SUMMARY
Necroptosis is a cell death pathway regulated by the receptor interacting protein kinase 3 (RIPK3) and the mixed lineage kinase domain-like (MLKL) pseudokinase. How MLKL executes plasma membrane rupture upon phosphorylation by RIPK3 remains controversial. Here, we characterize the hierarchical transduction of structural changes in MLKL that culminate in necroptosis. The MLKL brace, proximal to the N-terminal helix bundle (NB), is involved in oligomerization to facilitate plasma membrane targeting through the low-affinity binding of NB to phosphorylated inositol polar head groups of phosphatidylinositol phosphate (PIP) phospholipids. At the membrane, the NB undergoes a “rolling over” mechanism to expose additional higher-affinity PIP-binding sites responsible for robust association to the membrane and displacement of the brace from the NB. PI(4,5)P2 is the preferred PIP-binding partner. We investigate the specific association of MLKL with PIPs and subsequent structural changes during necroptosis.
Graphical Abstract

INTRODUCTION
Necroptosis, a mammalian form of programmed cell death, requires RIPK3 (Galluzzi et al., 2014) and MLKL (Murphy et al., 2013; Sun et al., 2012; Zhao et al., 2012). The activation of RIPK3 and MLKL yields the necrosome, the potential executioner of necroptosis. Upstream triggers of necroptosis activate RIPK3 via protein–protein interactions, mediated by its pronecrotic RIP homotypic interaction motif and regulated by RIPK1. Phosphorylated RIPK3 in turn phosphorylates and recruits MLKL to the necrosome. A chemical biology screen identified MLKL as a direct target of the necroptosis inhibitor necrosulfonamide (NSA) (Sun et al., 2012). Current models propose that MLKL causes plasma membrane permeabilization by binding to negatively charged phospholipids and pore formation (Dondelinger et al., 2014; Wang et al., 2014). Alternatively, MLKL may regulate ion channels at the plasma membrane, which in turn mediate osmolysis (Cai et al., 2014; Chen et al., 2014).
The crystal structure of full-length (FL) mouse MLKL has identified the NB separated by the brace region from the C-terminal psKD (Czabotar and Murphy, 2015; Murphy et al., 2013). The psKD interacts with the kinase domain of RIPK3 to allow phosphorylation of residues in the MLKL activation loop (Xie et al., 2013). This phosphorylation induces the monomer-to-oligomer transition of MLKL in the absence of membranes, which is required to activate necroptosis (Cai et al., 2014; Chen et al., 2014). The NB and brace (NBB) binds phospholipids, with phosphorylated phosphatidylinositols and cardiolipin binding most efficiently, whereas the psKD domain does not (Dondelinger et al., 2014; Wang et al., 2014). Enforced expression of NB, NBB, and FL MLKL leads to necroptosis in cells lacking a functional pathway, including mlkl−/− mouse embryonic fibroblasts (MEFs). Moreover, purified FL MLKL and truncated constructs (NB and NBB) can induce the permeabilization of large unilamellar vesicles (LUVs) (Dondelinger et al., 2014; Su et al., 2014; Wang et al., 2014). Further, several hotspots in helices 1 and 2 of NB are membrane-interacting regions with affinity for negatively charged phospholipids, and their mutagenesis can inhibit MLKL-mediated necroptosis (Dondelinger et al., 2014; Hildebrand et al., 2014; Su et al., 2014). A “plug release mechanism” has been proposed whereby the brace N-terminal helix inhibits the plasma membrane–permeabilizing NB domain (Su et al., 2014). Accordingly, the NB inserts into LUVs and is oligomerized to higher-order species, which resemble the active form of MLKL, suggesting that this is the effector of plasma membrane permeabilization during necroptosis.
In this study, we identify a stepwise activation mechanism of MLKL whereby the brace-mediated oligomerization enhances MLKL recruitment to the plasma membrane. Plasma membrane recruitment involves engagement of low-affinity phosphatidylinositol phosphate (PIP)–binding sites followed by a higher-affinity PIP-binding site to induce release of the inhibitory brace.
RESULTS
Brace-Mediated Oligomerization of NBB Increases Necroptotic Activity
The structure of MLKL identified the domain boundaries (Murphy et al., 2013). To delineate the functions of its individual domains in necroptosis (Figure S1), we generated constructs for inducible expression using the Tet-On 3G system, (Figures S1A–D). We produced NBB140, NBB182, psKD, and MLKL isoform 1 (MLKL, Figure S1B). Enforced expression of the four constructs in 3T3-SA MEFs led to robust production of three MLKL proteins detectable 4 h after doxycycline (Dox) induction (NBB140, psKD, MLKL, Figure S1C). We monitored necroptosis by flow cytometry of cells stained with propidium iodide (PI) 24 h after Dox induction (Figure S1D) and kinetically over 24 h by Incucyte imaging of Sytox Green–stained cells (Figure S1E). Although expressed at similar protein levels, psKD and NBB140 did not induce necroptosis in 3T3-SA MEFs (Figures S1D and S1E). MLKL and NBB182 induced necroptosis in about 15% and 55% of cells, respectively, and NBB182 had a 10-fold lower expression level compared to MLKL (Figures S1C and S1D). Addition of the caspase inhibitor qVD did not affect necroptosis, suggesting that apoptosis did not contribute to the observed cell death (Figure S1D). In contrast, the MLKL inhibitor NSA, which covalently attaches to Cys86 of NB (Sun et al., 2012), partially blocked MLKL-induced necroptosis for all constructs containing an intact brace, including NBB182 and MLKL. Interestingly, NSA slightly increased NBB140-mediated necroptosis, suggesting that the brace is required for inhibition (Figure S1D). Our findings suggest that psKD is dispensable for necroptosis. Moreover, the brace may exert a positive effect on the weak activity of NB, increasing it by at least an order of magnitude.
We characterized purified MLKL proteins to determine the difference in the ability of NBB140 and NBB182 to induce necroptosis (Figures 1A and S1D). Analytical ultracentrifugation (AUC) analysis of the oligomeric state of tagless NBB140 and NBB182 revealed that they form monomeric (M), and a mixture of monomeric (M 13%), dimeric (D 21%), and trimeric or tetrameric (T 66%) species, respectively (Figure 1B, Table S1).
Figure 1. Oligomerization of NB Is Essential for Necroptosis.
A. NBB182 and NBB140 purified proteins were >90% pure by Coomassie-staining. B. Analytical ultracentrifugation analysis of apo NBB140 and NBB182, and NSA-treated NBB140 and NBB182 identified their oligomeric profiles. M: monomer, D: dimer, T: trimer or tetramer. The experimentally derived MW is reported for each condition as in Table S1. C. Western blot analysis of Cerulean-MLKL141-182 (Cerulean-brace) oligomerization in the presence of DSS (disuccinimidyl suberate) crosslinker after 16 h of transient transfection in Hek-293 cell line. D. Kinetics and flow cytometry analyses of necroptosis induced by the 2xFV-Flag constructs during and after 12 h of protein expression. Error bars represent the SD from the mean of triplicate samples. Data are representative of n=3 independent experiments. E. Western blot analysis of spontaneous oligomerization of NBB182 after 4 h of protein expression using a BMH (bismaleimidohexane) crosslinker. *P<0.05, **P<0.01, ****P<0.0001, (Ordinary one-way ANOVA with Tukey multi-comparison post-test). See also Figure S1 and Table S1.
