Abstract
Vitamin D deficiency is implicated in multiple disease conditions and accumulating evidence supports that the variation in serum vitamin D (25(OH)D) levels, including deficiency, is under strong genetic control. However, the underlying genetic mechanism associated with vitamin 25(OH)D concentrations is poorly understood. We earlier reported a very high prevalence of vitamin D deficiency associated with an increased risk for type 2 diabetes and obesity in a Punjabi Sikh diabetic cohort as part of the Asian Indian diabetic heart study (AIDHS). Here we have performed the first genome-wide association study (GWAS) of serum 25(OH)D on 3538 individuals from this Punjabi Sikh population. Our discovery GWAS comprised of 1387 subjects followed by validation of 24 putative SNPs (P <10−4) using an independent replication sample (n = 2151) from the same population by direct genotyping. A novel locus at chromosome 20p11.21 represented by rs2207173 with minor allele frequency (MAF) 0.29, [β = −0.13, p = 4.47 × 10−9] between FOXA2 and SSTR4 was identified to be associated with 25(OH)D levels. Another suggestive association signal at rs11586313 (MAF 0.54) [β = 0.90; p = 1.36 × 10−6] was found within the regulatory region of the IVL gene on chromosome 1q21.3. Additionally, our study replicated 3 of 5 known GWAS genes associated with 25(OH)D concentrations including GC (p = 0.007) and CYP2R1 (p = 0.019) reported in Europeans and the DAB1 (p = 0.003), reported in Hispanics. Identification of novel association signals in biologically plausible regions with 25(OH)D metabolism will provide new molecular insights on genetic drivers of vitamin D status and its implications in health disparities.
Keywords: Vitamin D deficiency, 25(OH)D levels (ELISA), Type 2 diabetes, Genome-wide association study, Punjabi Sikhs
1. Introduction
Vitamin D deficiency is the most common medical condition in the world and is estimated to affect over one billion people worldwide [1]. Beyond its well established role in musculo-skeletal health, accumulating evidence suggests that vitamin D plays a crucial role in diverse physiological functions which include several commoncancers, cardiovascular disease, autoimmune diseases (e.g., type 1 diabetes (T1D), multiple sclerosis, Crohn’s disease and rheumatoid arthritis), Alzheimer’s disease, and Parkinson’s disease [2]. Clinical vitamin D deficiency causes rickets in children and osteomalacia in children and adults. Overwhelming evidence suggests that decreased vitamin D levels associate with increased age-related disorders including the development, progression, and severity of T2D [2]. Risk factors of vitamin D deficiency include living at higher latitudes (>35°N), dark skin, old age, sunscreen use, obesity, medication, chronic disease, and genetics.
Vitamin D status is known to be poor among Asian Indians [3], however, limited data exist to assess the implication of vitamin D insufficiency on cardiometabolic traits in Asian Indians. In 2012, we reported a strong association of serum 25(OH)D deficiency with T2D and obesity in 1765 Asian Indian participants enrolled in our Asian Indian diabetic heart study/Sikh diabetes study (AIDHS/SDS) [4]. A significantly higher percentage of T2D patients (83%) were vitamin D deficient compared to normoglycemic individuals (68%). Similarly, a significant linear increase (p <0.0001) in the frequency of vitamin D deficient individuals was observed (65.2%, 75.5%, 81.4%) as the BMI (kg/m2) increased (<23, 23–27.5, >27.5), respectively. There is accumulating evidence to support that variation in serum vitamin D levels, including deficiency, is under strong genetic control [4]. Results of family and twin studies suggest evidence for genetic influences on vitamin D metabolism and their association with a wide range of chronic complex diseases. 25(OH)D, the most abundant circulating metabolite, is the best indicator of vitamin D status and has a high heritability ranging from 28–80% [5–7]. Recent in vitro evidence also suggests that vitamin D modulates the expression of hundreds of genes, many of which belong to disease-associated pathways [8]. The global distribution of vitamin D receptors in the body controls nearly 3000 genes [8], which may have widespread health implications. However, the molecular mechanism by which vitamin D exerts its effects in diverse diseases remains incompletely understood. So far only a handful of Genome Wide Association Studies (GWAS) have been performed to identify genetic determinants of circulating 25(OH)D levels. Most of these studies (>80%) have been performed in European populations and the role of genetic variation in impacting vitamin D status among other ethnic groups remains poorly understood.
Two large GWAS and meta-analysis studies published in 2010 using 6722 [9] and 33,996 [10] participants of European descent revealed a significant association of serum 25(OH)D levels with several SNPs located on genes in the vitamin D metabolizing pathway, and have identified variants at five loci (GC, DHCR7/NADSYN1, CYP2R1, CYP2R2, CYP24A1) affecting 25(OH)D concentrations. Common variants in these five genes (GC [4p12; p = 1.9 × 10−109], DHCR7 [11q12; p = 2.1 × 10−27], CYP2R1 [11p15; 3.3 × 10−20], CYP2R2 [11p15; 2.9 × 10−17], and CYP24A1 [20q13; p = 6.0 × 10−10]) have appeared among the top hits in GWAS and meta-analysis studies, and revealed a significantly raised risk of vitamin D deficiency [9,10]. However, contribution of these loci in non-European populations is currently unknown, thus, additional GWAS and meta-analysis studies in non-European populations are required to confirm the contribution of previously known genes in affecting 25(OH)D concentrations. Moreover, the findings of these studies also suggest that additional genes contributing to the variability of serum vitamin D concentrations may exist and should be evaluated in other ethnic/racial populations. In this investigation, we have performed GWAS, replication and meta-analysis in 3538 participants enrolled in the AIDHS/SDS [11] to identify common genetic variants affecting vitamin D concentrations. To our knowledge, this is the first GWAS performed in a population from South Asian ancestry reporting the identification of new genetic variants affecting 25(OH)D concentrations.
