Abstract
Metabolic engineering is generally focused on static optimization of cells to maximize production of a desired product, though recently dynamic metabolic engineering has explored how metabolic programs can be varied over time to improve titer. However, these are not the only types of applications where metabolic engineering could make a significant impact. Here, we discuss a new conceptual framework, termed “precision metabolic engineering,” involving the design and engineering of systems that make different products in response to different signals. Rather than focusing on maximizing titer, these types of applications typically have three hallmarks: sensing signals that determine the desired metabolic target, completely directing metabolic flux in response to those signals, and producing sharp responses at specific signal thresholds. In this review, we will first discuss and provide examples of precision metabolic engineering. We will then discuss each of these hallmarks and identify which existing metabolic engineering methods can be applied to accomplish those tasks, as well as some of their shortcomings. Ultimately, precise control of metabolic systems has the potential to enable a host of new metabolic engineering and synthetic biology applications for any problem where flexibility of response to an external signal could be useful.
Keywords: Precision metabolic engineering, metabolic control, sensory systems, pathway regulation, product selectivity, synthetic biology
1. Introduction
Metabolic engineering efforts typically focus on designing an organism to maximize the final yield of a desired product, as this is usually the most commercially important goal. This approach has been successfully used to create and improve microbial production of high-demand products such as biofuels (Atsumi et al., 2008a, 2008b; Jin et al., 2005), pharmaceuticals (Alonso-Gutierrez et al., 2013; Ro et al., 2006), and commodity chemicals (Chemler et al., 2010; Raab et al., 2010). However, there are a number of applications where it is more important to design systems that can flexibly respond to different signals and tightly control output levels, rather than just produce as much of one single compound as possible. This class of problems can be referred to as “precision metabolic engineering”, and it is an area that will likely receive greater attention as metabolic engineering finds more diverse applications.
The fundamental goals and characteristics of precision metabolic engineering are the necessity for a singular output determined by the state of the system and an emphasis on maximizing product selectivity rather than just final titer. Based on the flexibility required for this approach, it is in some ways similar to the growing field of dynamic metabolic engineering (Brockman and Prather, 2015a; Venayak et al., 2015), in which metabolic flux is redirected as the system changes during the production process. The main contrast is that while the hallmark of dynamic metabolic engineering is control of metabolism as a function of time to maximize titer and productivity, precision metabolic engineering instead emphasizes having metabolic states that are completely and sharply switchable in response to a specific input (though temporal control could play a role in some precision metabolic engineering applications). While their objectives differ, many techniques used for dynamic control of metabolism can be used to implement precise control over metabolic systems. Precision metabolic engineering is relevant in any situation where a portable microbial cell factory capable of responding to external signals with multiple metabolic outputs could be useful.
One example of where precise control could be valuable is in the development of bacterial biosensors. Bacterial biosensors have been designed to sense and respond to the presence of harmful chemicals (Gil et al., 2000), radiation (Rosen et al., 2000), and heavy metals (Verma and Singh, 2005), though they often use fluorescent reporters or reporters that require addition of exogenous substrate for enzymatic reactions (e.g., luminescence), which limits utility in low-resource (electricity and equipment) environments. Precision metabolic engineering using only sugar substrates can be used to produce metabolite outputs (such as pigments) that are visible without the use of equipment, thus enabling such biosensors to be more widely used in low-resource environments. Extensive metabolic engineering efforts have already been made to increase microbial production of pigmented metabolites such as lycopene (Alper et al., 2005a; Farmer and Liao, 2000; Yoon et al., 2006) and violacein (Fang et al., 2015; Lee et al., 2013; Rodrigues et al., 2013). By tightly controlling conditions under which these metabolites are produced, they could be used as indicators for biosensors for diverse applications. One such application is biomedical: the development of a biosensor for a blood test to detect micronutrient deficiencies, which are most prevalent in low-resource areas and thus could gain substantially from having essentially equipment-free approaches for diagnosis. Recently our group (Watstein et al., 2015) reported engineered bacteria that produce visible pigments in response to different levels of zinc (an important mineral in human diet), which would enable their use as low-cost, point-of-care assays to detect micronutrient levels. In contrast to traditional metabolic engineering goals, the pigment-based biosensor was engineered not to maximize titer of the final product, but to instead maximize selectivity of pigment production based on the concentration of zinc, subject to the constraint of visible pigment production in a reasonable amount of time – a challenge of precision metabolic engineering. This whole-cell biosensor framework could also be extended and used to detect levels of other micronutrients or blood components.
Applications of precision metabolic engineering extend beyond diagnostics to pharmaceutical production. While pharmaceuticals are generally produced in tightly controlled industrial settings, there is a potentially significant value for flexible “on demand” drug production in certain situations. For example, because of the cost and logistical challenges of storing pharmaceuticals and protein therapeutics for portable use (e.g., by warfighters), DARPA established a battlefield medicine program to develop systems that can produce them in response to situations as they arise. Cells already designed to make pharmaceuticals could be further engineered to produce different therapeutics based on battlefield needs. Importantly, precision metabolic engineering would be needed to ensure that only the desired substance is produced in a certain condition to maximize product purity in the absence of industrial separation processes. Since the program’s initiation in 2012, four different drugs have met DARPA’s standards of being producible on demand (Choi and Ling, 2014), and incorporating selectivity and control into existing metabolic engineering techniques could contribute to the program’s success.
In addition to producing pharmaceuticals, microbes could also serve as specific drug delivery vehicles. The bacterium Salmonella typhimurium has been explored as novel anticancer vector (Pawelek et al., 1997) because of its protection from immune system clearance and its predilection for the hypoxic environment of a tumor (Brown and Wilson, 2004). Bacteria can be programmed to release cytotoxic agents, cytokines, or tumor antigens in the presence of the tumor, but suboptimal targeting efficiency and intrinsic bacterial toxicity have limited their use as clinical therapeutics (Forbes, 2010). Increasing metabolic control could help combat these challenges: cells could be engineered to release anti-cancer drugs only in the presence of cancer signaling molecules, and a population control mechanism could trigger a kill switch if bacteria accumulate in organs at dangerously high levels.