The difference in oligomeric states of purified NBB140 and NBB182 led us to hypothesize that oligomerization accounted for their ability to induce necroptosis, and that the brace may be responsible for oligomerization. A Cerulean-brace fusion construct, using the brace residues 141–182, induced high molecular weight species containing Cerulean, observed upon crosslinking (Figure 1C). This supports a role for the brace in oligomerization independent of other MLKL domains. To test how the brace oligomerization regulates necroptosis induction, we further engineered the Tet-On expression system with a tunable drug-inducible 1xFKBP (1xFV) dimerization or 2xFKBP (2xFV) oligomerization cassette introduced between MLKL and the C-terminal Flag or Venus tag (Figures S1A and S1F–K). Protein levels after 4 h of Dox-enforced expression of NBB140-2xFV and NBB182-2xFV were similar (Figure S1G). However, NBB140-2xFV was inactive and NBB182-2xFV was active when assessed kinetically by imaging over 12 h and by FACS analysis at 12 h (Figure 1D). Addition of a dimerizer (Dim) to oligomerize the NBB140-2xFV and NBB182-2xFV fusions led to induction of necroptosis by NBB140-2xFV with the same kinetics and extent as for NBB182-2xFV (Figure 1D). Dim-mediated oligomerization of the NBB182-2xFV fusion led to earlier onset of necroptosis (Figure 1D), supporting the idea that oligomeric MLKL hubs are active species in necroptosis. NSA decreased the rate of NBB182-2xFV–induced necroptosis but did not affect NBB140-2xFV–induced necroptosis, corroborating the suggestion that the brace is required for the inhibitory activity of NSA.
We trapped NBB182 oligomers using the cysteine-directed crosslinking agent bismaleimidohexane. Although the brace does not contain cysteines, using the same conditions, we did not trap the NBB140 oligomers (Figure 1E), which suggests that spontaneous oligomerization of NBB182 brings cysteines in the NB into proximity for crosslinking. NSA addition stabilized monomeric NBB182, suggesting that NSA may interfere with NBB oligomerization (Figure 1E). AUC analysis after adding NSA to purified NBB182 disfavored the production of intermediate dimeric species (Figures 1A and 1B, Table S1). Nonetheless, NSA did not disrupt oligomeric T species, which may explain the level of residual necroptosis with NBB182 plus NSA in cell death assays (Figures 1D and S1D). We therefore tested if dimerization of NBB140 was sufficient to induce necroptosis. Indeed, drug-inducible dimeric NBB140-1xFV induced necroptosis with similar kinetics as the oligomeric NBB140-2xFV (Figures S1H and S1I).
Thus, potent activation by addition of Dim to the 1xFV or 2xFV cassettes restores the ability of monomeric NBB140 to induce necroptosis, supporting the idea that brace oligomerization is essential for NBB182-mediated necroptosis.
NB, the Effector Core of MLKL, Localizes to the Plasma Membrane
If MLKL is the effector of necroptosis, it should localize to the plasma membrane for permeabilization. We visualized plasma membrane localization in real time using C-terminal Venus fusions of NBB140-2xFV and NBB182-2xFV. Upon Dox-mediated induction, the two constructs had similar expression in mlkl−/− MEFs (Figures S2A and S2B) and the potential to induce necroptosis was similar to that for C-terminal Flag fusions (Figures 1D and S2C). As expected, the NBB140-2xFV-Venus fusion was inactive upon Dox-induced expression but became maximally potent when Dox induction was followed by Dim-induced 2xFV oligomerization (Figures 2A and 2B). The NBB182-2xFV-Venus fusion was potently active under both conditions (Figure S2C). NSA inhibited NBB182-2xFV-Venus–induced necroptosis (Figure S2C). Live-cell confocal fluorescence microscopy of the NBB140-2xFV-Venus fusion showed homogeneous cytosolic and nuclear localization upon Dox-induced expression and before Dim-induced 2xFV oligomerization (Figure 2C, 0 min). Analysis of the NBB182-2xFV-Venus fusion was precluded due to the rapid induction of necroptosis (Figures S2A and S2C). We monitored the kinetics of NBB140-2xFV-Venus recruitment to the plasma membrane and used Lymphocyte-Specific Protein Tyrosine Kinase tagged at the C terminus with red fluorescent protein (LCK-C-RFP) as a plasma membrane marker. Upon addition of Dim to induce oligomerization of the 2xFV cassette, NBB140-2xFV-Venus translocated from the cytosol to puncta associated with the plasma membrane in as early as 5 min and colocalized with the LCK-C-RFP staining (Figure 2C, Movie S1).
Figure 2. NB Oligomerization Drives Plasma Membrane Localization Before Rupture.
A. Fast kinetics of necroptosis in mlkl−/− MEFs induced by oligomerization of NBB140-2xFV-Venus after 12 h of Dox pretreatment. Error bars represent the SD from the mean of triplicate samples. Data are representative of n=3 independent experiments. B. Corresponding flow cytometry analysis after 120 min. Error bars represent the SD from the mean of triplicate samples. Data are representative of n=2 independent experiments. C. Live confocal microscopy revealed rapid translocation of NBB140-2xFV-Venus from the cytosol to the plasma membrane upon Dim-induced oligomerization in mlkl−/− MEFs pretreated with Dox for 12 h. Yellow: NBB140-2xFV-Venus, red: LCK-C-RFP. Bar = 10 μm. Data show one representative of n=3 independent experiments. D. Total internal reflection fluorescence of mlkl−/− MEFs expressing NBB140-2xFV-Venus and NBB140(9A)-2xFV-Venus upon 12 h Dox incubation and Dim-mediated oligomerization. Bar = 10 μm. Data show one representative of n=3 independent experiments per condition. E. Magnified view of the 4 min panel shown in D. The red arrows show protein aggregations on the cell surface. F. Summary of the hallmarks of necroptosis as seen by electron microscopy after Dim-induced oligomerization of mlkl−/− MEFs and 12 h of Dox-enforced expression of NBB140-2xFV-Venus. G. Representative 3D-SEM reconstruction of mlkl−/− MEFs expressing NBB140-2xFV-Venus after a 12-h Dox treatment at 0 and 10 min after Dim treatment. See also Figure S2 and Movies S1–S5.
Total internal reflection fluorescence (TIRF) microscopy was performed to determine if NBB140-2xFV-Venus accumulated at the plasma membrane. TIRF microscopy can distinguish molecular events at cellular surfaces (Axelrod, 1981). We observed plasma membrane accumulation of NBB140-2xFV-Venus in puncta as quickly as 4 min after addition of Dim (Figures 2D and 2E, Movie S2). Interestingly, puncta formation preceded membrane permeabilization, quantified as Sytox positive cells, which appeared 20 min after Dim addition (Figure 2A).
Ala substitutions in NBB140- and NBB182-2xFV-Venus at nine Arg, Lys, and His residues in helices 1 and 2 (9A) can inhibit MLKL-mediated necroptosis (Dondelinger et al., 2014). As expected, NBB140(9A)- and NBB182(9A)-2xFV-Venus did not mediate necroptosis, although their expression levels were similar to those of wild-type (WT) counterparts (Figure S2D and S2E, Movie S3). NBB140(9A)-2xFV-Venus TIRF microscopy revealed that Venus was not recruited to the plasma membrane. Our results suggest NBB140-mediated plasma membrane targeting is required for necroptosis (Figures 2D and 2E).
To characterize the morphology of necroptosis, we imaged NBB140-2xFV-Venus mlkl−/− MEFs by three-dimensional scanning electron microscopy (3D SEM) at different time points after addition of Dim (Figures 2F, 2G, S2F, and S2G; Movies S4 and S5). We scored progression from normal to necrotic cell morphology. At 0 min, cells showed a normal plasma membrane with numerous microvilli and intact cell architecture. At 5 and 10 min, 75% and 88% of cells, respectively, showed pre-necrotic phenotypes, including major plasma membrane modifications such as the disappearance of microvilli, flattening, micro and macro ruptures ranging from 10 nm to 0.5–1 μm, and organelle depletion defined as large cytosolic area, at least 30–40% of the whole cell area, devoid of organelles (Figures S2F and S2G). By 20 min, most cells were pre-necrotic (52%) or necrotic (45%).