2. Research design and methods
2.1. Study subjects
We carried out a two stage GWAS of 25(OH)D levels in a total of 3538 subjects comprised of 1387 individuals in a ‘discovery sample set’ and 2151 individuals in an independent ‘replication sample set’ from the AIDHS/SDS, described earlier [11,12]. All individuals in this study were recruited from the single geographical location of the state of Punjab, India. The details of the clinical characteristic of the study subjects are shown in Supplementary Table 1 and recruitment details are described elsewhere [11,12]. Briefly, the diagnosis of T2D was confirmed in all the case participants by reviewing medical records for symptoms, use of medications, and measuring fasting glucose levels following the guidelines of the American Diabetes Association [13]. Subjects with T1D, or those having a family member with T1D, or rare forms of T2D subtypes maturity onset diabetes of the young (MODYs), or secondary diabetes (from e.g. hemochromatosis, pancreatitis) were excluded from the study. The selection of controls was based on fasting glucose levels <100.8 mg/dL or a 2 h glucose <141.0 mg/dL, and subjects were excluded if they had impaired fasting glucose (IFG) or impaired glucose tolerance (IGT). Body mass index (BMI) was calculated as weight (kg)/height (m2), and obesity was defined using the World Health Organization’s BMI recommendations for Asian populations as described earlier [14,15]. All blood samples were obtained at the baseline visits. All participants provided a written informed consent, and the study was reviewed and approved by the University of Oklahoma Health Sciences Center Institutional Review Board, as well as the Human Subject Protection Committee at the participating hospitals and institutes in India.
2.2. Serum vitamin D measurements
Serum was obtained from fasting blood samples to quantify 25(OH)D levels according to PhenX protocol (#051100) in the entire AIDHS/SDS cohort (n = 3538) using standard monoclonal antibody based ELISA kits from ALPCO Diagnostics (Salem, NH, USA) as described previously [4]. Samples were blinded for 25 (OH)D measurements and each specimen was run in duplicate following the manufacturer’s instructions. A standard curve was used with a range of concentrations (2-fold dilutions) and mixing multiple samples during initial optimization. Any sample that fell out of range was repeated. To minimize batch effect and inter-assay variation across cohorts, samples from both discovery and replication cohorts were quantified using ELISA kits from one manufacturer (ALPCO Diagnostics, Salem, NH, USA) and using one instrument (Tecan Infinite 200 PRO microplate reader).
2.3. Genotyping and quality control
Genomic DNA was extracted from buffy coats using QiaAmp blood kits (Qiagen, Chatsworth, CA) or by the salting-out procedure as described earlier [16,17]. We genotyped the discovery set using Human 660 W Quad BeadChip panel (Illumina, Inc., San Diego, CA) as described previously [11,12]. We assessed population stratification in the discovery set using principal-components analysis that revealed little population structure (Supplementary Fig. 1). We also performed pairwise identity-by-state (IBS) clustering in PLINK across all individuals to assess population substructure due to cryptic relatedness and to remove outliers. Related individuals with a pi-hat >0.3, samples with <93% call rate, SNPs with <95% call rate, SNPs with Hardy–Weinberg Equilibrium (HWE) of p <10−6 in controls, or SNPs with minor allele frequency (MAF) of <1% were excluded from analysis. After stringent quality control, there were 474,231 directly genotyped SNPs in 1616 subjects available for further analysis and used for the basis of imputation. To increase genome coverage, imputation was performed using IMPUTE2 with the reference panel of 1092 worldwide subjects from 1000 Genomes Project Phase I integrated variant set (March 2012 release) in NCBI Build 37 (hg19) coordinates. Quality control for the imputed SNPs included removal of variants with an imputation certainty (info score) <0.5, MAF <0.05, and SNPs significantly deviated from HWE in controls (p <1 × 10−6). Of a total of 6,378,483 variants, 5,904,251 variants with MAF > 5% were used in association analysis. A total of 1387 subjects with available measures of 25(OH)D levels were used in final association analysis. Genotyping of our top 24 SNPs selected for validation in the replication set (n = 2151) was performed using TaqMan assay (Applied Biosystems, Foster City, CA) and the KASPAR technology (LGC Genomics, Boston, MA).
2.4. Statistical analysis
2.4.1. Association testing for discovery
To test the associations of SNPs with circulating 25(OH)D levels, a linear regression and an additive genetic model implemented in SNPTEST V2.3.0 were used with the natural log transformed 25 (OH)D level adjusted with age, gender, BMI and T2D status. Population structure was assessed by principal component analysis and we included five principal components as covariates to account for population stratification. These principal components used for this correction were estimated from our Sikh population sample, due to a lack of Sikh reference data in the existing HapMap2, HapMap3, or 1000 Genomes panels.
2.4.2. Stage 1 replication
To identify 25(OH)D associated signals, we analyzed the top 24 SNPs from 18 independent signals (p <10−4) derived from the discovery set in an additional 2151 individuals (1258 T2D cases and 893 controls) from the AIDHS study cohorts. Associations between the 24 selected SNPs and natural log transformed 25(OH)D levels were analyzed using a mixed linear regression model implemented in SVS software (Golden Helix, MT, USA), with SNPs coded in an additive genetic model adjusted for gender, age, BMI, and T2D status.
2.4.3. Meta-analysis
To combine the results from the discovery and replication stages, a fixed effect, inverse variance meta-analysis implemented in METAL was performed. The 2-tailed p values lower than 5 × 10−8 were considered as genome-wide significant. Assessment of heterogeneity across the two studies was executed by evaluating the p values from chi-squared statistic in a heterogeneity test implemented in METAL and p-values <0.001 were considered significant. The LocusZoom standalone tool was used to generate a regional association plot for each locus and Forest Plot Viewer (http://ntp.niehs.nih.gov/ntp/ohat/forestplot/) was used to generate the forest plots.