In this review, we will discuss the characteristics of precision metabolic engineering and ways that existing metabolic engineering techniques can be used in applications that require precise metabolic control. For metabolic engineering to be used in situations where tight control is necessary, a system must have the machinery to sense signals that determine the desired metabolic target, must completely direct metabolic flux based on those signals, and must produce sharp responses based on specific signal thresholds (Figure 1). First, we will describe sensory systems that can respond to autonomous and exogenous signals and the ways that protein engineering can enhance selectivity and sensitivity of a sensor. Next, we will describe the ways that different types of transcriptional and post-transcriptional regulation can be used, often in combination, to direct metabolic flux to only the desired pathway. Finally, we will describe methods that can be used to make the system respond sharply to the signal thresholds dictated by the application. Since dynamic metabolic engineering and precision metabolic engineering do have a bit in common and much more work has been done in dynamic than precision metabolic engineering, we will often first refer to applications of specific techniques in a dynamic metabolic engineering context before evaluating their relevance for direct application to precision metabolic engineering.
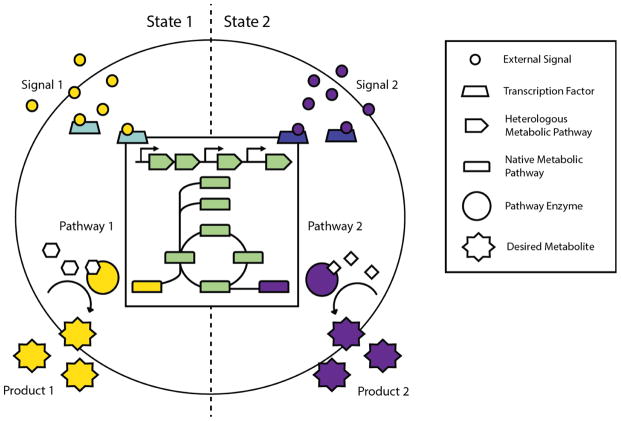
Figure 1. Schematic of precision metabolic engineering.
For precision metabolic engineering, cells must be capable of sensing different signals and, through robust cell regulation of native and heterologous metabolic pathways, ensure that only the desired products are made. In this example, external signals (small circles) enter the cell and bind transcription factors (trapezoids). The activated transcription factors control cell metabolism (central schematic in box) by affecting transcription from either native or heterologous metabolic pathways. Pathway enzymes (large circles) produce the desired metabolites (starburst shapes), and control mechanisms ensure high product selectivity over metabolites of competing pathways.
2. Sensory mechanisms to determine the state of the cell
A key characteristic of precision metabolic engineering is the production of a signal-specific metabolic response; to produce a specific response, a cell must first sense that signal. Whole cell biosensors have been designed to sense environmental signals and specific chemicals (Zhang and Keasling, 2011), and recent reviews have highlighted how dynamic metabolic engineering systems have been designed to similarly sense small molecules and metabolites, responding to inducers added at specific points in the production process or to a specific concentration of a naturally occurring molecule that changes over time (Brockman and Prather, 2015a; Venayak et al., 2015). Though these sensors are explicitly designed to be activated at a time point within the process, the same design approaches can be used to make sensors that do not necessarily effect changes in time.
2.1 Small molecule inducers
Exogenous inducers have been used in several dynamic systems to initiate a switch from a growth phase to a production phase (Brockman and Prather, 2015b; Davis et al., 2011; Soma et al., 2014; Torella et al., 2013). Small molecules like allolactose analogs (IPTG) and anhydrotetracycline have well-characterized interactions with promoter-repressor pairs and are widely used. Genetic circuits can be designed such that genes critical to growth but detrimental to production of a commercial target are expressed during the growth phase and repressed upon addition of an inducer. Optimal inducer concentration and time of induction can be determined to maximize titer and productivity.
The use of exogenous inducers could be useful in precision applications in which the user knows what the system should be producing. For example, microbes capable of producing a range of pharmaceuticals based on demand could produce specific pharmaceuticals in response to specific exogenous inducers. Exogenous inducers have less utility in biosensor applications, since the system must respond to a specific environmental signal. However, they could be used to activate or repress a sensory system: if sensory systems need to be off during the mass production of biosensor cells for later use in assays, then the sensory and response systems could be repressed through small molecule-responsive promoters and repressors, to be turned on only when desired.
2.2 Autonomous sensory systems
Cells have many natural mechanisms to sense and respond to their surroundings, and dynamic metabolic engineering has begun to harness this cellular machinery for use in controlled autonomous regulation (Carter et al., 2012). One commonly used mechanism is quorum sensing, the process through which cells secrete and sense autoinducers to sense their population density. E. coli have been engineered to sense and respond to quorum sensing molecules of their own and of different species (Kobayashi et al., 2004; Tsao et al., 2010) by expressing genes under quorum-responsive promoters. Similarly, the nitrogen regulatory system in E. coli was rewired to initiate the production of heterologous proteins upon sensing of acetyl-phosphate (Farmer and Liao, 2000) and acetate (Bulter et al., 2004), signals of excess metabolic flux that indicated cells were approaching the late exponential growth phase.
Natural methods to sense population density could replace small molecule induction in industrial processes where redirection of metabolic flux is required upon reaching a certain cell density. In the general case of precision metabolic engineering, in which production does not necessarily change over time, quorum sensing may have limited utility. However, quorum sensing could be useful in a subset of problems in which cells must modulate their response based on the number of cells present. For example, as discussed earlier, one challenge to using microbes to treat cancer is the inherent toxicity of bacteria and infections that can occur if bacterial growth cannot be controlled (Forbes, 2010). Quorum sensing could be used to trigger bacterial self-destruction upon reaching a threshold cell density, in this case enforcing precision on the dose of therapeutic cells in addition to the synthesis of their pharmaceutical products.
The same methods used to engineer specific quorum responses can be used to harness any natural transcriptional regulatory mechanism of a cell. Cells have transcriptional regulatory mechanisms to sense and respond to relative concentrations of different nutrients, ions, and sugars present in the cell (Chantranupong et al., 2015), and these sensory systems could be engineered to differentially regulate metabolism in response to different levels of analytes in the system.