Our results pinpoint NBB140 as the plasma membrane targeting core of MLKL, whose recruitment and accumulation at the plasma membrane is induced by oligomerization and precedes plasma membrane rupture. Moreover, NBB140-induced morphological changes resemble necrotic phenotypes, including plasma membrane rupture with extravasation of cytosol and organelles.
Weak Binding of Phosphorylated Inositols Supports NB Binding to PIPs
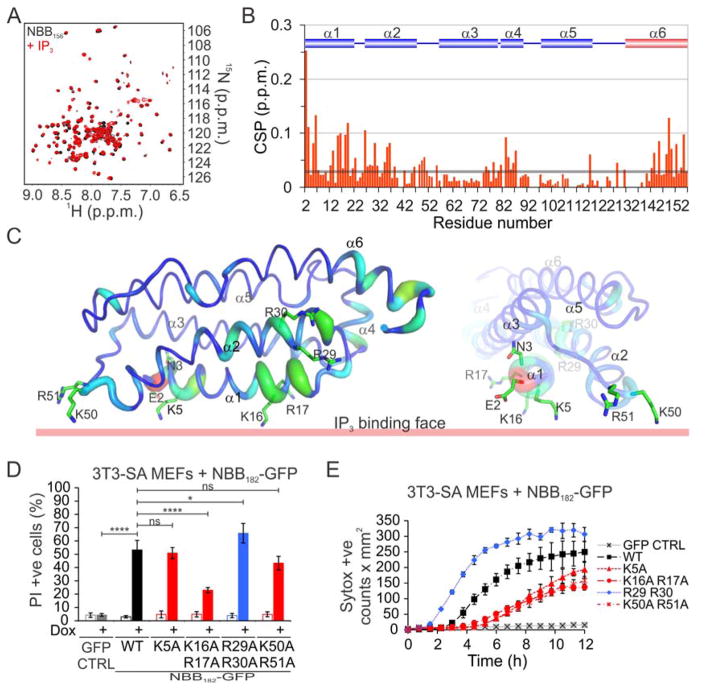
Our finding that NBB140 is recruited to the plasma membrane before plasma membrane rupture prompted us to characterize the molecular basis of its interaction with plasma membrane phospholipids. Previous studies showed that NB associates with membranes in liposome co-flotation assays, and parts of NBB2-154 may become buried in the lipid bilayer according to the differential sensitivity of a site-labeled fluorescent probe to aqueous and hydrophobic environments (Su et al., 2014). Moreover, PIPs and cardiolipin bind to MLKL NB (Wang et al., 2014), presumably through the interaction of negatively charged phospholipid polar head groups (inositol phosphates and phosphate, respectively) and positively-charged patches in NB (Dondelinger et al., 2014). Given that the plasma membrane does not contain cardiolipin, we focused on the interaction of PIPs with MLKL. PIPs can be deconstructed into diacylglycerol (the hydrophobic tail) and inositol phosphates (polar head group). We tested the ability of inositol polar head groups myoinositol, inositol-1-phosphate [IP(1)], inositol-1,4-diphosphate [IP2(1,4)], inositol-1,5-diphosphate [IP2(1,5)], inositol-1,4,5-triphosphate [IP3(1,4,5)], and inositol-1,3,4,5-tetrakisphosphate [IP4(1,3,4,5)] to bind to the NBB by NMR spectroscopy. We performed preliminary NMR analysis on NBB156, an N-terminal construct of MLKL that exhibits well-dispersed NMR spectra, as previously reported for a similar construct (NBB2-154) (Su et al., 2014). We assigned the backbone amide resonances for NBB156, which were similar to those of NBB2-154 (Su et al., 2014). We performed titrations of 15N-labeled NBB156 with inositols and monitored perturbations in [15N, 1H] NMR spectra. These revealed the weak millimolar affinity and binding interface of phosphorylated inositols. The chemical shift perturbation (CSP) observed in NMR titrations enabled us to rank the affinity of inositols for NBB156: myoinositol at 20 mM and IP(1) at 2 mM had no detectable binding; IP2(1,4) and IP2(1,5) at 2.7 mM had intermediate binding, and IP3(1,4,5) and IP4(1,3,4,5) at 1.4 mM had the highest binding (Figure S3A).
Because IP3 induced more chemical shifts at the N terminus of NBB156 (residues E2 and N3) than IP4, we further characterized the NBB156–IP3 interaction. Although the weak-affinity binding manifested as fast exchange by NMR and the CSP was traced during titration, we reassigned the backbone N-H resonances of NBB156 at 5 mM IP3 to prevent assignment ambiguities due to overlaps (Figures 3A and 3B). The CSP induced by IP3 mapped extensively to helices 1 and 2 and also to the beginning of helices 3 and 4 and the end of helix 6 (Figures 3B and 3C). These sites shared Arg, Lys, and His residues, all of which may interact with the negatively charged phosphates in IP3. Due to weak interactions, there were no intermolecular nuclear Overhauser effects between IP3 and any of the residues exhibiting CSP in NBB156; therefore, we performed titration analysis to determine the differential affinity of binding across the NBB156 structure. The CSP-based affinity (Figure S3B) ranged from low (~1–2 mM, Arg 17, Lys26, Gln27, Lys50, and Arg51) to intermediate millimolar (~5–10 mM, Glu2, Asn3, Lys5, Arg30).
Figure 3. NB Binds to Isolated Polar Head Groups of PIPs.
A. Overlaid [15N-1H] TROSY spectra of 200 μM 15N-NBB156 ± 5 mM IP3. B. CSP of NBB156 induced by 5 mM IP3 as a function of residue number. C. Mapping of IP3-binding residues onto the NMR structure of NBB2-152 (PDB ID 2MSV). The thickness of the tubes represent the magnitude of the CSP in panel B. D. Flow cytometry analysis of necroptosis induced after 12 h by Dox-enforced expression of WT and IP3-binding mutant NBB182-GFP constructs. Data are the average and SD of n=3 independent experiments in triplicate. E. Kinetics of necroptosis induced by 12 h of Dox-enforced expression of the constructs in panel D. Error bars represent the SD from the mean of triplicate samples. *P<0.05, ****P<0.0001, (Ordinary one-way ANOVA with Tukey multi-comparison post-test). See also Figure S3.
We then examined enforced expression-induced necroptosis in 3T3-SA MEFs, using NBB182-GFP with Ala substitutions at Lys5, Lys16 Arg17, and Lys50 Arg51, because these residues face one side of the NB opposite the brace (Figure 3C). Mutagenesis of higher-affinity sites had the most pronounced negative effect on NBB182-GFP activity (Figures 3D, 3E, and S3C). In contrast, the Ala substitution at Arg29 Arg30 slightly increased the activity of NBB182-GFP compared to WT (discussed in more detail below). Together, NMR and mutagenesis analyses show that positively-charged clusters on the surface of NBB156 bind to inositol phosphates with weak affinity and mediate early interactions with the plasma membrane, most likely through PIPs (Dondelinger et al., 2014; Wang et al., 2014).