3. Results
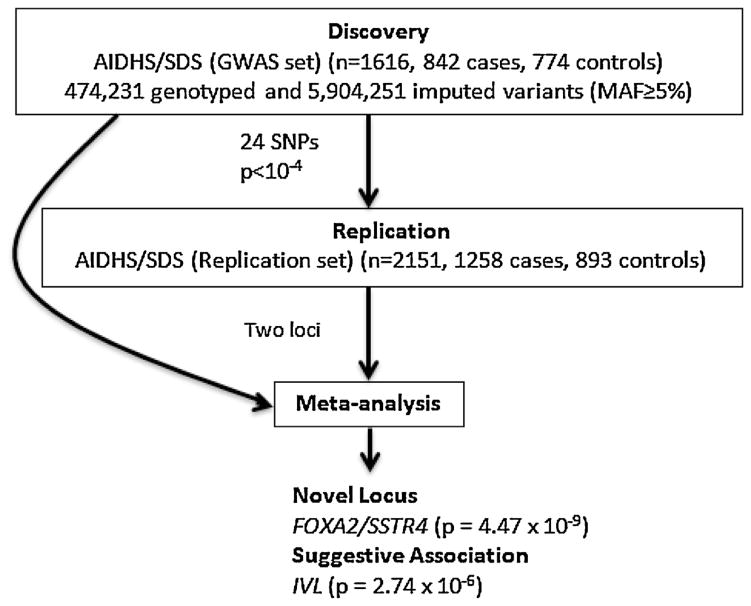
To identify genetic variants associated with the circulating 25(OH)D levels, we conducted the two stage GWAS in a total of 3538 subjects of Punjabi Sikhs from the AIDHS [11]: discovery GWAS (n = 1387) and replication (n = 2151). Details on clinical characteristics of GWAS discovery and replication samples are summarized in Supplementary Table 1. The study design is outlined in Fig. 1.
Fig. 1.
Summary of study design and key outcomes.
3.1. GWAS discovery
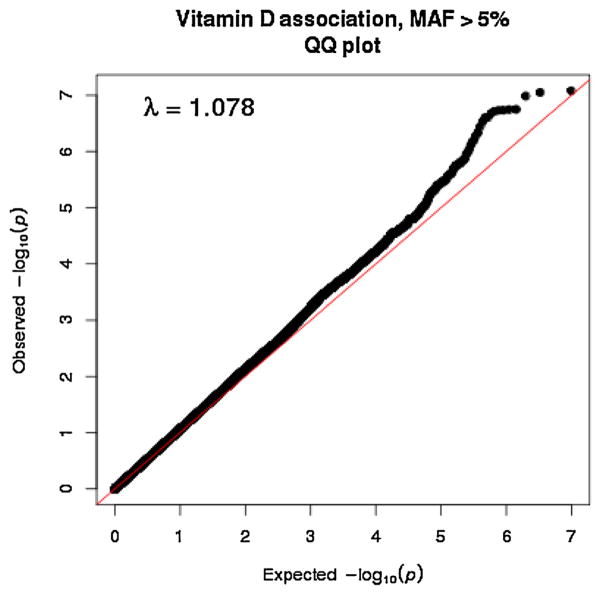
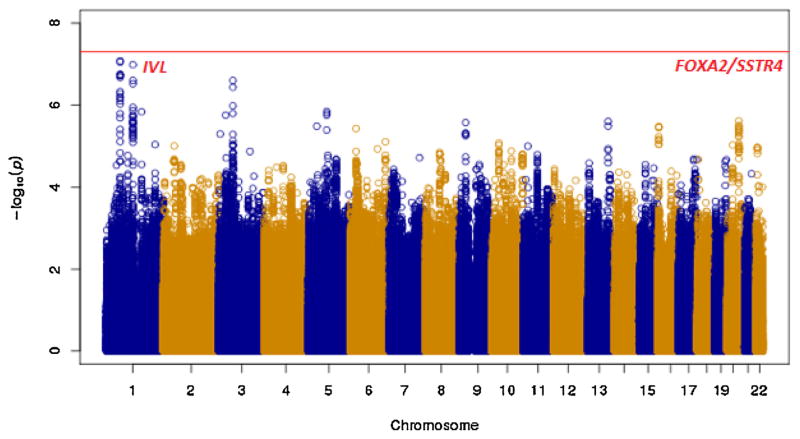
After stringent quality control criteria as described earlier (Saxena et al.) [12], a total of 474,231 directly genotyped SNPs (MAF ≥ 5%) were analyzed. Principal components analysis revealed the close proximity of the Sikh GWAS population to the Gujarati Indians in Houston, Texas, USA (GIH) and Utah residents with ancestry from Northern and Western Europe (CEU) populations, and the close matching of cases and controls of the Sikh GWAS discovery with little population substructure (Supplementary Fig. 1). A genome-wide association analysis was performed on 5,904,251 directly genotyped and high quality imputed variants with MAF ≥ 5% to identify gene variants associated with 25(OH)D concentrations using multiple linear regression. The analysis was adjusted for age, gender, BMI, T2D status, and five principal components of ancestry. Quantile–Quantile (Q–Q) plots revealed no deviation under the expected null distribution of the observed p values except at the tail end of the distribution (Fig. 2). The genomic inflation factor was λ = 1.078 which suggested insignificant inflation by population stratification, relatedness and other possible artifacts. In discovery GWAS analysis, no association signals exceeded conventional genome-wide significance level (p <5 × 10−8), but several strong signals (p <10−5) were observed as shown in the Manhattan plot of the distribution of p values (inverse log10 scale) after adjusting for covariates and principal components of ancestry (Fig. 3).
Fig. 2.
Quantile–Quantile (Q–Q) plot of Vitamin D GWAS of the AIDHS discovery set after quality control of directly genotyped and imputed variants (MAF >0.05) from the 1000 Genome (March 2012) reference panel.
Fig. 3.
Manhattan plot showing 25(OH)D GWAS analysis of the AIDHS/SDS discovery cohort using directly genotyped and imputed SNPs on X-axis and–log p-values on the Y-axis.