2.3 Protein engineering to alter receptor-ligand affinity and selectivity
In the presence of known regulator-promoter interactions, protein engineering can be used to affect a sensor’s dynamic range by modulating regulator-substrate affinity (Cobb et al., 2013). Transcription factors involved in quorum sensing have been the target of extensive protein engineering efforts to accomplish such goals (Collins et al., 2005; Hawkins et al., 2007; Kambam et al., 2008, 2009). For example, by screening a large library of LuxR mutants, a library of transcription factors with up to a 100 fold increase in sensitivity to specific molecules was developed (Collins et al., 2005). Similar approaches have been successful in changing the dynamic range of other ligand-regulated transcription factors (Michener et al., 2012). Protein engineering can also change substrate specificity, allowing the construction of entirely new ‘sensors’ from fairly well-characterized scaffolds (Cobb et al., 2013). As an alternative to high-throughput screening of a mutant library if the protein structure is well-characterized, rational, directed mutagenesis of amino acids can effectively change receptor-ligand affinity and selectivity (Khan et al., 2002).
Protein engineering thus has significant potential to enhance responses to specific signals so that the dynamic sensing range corresponds with relevant concentrations of analyte (see also Section 4). Combined with the broad range of natural sensory mechanisms available across different organisms, this could allow a metabolic system to respond to an increasing number of external signals, whether exogenous or endogenous to the system.
3. Tight control of signal transduction through metabolic pathways
Another hallmark of precision metabolic engineering is tight control of metabolic state based on the signals that are sensed. A recently growing approach in metabolic engineering is the use of metabolite valves, which redirect metabolic flux from central carbon metabolic pathways to competing production pathways (Solomon et al., 2012; Soma et al., 2014). Though these valves shift the bulk of metabolic flux, some baseline expression through competing pathways can be permitted, which is often problematic in precision applications. A similar approach can be applied in a precision metabolic engineering context, where instead of just diverting flux, competing valves must be completely closed upon determination of state (Figure 2).
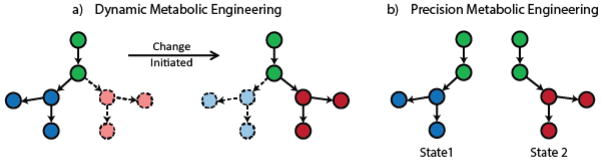
Figure 2. Direction of flux in dynamic and precision metabolic engineering.
A) Dynamic control redirects flux based on a change in the system. The pathway flux distribution changes, but flux may still be permitted through competing pathways. Initially, the majority of the metabolic flux is through the left branch of the pathway, with some flux permitted through the right branch. Upon initiation of a change, the flux shifts so that the majority is through the right branch. Circles represent metabolites in the pathway, with solid circles indicating significant production of a metabolite and dotted circles indicating partial repression of its production.
B) Rather than emphasizing the ability to dynamically switch between metabolic states, precision metabolic engineering emphasizes the completeness of the switch in metabolic state. Competing pathways are completely repressed so that only the desired metabolic pathway is active.
In dynamic metabolic engineering, leaky expression of enzymes vital to growth can enable mild growth during the production phase (Soma et al., 2014), which could in some situations be advantageous. However, in situations where complete control of metabolic flux is required, even minimal leakage can cause problems, and layers of regulation will likely be necessary to ensure that flux is completely redirected from competing pathways. Transcriptional, translational, and post-translational methods can be used, often in combination, to completely control flux through pathways.
3.1 Promoter engineering to tighten transcriptional regulation
E. coli has numerous known transcriptional regulatory systems in which the interaction of a transcription factor with an external signal can change the rate of transcription of different genes. However, the regulators may not be sufficiently strong to tightly control (induce or repress transcription of) engineered pathways. Promoter engineering can be used to enable transcriptional regulators to more strongly affect transcription. For example, promoters were designed to increase the regulatory capabilities of FadR (Zhang et al., 2012) and FapR (Xu et al., 2014a), transcription factors that respond to the fatty acid precursors acyl-CoA and malonyl-CoA, respectively. Zhang et al. integrated the FadR operator into two locations in the phage lambda and phage T7 promoters and demonstrated a 60 fold increase in the system’s response to acyl-CoA (Zhang et al., 2012). Xu et al. had similar success integrating the FapR operator into a T7 promoter to increase its response to malonyl-CoA (Xu et al., 2014a).
In designing promoters to be more strongly repressible, predicting regulator-polymerase interactions is difficult. When trying to make a second promoter that would also be active in the presence of malonyl-CoA, Xu et al. (Xu et al., 2014b) designed a promoter that was in fact repressed upon malonyl-CoA binding to the transcription factor. This ultimately proved beneficial—the two promoters with opposite responses to malonyl-CoA could differentially regulate genes in the presence of malonyl-CoA—but demonstrates the unpredictability of the effects of different regulators. Adding operators to different promoters can increase transcriptional regulation, but it cannot change the affinity with which the regulator binds DNA; without protein engineering, this method may not be effectively extrapolated to transcriptional regulators that interact weakly with DNA.
To help reduce leaky expression, both groups designed hybrid promoters that were repressed by the native transcription factor and LacI. This added an extra layer of control to the system: expression from the promoter would only occur if both the fatty acid precursor and IPTG were present in the system. This created a transcriptional AND gate, which is of importance in synthetic biology and biological computing and could have an increasingly important role in metabolic engineering as systems are designed to specifically respond to different combinations of signals. For example, in drug delivery applications, requiring two signaling molecules to be present instead of one could decrease nonselective delivery of a toxic drug.
While promoter engineering has significant potential to help redirect flux, limitations imposed by desired regulators could render this strategy alone insufficient. In these cases, regulation through other mechanisms would be necessary.