Mutagenesis of Inositol Phosphate–Binding Sites in NB Impairs Necroptosis
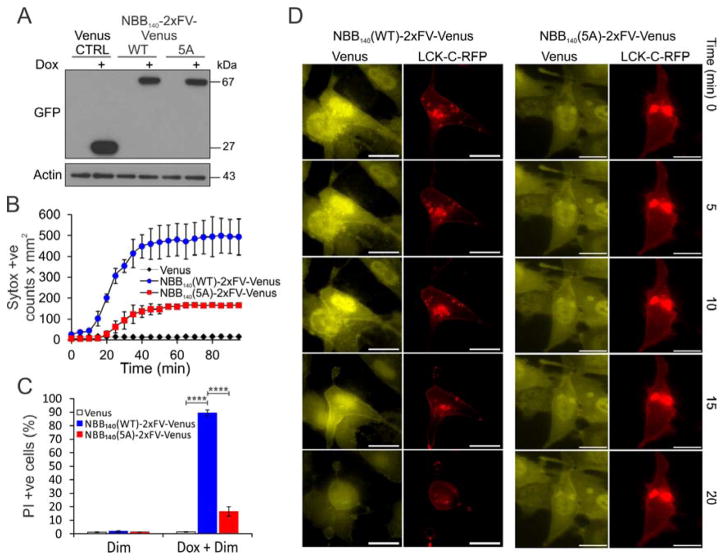
We explored membrane recruitment of IP3-binding mutants and combined the 5 Ala mutations of Lys5, Lys16 Arg17, and Lys50 Arg51 clusters in the context of NBB140- or NBB182-2xFV-Venus enforced expression (Figure 3E). We refer to this as the 5A mutant. Compared to the WT, the 5A mutant showed delayed kinetics of necroptosis in mlkl−/− MEFs measured by imaging over 12 h (Figure S4A). This delay resulted in about 30% of cells undergoing necroptosis after 12 h for the 5A mutant compared with 80%–85% for the WT (Figure S4B) despite similar protein levels (Figure S4C). A more pronounced ablation of necroptosis occurred after 12 h enforced expression of the NBB140-2xFV-Venus 5A mutant in mlkl−/− MEFs, followed by 2 h incubation with Dim (Figures 4A–C). Unlike the WT, the NBB140-2xFV-Venus 5A mutant did not show punctate staining at the plasma membrane, instead maintaining a cytosolic distribution at 20 min, when most of the cells expressing the WT construct were dead (Figure 4D). The 5A mutant did not inhibit the ability of NBB182 to oligomerize, underscoring its defect in membrane recruitment (Figure S4D).
Figure 4. IP3-Binding Sites Recruit MLKL to Plasma Membrane.
A. Western blot analysis after 12 h of Dox-enforced expression of WT and 5A mutant NBB140-2xFV-Venus constructs in mlkl−/− MEFs.
B. Fast kinetics of necroptosis induced at 120 min of Dim-induced oligomerization of WT and 5A mutant NBB140-2xFV-Venus constructs after 12 h of Dox-enforced expression in mlkl−/− MEFs. Error bars represent the SD from the mean of triplicate samples. Data are representative of n=3 independent experiments. C. Corresponding flow cytometry analysis after 120 min. Data are the average and SD of n=3 independent experiments in triplicate. D. Live microscopy of early time points of necroptosis induced by Dim-mediated oligomerization in mlkl−/− MEFs after 12 h of Dox-enforced expression of WT and 5A mutant NBB140-2xFV-Venus. Yellow: Venus, Red: LCK-C-RFP. Bar = 20 μm. Data show one representative of n=3 independent experiments per condition.****P<0.0001, (Ordinary one-way ANOVA with Tukey multi-comparison post-test). See also Figure S4.
Human MLKL is not phosphorylated by murine RIPK3 (Chen et al., 2013) and therefore not activated by necroptotic signals in murine cells. Thus, to study the behavior of FL hMLKL, we developed a coexpression strategy involving Dim-induced oligomerization of Cerulean-2xFV-hRIPK3 to activate hMLKL-Venus, to test the plasma membrane recruitment of the deficient 5A FL mutant expressed in ripk3−/− mlkl−/− MEFs (Figures 5 and S5). We found low-level necroptosis (~10% PI +ve cells) upon Dox-induced expression of MLKL-Venus, which was augmented by RIPK3 oligomerization (~30% PI +ve cells, Figure 5B). Necroptosis increased dramatically when the caspase inhibitor zVAD was added (~80% PI +ve cells), presumably because oligomerized RIPK3 recruited the murine RIPK1/FADD/caspase-8–FLIP complex to inhibit the necrosome (Orozco et al., 2014). Necroptosis was inhibited by NSA, and was resistant to the RIPK1 inhibitor Nec-1s (Figure 5B), which is consistent with a study involving murine RIPK3 (Orozco et al., 2014).
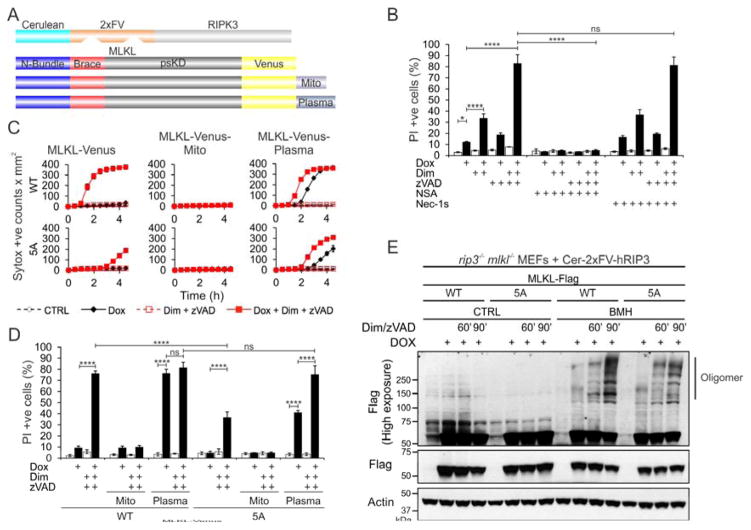
Figure 5. Plasma Membrane Targeting Bypasses Requirement for Weak-Affinity Recruitment.
A. Schematic representation of the co-expressed chimeric RIPK3 and MLKL proteins. B. Flow cytometry analysis of necroptosis after 5 h of Dox-enforced expression of Cer-2xFV-hRIPK3 in ripk3−/−mlkl−/− MEFs that express the MLKL-Venus constructs. Error bars represent the SD from the mean of triplicate samples. Data are representative of n=3 independent experiments. C. Kinetics of necroptosis after Dox-enforced expression of RIPK3 in ripk3−/−mlkl−/− MEFs expressing the WT and 5A MLKL constructs from panel A. Error bars represent the SD from the mean of triplicate samples. Data are representative of n=3 independent experiments. D. Flow cytometry analysis of necroptosis for the cells in panel C. E. Western blot analysis of WT and 5A mutant MLKL-Venus constructs in ripk3−/− mlkl−/− MEFs upon activation by RIPK3. Data are the average and SD of n=3 independent experiments in triplicate. *P<0.05, ****P<0.0001, (Ordinary one-way ANOVA with Tukey multi-comparison post-test). See also Figure S5.
We designed mitochondrial- and plasma membrane–targeted constructs of MLKL-Venus by adding at the C terminus the BCL-xL transmembrane-targeting tail (Mito), and HRas25 plasma membrane–targeting (Plasma) sequences, respectively (Figures 5A, S5B and S5C) (Llambi et al., 2011; Sancak et al., 2010). MLKL-Venus-Mito did not induce necroptosis even after RIPK3 activation in the presence of zVAD, supporting our previous observation that mitochondria are dispensable for necroptosis (Figures 5C and 5D) (Tait et al., 2013). In contrast, MLKL-Venus-Plasma caused necroptosis upon Dox-induced expression, indicating that plasma membrane targeting is sufficient to activate MLKL. Necroptosis was accelerated by RIPK3 oligomerization in the presence of zVAD.
