sensitized background, like the cikA-null strain used in (73), or in double knockout screens. The effects of clock perturbation on the broader cellular context also need to be characterized in greater detail. Assays that measure systems-level properties (like gene expression levels or metabolic profile), are becoming cheaper and easier to use. Future mechanistic studies should use these assays to investigate the impact of altered clock protein dynamics on the cell as a whole. These data will help assign a deeper biological significance to individual molecular interactions, by directly connecting them to cellular consequences. By making these connections, we can begin to truly understand the value of the clock to the cell.
SUMMARY POINTS.
Input and output functions of the circadian clock are products of highly connected network interactions among clock components.
A single state of the Kai oscillator, corresponding to the pST phosphostate of KaiC, is responsible for signaling a negative output on transcription on the majority of phase class 1 genes.
Clock output is integrated with environmentally-sensitive signaling pathways to provide modulation of transcriptional responses.
The transcription factor RpaA is a nexus for circadian control of global transcription and controls genes for metabolic processes at night.
The clock provides an adaptive fitness advantage during LD cycles.
ELIMINATING RHYTHMS WHILE KEEPING THE CLOCK.
It has become clear that distinct “clockless” mutants are not uniform but exhibit a spectrum of global attributes. Deletions of kaiA, kaiB, or kaiC individually render cellular processes arrhythmic, but each results in distinct global transcriptional profiles, and in the case of kaiB-null strains, a severe cell division defect resulting in hyper-elongated cells. A deletion of rpaA, while causing arrhythmic transcription, has severe pleiotropic consequences on cell growth and metabolic regulation. The recent identification of a transposon insertion strain, crm1, that lacks rhythms of both transcription and KaiC phosphorylation, yet has WT levels of RpaA, provides a background that may be preferred for studies in which an arrhythmic strain is desired that retains a complete circadian clock and whose transcription profile is not as lop-sided as an rpaA-null strain (6). The transposon insertion is located immediately upstream of the rpaA gene in a short ORF encoding a 62-residue polypeptide, dubbed crm for circadian rhythmicity modulator. Expression from the kaiBC promoter is locked at the WT trough level and is complemented by ectopic expression of the crm ORF. Unlike an rpaA deletion mutant, the crm1-null strain does not exhibit a growth defect in LD cycles.
KAI GENES IN NON-CYANOBACTERIAL SPECIES.
There are many putative homologs of the kai genes present in prokaryotes outside the cyanobacteria. Archaea lack homologs of kaiA and kaiB but possess several kaiC-like genes; the cir genes from the halophilic Archaeon Haloferax volcanii are rhythmically expressed in LD conditions (54). Legionella pneumophila harbors kaiBC homologs that may function in stress response (49). As yet, no non-cyanobacterial prokaryote has a confirmed bona fide circadian clock that fulfills the criteria observed in eukaryotes, but as functional studies penetrate the rapidly expanding wealth of genomic data, the future promises to yield fascinating insights into the roles of clock-like genes in non-circadian cellular processes.
ACKNOWLEDGMENTS
We thank Mark Paddock for helpful comments. This work was supported by the W. M. Keck Foundation and by Army Research Office Grant W911NF-13-1-0097 (to R.J.G.); National Science Foundation Grant MCB1244108 and National Institute of Health Cell and Molecular Genetics Training Grant T32GM007240 (to S.D.); and National Institutes of Health Grant R01GM062419 (to S.S.G.).
Glossary
- KaiC phosphorylation states
KaiC cycles through four possibilities of Ser431 and Thr432 phosphorylation: ST (none), SpT (Thr-phos), pSpT (both), pST (Ser-phos).
- CLASS 1 PROMOTER
Peak expression level is at dusk.
- CLASS 2 PROMOTER
Peak expression level is at dawn.
- Kai-complex Output Activity (KOA)
The difference in gene expression between a clockless state and one where KaiC is locked in a given phosphostate.