3.2 CRISPR Transcriptional Regulation
Recently, the CRISPR/Cas system was demonstrated to be a powerful tool for genome engineering (Cong et al., 2013; Mali et al., 2013). With some modifications, CRISPR/Cas systems can also be used for metabolic regulation. CRISPR interference (CRISPRi) has been used as a gene knock down tool: catalytically inactive Cas9 (dCas9) brings a guide RNA to the chromosome and unwinds the DNA through helicase activity. Upon reaching the gene of interest, the guide RNA binds to its complementary DNA strand, and dCas9 sterically interferes to prevent transcription of the gene (Qi et al., 2013). Transcriptional effectors can be fused to dCas9 to further repress or activate transcription from adjacent genes (Gilbert et al., 2013). Alternatively, guide RNA can also be extended with modular RNA domains to recruit transcriptional regulators via RNA-binding modules so that targeted genes can be differentially regulated (Zalatan et al., 2015). Generally, inducing the expression of dCas9 can turn on all parts of CRISPR regulation and tightly control the enzymatic activity of complex systems. Orthogonal dCas9 proteins that recognize different RNA sequences (Esvelt et al., 2013) could be induced under different conditions to differentially regulate genes (Zalatan et al., 2015).
Regulation with CRISPR/Cas9 could be useful in precisely directing metabolic flux because of its high selectivity for its target gene, use of orthogonal Cas9 proteins as master regulators, and limited interference with natural cellular pathways. CRISPR/Cas9 regulation completely repressed competing pathways in the branched violacein pathway, demonstrating its ability to tightly control which products are produced (Zalatan et al., 2015). Since CRISPR/Cas9 enables robust genomic regulation, it can also provide an alternative to strictly plasmid-based heterologous expression and regulation, which has the disadvantages of metabolic burden and unstable propagation characteristics (Anthony et al., 2009; Bentley et al., 1990; Diaz Ricci and Hernández, 2000).
3.3 Synthetic biology methods to control transcription
The field of synthetic biology has many examples of novel methods to turn cellular responses on and off, many of which have been used in metabolic engineering. The development of more tightly controlled synthetic biology devices could enable more precise control of metabolism. The genetic toggle switch (Gardner et al., 2000) has been used in dynamic metabolic engineering applications to generate “on” and “off” states through the use of well-characterized promoter-regulator interactions (Soma et al., 2014), and more elaborate switches can be made by layering logic gates (Moon et al., 2012). In a basic synthetic biology circuit, toggle switches can enable nearly complete shifts between states, but when controlling more complex circuits, toggle switches are not always effective at eliminating leaky expression and completely repressing alternative pathways (Soma et al., 2014). Applying these techniques to existing sensors and regulators could help enable more complete switch-like behavior, but their limitations would need to be addressed for direct application in precision metabolic engineering.
Riboregulators have shown the potential to regulate metabolic flux more precisely than toggle switches. The Collins lab has designed riboregulators that have sequence complementarity to a ribosomal binding site in the untranslated region. Upon transcription, this sequence binds to the RBS and forms a hairpin to prevent RNA translation. A short noncoding RNA sequence, expressed from a different promoter, interacts with the interfering RNA to expose the RBS and allow protein translation (Isaacs et al., 2004). They demonstrated some of this system’s major advantages: leakage minimization, tunable gene expression, fast response time, and independent regulation of multiple genes (Callura et al., 2010), all of which make this a very attractive regulatory mechanism to precisely control metabolism. A layer of complexity was added to riboregulator regulation through a switchboard that can detect and produce unique responses to a theoretically infinite number of signals; they demonstrated its ability to control metabolic flux through a branched pathway and prevent unwanted enzyme activity (Callura et al., 2012). Practically, this approach is limited by the small number of characterized natural receptor-promoter pairs, but it still provides an excellent framework for tight, synthetic biology-based control of transcription.
3.4 Post-transcriptional regulation methods
While transcriptional control is useful, control of translation is an equally important way to control cellular state, since it is ultimately enzyme activity that controls cellular metabolism. Dynamic metabolic engineering efforts have previously used interfering RNA (Williams et al., 2015) to prevent translation of specific genes. Precision metabolic engineering must go beyond preventing translation of unwanted proteins, to controlling exactly how much individual proteins are translated. Tools used for flux balancing and in synthetic biology can be used to accomplish this.
To effectively control protein concentration, several levels of post-transcriptional control will likely be necessary. Altering the ribosomal binding site of individual proteins can change relative protein translation efficiency (Salis et al., 2009), changing gene order within an operon can change relative translation rates of different proteins (Lee et al., 2013), and changing the 3′ untranslated region of messenger RNA can affect RNA stability (Lu et al., 2011; Zhao et al., 2013). Varying tunable intergenic regions between genes can alter RNA stability and ribosomal binding site availability independent of RBS strength (Pfleger et al., 2006). Protein expression can be altered after translation by adding different degradation tags to change the rate at which specific proteins are degraded (McGinness et al., 2006). Taken together, these constitute a powerful suite of tools for post-transcriptional control in precision metabolic engineering.
3.5 Dynamic protein degradation
While degradation tags are usually used to permanently target proteins for degradation, inducible expression of the degradation machinery can be used to selectively enhance protein degradation only in certain system (signal) states. This is a useful tool in dynamic metabolic engineering, as it can increase the rate at which enzymes critical for growth are degraded so that metabolic flux can be directed to production pathways. Knockout of sspB (the gene responsible for tethering tagged proteins to E. coli’s ClpXP degradation system) can prevent protein degradation, so inducible complementation of sspB expression at a specific time can allow for temporally controlled degradation of tagged proteins (Brockman and Prather, 2015b; Davis et al., 2011; Torella et al., 2013). Since knockout and induction of sspB could have deleterious effects on cell metabolism, as it disrupts natural protein degradation functions, inducible heterologous protein degradation systems have also been explored. An ssrA tag and protease from M. florum have been successfully used to orthogonally control protein degradation in E. coli (Cameron and Collins, 2014): E. coli’s ClpXP system does not recognize the heterologous tag, and the heterologous protease does not disrupt E. coli’s native ClpXP-ssrA degradation system (Cameron and Collins, 2014).
As noted above, inducible protein degradation has been used in dynamic metabolic engineering as a stand-alone method to decrease the concentration of pivotal enzymes (Brockman and Prather, 2015b; Davis et al., 2011; Torella et al., 2013). However, tag-based degradation (whether induced or constitutive) is never complete (there is always some recently translated protein having some potentially undesirable enzymatic activity), and expressing an unwanted protein just to quickly degrade it can place a significant metabolic burden on the cell. Thus, in precision metabolic engineering, protein degradation (through either native or heterologous systems) might instead be used as a secondary level of control. Transcription of competing pathways could be repressed, and if leaky expression of genes is problematic, the unwanted proteins could be degraded to reduce unwanted enzyme expression.