3.2. Replication and Meta-analysis
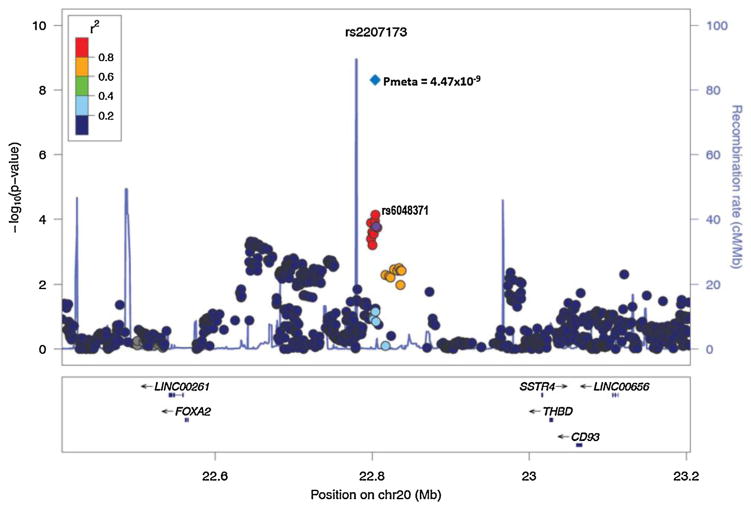
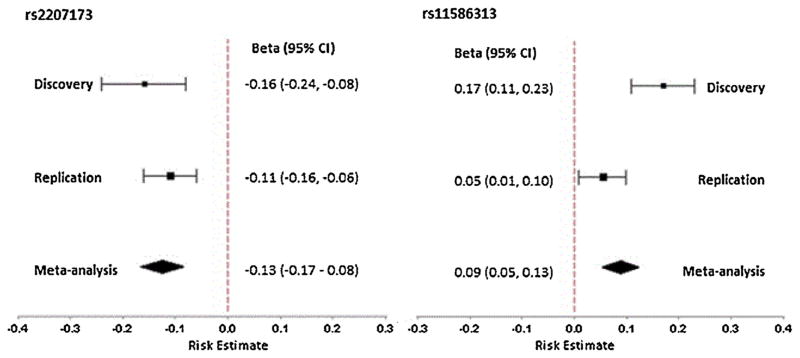
Twenty four top SNPs from 18 independent signals (p <10−4) (selected based on putative novel signals and possible candidates with biological relevance) were directly genotyped in an additional 2151 subjects (1258 T2D cases and 893 controls) from the AIDHS/SDS cohort for replication studies (Supplementary Tables 2 and 3). Association of two out of 24 SNPs was confirmed in the replication sample after adjustment for gender, age, BMI, and T2D status (Table 1). In combined meta-analysis of our discovery and replication, we identified a robust novel association signal at chromosome 20p11.21 represented by imputed SNP rs2207173 (info = 0.98; β = −0.13, p = 4.47 × 10−9) and genotyped SNP rs6048371 (β = −0.13; p = 4.82 × 10−9) located in the intergenic region between the FOXA2/SSTR4 genes (Fig. 4 and 5). The other suggestive association signal was discovered at chromosome 1q21.3 for the IVL gene (directly genotyped SNP rs11586313; β = 0.09, p = 1.36 × 10−6) that could not achieve conventional genome-wide significance (p <5 × 10−8) (Fig. 5 and Supplementary Fig. 2). Of the five regions identified previously in European and Hispanic GWA studies, two SNPs, including the top associated variant in Europeans (rs2282679) from the GC gene (p = 0.007) and a variant at chromosome 1p32.1 represented by rs6680429 in the DAB1 gene (p = 0.003) showed statistically significant association with vitamin D concentrations in our sample. Both SNPs showed the allelic effects in the same direction despite differences in allele frequency in the DAB1 gene [0.49 Sikhs vs. 0.42 Hispanic]. However, the association of rs12794714 in the CYP2R1 gene was only marginally significant (β = −0.08; p = 0.019) in our sample (Supplementary Table 4).
Table 1.
GWAS, replication and meta-analysis results of 25(OH)D loci identified in AIDHS/SDS.
| SNP | Chromosome position | Nearest gene | Effect/other allele | Effect allele frequency
|
Discovery GWAS | Replication | Combined meta-analysis | Heterozygosity P value |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Sikh population
|
1000G Global frequency | Beta (95%CI) | Beta (95%CI) | Beta (95%CI) | ||||||
| Discovery | Replication | P value | P value | P value | ||||||
| rs2207173 | 20 | FOXA2/SSTR4 | G/A | 0.28 | 0.29 | 0.30 | −0.16 (−0.24, −0.08) | −0.11 (−0.16, −0.06) | −0.13 (−0.17, −0.08) | 0.55 |
| 22805061 | 3.70E-05 | 2.74E-05 | 4.47E-09 | |||||||
| rs6048371 | 20 | FOXA2/SSTR4 | T/C | 0.28 | 0.29 | 0.30 | −0.16 (−0.24, −0.08) | −0.11 (−0.16, −0.06) | −0.13 (−0.17, −0.08) | 0.54 |
| 22804107 | 3.39E-05 | 2.74E-05 | 4.82E-09 | |||||||
| rs11586313 | 1 | IVL | G/A | 0.54 | 0.55 | 0.48 | 0.17 (0.11, 0.23) | 0.05 (0.01, 0.10) | 0.09 (0.05, 0.13) | 0.02 |
| 152890470 | 1.45E-06 | 0.02 | 1.36E-06 | |||||||
GWAS: genome-wide association study, CI: confidence interval.
Fig. 4.
Regional association plot showing the novel locus (rs2207173) representing the nearby genes FOXA2/SSTR4 in discovery GWAS and in combined analysis of GWAS and replication studies. Meta-analysis p value is represented in blue diamond. At the bottom of the plot, the locations of known genes in the region are shown. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Forest plots showing association of lead SNP (rs2207173) near FOXA2/SSTR4 (left) and rs11586313 within IVL (right) with serum 25(OH)D concentrations. The size of the square represents weight number. Diamonds represent summary estimate of the effect allele combining the study-specific estimates with a fixed-effects model and the dashed vertical line represents the beta (risk estimate) = 0, and CI (confidence interval).