- GLYCOGEN
The primary carbon storage polymer in cyanobacteria.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Arita K, Hashimoto H, Igari K, Akaboshi M, Kutsuna S, et al. Structural and biochemical characterization of a cyanobacterium circadian clock-modifier protein. Journal of Biological Chemistry. 2007;282:1128–1135. doi: 10.1074/jbc.M608148200. [DOI] [PubMed] [Google Scholar]
- 2.Ashby MK, Mullineaux CW. Cyanobacterial ycf27 gene products regulate energy transfer from phycobilisomes to photosystems I and II. FEMS microbiology letters. 1999;181:253–260. doi: 10.1111/j.1574-6968.1999.tb08852.x. [DOI] [PubMed] [Google Scholar]
- 3.Axmann IM, Dühring U, Seeliger L, Arnold A, Vanselow JT, et al. Biochemical evidence for a timing mechanism in Prochlorococcus. J Bacteriol. 2009;191:5342–5347. doi: 10.1128/JB.00419-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baca I, Sprockett D, Dvornyk V. Circadian input kinases and their homologs in cyanobacteria: evolutionary constraints versus architectural diversification. J Mol Evol. 2010;70:453–465. doi: 10.1007/s00239-010-9344-0. [DOI] [PubMed] [Google Scholar]
- 5.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nature Reviews Genetics. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd JS, Bordowitz JR, Bree AC, Golden SS. An allele of the crm gene blocks cyanobacterial circadian rhythms. Proc Natl Acad Sci USA. 2013;110:13950–13955. doi: 10.1073/pnas.1312793110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang YG, Cohen SE, Phong C, Myers WK, Kim YI, et al. Protein fold switching is the linchpin joining the circadian oscillator to clock output in cyanobacteria. Science. 2015 doi: 10.1126/science.1260031. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang YG, Tseng R, Kuo NW, LiWang A. Rhythmic ring–ring stacking drives the circadian oscillator clockwise. Proceedings of the National Academy of Sciences. 2012;109:16847–16851. doi: 10.1073/pnas.1211508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond S, Jun D, Rubin BE, Golden SS. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proceedings of the National Academy of Sciences. 2015 doi: 10.1073/pnas.1504576112. 201504576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 11.Doi R, Oishi K, Ishida N. CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. Journal of Biological Chemistry. 2010;285:22114–22121. doi: 10.1074/jbc.M110.110361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doolittle WF, Singer RA. Mutational analysis of dark endogenous metabolism in the blue-green bacterium Anacystis nidulans. Journal of Bacteriology. 1974;119:677–683. doi: 10.1128/jb.119.3.677-683.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: biological timekeeping. Sinauer Associates; 2004. [Google Scholar]
- 14.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egli M. Intricate protein-protein interactions in the cyanobacterial circadian clock. Journal of Biological Chemistry. 2014;289:21267–21275. doi: 10.1074/jbc.R114.579607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espinosa J, Boyd JS, Cantos R, Salinas P, Golden SS, Contreras A. Cross-talk and regulatory interactions between the essential response regulator RpaB and cyanobacterial circadian clock output. Proceedings of the National Academy of Sciences. 2015 doi: 10.1073/pnas.1424632112. 201424632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garces RG, Wu N, Gillon W, Pai EF. Anabaena circadian clock proteins KaiA and KaiB reveal a potential common binding site to their partner KaiC. The EMBO journal. 2004;23:1688–1698. doi: 10.1038/sj.emboj.7600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gontero B, Maberly S. An intrinsically disordered protein, CP12: jack of all trades and master of the Calvin cycle. Biochemical Society Transactions. 2012;40:995. doi: 10.1042/BST20120097. [DOI] [PubMed] [Google Scholar]
- 19.Gonze D, Roussel M, Goldbeter A. A model for the enhancement of fitness in cyanobacteria based on resonance of a circadian oscillator with the external light–dark cycle. Journal of theoretical biology. 2002;214:577–597. doi: 10.1006/jtbi.2001.2476. [DOI] [PubMed] [Google Scholar]