4. Engineering response hypersensitivity to specified signal concentrations
For a system to properly respond to a specific input, it must be calibrated so that it responds to the correct concentration of signal in a sharp, switch-like way. The system should ideally respond with either one metabolic mode or another, rather than with intermediate or gradient-like responses that are often present in natural biological and metabolic systems. Biosensor and drug delivery devices require sharp switch-like behavior to prevent ambiguous sensor readings and unwanted delivery of toxic drugs. Many of the transcriptional and post-transcriptional methods described in Section 3 can be used to tune the set point of the response and to facilitate sharper responses; those will not be rehashed in this section. Instead, we will focus on additional approaches that are useful for tuning the sharpness and set point of the response, including precursor availability and gene dosage.
4.1 Tuning precursor availability
Since the rate at which an enzyme converts metabolites often scales with the concentration of metabolite present, metabolic flux can be adjusted by modulating the concentration of precursors. This approach could then increase the sharpness or signaling set point of a response.
Controlling precursor concentration is not a new idea in metabolic engineering. Increasing precursor availability is an effective approach used widely across the field of metabolic engineering to increase titer for pathways that are not already saturated with flux. In addition, it has been used in balancing flux to prevent accumulation of toxic intermediates. Tunable intergenic regions (Pfleger et al., 2006), protein scaffolds (Dueber et al., 2009), addition of pathways (Pitera et al., 2007), and pathway balancing with metabolite responsive promoters (Dahl et al., 2013) have all been used to minimize the build-up of toxic metabolites.
A prototypical example demonstrating this concept for precision metabolic engineering is our recently described pigment-based biosensor (Watstein et al., 2015). In this work, two pigments from the same pathway (lycopene and β-carotene) were used as indicators, with conversion between them by the gene crtY controlled by a signal-responsive transcription factor. However, flux to the entry point of the pathway was low, which meant that slightly leaky expression of crtY (in what should have been a lycopene-on, β-carotene-off state) resulted in the reaction of all lycopene to β-carotene. By supplementing with the mevalonate pathway, total flux to the pathway was increased, rendering the low crtY expression negligible, and allowing the lycopene-producing state to be accessible. Moreover, for circuit configurations already with switch-like behavior, manipulating precursor availability tuned the set point for the switch relative to sensor input.
4.2 Regulating gene dosage
In addition to modulation of precursor levels, fluxes can also be regulated by changing enzyme concentration through selection of plasmid copy number for DNA elements that are not chromosomally integrated. Modular metabolic engineering has successfully used this method to eliminate flux imbalances by grouping pathway enzymes with a lower turnover rate on a higher copy plasmid to make the turnover rates of all enzymes more equal (Yadav et al., 2012). By varying plasmid copy number of different modules and adjusting promoter strengths, Ajikumar et al. achieved a 15,000 fold greater titer of taxane than through previous production methods (Ajikumar et al., 2010). This approach can also be useful for plasmid-based systems with regulatory elements (cis or trans) that are not orthogonal to the host: cross-talk interactions that are detrimental to the host or interfere with the metabolic program being added can be titrated out by changing the relative contributions of native versus plasmid-based regulatory elements.
4.3 Rational optimization methods
Tuning systems to sharply respond to a specific concentration of signal will likely require multiple levels of regulation, and determining the optimum combination of parts can be very difficult. It is often possible to create combinatorial libraries of regulated constructs, but assessing the performance of such libraries typically entails the use of high-throughput screens, which are not necessarily available for arbitrary phenotypes. Lee et al. used limited testing of a library combined with regression algorithms to predict the optimal combination of regulatory elements for a given phenotype, eliminating the need for a high-throughput screen (Lee et al., 2013) (though they still had to create the library).
As an alternative, multivariate modular metabolic engineering (MMME) has been used to maximize yield of a desired product by breaking a complicated system into modules and using transcriptional, translational, and post-translational modification methods to determine the optimum expression levels of each module. By treating a complicated system as a series of simpler, independent subsystems, a multivariate statistical analysis can be used to determine the optimal configuration so that extensive combinations of parts do not have to be made and tested (Yadav et al., 2012).
This multivariate approach could also be used to enhance precise control of metabolic systems. Rational design of individual modules could remove the need for high-throughput screening and enable more efficient combination of optimized modules. Variables would not be used to eliminate bottlenecks and balance flux, but rather to eliminate unwanted protein expression, increase sharpness of the response, and tune the set point (Figure 3).
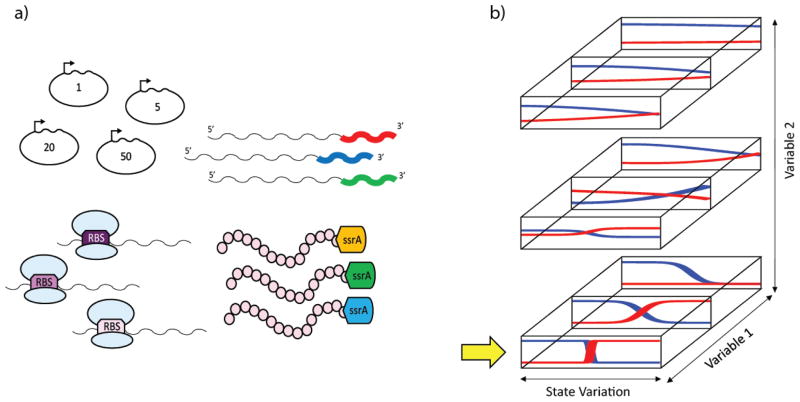
Figure 3. Multivariate optimization for precision metabolic engineering.
Adapted from Yadav et al. (Yadav et al., 2012)
A) Variables in precision metabolic engineering could include plasmid copy number, mRNA stabilizing regions, RBS strength, and protein degradation tags of the modules to be tuned
B) A two-variable optimization scheme. Variables 1 and 2 are changed to optimize the state-based selectivity and sharpness of the system response. The optimum combination, indicated by the yellow arrow, sharply responds to a change in state and has a switch point in an appropriate region.