The association of our key SNPs for the FOXA2/SSTR4 (rs2207173 and rs6048371) and the IVL gene (rs11586313) remained strongly significant when analyzed separately in the data stratified by gender (Supplementary Table 5). We further examined the relationship of our top variants with T2D (yes/no) and cardiometabolic traits including fasting glucose, BMI, waist circumference, serum lipids, and blood pressure. None of these variants revealed any significant associated with T2D (Supplementary Table 6) and with cardiometabolic traits (data not shown).
To investigate whether genetic data through statistical associations provide further insights on the pathways through which these loci may exert their effect on vitamin D metabolism, we performed in silico analysis using multiple bioinformatics analysis tools. Ingenuity pathway analysis (IPA) revealed the relationship between FOXA2 and other genes in the vitamin D pathway, T2D and cardiovascular disease (Supplementary Fig. 3). Cis eQTL analysis using publically available MuTHER data in the region surrounding LCE1E-IVL genes (1q21.3) revealed a significantly elevated expression of IVL and LCE1E mRNA (eQTL <10−3) in skin tissues but not in adipose and lymphoblastiod cell lines (Supplementary Fig. 4). Next we performed pathway analysis using DEPICT, a tool that uses predicted gene functions to prioritize causal genes, highlight enriched pathways, and identify causal tissues/cell types with high expression at GWAS loci. The pathway analysis using DEPICT not only validated our top association signal near FOXA2/SSTR4, but also significantly confirmed several other independent hits in our discovery GWAS (including C1orf187, FHIT and IVL) (Supplementary Table 7).
4. Discussion
There is compelling evidence suggesting that vitamin D deficiency is increasing in conjunction with T1D, T2D, obesity, and cardiovascular disease [18,19], and that vitamin D deficiency accelerates the progression from prediabetes to T2D [20]. We earlier reported a very high prevalence of vitamin D deficiency in this Punjabi diabetic cohort originating from Northern India; showing a strong predisposition to T2D and cardiovascular disease risk [4]. Ethnic, cultural, and genetic differences in vitamin D metabolism may affect the vitamin D status of different populations including Chinese [21], Asians Indians [22], and African Americans [23]. According to Lo et al. [24], the people from the Indian subcontinent require twice as much UVB exposure to produce 25(OH)D levels equal to Caucasians due to increased skin pigmentation. In addition, a cultural tendency to avoid direct sunlight, sunscreen usage, and customary dress that covers most of the body, may contribute to suboptimal vitamin D status despite the climate in India and Middle Eastern countries is sunny throughout the year. Since Punjabis are known to be susceptible to metabolic syndrome, T2D, and early cardiovascular disease, vitamin D deficiency may put them at an additional risk, making a strong case for exploring genetic mechanisms underlying the insufficiency and its implications in increased cardiometabolic risk.
Here we have performed the first GWAS of serum 25(OH)D in this Sikh population followed by validation of 24 putative SNPs using an independent replication cohort from the same Punjabi population by direct genotyping. A novel locus at chromosome 20p11.21 represented by rs2207173, [β = −0.13, p = 4.47 × 10−9] within the intergenic region between FOXA2 (also named HNF3B) and SSTR4 was identified to be associated with 25(OH)D concentrations. Additionally, a suggestive association signal represented by rs11586313 [β = 0.90; p = 1.36 × 10−6] was found at IVL gene located at chromosome 1q21.3. The top associated variant (rs2282679) in the GC gene in Europeans was confirmed in our sample (β = −0.10, p = 0.007 for the G allele) and a modest replication of CYP2R1 (β = −0.08; p = 0.019 for the A allele). We also confirmed significant association of another locus at chromosome 1p32.1 in the DAB1 gene represented by rs6680429 (β = 0.096, p = 0.003 for the A allele) with vitamin D concentrations in Hispanics [25]. No other loci including previously known candidate gene associations influencing 25(OH)D concentrations could be confirmed in our study.
Our novel signal is located near the FOXA2 (or HNF3B) gene which encodes hepatocyte nuclear factor 3B. It belongs to a family of DNA-binding proteins that are transcriptional activators for liver-specific genes [26,27]. FOXA2 controls the expression of genes important for glucose sensing in pancreatic beta-cells including MODY genes (PDX-1 and HNF4A) and regulates genes involved in glucose homeostasis and fat metabolism by interacting with the cis-acting regulatory regions of these genes. The targeted knockout of pancreatic β-cell-specific FOXA2 in mice results in postnatal death due to severe hyperinsulinemic hypoglycemia, and the down-regulation of ATP-sensitive potassium channel subunits Sur1 and Kir6.2 [28]. Accordingly to Lamba et al. [29], a trinucleotide (CCT) repeat polymorphism in FOXA2 downregulates expression of CYP3A4 in hepatocytes. The CYP3A4 is a known hepatic microsomal vitamin 25-hydroxylase and is involved in the hydrolysis of vitamin D. The exact mechanism underlying the observed association of FOXA2 with vitamin D levels is currently unknown. However, from the study of Lamba et al. [29], it is possible that the genetic variants in the FOXA2 gene may impact/dysregulate the expression of CYP3A4 in hepatocytes which may in turn influence 25(OH)D concentration and its metabolism. Moreover, a CCT trinucleotide repeat polymorphism in CYP3A4 was associated with risk of T2D in North Indians [30] making this a strong candidate for further investigation regarding its T2D associated risk in conjunction with the altered CYP3A4-mediated effect on 25(OH)D levels.
Additionally, FOXA2 has been shown to act as a master regulator of key pathways critical to endocrine traits in a genome-wide analysis of Chip-Seq by Johnson et al. [31]. Our findings are also in line with previous studies that suggest the emerging function of the skeleton physiology as a regulator of energy metabolism [32,33]. A study conducted by Yoshikawa et al., [34] using murine model showed how osteoblast ablation in adult mice resulted into insulin resistance and hyperglycemia through down-regulation of FOXA2. Another recent study of global gene expression profiling of pancreatic islets for evaluating the effects of treptozotocin (STZ) on pancreatic beta cells in mice showed that the STZ treatment not only induced impaired insulin expression but also significantly suppressed expression of a wide range of genes including FOXA2, VDR, PDX 1, and SLC30A8 linked with key β-cell functions or progression of diabetes development [35]. These findings further substantiate functional role of FOXA2 in pancreatic beta cells.