We used the RIPK3 MLKL coexpression system to monitor necroptosis for the WT and 5A mutant. Although oligomerization was not impaired in the WT and 5A mutant (Figure 5E), the 5A mutation affected the kinetics and extent of cell death. Necroptosis began within the first 30 min of RIPK3 oligomerization in WT MLKL-Venus, followed by a steep increase in rate. By 3 h, the Sytox positive counts reached a plateau with the WT, as most cells had undergone necroptosis (Figure 5C). In contrast, necroptosis was first detected at 3 h in the 5A mutant, and the death rate was much slower subsequently. At 4.5 h, only ~35% of cells had undergone necroptosis (Figure 5D). 5A MLKL-Venus-Plasma was more active than the untargeted mutant, showing a delay in onset of only 1 h compared to the WT and induction of necroptosis with or without RIPK3 oligomerization. Therefore, impaired plasma membrane recruitment and cell death in the 5A MLKL was overcome by plasma membrane targeting. We conclude that positively-charged clusters in NB mediate the weak-affinity recruitment to plasma membrane downstream of brace oligomerization.
PIP-Induced Displacement of Brace from NB Mediates Necroptosis
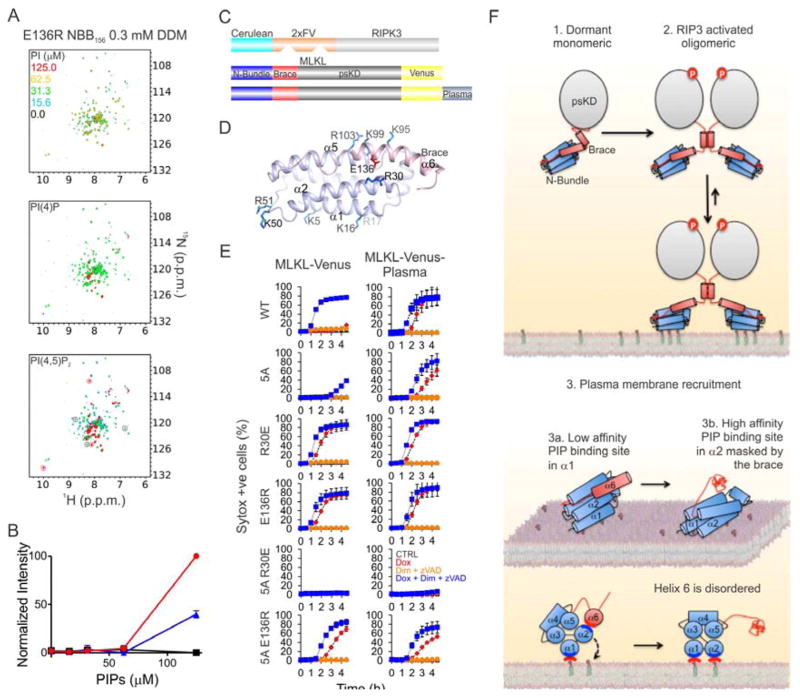
If MLKL is the effector of plasma membrane permeabilization, it may undergo a conformational change at the membranes. To mimic association with membranes we used detergent micelles. Nonionic detergents, including dodecyl maltoside (DDM), nonyl maltoside (NM), IGEPAL, and others, even above their critical micellar concentration (CMC), did not induce a micelle-associated conformation of NBB156, which remained monomeric (Figure 6B, S6A and S6B).
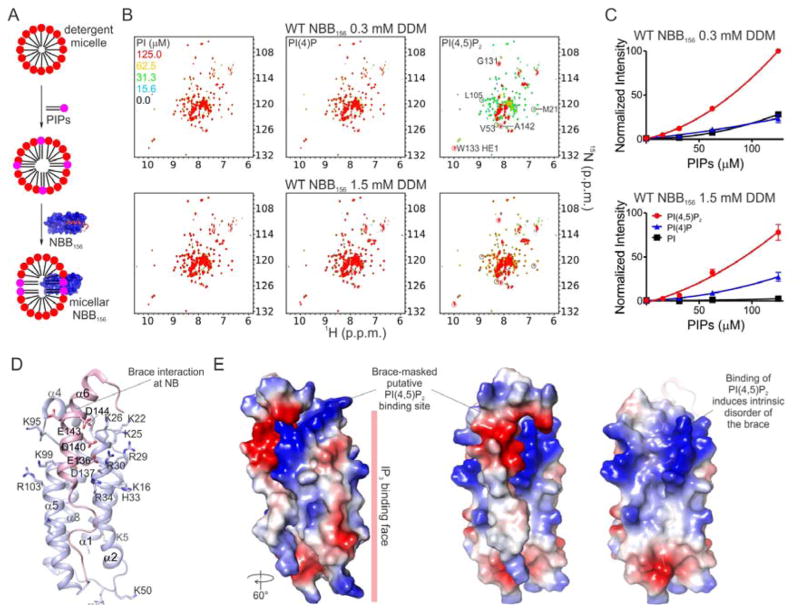
Figure 6. PI(4,5)P2 Binding Displaces the Brace from NB.
A. Detergent micelles were mixed with PIPs followed by NBB156 to generate micellar NBB156. B. Overlaid [15N-1H] TROSY spectra of 15N-NBB156 in micellar DDM containing PIP as indicated. The free and micelle-associated NBB156 resonances used to normalize intensities between the different concentration of detergent and PIPs are labeled and circled grey and red, respectively. C. PIP-dependent induction of intrinsic disorder in the brace was quantified for the micelle-associated resonances circled red in panel B. The plot shows average and standard deviation of three normalized intensities. Details of intensity normalization are provided in Figure S6 and Experimental Procedures. D. The unstructured region corresponding to the C terminus of NBB156 was mapped onto NBB2-152 (pink PDB ID 2MSV). Basic and acidic residues involved in the interaction with IP3 and the brace are represented as sticks. E. Surface representation of NBB2-154 highlighting the acidic nature of the brace (left and center), which masks the putative basic PI(4,5)P2-binding site (right). See also Figure S6.
We supplemented micellar DDM (0.3 mM and 1.5 mM), NM (12 mM), and IGEPAL (0.5%) with phosphatidylinositols, including PI, PI(4)P, and PI(4,5)P2 up to 125 μM, and monitored NBB156 conformation by NMR (Figure 6A). Remarkably, we observed PIP-dependent interaction of NBB156 with all detergent micelles (Figures 6B, S6A and S6B). Under these conditions two species were detected by TROSY NMR analysis, including the apo monomer and the putative micelle-associated NBB156 (micellar NBB156). At low and high PIP concentration the monomer and micellar NBB156 species dominated, respectively. A mixture of the two species was observed at intermediate PIP concentration. Micelle association was accompanied by PIP-dependent line broadening and disappearance of about 80% of the apo monomer resonances and the appearance of new resonance (Figures 6B, S6A and S6B). The NBB156 resonances induced by high PIP concentration exhibited intrinsic disorder character, suggested by their narrow chemical shift dispersion (7.7 and 8.3 p.p.m.). At the highest PIP concentration in 0.3 mM DDM (~2x CMC), intrinsic disorder in NBB156 was fully induced by PI(4,5)P2 (100% micellar) and partially induced by PI(4)P or PI (~25% micellar, Figures 6B and 6C). At 1.5 mM DDM (~10x CMC), the apparent PIPs concentration was decreased through dilution within excess micelles, thus lowering the ability of PI(4,5)P2, PI(4)P, and PI to engage NBB156 in interactions with micelles to ~80, 25, and 5%, respectively (Figures 6B and 6C). Similar interactions were observed with the detergents NM and IGEPAL (Figures S6B and S6C).