While the main focus of MMME applications has typically been to identify optimal promoter strength for different parts, variation of promoters is not always an option. When promoter-regulator interactions are fixed, it may be impossible to alter promoter strength without losing regulator effectiveness (Xu et al., 2013). In these cases, post-transcriptional regulation is necessary, and variables to be optimized can potentially include copy number, ribosomal binding site sequence, protein degradation tag, and tunable intergenic regions.
More tools for MMME are continuously being developed. Libraries of characterized parts (Alper et al., 2005b; Pfleger et al., 2006; Zaslaver et al., 2006) have decreased the number of combinations required to be tested for effective MMME, and predictive algorithms (Salis, 2011; Salis et al., 2009) can further decrease the range of variation required to achieve optimal expression. A recent review (Biggs et al., 2014) discusses the need to expand and further characterize the tools available for tuning in MMME. As the MMME toolset expands, precision metabolic engineering will be able to be more easily applied to other systems.
5. Conclusions
By using existing techniques in new ways, metabolic engineering can expand its applications to produce flexible systems capable of sensing their environment and responding to different stimuli with distinct metabolic programs. In applications that require extreme product selectivity, tight control mechanisms must be in place to ensure that only the desired product is made. This is similar to (but a departure from) dynamic metabolic engineering, which focuses on temporarily diverting flux from, but not necessarily shutting down, competing pathways. While exogenous small molecule induction that is useful in dynamic applications has limited utility in precision metabolic engineering, autonomous sensory mechanisms and methods used to enhance their dynamic range and selectivity can be used to develop sensors for precision metabolic engineering applications. Transcriptional and post-transcriptional methods of controlling metabolic flux can be used in combination to ensure complete pathway selectivity. Of course, the goal of 100% complete elimination of anything in a biological system may be an impractical ideal; in the context of precision metabolic engineering, the point is that the 10% or 1% leakiness that is easy to achieve and is permissible in many applications can be quite problematic, requiring much tighter control than is typically pursued in metabolic engineering or synthetic biology applications.
Precision metabolic engineering will almost always require extensive tuning of metabolic responses. Standard methods of promoter engineering to change transcription rate are not viable options when the promoter to be used is dictated by sensor-regulator interactions. Protein engineering can be used to increase regulator affinity and dynamic range, but more typically post-transcriptional regulation would be used to control translation rate and protein degradation. Multivariate modular metabolic engineering provides a potentially valuable framework for the rational variation of multiple components to efficiently optimize systems.
There are a number of ways that future technological developments would address the current challenges of precision metabolic engineering. One of the main limits on the application of precision metabolic engineering is the limited number of well-characterized sensory-regulator systems available, such that developing new sensory systems would be extremely valuable. Aptamers, which are DNA or RNA sequences that can specifically bind small molecules and can be coupled with other elements to selectively induce enzymatic or regulatory activity, show significant potential in helping combat this challenge. In particular, aptamers used in combination with riboregulators could create tightly controlled systems that could specifically respond to different molecular signals. To date, the effort and difficulty associated with creating aptamers that bind specifically to small molecules of interest has limited their wide-spread use. However, recent improvements on high-throughput technologies to develop sensitive and specific aptamers (Cho et al., 2013; Wang et al., 2014) will likely hasten creation of novel aptamers, potentially broadening the scope of who can reasonably develop new aptamers and increasing the use of aptamer-based regulation. Other significant challenges to be addressed for precision metabolic engineering include reducing the experimental burden of tuning the system to respond in a switch-like way at a specified signal threshold. Computational models have been developed in dynamic metabolic engineering to predict the ways that changing time of induction and other controllable parameters would affect productivity (Anesiadis et al., 2008, 2013; Gadkar et al., 2005), and similar models could be developed to predict the ways that the switching point and switch-like behavior of a system could be optimized.
We expect the number of precision metabolic engineering applications to increase as scientists develop more and more complex tasks for microbes to complete. While existing examples of precision metabolic engineering – including using microbes as micronutrient biosensors, portable pharmaceutical platforms, and drug delivery vehicles – are exciting and have great potential, they are likely just the beginning of ways that metabolic engineers will look to precisely control cell state and function.
Table 1.
Comparison of precision metabolic engineering, dynamic metabolic engineering, and general metabolic engineering approaches
| Characteristic | General Metabolic Engineering | Dynamic Metabolic Engineering | Precision Metabolic Engineering |
|---|---|---|---|
| Engineering optimization goal | Product titer, yield, productivity | Product titer, yield, productivity | State-based selectivity and response sensitivity |
| Temporal behavior | None; optimization through static manipulations | System behavior necessarily changes over time | System behavior may (but does not necessarily) change over time |
| Number of possible states | One; static optimization produces singular desired state | Two; change between states initiated at time point during production | Many; states could change over time or be determined by initial concentration of sensory molecule |
| Signal that dictates metabolic state | None; state of system predetermined | Exogenous molecules, naturally produced metabolites | Exogenous molecules, naturally produced metabolites |
| Methods used to change metabolic state | None; state of system predetermined | Inducible transcriptional, post-transcriptional regulation | Inducible transcriptional, post-transcriptional regulation |
| Necessary degree of control between multiple desired metabolic states | None; state of system predetermined | Majority of metabolic flux must be through desired pathways, but leakiness in transcription and enzymatic activity may be permitted | All measurable metabolic flux must be through desired pathways, leakiness in transcription and enzymatic activity not permitted |
| Examples | Industrial production of biofuels, pharmaceuticals, commodity chemicals | Industrial production in which titer and productivity can be increased either by shifting from a growth phase to a production phase or by balancing fluxes to reduce toxic intermediates | Metabolite biosensors, portable pharmaceutical production, targeted drug delivery |
Highlights.
Metabolic engineering has potential for applications beyond maximization of titer.
These new “precision” applications have different goals and characteristics.
Many existing techniques can be applied for precision metabolic engineering.