- 20.Gründel M, Scheunemann R, Lockau W, Zilliges Y. Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology. 2012;158:3032–3043. doi: 10.1099/mic.0.062950-0. [DOI] [PubMed] [Google Scholar]
- 21.Gutu A, OShea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanaoka M, Takai N, Hosokawa N, Fujiwara M, Akimoto Y, et al. RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. Journal of Biological Chemistry. 2012;287:26321–26327. doi: 10.1074/jbc.M111.338251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanaoka M, Tanaka K. Dynamics of RpaB–promoter interaction during high light stress, revealed by chromatin immunoprecipitation (ChIP) analysis in Synechococcus elongatus PCC 7942. The Plant Journal. 2008;56:327–335. doi: 10.1111/j.1365-313X.2008.03600.x. [DOI] [PubMed] [Google Scholar]
- 24.Hellweger FL. Resonating circadian clocks enhance fitness in cyanobacteria in silico. Ecological Modelling. 2010;221:1620–1629. [Google Scholar]
- 25.Hitomi K, Oyama T, Han S, Arvai AS, Getzo ED. Tetrameric architecture of the circadian clock protein KaiB a novel interface for intermolecular interactions and its impact on the circadian rhythm. Journal of Biological Chemistry. 2005;280:19127–19135. doi: 10.1074/jbc.M411284200. [DOI] [PubMed] [Google Scholar]
- 26.Holtman CK, Chen Y, Sandoval P, Gonzales A, Nalty MS, et al. High-throughput func-tional analysis of the Synechococcus elongatus PCC 7942 genome. DNA research. 2005;12:103–115. doi: 10.1093/dnares/12.2.103. [DOI] [PubMed] [Google Scholar]
- 27.Holtzendor J, Partensky F, Mella D, Lennon JF, Hess WR, Garczarek L. Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J Biol Rhythms. 2008;23:187–199. doi: 10.1177/0748730408316040. [DOI] [PubMed] [Google Scholar]
- 28.Hosokawa N, Kushige H, Iwasaki H. Attenuation of the posttranslational oscillator via transcription–translation feedback enhances circadian-phase shifts in Synechococcus. Proceedings of the National Academy of Sciences. 2013;110:14486–14491. doi: 10.1073/pnas.1302243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson C, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 30.Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proceedings of the National Academy of Sciences. 2009;106:14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: a component of the circadian clock senses redox state of the cell. The EMBO journal. 2005;24:1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci USA. 2006;103:17468–17473. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwasaki H, Williams SB, Kitayama Y, Ishiura M, Golden SS, Kondo T. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 34.Iwase R, Imada K, Hayashi F, Uzumaki T, Morishita M, et al. Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex. Journal of Biological Chemistry. 2005;280:43141–43149. doi: 10.1074/jbc.M503360200. [DOI] [PubMed] [Google Scholar]
- 35.Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Annual review of biophysics. 2011;40:143. doi: 10.1146/annurev-biophys-042910-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katayama M, Kondo T, Xiong J, Golden SS. ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. Journal of Bacteriology. 2003;185:1415–1422. doi: 10.1128/JB.185.4.1415-1422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katayama M, Tsinoremas NF, Kondo T, Golden SS. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J Bacteriol. 1999;181:3516–3524. doi: 10.1128/jb.181.11.3516-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato H, Watanabe S, Nimura-Matsune K, Chibazakura T, Tozawa Y, Yoshikawa H. Exploration of a possible partnership among orphan two-component system proteins in cyanobacterium Synechococcus elongatus PCC 7942. Bioscience, biotechnology, and biochemistry. 2012;76:1484–1491. doi: 10.1271/bbb.120172. [DOI] [PubMed] [Google Scholar]