We assigned backbone amide resonances for the intrinsically disordered region, which maps to the C-terminal region of NBB156, starting with residue 122 and stretching to the C terminus (Figures S6D–S6F). The observed Cα chemical shifts suggest that this region is unstructured compared to the free form (Figure S6G). The NBB2-154 NMR structure suggested that the PI(4,5)P2 binding site may involve clusters of positively charged Arg and Lys residues that interact with the brace (Figure 6D). Their engagement presumably resulted in brace displacement from the NB (Figure 6E). Addition of IP3 to compete PI(4,5)P2 binding at the putative high-affinity site in NB did not recover NBB156 resonances (Figure S6D). We speculate that PIP engagement of the putative PI(4,5)P2-binding site, masked by the brace, may trigger the release of the brace from the NB in the presence of membrane. Presumably, because the NB interacts with PI(4,5)P2 associated with detergent micelles, it tumbles too slowly and relaxes too fast to be detected on the NMR time scale. We therefore could not further explore the structure of NBB156 in micellar detergent supplemented with PIPs by solution NMR.
Modulating Necroptosis Through Mutations in MLKL Inositol-Binding Sites
We introduced Ala or Glu substitutions at Arg30 of helix α2 and at Lys95, Lys99, and Arg103 of helix α5 of the NB to test how these electrostatic residues masked by the brace may contribute to the putative interaction with the negatively charged polar head group of PI(4,5)P2 (Figures 6D and 6E). Using TROSY NMR analysis of NBB156, we showed that all tested mutants were able to bind micellar PI(4,5)P2 to induce disorder in the brace, albeit the 5A mutant, Arg30Glu, and Arg103Ala exhibited diminished potency (Figures S7A and S7B), suggesting roles in the interaction with PI(4,5)P2. Arg30 Ala or Glu substitutions in FL MLKL-Venus, alone or in combination with Ala or Arg substitutions at Arg29 and Glu136 (the salt bridge partner to Arg30 in the brace), respectively, induced necroptosis better than WT, at similar expression levels (Figures S7C and S7D). The related Glu136Ala and Glu136Arg mutants induced similar Dox-induced necroptosis (Figures S7C and S7D). TROSY NMR analysis of Glu136Arg NBB156 at 0.3mM DDM indicated that, similar to the WT (Figures 6B and 6C), PI(4,5)P2 is absolutely essential to fully induce intrinsic disorder in the brace, presumably by engaging the putative, unaltered PIP binding site in NB (Figures 7A and 7B). Our results underscore the strict requirement for PI(4,5)P2 in triggering the micelle-associated conformation of NBB156, challenging the notion that the Arg30–Glu136 salt bridge is the main molecular determinant of the putative plug release mechanism (Su et al., 2014). Interestingly, all of the mutants were more resistant than the WT to NSA-mediated inhibition, and the Arg30Glu, Glu136Arg, and Arg30Glu Glu136Arg mutants were completely resistant (Figure S7C). The resistance to NSA inhibition was more apparent for Dox-induced NBB182-2xFV-Venus–mediated necroptosis (Figure S7E and S7F). Our results suggest that NSA-mediated inhibition takes advantage of an intact interaction between the brace and NB.
Figure 7. Sequential PI(4,5)P2 Binding Through a “Rolling Over” Mechanism Induces Brace Displacement.
A. Overlaid [15N-1H] TROSY spectra of Glu136Arg 15N-NBB156 in micellar DDM containing PIP as indicated. The free and micelle-associated NBB156 resonances used to normalize intensities between the different PIP concentrations are labeled and circled grey and red, respectively. B. PIP-dependent induction of intrinsic disorder in the brace was quantified for the micelle-associated resonances circled red in panel A. C. Schematic representation of the co-expressed RIPK3 and MLKL chimeric proteins. D. The mutated basic residue are highlighted on the cartoon representation of NBB2-154. E. Kinetics of necroptosis induced by Dim-mediated oligomerization of Cer-2xFV-RIPK3 after 5 h of Dox-enforced expression of WT and mutant MLKL-Venus constructs in ripk3−/− mlkl−/− MEFs. Error bars represent the SD from the mean of triplicate samples. Data are representative of n=3 independent experiments. F. Our revised mechanism of MLKL-mediated necroptosis involves RIPK3-mediated phosphorylation of MLKL inducing a poorly understood transition from the cytosolic monomeric conformation, elucidated by the crystal structure of FL mMLKL (PDB ID 4BTF), to a transient brace-mediated oligomeric conformation. The brace-mediated oligomerization may be the site of action for the MLKL inhibitor NSA. Brace-mediated oligomerization facilitates recruitment to the plasma membrane via low-affinity binding of Lys16 Arg17 of helix α1 to the polar head group of PI(4,5)P2. We propose that after the initial plasma membrane recruitment, there is a “rolling over” mechanism involving more extensive interactions of basic residues in helix α2 with PI(4,5)P2, resulting in displacement of the brace and its intrinsic disordering. Our study does not address steps downstream of PI(4,5)P2 binding, but others (Dondelinger et al., 2014; Su et al., 2014; Wang et al., 2014) have demonstrated that MLKL can permeabilize synthetic liposomes, supporting its role as the effector of plasma membrane permeabilization. See also Figure S7.
We next tested the effect of Arg30Glu and Glu136Arg mutations, alone and in combination with the 5A mutant, on necroptosis by RIPK3, MLKL coexpression in ripk3−/− mlkl−/− MEFs. This was performed for MLKL-Venus or MLKL-Venus-Plasma constructs (Figures 7C–E and S7G). For both constructs, the Arg30Glu, Glu136Arg and the combined 5A Glu136Arg mutant induced potent activation of MLKL in the absence of RIPK3 oligomerization, although the latter showed a delay in onset and a slower rate of cell death (Figure 7E). There was an increase in necroptosis upon RIPK3 oligomerization for all mutants (Figure 7E) except for the combined 5A, Arg30Glu mutant (Figure 7E). This mutant was completely inactive and neither plasma membrane targeting nor RIPK3 oligomerization induced its activity (Figure 7E). Lys95Ala, Lys99Ala, and Arg103Ala mutations in helix 5 of MLKL-Venus showed decreased ability to induce necroptosis due to slower kinetics compared to the WT in the RIPK3 MLKL coexpression system (Figures S7H and S7I). We therefore identified positively-charged clusters in NB that cooperate to mediate membrane recruitment via PIP binding, which leads to release of the brace from NB as a major mechanistic step in MLKL-mediated necroptosis.
Discussion
During necroptosis, RIPK3 phosphorylates the MLKL activation loop (Cai et al., 2014; Murphy et al., 2013; Sun et al., 2012; Wang et al., 2014). The binding of the RIPK3 kinase domain to the psKD of MLKL positions the active site of the former close to the activation loop of the latter (Xie et al., 2013), and mutations at this interface disrupt MLKL phosphorylation. Phosphomimetic mutations of RIPK3 phosphorylation sites in MLKL activate the pseudokinase to induce a monomer-to-oligomer transition of MLKL in vitro and in cells. Mounting evidence, including that presented herein, suggests that NB is the minimal region of MLKL active in necroptosis (Chen et al., 2014; Dondelinger et al., 2014; Hildebrand et al., 2014). Therefore, phosphorylation by RIPK3 at the activation loop likely triggers the active form of NB. The activated NB of MLKL binds phospholipids, especially those that are negatively charged, which brings MLKL to the plasma membrane and potentially to other intracellular membranes. NB can be recruited to and permeabilizes liposomes, and the interaction between the brace and NB likely acts as an inhibitory plug that must be released to unleash the pore-forming activity of NB (Dondelinger et al., 2014; Su et al., 2014; Wang et al., 2014).