The second suggestive association signal at chromosome1q21.3 represented by rs11586313 (β = 0.09; p = 1.36 × 10−6) is located within the IVL gene encoding involucrin. Although the association of our index SNP could not achieve a genome-wide empirical p-value, this signal appears to have strong biological relevance in vitamin D metabolism. Involucrin is a major protein involved in keratinocyte differentiation in the epidermis and maintains the structural integrity of skin [36]. Vitamin D (25(OH)D) upregulates the transcription of IVL, increasing gene expression and the mRNA and protein levels for involucrin in keratinocytes [36,37]. When 7-dehydrocholesterol is converted into vitamin D3 in the presence of sunlight, it is transported out of the keratinocyte to the liver through the vitamin D receptors in the epidermis where it is converted to 25(OH)D3 through hydroxylation by cytochrome P450 enzymes (CYP27A1, CYP3A4) [37,38]. Both 25(OH)D and calcium regulate differentiation in keratinocytes through the up-regulation of IVL gene [37]. Studies suggest the presence of calcium and vitamin D response elements(VDRE) in the IVL promoter play an important role in mediating the action of 25(OH)D on IVL expression. Future sequencing, and functional studies will provide further insight on the discovery of putative functional variants in IVL, and how these may impact binding of VDRE or how calcium response elements regulate the induction of IVL in keratinocytes.
In addition to identifying novel loci associated with vitamin D concentrations in Sikhs, our study confirmed 3 of 5 previously identified GWAS loci, validating our vitamin D measures and at least partial overlap of causal vitamin D loci between European, Hispanic and Sikh populations. The reason for the non-replication of the two other previously known GWAS regions in this study could be the differences in linkage disequilibrium between marker and causal SNPs between the populations, though, the allele frequencies in Sikhs did not vary significantly with respect to Caucasians(Supplementary Table 4). Of note, our study did not detect any association between VDR gene variants and 25(OH)D concentrations in this population. Genetic polymorphisms in the VDR gene (BsmI, FokI, ApaI, and TaqI) have been extensively studied for assessing VDR-disease association in multiple conditions, and results have been conflicting across studies [39]. Recent meta-analyses studies suggest association between VDR variants with cancer [40], Fractures [41], tuberculosis [42], and rheumatoid arthritis [43]. However, contrarily to expectations, SNPs in VDR have not been associated with 25(OH)D concentrations in most GWA studies. Even a large meta-analysis study by Wang et al. [10], could not identify the association of VDR locus with 25(OH)D concentrations.
Strengths of our study include the use of a well-characterized, relatively homogenous population with high predisposition to diabetes and cardiovascular disease. Both discovery and replication data sets are from one geographic location from the Punjab province of India. We also minimized inter-study variability of discovery and replication cohort by measuring 25(0H)D levels using immunoassay kit from one source and using one instrument. None of our study subjects were on vitamin supplementation which adds further strength to the study. We understand that the lack of availability of seasonal data to adjust seasonal changes which may influence vitamin D concentrations, is a limitation in our study. However, literature suggests that people living above 35°N and below 35°S latitude do not receive adequate UV radiations in the winter months to produce adequate vitamin D in these months [44]. Conversely, India is below 35°N and has tropical climate with availability of sunlight through most part of the year would make Indian people to be less affected by seasonal variation [44]. The other limitation includes our relatively small sample size (n = 3538). Despite the lack of sufficient power for capturing small effect sizes for the loci discovered in a large study exceeding 33,000 individuals from the SUNLIGHT consortium [10], our study was able to replicate variants in 3 of 5 known genes including GC (DBP) and CYP2R1 reported in Europeans and the DAB1 reported in Hispanics with a much smaller sample size. Additionally, no other South Asian study was available with serum vitamin D measures to expand this analysis.
5. Conclusions
Our study identified a novel locus and one suggestive gene region associated with circulating 25(OH)D concentrations in a diabetic case-control cohort of Punjabi Sikh ancestry. Thus far, no GWAS has been conducted for circulating vitamin D concentration in populations from South Asia. Detection of novel association signals in biologically plausible regions in vitamin D metabolism will provide new molecular insights on the genetic drivers for vitamin D status and its implications in health disparities.
Supplementary Material
Acknowledgments
This work was supported by NIH grants -R01DK082766 funded by the National Institute of Health (NIDDK) and NOT-HG-11-009 funded by NHGRI, and a VPR Bridge Grant from University of Oklahoma Health Sciences Center. Technical support provided by Timothy Braun and Praveen Natt is duly acknowledged. Authors thank all the participants of AIDHS/SDS and PhenX RISING consortium (NHGRI), and are grateful for their contribution in this study.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsbmb.2015.12.014.
Footnotes
Conflict of interest
We declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author contributions
Conceived and designed the experiments D.K.S., P.R.B.; Data collection sample recruitment: D.K.S., S.R., G.S.W., N.K.M., and J.R. S.; Data analysis: B.R.S., A.B., R.H., and R.S.; Manuscript preparation: D.K.S. and R.S. All authors read and provided critical comment on the manuscript.
URLs
IMPUTE http://mathgen.stats.ox.ac.uk/impute/impute.html;
METAL http://www.sph.umich.edu/csg/abecasis/Metal;
LocusZoom http://csg.sph.umich.edu/locuszoom/;
Forestplot http://ntp.niehs.nih.gov/ntp/ohat/forestplot;
IPA http://www.ingenuity.com/products/ipa;
DEPICT http://www.broadinstitute.org/mpg/depict/contact.html;
MuTHER http://www.muther.ac.uk/;
PLINK http://pngu.mgh.harvard.edu/~purcell/plink/;
SNPTEST https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html.