Acknowledgments
The authors acknowledge the Bill & Melinda Gates Foundation for funding support (OPP1046289), as well as the National Science Foundation (1254382). MPM was supported by an NIH training grant (T32-EB0064343).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ajikumar PK, Xiao WH, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid Pathway Optimization for Taxol Precursor Overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Gutierrez J, Chan R, Batth TS, Adams PD, Keasling JD, Petzold CJ, Lee TS. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab Eng. 2013;19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol. 2005a;23:612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci U S A. 2005b;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anesiadis N, Cluett WR, Mahadevan R. Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng. 2008;10:255–266. doi: 10.1016/j.ymben.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Anesiadis N, Kobayashi H, Cluett WR, Mahadevan R. Analysis and Design of a Genetic Circuit for Dynamic Metabolic Engineering. ACS Synth Biol. 2013;2:442–452. doi: 10.1021/sb300129j. [DOI] [PubMed] [Google Scholar]
- Anthony JR, Anthony LC, Nowroozi F, Kwon G, Newman JD, Keasling JD. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab Eng. 2009;11:13–19. doi: 10.1016/j.ymben.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJY, Hanai T, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng. 2008a;10:305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008b;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Bentley WE, Mirjalili N, Andersen DC, Davis RH, Kompala DS. Plasmid-encoded protein: The principal factor in the “metabolic burden” associated with recombinant bacteria. Biotechnol Bioeng. 1990;35:668–681. doi: 10.1002/bit.260350704. [DOI] [PubMed] [Google Scholar]
- Biggs BW, De Paepe B, Santos CNS, De Mey M, Kumaran Ajikumar P. Multivariate modular metabolic engineering for pathway and strain optimization. Curr Opin Biotechnol. 2014;29:156–162. doi: 10.1016/j.copbio.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Brockman IM, Prather KLJ. Dynamic metabolic engineering: New strategies for developing responsive cell factories. Biotechnol J. 2015a doi: 10.1002/biot.201400422. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman IM, Prather KLJ. Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites. Metab Eng. 2015b;28:104–113. doi: 10.1016/j.ymben.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- Bulter T, Lee SG, Wong WW, Fung E, Connor MR, Liao JC. Design of artificial cell-cell communication using gene and metabolic networks. Proc Natl Acad Sci U S A. 2004;101:2299–2304. doi: 10.1073/pnas.0306484101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, Collins JJ. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc Natl Acad Sci. 2010;107:15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callura JM, Cantor CR, Collins JJ. Genetic switchboard for synthetic biology applications. Proc Natl Acad Sci. 2012;109:5850–5855. doi: 10.1073/pnas.1203808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DE, Collins JJ. Tunable protein degradation in bacteria. Nat Biotechnol. 2014;32:1276–1281. doi: 10.1038/nbt.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KK, Valdes JJ, Bentley WE. Pathway engineering via quorum sensing and sRNA riboregulators-interconnected networks and controllers. Metab Eng. 2012;14:281–288. doi: 10.1016/j.ymben.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Sabatini DM. Nutrient-Sensing Mechanisms across Evolution. Cell. 2015;161:67–83. doi: 10.1016/j.cell.2015.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemler JA, Fowler ZL, McHugh KP, Koffas MAG. Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metab Eng. 2010;12:96–104. doi: 10.1016/j.ymben.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Gil G, Mitchell RJ, Tai Chang S, Bock Gu M. A biosensor for the detection of gas toxicity using a recombinant bioluminescent bacterium. Biosens Bioelectron. 2000;15:23–30. doi: 10.1016/s0956-5663(99)00074-3. [DOI] [PubMed] [Google Scholar]
- Cho M, Soo Oh S, Nie J, Stewart R, Eisenstein M, Chambers J, Marth JD, Walker F, Thomson JA, Soh HT. Quantitative selection and parallel characterization of aptamers. Proc Natl Acad Sci U S A. 2013;110:18460–18465. doi: 10.1073/pnas.1315866110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EJ, Ling GSF. Battlefield Medicine: Paradigm Shift for Pharmaceuticals Manufacturing. PDA J Pharm Sci Technol. 2014;68:312–312. doi: 10.5731/pdajpst.2014.01002. [DOI] [PubMed] [Google Scholar]
- Cobb RE, Sun N, Zhao H. Directed evolution as a powerful synthetic biology tool. Methods San Diego Calif. 2013;60:81–90. doi: 10.1016/j.ymeth.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CH, Arnold FH, Leadbetter JR. Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol Microbiol. 2005;55:712–723. doi: 10.1111/j.1365-2958.2004.04437.x. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, et al. Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol. 2013;31:1039–1046. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- Davis JH, Baker TA, Sauer RT. Small-molecule control of protein degradation using split adaptors. ACS Chem Biol. 2011;6:1205–1213. doi: 10.1021/cb2001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Ricci JC, Hernández ME. Plasmid effects on Escherichia coli metabolism. Crit Rev Biotechnol. 2000;20:79–108. doi: 10.1080/07388550008984167. [DOI] [PubMed] [Google Scholar]
- Dietrich JA, McKee AE, Keasling JD. High-Throughput Metabolic Engineering: Advances in Small-Molecule Screening and Selection. Annu Rev Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KLJ, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–U107. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang MY, Zhang C, Yang S, Cui JY, Jiang PX, Lou K, Wachi M, Xing XH. High crude violacein production from glucose by Escherichia coli engineered with interactive control of tryptophan pathway and violacein biosynthetic pathway. Microb Cell Factories. 2015;14:8. doi: 10.1186/s12934-015-0192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18:533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10:785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadkar KG, Doyle FJ, III, Edwards JS, Mahadevan R. Estimating optimal profiles of genetic alterations using constraint-based models. Biotechnol Bioeng. 2005;89:243–251. doi: 10.1002/bit.20349. [DOI] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins AC, Arnold FH, Stuermer R, Hauer B, Leadbetter JR. Directed Evolution of Vibrio fischeri LuxR for Improved Response to Butanoyl-Homoserine Lactone. Appl Environ Microbiol. 2007;73:5775–5781. doi: 10.1128/AEM.00060-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- Jin YS, Alper H, Yang YT, Stephanopoulos G. Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl Environ Microbiol. 2005;71:8249–8256. doi: 10.1128/AEM.71.12.8249-8256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambam PKR, Sayut DJ, Niu Y, Eriksen DT, Sun L. Directed evolution of LuxI for enhanced OHHL production. Biotechnol Bioeng. 2008;101:263–272. doi: 10.1002/bit.21901. [DOI] [PubMed] [Google Scholar]