- 40.Kim YI, Vinyard DJ, Ananyev GM, Dismukes GC, Golden SS. Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc Natl Acad Sci USA. 2012;109:17765–17769. doi: 10.1073/pnas.1216401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knowles VL, Plaxton WC. From genome to enzyme: analysis of key glycolytic and oxidative pentose-phosphate pathway enzymes in the cyanobacterium Synechocystis sp. PCC 6803. Plant and Cell Physiology. 2003;44:758–763. doi: 10.1093/pcp/pcg086. [DOI] [PubMed] [Google Scholar]
- 42.Kondo T, Ishiura M. Circadian rhythms of cyanobacteria: monitoring the biological clocks of individual colonies by bioluminescence. J Bacteriol. 1994;176:1881. doi: 10.1128/jb.176.7.1881-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondo T, Mori T, Lebedeva NV, Aoki S, Ishiura M, Golden SS. Circadian rhythms in rapidly dividing cyanobacteria. Science. 1997;275:224–227. doi: 10.1126/science.275.5297.224. [DOI] [PubMed] [Google Scholar]
- 44.Kondo T, Strayer C, Kulkarni R, Taylor W, Ishiura M, et al. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kondo T, Tsinoremas N, Golden S, Johnson C, Kutsuna S, Ishiura M. Circadian clock mutants of cyanobacteria. Science. 1994;266:1233. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- 46.Kucho Ki, Okamoto K, Tsuchiya Y, Nomura S, Nango M, et al. Global analysis ofcircadian expression in the cyanobacterium Synechocystis sp. strain PCC 6803. Journal of Bacteriology. 2005;187:2190–2199. doi: 10.1128/JB.187.6.2190-2199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kutsuna S, Kondo T, Aoki S, Ishiura M. A period-extender gene, pex, that extends the period of the circadian clock in the cyanobacterium Synechococcus sp. strain PCC 7942. Journal of Bacteriology. 1998;180:2167–2174. doi: 10.1128/jb.180.8.2167-2174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, et al. Circadian orchestration of gene expression in cyanobacteria. Genes & development. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 49.Loza-Correa M, Sahr T, Rolando M, Daniels C, Petit P, et al. The Legionella pneumophila kai operon is implicated in stress response and confers fitness in competitive environments. Environmental microbiology. 2014;16:359–381. doi: 10.1111/1462-2920.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma P, Woelfle MA, Johnson CH. An evolutionary fitness enhancement conferred by the circadian system in cyanobacteria. Chaos, Solitons & Fractals. 2013;50:65–74. doi: 10.1016/j.chaos.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mackey SR, Choi JS, Kitayama Y, Iwasaki H, Dong G, Golden SS. Proteins found in a CikA interaction assay link the circadian clock, metabolism, and cell division in Synechococcus elongatus. Journal of Bacteriology. 2008;190:3738–3746. doi: 10.1128/JB.01721-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mackey SR, Ditty JL, Clerico EM, Golden SS. Circadian Rhythms. Springer; 2007. pp. 115–129. [DOI] [PubMed] [Google Scholar]
- 53.Mackey SR, Golden SS, Ditty JL. The itty-bitty time machine: genetics of the cyanobacterial circadian clock. Advances in genetics. 2011;74:13. doi: 10.1016/B978-0-12-387690-4.00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maniscalco M, Nannen J, Sodi V, Silver G, Lowrey PL, Bidle KA. Light-dependent expression of four cryptic archaeal circadian gene homologs. Frontiers in microbiology. 2014;5 doi: 10.3389/fmicb.2014.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Markson JS, Piechura JR, Puszynska AM, OShea EK. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell. 2013;155:1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moronta-Barrios F, Espinosa J, Contreras A. In vivo features of signal transduction by the essential response regulator RpaB from Synechococcus elongatus PCC 7942. Microbiology. 2012;158:1229–1237. doi: 10.1099/mic.0.057679-0. [DOI] [PubMed] [Google Scholar]