Role of the funding source
The sponsors of this study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data, and takes full responsibility for the integrity of data and the accuracy of data analysis.
References
- 1.Hollick MF, Chen TC. Vitamin D deficiency a worldwide problem with health consequences. Am J Clin Nutr. 2008;87:10805–10868. doi: 10.1093/ajcn/87.4.1080S. [DOI] [PubMed] [Google Scholar]
- 2.Autier P, Boniol M, Pizot C, Mullie P. Vitamin D status and ill health: a systematic review. Lancet Diabetes Endocrinol. 2014;2(1):76–89. doi: 10.1016/S2213-8587(13)70165-7. [DOI] [PubMed] [Google Scholar]
- 3.Harinarayan CV, Ramalakshmi T, Venkataprasad U. High prevalence of low dietary calcium and low vitamin D status in healthy south Indians. Asia Pac J Clin Nutr. 2004;13(4):359–364. [PubMed] [Google Scholar]
- 4.Braun TR, Been LF, Blackett PR, Sanghera DK. Vitamin D deficiency and cardio-metabolic risk in a North Indian Community with highly prevalent type 2 diabetes. J Diabetes Metab. 2012;3:213. doi: 10.4172/2155-6156.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wjst M, Altmuller J, Braig C, Bahnweg M, Andre E. A genome-wide linkage scan for 25-OH-D(3) and 1, 25-(OH)2-D3 serum levels in asthma families. J Steroid Biochem Mol Biol. 2007;103(3–5):799–802. doi: 10.1016/j.jsbmb.2006.12.053. [DOI] [PubMed] [Google Scholar]
- 6.Bu FX, Armas L, Lappe J, Zhou Y, Gao G, Wang HW, et al. Comprehensive association analysis of nine candidate genes with serum 25-hydroxy vitamin D levels among healthy Caucasian subjects. Hum Genet. 2010;128(5):549–556. doi: 10.1007/s00439-010-0881-9. [DOI] [PubMed] [Google Scholar]
- 7.Hunter D, De Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, et al. Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. J Bone Miner Res. 2001;16(2):371–378. doi: 10.1359/jbmr.2001.16.2.371. [DOI] [PubMed] [Google Scholar]
- 8.Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20(10):1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19(13):2739–2745. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376(9736):180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena R, Saleheen D, Been LF, Garavito ML, Braun T, Bjonnes A, et al. Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes. 2013;62(5):1746–1755. doi: 10.2337/db12-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saxena R, Bjonnes A, Prescott J, Dib P, Natt P, Lane J, et al. Genome-wide association study identifies variants in casein kinase II (CSNK2A2) to be associated with leukocyte telomere length in a Punjabi Sikh diabetic cohort. Circ Cardiovasc Genet. 2014;7(3):287–295. doi: 10.1161/CIRCGENETICS.113.000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guidlines ADA. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2004;27(Suppl 1):S5–S10. doi: 10.2337/diacare.27.2007.s5. [DOI] [PubMed] [Google Scholar]
- 14.Panel WE. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. 2004;363(9403):157–163. doi: 10.1016/S0140-6736(03)15268-3. [DOI] [PubMed] [Google Scholar]
- 15.Been LF, Hatfield JL, Shankar A, Aston CE, Ralhan S, Wander GS, et al. A low frequency variant within the GWAS locus of MTNR1B affects fasting glucose concentrations: genetic risk is modulated by obesity. Nutr Metab Cardiovasc Dis. 2012;22(11):944–951. doi: 10.1016/j.numecd.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, et al. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet. 2008;9:59. doi: 10.1186/1471-2350-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanghera DK, Demirci FY, Been L, Ortega L, Ralhan S, Wander GS, et al. PPARG and ADIPOQ gene polymorphisms increase type 2 diabetes mellitus risk in Asian Indian Sikhs: Pro12Ala still remains as the strongest predictor. Metabolism. 2010;59(4):492–501. doi: 10.1016/j.metabol.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahn HS, Morgan TM, Case LD, Dabelea D, Mayer-Davis EJ, Lawrence JM, et al. Association of type 1 diabetes with month of birth among U.S. youth: the search for diabetes in youth study. Diabetes Care. 2009;32(11):2010–2015. doi: 10.2337/dc09-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vacek JL, Vanga SR, Good M, Lai SM, Lakkireddy D, Howard PA. Vitamin D deficiency and supplementation and relation to cardiovascular health. Am J Cardiol. 2012;109(3):359–363. doi: 10.1016/j.amjcard.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 20.Deleskog A, Hilding A, Brismar K, Hamsten A, Efendic S, Ostenson CG. Low serum 25-hydroxyvitamin D level predicts progression to type 2 diabetes in individuals with prediabetes but not with normal glucose tolerance. Diabetologia. 2012;55(6):1668–1678. doi: 10.1007/s00125-012-2529-x. [DOI] [PubMed] [Google Scholar]
- 21.Yan L, Zhou B, Wang X, D’Ath S, Laidlaw A, Laskey MA, et al. Older people in China and the United Kingdom differ in the relationships among parathyroid hormone, vitamin D, and bone mineral status. Bone. 2003;33(4):620–627. doi: 10.1016/s8756-3282(03)00216-3. [DOI] [PubMed] [Google Scholar]
- 22.Awumey EM, Mitra DA, Hollis BW, Kumar R, Bell NH. Vitamin D metabolism is altered in Asian Indians in the southern United States: a clinical research center study. J Clin Endocrinol Metab. 1998;83(1):169–173. doi: 10.1210/jcem.83.1.4514. [DOI] [PubMed] [Google Scholar]
- 23.Harris SS. Vitamin D and African Americans. J Nutr. 2006;136(4):1126–1129. doi: 10.1093/jn/136.4.1126. [DOI] [PubMed] [Google Scholar]