Our study reveals insights into MLKL oligomerization outside of membranes, plasma membrane recruitment, and phospholipid binding. NBB182 was active whereas NBB140 was inactive in mediating necroptosis, which may be due to their difference in oligomerization. Purified NBB140 and NBB156 are monomeric, whereas purified NBB182 is a mixture of monomeric, dimeric, and mostly oligomeric species. Forced oligomerization of NBB140 using the 2xFV cassette restored its necroptotic activity, supporting the notion that oligomerization is needed for function. Even dimerization of NBB140 using the 1xFV cassette induced necroptosis, suggesting that NBB140 dimers may be the minimal core required for MLKL-induced necroptosis. Surprisingly, NSA, which attaches to the Cys86 in helix 4 of NB, inhibits only NBB182 but not NBB140, suggesting that the C-terminal region of the brace is required for NSA-mediated inhibition. Helix 4 is positioned in NB adjacent to Phe148 of the brace, and given the extended conformation of NSA we speculate that it may interact with the C-terminal portion of the brace. AUC analysis indicated that NSA disrupts the monomer-to-oligomer equilibrium of NBB182 in favor of the monomer by preferentially breaking down dimers over oligomers. Remarkably, mutations that disrupt a critical contact between the brace and NB, the Arg30–Glu136 salt bridge, and potentially the overall architecture of NBB182, become refractory to NSA inhibition. Our analysis suggests that the brace contains an oligomerization cassette whose engagement leads to some of the observed soluble higher-order MLKL species and necroptosis. Preliminary sequence analysis suggested that the brace may form β-strand aggregates, reminiscent of the hetero- and homotypic interactions governing RIPK1 and RIPK3 oligomerization (Li et al., 2012).
Nine positively-charged residues are found in helices 1 and 2 of NB, and their substitution prevents plasma membrane recruitment and necroptosis (Dondelinger et al., 2014). Our NMR analysis deconstructed the interaction of PIP with NBB156. Our per residue NMR titrations using IP3 identified multiple basic clusters across the NB surface that show different binding affinities. The Lys16 Arg17, Lys26 Gln27, and the Lys50 Arg51 clusters had the highest affinities for IP3. Combined Ala substitution of Lys5, Lys16 Arg17, and Lys50 Arg51 clusters (5A mutant) had an additive effect on MLKL activity inhibiting plasma membrane recruitment and necroptosis, but this effect was overcome by enforced recruitment of MLKL to the plasma membrane. We propose that the weak-affinity binding to inositol polar head groups of PIPs, with preference for the more negatively charged PI(4,5)P2, induces initial MLKL recruitment to the plasma membrane through a helix α1-mediated interaction. Another inositol phosphate-binding site involves clusters of basic residues in helix α2, which also mediate the interaction with the brace. Nonetheless, in the context of micellar detergents, which otherwise do not interact with NBB, this site engages PIP phospholipids to induce intrinsic disorder in the brace and the region N terminal to the brace. Moreover, we saw a correlation between the binding preference for inositol phosphates and their corresponding phospholipids, the IP3 polar head group and its parent PI(4,5)P2 phospholipid being preferred MLKL ligands. However, while PIP2 induced unfolding of MLKL in micelles, soluble IP3 did not.
On the basis of our analysis, we propose a revised mechanism of MLKL activation (Figure 7F). MLKL is a dormant monomeric protein that resides largely in the cytosol. Upon RIPK3 phosphorylation of the activation loop, MLKL undergoes unfolding and release of the brace from psKD by a poorly understood mechanism. After release, the brace propensity to oligomerize through residues 157–182 likely contributes to an overall increase in avidity of NB for the plasma membrane. Initially, NB recruitment to the plasma membrane is through low-affinity PIP-binding sites made up of clusters of basic residues. It is likely that the Lys16 Arg17 cluster in helix α1 and Lys50 Arg51 in the loop between helices α2 and α3 are engaged initially opposite to the brace. This initial targeting is followed by a rolling mechanism whereby helix α2 is engaged to bring the brace in proximity to the plasma membrane. Rearrangement of the salt bridge network involved in the interaction of basic residues in NB with acidic residues in the brace neutralizes the negatively charged polar head group of PIPs. The ability to overcome the deficiency in the 5A mutant by enforced localization supports that the primary function of the exposed basic residues in helix 1 is to bind to the plasma membrane. However, basic residues of helix α2 then become critical, as mutation of even 1 residue is sufficient to completely inactivate the plasma membrane–targeted 5A mutant. This rearrangement leads to displacement of the brace, which becomes intrinsically disordered. The interaction of NB with PIPs induces strong association of NB with the plasma membrane, and we suspect that one putative PIP binding site may be masked by the brace. This was supported by our mutagenesis analysis of Lys and Arg residues in helix α5, which inhibited PI(4,5)P2 binding and necroptosis. In contrast, substitutions at Arg30 in helix α2 resulted in hyperactive MLKL, supporting its role in the plug release mechanism (Su et al., 2014). Unfortunately, the low yield of the mutant Arg30 recombinant proteins precluded a thorough analysis of their PIP binding preference. Therefore, we do not exclude the possibility of the charge at Arg30 being a critical determining of phospholipid binding preference.
Our model does not discriminate between direct or indirect involvement of MLKL in plasma membrane rupture, but is consistent with findings from our structural, biochemical, and cell-based studies and identifies critical steps in the mechanism of MLKL-mediated necroptosis. As executioner (Sun and Wang, 2014), characterization of the PIP-mediated micelle-associated conformation becomes paramount as it may reflect the active MLKL structure associated with the plasma membrane. Indirectly, plasma membrane associated MLKL may cooperate with previously reported ion channels to mediate necroptosis by deregulation of sodium and calcium homeostasis leading to osmolysis (Cai et al., 2014; Chen et al., 2014).
EXPERIMENTAL PROCEDURES
Cell Lines
Primary MEFs were generated as previously described (Dillon et al., 2014; Rodriguez et al., 2015). MEFs were maintained in DMEM supplemented with 10% FBS.
Plasmid and Retroviral Transduction
Human MLKL and RIPK3 constructs were cloned into the Dox-inducible vector pRetroX-TRE3G (631188, Clontech) or the pC4M-FV2E vector (Ariad Pharmaceuticals) in frame with several N- and C-terminal fusion partners. Retroviral transduction of target cells was performed according to standard protocols.
Western Blotting and Antibodies
Protein extracts were resolved by SDS PAGE, transferred to nitrocellulose membrane and probed with the following specific antibodies: hMLKL (M6697) and flagM2 (F1804, F7425, Sigma); histone H4 (ab10158), hMLKL 3H1 (ab10158), MLKL phospo S358 (EPR9514/ab187091, Abcam); GFP (sc8334), β-actin C4 (sc47778), hRIPK3 B2 (sc374639, Santa Cruz Biotechnology).
Cell Death Assay
Necroptosis was induced with a combination of Dox, Dim and other drugs as specified. Necroptosis was monitored with cell impermeable dyes that bind nucleic acids by imaging using Incucyte FLR or Zoom imaging system (Essen Bioscience) and by flow cytometry using FACScan and FACsCalibur systems (BD Biosciences).