- 57.Moronta-Barrios F, Espinosa J, Contreras A. Negative control of cell size in the cyanobacterium Synechococcus elongatus PCC 7942 by the essential response regulator RpaB. FEBS letters. 2013;587:504–509. doi: 10.1016/j.febslet.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 58.Nakahira Y, Katayama M, Miyashita H, Kutsuna S, Iwasaki H, et al. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 60.Nishiwaki T, Satomi Y, Nakajima M, Lee C, Kiyohara R, et al. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13927–13932. doi: 10.1073/pnas.0403906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ouyang Y, Andersson C, Kondo T, Golden S, Johnson C. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paddock ML, Boyd JS, Adin DM, Golden SS. Active output state of the Synechococcus Kai circadian oscillator. Proceedings of the National Academy of Sciences. 2013;110:E3849–E3857. doi: 10.1073/pnas.1315170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paranjpe D, Sharma V. Evolution of temporal order in living organisms. Journal of Circadian Rhythms. 2005;3:7. doi: 10.1186/1740-3391-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pattanayak GK, Phong C, Rust MJ. Rhythms in Energy Storage Control the Ability of the Cyanobacterial Circadian Clock to Reset. Current Biology. 2014:1–5. doi: 10.1016/j.cub.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pattanayek R, Williams DR, Pattanayek S, Xu Y, Mori T, et al. Analysis of KaiA–KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods. The EMBO journal. 2006;25:2017–2028. doi: 10.1038/sj.emboj.7601086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petit JM, Burlet-Godinot S, Magistretti PJ, Allaman I. Glycogen metabolism and the homeostatic regulation of sleep. Metabolic brain disease. 2015;30:263–279. doi: 10.1007/s11011-014-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7:e1000062. doi: 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rust MJ, Golden SS, OShea EK. Light-driven changes in energy metabolism directlyentrain the cyanobacterial circadian oscillator. Science. 2011;331:220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scanlan DJ, Sundaram S, Newman J, Mann NH, Carr NG. Characterization of a zwf mutant of Synechococcus sp. strain PCC 7942. Journal of Bacteriology. 1995;177:2550–2553. doi: 10.1128/jb.177.9.2550-2553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophy-tochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 72.Shestakov S, Khyen NT. Evidence for genetic transformation in blue-green alga Anacystis nidulans. Molecular and General Genetics MGG. 1970;107:372–375. doi: 10.1007/BF00441199. [DOI] [PubMed] [Google Scholar]
- 73.Shultzaberger RK, Boyd JS, Katsuki T, Golden SS, Greenspan RJ. Single mutations in sasA enable a simpler cikA gene network architecture with equivalent circadian properties. Proceedings of the National Academy of Sciences. 2014a;111:E5069–E5075. doi: 10.1073/pnas.1419902111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shultzaberger RK, Paddock ML, Katsuki T, Greenspan RJ, Golden SS. High-throughput and quantitative approaches for measuring circadian rhythms in cyanobacteria using bioluminescence. Methods in Enzymology. 2014b doi: 10.1016/bs.mie.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annual review of biochemistry. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki E, Umeda K, Nihei S, Moriya K, Ohkawa H, et al. Role of the GlgX protein in glycogen metabolism of the cyanobacterium, Synechococcus elongatus PCC 7942. Biochimica et Biophysica Acta (BBA) - General Subjects. 2007;1770:763–773. doi: 10.1016/j.bbagen.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 77.Sweeney BM, Borgese MB. A circadian rhythm in cell division in a prokaryote, the cyanobacterium Synechococcus WH78031. Journal of phycology. 1989;25:183–186. [Google Scholar]
- 78.Takai N, Ikeuchi S, Manabe K, Kutsuna S. Expression of the circadian clock-related gene pex in cyanobacteria increases in darkness and is required to delay the clock. Journal of biological rhythms. 2006a;21:235–244. doi: 10.1177/0748730406289400. [DOI] [PubMed] [Google Scholar]
- 79.Takai N, Nakajima M, Oyama T, Kito R, Sugita C, et al. A KaiC-associating SasA– RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proceedings of the National Academy of Sciences. 2006b;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD (H)/NADP (H) ratio under light/dark conditions. The Plant Journal. 2005;42:504–513. doi: 10.1111/j.1365-313X.2005.02391.x. [DOI] [PubMed] [Google Scholar]