- 24.Lo CW, Paris PW, Holick MF. Indian and Pakistani immigrants have the same capacity as Caucasians to produce vitamin D in response to ultraviolet irradiation. Am J Clin Nutr. 1986;44(5):683–685. doi: 10.1093/ajcn/44.5.683. [DOI] [PubMed] [Google Scholar]
- 25.Engelman CD, Meyers KJ, Ziegler JT, Taylor KD, Palmer ND, Haffner SM, et al. Genome-wide association study of vitamin D concentrations in Hispanic Americans: the IRAS family study. J Steroid Biochem Mol Biol. 2010;122(4):186–192. doi: 10.1016/j.jsbmb.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen J. Gene regulatory factors in pancreatic development. Dev Dyn. 2004;229(1):176–200. doi: 10.1002/dvdy.10460. [DOI] [PubMed] [Google Scholar]
- 27.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435(7044):944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 28.Lantz KA, Vatamaniuk MZ, Brestelli JE, Friedman JR, Matschinsky FM, Kaestner KH. Foxa2 regulates multiple pathways of insulin secretion. J Clin Invest. 2004;114(4):512–520. doi: 10.1172/JCI21149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamba V, Panetta JC, Strom S, Schuetz EG. Genetic predictors of interindividual variability in hepatic CYP3A4 expression. J Pharmacol Exp Ther. 2010;332(3):1088–1099. doi: 10.1124/jpet.109.160804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabassum R, Chavali S, Dwivedi OP, Tandon N, Bharadwaj D. Genetic variants of FOXA2: risk of type 2 diabetes and effect on metabolic traits in North Indians. J Hum Genet. 2008;53(11–12):957–965. doi: 10.1007/s10038-008-0335-6. [DOI] [PubMed] [Google Scholar]
- 31.Johnson ME, Schug J, Wells AD, Kaestner KH, Grant SF. Genome-wide analyses of ChIP-Seq derived FOXA2 DNA occupancy in liver points to genetic networks underpinning multiple complex traits. J Clin Endocrinol Metab. 2014;99(8):E1580–E1585. doi: 10.1210/jc.2013-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fulzele K, Riddle RC, DiGirolamo DJ, Cao X, Wan C, Chen D, et al. Insulin receptor signaling in osteoblasts regulates postnatal bone acquisition and body composition. Cell. 2010;142(2):309–319. doi: 10.1016/j.cell.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferron M, Wei J, Yoshizawa T, Del Fattore A, DePinho RA, Teti A, et al. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell. 2010;142(2):296–308. doi: 10.1016/j.cell.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshikawa Y, Kode A, Xu L, Mosialou I, Silva BC, Ferron M, et al. Genetic evidence points to an osteocalcin-independent influence of osteoblasts on energy metabolism. J Bone Miner Res. 2011;26(9):2012–2025. doi: 10.1002/jbmr.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonne JM, Sakuma T, Deeds MC, Munoz-Gomez M, Barry MA, Kudva YC. Global gene expression profiling of pancreatic islets in mice during streptozotocin-induced beta-cell damage and pancreatic Glp-1 gene therapy. Dis Model Mech. 2013;6(5):1236–1245. doi: 10.1242/dmm.012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hosomi J, Hosoi J, Abe E, Suda T, Kuroki T. Regulation of terminal differentiation of cultured mouse epidermal cells by 1 alpha, 25-dihydroxyvitamin D3. Endocrinology. 1983;113(6):1950–1957. doi: 10.1210/endo-113-6-1950. [DOI] [PubMed] [Google Scholar]
- 37.Bikle DD, Ng D, Oda Y, Hanley K, Feingold K, Xie Z. The vitamin D response element of the involucrin gene mediates its regulation by 1, 25-dihydroxyvitamin D3. J Invest Dermatol. 2002;119(5):1109–1113. doi: 10.1046/j.1523-1747.2002.19508.x. [DOI] [PubMed] [Google Scholar]
- 38.Stumpf WE, Sar M, Reid FA, Tanaka Y, DeLuca HF. Target cells for 1,25-dihydroxyvitamin D3 in intestinal tract, stomach, kidney, skin, pituitary, and parathyroid. Science. 1979;206(4423):1188–1190. doi: 10.1126/science.505004. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Li W, Liu J, Wu W, Ouyang H, Zhang Q. Polymorphisms in the vitamin D receptor gene and type 1 diabetes mellitus risk: an update by meta-analysis. Mol Cell Endocrinol. 2012;355(1):135–142. doi: 10.1016/j.mce.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 40.Randerson-Moor JA, Taylor JC, Elliott F, Chang YM, Beswick S, Kukalizch K, et al. Vitamin D receptor gene polymorphisms, serum 25-hydroxyvitamin D levels, and melanoma: UK case-control comparisons and a meta-analysis of published VDR data. Eur J Cancer. 2009;45(18):3271–3281. doi: 10.1016/j.ejca.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji GR, Yao M, Sun CY, Li ZH, Han Z. TaqI BsmI, ApaI and FokI polymorphisms in the vitamin D receptor (VDR) gene and risk of fracture in Caucasians: a meta-analysis. Bone. 2010;47(3):681–686. doi: 10.1016/j.bone.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 42.Gao L, Tao Y, Zhang L, Jin Q. Vitamin D receptor genetic polymorphisms and tuberculosis: updated systematic review and meta-analysis. Int J Tuberc Lung Dis. 2010;14(1):15–23. [PubMed] [Google Scholar]
- 43.Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Associations between vitamin D receptor polymorphisms and susceptibility to rheumatoid arthritis and systemic lupus erythematosus: a meta-analysis. Mol Biol Rep. 2011;38(6):3643–3651. doi: 10.1007/s11033-010-0477-4. [DOI] [PubMed] [Google Scholar]
- 44.Webb AR, Kline L, Holick MF. Influence of season and latitude on the cutaneous synthesis of vitamin D3: exposure to winter sunlight in Boston and Edmonton will not promote vitamin D3 synthesis in human skin. J Clin Endocrinol Metab. 1988;67(2):373–378. doi: 10.1210/jcem-67-2-373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.