Cell Imaging
Live-cell microscopy of MLKL-mediated necroptosis was performed using a Marianas spinning disc laser scanning confocal system (Intelligent Imaging Innovations). Time-lapsed TIRF microscopy was performed using the AxioObserverZ.1 microscope equipped with an automated TIRF slider (Carl Zeiss Microscopy). Time-lapsed imaging using wide-field illumination was performed using a Nikon Ti-E microscope.
Electron Microscopy
Samples were fixed and dehydrated using standard protocols, embedded in epoxy resin and polymerized prior to EM analysis. The 3D EM images were collected on a FEI Helios Nanolab 660 Dualbeam system using Auto Slice and View software and processed with Amira 6 software. For the quantification analysis, a low-magnification image of an Epon block surface with exposed cells was imaged at 5keV using the NanoLab 660 SEM. The Maps software was used to create a high-resolution montage containing about 100 cells.
Bacterial Expression and Purification
Human MLKL constructs were expressed using the pNic28Bsa4 vector (Savitsky et al., 2010) in BL21* E. coli (Invitrogen). Protein was purified by standard chromatography.
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted in 20 mM HEPES pH 7.5, 100 mM NaCl, and 0.22 mM DTT in a ProteomeLab XL-I analytical ultracentrifuge (Beckman Coulter) following standard protocols (Schuck, 2000; Zhao et al., 2013).
NMR Assignment and Titrations
The backbone of 13C/15N-labeled NBB156 with or without IP3 and with micellar detergents supplemented with PI(4,5)P2 was assigned using standard triple-resonance experiments (Sattler, 1999). Backbone assignment experiments and NMR titrations were acquired in 20 mM phosphate buffer pH 6.8 and 10% D2O at 298K on Bruker Avance 600 MHz spectrometers equipped with cryogenically cooled triple resonance z-gradient probes. Data were processed in TopSpin (Bruker BioSpin) and spectral analysis was done in Cara (Keller, 2004).
Supplementary Material
Highlights.
MLKL activation is governed by a hierarchical transduction of structural changes
The brace undergoes oligomerization prior to plasma membrane recruitment
Low affinity NB binding to PIP mediates plasma membrane targeting
High affinity PIP binding to NB displaces the brace for robust membrane association
Acknowledgments
We thank Dr. Christopher P. Dillon for providing mlkl−/− and ripk3−/− mlkl−/− MEFs and Patrick Fitzgerald for extensive technical assistance. G.Q. thanks Drs. Nazzareno Capitanio, Claudia Piccoli, and Ricardo Weinlich for helpful discussions and advice. This work was supported by grants from the U.S. National Institutes of Health and ALSAC.
Footnotes
AUTHOR CONTRIBUTIONS
G.Q., T.M. and D.R.G. conceived the study. G.Q. and T.M. designed, executed and analyzed the majority of cell-based and in vitro experiments, respectively. C.S.G. performed the confocal microscopy analysis, F.L. and D.A.R. some of the biochemical oligomerization assays, R.W. and S.F. the EM analysis, C.R.G. some of the NMR analyses and A.N. the AUC analysis. G.Q., T.M. and D.R.G. wrote and edited the manuscript with input from all co-authors.
Supplemental Information includes Supplemental Experimental Procedures, four figures, and five tables and can be found with this article online at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol. 1981;89:141–145. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Jitkaew S, Zhao J, Chiang HC, Choksi S, Liu J, Ward Y, Wu LG, Liu ZG. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol. 2014;16:55–65. doi: 10.1038/ncb2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhou Z, Li L, Zhong CQ, Zheng X, Wu X, Zhang Y, Ma H, Huang D, Li W, et al. Diverse sequence determinants control human and mouse receptor interacting protein 3 (RIP3) and mixed lineage kinase domain-like (MLKL) interaction in necroptotic signaling. J Biol Chem. 2013;288:16247–16261. doi: 10.1074/jbc.M112.435545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li W, Ren J, Huang D, He WT, Song Y, Yang C, Li W, Zheng X, Chen P, et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014;24:105–121. doi: 10.1038/cr.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Murphy JM. A tale of two domains - a structural perspective of the pseudokinase, MLKL. FEBS J. 2015 doi: 10.1111/febs.13504. [DOI] [PubMed] [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon CP, Weinlich R, Rodriguez DA, Cripps JG, Quarato G, Gurung P, Verbist KC, Brewer TL, Llambi F, Gong YN, et al. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell. 2014;157:1189–1202. doi: 10.1016/j.cell.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondelinger Y, Declercq W, Montessuit S, Roelandt R, Goncalves A, Bruggeman I, Hulpiau P, Weber K, Sehon CA, Marquis RW, et al. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 2014;7:971–981. doi: 10.1016/j.celrep.2014.04.026. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Kroemer G. MLKL regulates necrotic plasma membrane permeabilization. Cell Res. 2014;24:139–140. doi: 10.1038/cr.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JM, Tanzer MC, Lucet IS, Young SN, Spall SK, Sharma P, Pierotti C, Garnier JM, Dobson RC, Webb AI, et al. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc Natl Acad Sci. 2014;111:15072–15077. doi: 10.1073/pnas.1408987111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. The computer aided resonance assignment tutorial. CANTINA Verlag; Goldau, Switerland: 2004. [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S, Lewis R, Lalaoui N, Metcalf D, Webb AI, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- Orozco S, Yatim N, Werner MR, Tran H, Gunja SY, Tait SW, Albert ML, Green DR, Oberst A. RIPK1 both positively and negatively regulates RIPK3 oligomerization and necroptosis. Cell Death Differ. 2014;21:1511–1521. doi: 10.1038/cdd.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez AJ, Seyedhosseini M, Deerinck TJ, Bushong EA, Panda S, Tasdizen T, Ellisman MH. A workflow for the automatic segmentation of organelles in electron microscopy image stacks. Front Neuroanatomy. 2014;8:126. doi: 10.3389/fnana.2014.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez DA, Weinlich R, Brown S, Guy C, Fitzgerald P, Dillon CP, Oberst A, Quarato G, Low J, Cripps JG, et al. Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 2015;23:76–88. doi: 10.1038/cdd.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectr. 1999;34:66. [Google Scholar]
- Savitsky P, Bray J, Cooper CD, Marsden BD, Mahajan P, Burgess-Brown NA, Gileadi O. High-throughput production of human proteins for crystallization: the SGC experience. J Struct Biol. 2010;172:3–13. doi: 10.1016/j.jsb.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Quade B, Wang H, Sun L, Wang X, Rizo J. A plug release mechanism for membrane permeation by MLKL. Structure. 2014;22:1489–1500. doi: 10.1016/j.str.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang X. A new kind of cell suicide: mechanisms and functions of programmed necrosis. Trends Biochem Sci. 2014;39:587–593. doi: 10.1016/j.tibs.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Tait SW, Oberst A, Quarato G, Milasta S, Haller M, Wang R, Karvela M, Ichim G, Yatim N, Albert ML, et al. Widespread mitochondrial depletion via mitophagy does not compromise necroptosis. Cell Rep. 2013;5:878–885. doi: 10.1016/j.celrep.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, Wang FS, Wang X. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell. 2014;54:133–146. doi: 10.1016/j.molcel.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Xie T, Peng W, Yan C, Wu J, Gong X, Shi Y. Structural insights into RIP3-mediated necroptotic signaling. Cell Rep. 2013;5:70–78. doi: 10.1016/j.celrep.2013.08.044. [DOI] [PubMed] [Google Scholar]
- Zhao H, Brautigam CA, Ghirlando R, Schuck P. Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. In: Coligan John E, et al., editors. Curr Prot Prot Sci/editorial board. Unit 20. Chapter 20. 2013. p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J, Liu ZG. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci. 2012;109:5322–5327. doi: 10.1073/pnas.1200012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.