- Kambam PKR, Eriksen DT, Lajoie J, Sayut DJ, Sun L. Altering the Substrate Specificity of RhlI by Directed Evolution. Chem Bio Chem. 2009;10:553–558. doi: 10.1002/cbic.200800636. [DOI] [PubMed] [Google Scholar]
- Khan S, Brocklehurst KR, Jones GW, Morby AP. The functional analysis of directed amino-acid alterations in ZntR from Escherichia coli. Biochem Biophys Res Commun. 2002;299:438–445. doi: 10.1016/s0006-291x(02)02660-8. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Kærn M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: Interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ME, Aswani A, Han AS, Tomlin CJ, Dueber JE. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 2013;41:10668–10678. doi: 10.1093/nar/gkt809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Tang J, Liu Y, Zhu X, Zhang T, Zhang X. Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl Microbiol Biotechnol. 2011;93:2455–2462. doi: 10.1007/s00253-011-3752-y. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Mol Cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Michener JK, Thodey K, Liang JC, Smolke CD. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metab Eng. 2012;14:212–222. doi: 10.1016/j.ymben.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491:249–253. doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelek JM, Low KB, Bermudes D. Tumor-targeted Salmonella as a Novel Anticancer Vector. Cancer Res. 1997;57:4537–4544. [PubMed] [Google Scholar]
- Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- Pitera DJ, Paddon CJ, Newman JD, Keasling JD. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng. 2007;9:193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab AM, Gebhardt G, Bolotina N, Weuster-Botz D, Lang C. Metabolic engineering of Saccharomyces cerevisiae for the biotechnological production of succinic acid. Metab Eng. 2010;12:518–525. doi: 10.1016/j.ymben.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Rodrigues AL, Trachtmann N, Becker J, Lohanatha AF, Blotenberg J, Bolten CJ, Korneli C, de Souza Lima AO, Porto LM, Sprenger GA, et al. Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metab Eng. 2013;20:29–41. doi: 10.1016/j.ymben.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Rosen R, Davidov Y, LaRossa RA, Belkin S. Microbial sensors of ultraviolet radiation based on recA’::Lux fusions. Appl Biochem Biotechnol - Part Enzyme Eng Biotechnol. 2000;89:151–160. doi: 10.1385/abab:89:2-3:151. [DOI] [PubMed] [Google Scholar]
- Salis HM. Chapter two - The Ribosome Binding Site Calculator. In: Voigt C, editor. Methods in Enzymology. Academic Press; 2011. pp. 19–42. [DOI] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–U112. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon KV, Sanders TM, Prather KLJ. A dynamic metabolite valve for the control of central carbon metabolism. Metab Eng. 2012;14:661–671. doi: 10.1016/j.ymben.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Soma Y, Tsuruno K, Wada M, Yokota A, Hanai T. Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metab Eng. 2014;23:175–184. doi: 10.1016/j.ymben.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Torella JP, Ford TJ, Kim SN, Chen AM, Way JC, Silver PA. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc Natl Acad Sci U S A. 2013;110:11290–11295. doi: 10.1073/pnas.1307129110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao CY, Hooshangi S, Wu HC, Valdes JJ, Bentley WE. Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metab Eng. 2010;12:291–297. doi: 10.1016/j.ymben.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Venayak N, Anesiadis N, Cluett WR, Mahadevan R. Engineering metabolism through dynamic control. Curr Opin Biotechnol. 2015;34:142–152. doi: 10.1016/j.copbio.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Verma N, Singh M. Biosensors for heavy metals. Biometals. 2005;18:121–129. doi: 10.1007/s10534-004-5787-3. [DOI] [PubMed] [Google Scholar]
- Wang J, Gong Q, Maheshwari N, Eisenstein M, Arcila ML, Kosik KS, Soh HT. Particle display: a quantitative screening method for generating high-affinity aptamers. Angew Chem Int Ed Engl. 2014;53:4796–4801. doi: 10.1002/anie.201309334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watstein DM, McNerney MP, Styczynski MP. Precise metabolic engineering of carotenoid biosynthesis in Escherichia coli towards a low-cost biosensor. Metab Eng. 2015 doi: 10.1016/j.ymben.2015.06.007. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TC, Averesch NJH, Winter G, Plan MR, Vickers CE, Nielsen LK, Krömer JO. Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metab Eng. 2015;29:124–134. doi: 10.1016/j.ymben.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Xu P, Gu Q, Wang W, Wong L, Bower AGW, Collins CH, Koffas MAG. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun. 2013;4:1409. doi: 10.1038/ncomms2425. [DOI] [PubMed] [Google Scholar]
- Xu P, Wang W, Li L, Bhan N, Zhang F, Koffas MAG. Design and Kinetic Analysis of a Hybrid Promoter–Regulator System for Malonyl-CoA Sensing in Escherichia coli. ACS Chem Biol. 2014a;9:451–458. doi: 10.1021/cb400623m. [DOI] [PubMed] [Google Scholar]
- Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci. 2014b;111:11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VG, De Mey M, Giaw Lim C, Kumaran Ajikumar P, Stephanopoulos G. The future of metabolic engineering and synthetic biology: Towards a systematic practice. Metab Eng. 2012;14:233–241. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SH, Lee YM, Kim JE, Lee SH, Lee JH, Kim JY, Jung KH, Shin YC, Keasling JD, Kim SW. Enhanced lycopene production in Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate. Biotechnol Bioeng. 2006;94:1025–1032. doi: 10.1002/bit.20912. [DOI] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- Zhang F, Keasling J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 2011;19:323–329. doi: 10.1016/j.tim.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- Zhao J, Li Q, Sun T, Zhu X, Xu H, Tang J, Zhang X, Ma Y. Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metab Eng. 2013;17:42–50. doi: 10.1016/j.ymben.2013.02.002. [DOI] [PubMed] [Google Scholar]