- 81.Taniguchi Y, Katayama M, Ito R, Takai N, Kondo T, Oyama T. labA: a novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes & development. 2007;21:60–70. doi: 10.1101/gad.1488107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taniguchi Y, Nishikawa T, Kondo T, Oyama T. Overexpression of lalA, a paralog of labA, is capable of affecting both circadian gene expression and cell growth in the cyanobacterium Synechococcus elongatus PCC 7942. FEBS letters. 2012;586:753–759. doi: 10.1016/j.febslet.2012.01.035. [DOI] [PubMed] [Google Scholar]
- 83.Teng SW, Mukherji S, Mo tt, JR, De Buyl S, Oshea EK. Robust circadian oscillations in growing cyanobacteria require transcriptional feedback. Science. 2013;340:737–740. doi: 10.1126/science.1230996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Terauchi K, Kitayama Y, Nishiwaki T, Miwa K, Murayama Y, et al. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proceedings of the National Academy of Sciences. 2007;104:16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 86.Tseng R, Chang YG, Bravo I, Latham R, Chaudhary A, et al. Cooperative KaiA–KaiB– KaiC interactions affect KaiB/SasA competition in the circadian clock of cyanobacteria. J Mol Biol. 2014;426:389–402. doi: 10.1016/j.jmb.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Umetani M, Hosokawa N, Kitayama Y, Iwasaki H. Hypersensitive photic responses and intact genome-wide transcriptional control without the KaiC phosphorylation cycle in the Synechococcus circadian system. Journal of Bacteriology. 2014;196:548–555. doi: 10.1128/JB.00892-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vijayan V, Zuzow R, O’Shea E. Oscillations in supercoiling drive circadian gene expressionin cyanobacteria. Proc Natl Acad Sci USA. 2009;106:22564. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wiegard A, Dörrich AK, Deinzer HT, Beck C, Wilde A, et al. Biochemical analysis of three putative KaiC clock proteins from Synechocystis sp. PCC 6803 suggests their functional divergence. Microbiology. 2013;159:948–958. doi: 10.1099/mic.0.065425-0. [DOI] [PubMed] [Google Scholar]
- 90.Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proceedings of the National Academy of Sciences. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Woelfle MA, Johnson CH. Bacterial Circadian Programs. Springer; 2009. pp. 205–221. [Google Scholar]
- 92.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Current Biology. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 93.Woelfle MA, Xu Y, Qin X, Johnson CH. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proceedings of the National Academy of Sciences. 2007;104:18819–18824. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wood TL, Bridwell-Rabb J, Kim YI, Gao T, Chang YG, et al. The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proceedings of the National Academy of Sciences. 2010;107:5804–5809. doi: 10.1073/pnas.0910141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu Y, Mori T, Pattanayek R, Pattanayek S, Egli M, Johnson CH. Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13933–13938. doi: 10.1073/pnas.0404768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu Y, Weyman PD, Umetani M, Xiong J, Qin X, et al. Circadian yin-yang regulation and its manipulation to globally reprogram gene expression. Current Biology. 2013;23:2365–2374. doi: 10.1016/j.cub.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang C, Hua Q, Shimizu K. Integration of the information from gene expression and metabolic fluxes for the analysis of the regulatory mechanisms in Synechocystis. Applied Microbiology and Biotechnology. 2002;58:813–822. doi: 10.1007/s00253-002-0949-0. [DOI] [PubMed] [Google Scholar]
- 98.Yen UC, Huang TC, Yen TC. Observation of the circadian photosynthetic rhythm in cyanobacteria with a dissolved-oxygen meter. Plant Science. 2004;166:949–952. [Google Scholar]
- 99.Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, et al. Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS One. 2009;4:e5135. doi: 10.1371/journal.pone.0005135. [DOI] [PMC free article] [PubMed] [Google Scholar]