Abstract
The conversion of grape must into wine involves the development and succession of yeast populations differing in species composition. The initial population is formed by vineyard strains which are washed into the must from the crushed grapes and then completed with yeasts coming from the cellar environment. As the origin and natural habitat of the vineyard yeasts are not fully understood, this study addresses the possibility, that grape yeasts can be preserved in berries left behind on vines at harvest until the spring of the next year. These berries become mummified during the winter on the vines. To investigate whether yeasts can survive in these overwintering grapes, mummified berries were collected in 16 localities in the Tokaj wine region (Hungary-Slovakia) in early March. The collected berries were rehydrated to recover viable yeasts by plating samples onto agar plates. For the detection of minority species which would not be detected by direct plating, an enrichment step repressing the propagation of alcohol-sensitive yeasts was also included in the process. The morphological, physiological, and molecular analysis identified 13 basidiomycetous and 23 ascomycetous species including fermentative yeasts of wine-making relevance among the 3879 isolates. The presence of viable strains of these species demonstrates that the grapes mummified on the vine can serve as a safe reservoir of yeasts, and may contribute to the maintenance of grape-colonizing yeast populations in the vineyard over years, parallel with other vectors and habitats. All basidiomycetous species were known phylloplane yeasts. Three Hanseniaspora species and pigmented Metschnikowia strains were the most frequent ascomycetes. Other fermentative yeasts of wine-making relevance were detected only in the enrichment cultures. Saccharomyces (S. paradoxus, S. cerevisiae, and S. uvarum) were recovered from 13% of the samples. No Candida zemplinina was found. The isolates with Aureobasidium morphology turned out to belong to Aureobasidium subglaciale, Kabatiella microsticta, or Columnosphaeria fagi. The ascomyceteous isolates grew at high concentrations of sugars with Wickerhamomyces anomalus being the most tolerant species. Complex interactions including antagonism (growth inhibition, contact inhibition, competition for nutrients) and synergism (crossfeeding) among the isolates and with Botrytis cinerea shape the composition of the overwintering communities.
Keywords: yeasts, grape, Tokaj, molecular taxonomy, diversity, antagonism, wine
Introduction
Wine is the product of the activity of complex microbial communities in which fermentative yeasts and bacteria play the major roles. It has been demonstrated by numerous studies, that the vineyard flora is the primary source of inoculation of the grape must with yeasts (for a review, see Fleet et al., 2002). When the fermentation takes place in wineries regularly used for wine-making, the population of the grape yeasts is supplemented with residential winery yeasts. In spontaneous fermentation, these yeasts drive the fermentation process with successive dominance of less and more alcohol tolerant species. Although fermentative yeasts, including Saccharomyces, can occur on grapes (e.g., Combina et al., 2005; Schuller et al., 2005; Valero et al., 2007; Cordero-Bueso et al., 2011; Setati et al., 2012; Bokulich et al., 2014; Taylor et al., 2014), the winery-borne yeasts usually overgrow the grape-borne strains in advanced stages of fermentation performed in cellars and can dominate the process until its completion (e.g., Rosini, 1984; Martini, 1993; Vaughan-Martini and Martini, 1995; Egli et al., 1998; Gutierrez et al., 1999; Ciani et al., 2004; Santamarıa et al., 2005; Mercado et al., 2007; Di Maio et al., 2012).
The grape berries are colonized in the vineyard by both ascomyceteous and basidiomyceteous yeasts, and their communities change over time depending on the stage of ripening (for reviews, see Fleet et al., 2002; Bisson and Joseph, 2009; Barata et al., 2012). In a comprehensive study, Bourret et al. (2013) identified 53 yeast species on grapes in a Washington State vineyard. Other laboratories have found most of these species in many other wine-growing regions of the globe and found numerous additional species (e.g., Yanagida et al., 1992; Zahavi et al., 2002; Antunovics et al., 2003; Prakitchaiwattana et al., 2004; Raspor et al., 2006; Sipiczki, 2006; Renouf et al., 2007; Chavan et al., 2009; Brysch-Herzberg and Seidel, 2015; Nemcová et al., 2015; Setati et al., 2015).
The main factor determining the composition of the yeasts communities on the grape appears to be nutrient availability on the berry surface which increases with ripening. During the growth of the grape, before the unset of ripening, the surface yeast flora is dominated by basidiomyceteous genera (e.g., Cryptococcus, Rhodosporidium, Rhodotorula, Sporobolomyces) and the dimorphic ascomyceteous genus Aureobasidium (reviewed in Bisson and Joseph, 2009; Barata et al., 2012). These yeasts capable of growth in the nutrient-poor surface of the developing berries are also present on other parts of the grapevine and on the phylloplane of other plants (for a review, see Fonseca and Inacio, 2006). When the fruit begins to ripen, yeasts belonging to ascomyceteous genera (e.g., Hanseniaspora, Metschnikowia, Candida) start proliferating on the grape skin, probably due to nutrients leaking out through the thinning skin. Interestingly, Saccharomyces, the major wine yeast is not ubiquitous on the ripening grape and if present, only constitutes very small fractions of the yeast communities (Setati et al., 2012; Bokulich et al., 2014; Taylor et al., 2014). Saccharomyces strains were more frequently isolated from heavily damaged grapes (e.g., Mortimer and Polsinelli, 1999), where the juice of the grape became accessible to the yeasts through the skin lesions.
All yeasts present on the grape at harvest are washed into the must at crush. However, not all grape-borne yeasts are equally important for the process of turning the grape must into wine (vinification). The basidiomyceteous species are the least relevant group because they die off very quickly in the must due to their inability to ferment the juice sugars. The ascomyceteous yeast-like Aureobasidium does not survive in the wine either (Renouf et al., 2007). Among the fermentative species, S. cerevisiae and S. uvarum (S. bayanus var. uvarum) are the most important yeasts because they drive the alcoholic fermentation and release the most important metabolites into the fermenting wine. The non-Saccharomyces yeasts usually play secondary roles by fine-tuning the wine character or act as spoilage microorganisms producing off-flavors (for reviews, see Loureiro and Malfeito-Ferreira, 2003; Jolly et al., 2014).
Some important questions are still to be answered about how the fermentative non-phylloplane yeasts show up on the grapes. Insects attracted by damaged berries have been implicated in the dispersal of the yeasts, with honey bees (Goddard et al., 2010), wasps (Stefanini et al., 2012), and the fruit fly Drosophila assumed to act as vectors (Lam and Howell, 2015) and perhaps also to preserve the yeasts in their (hibernated or dehydrated) bodies over the winter until the next spring. It is pertinent to note here, that various yeasts species have been isolated from various Drosophila species collected in various habitats such as tree exudates, rotting cacti, rain forests, oak-pine forests, etc. (e.g., Dobzhansky et al., 1956; Phaff and Knapp, 1956; Starmer et al., 1976; Gilbert, 1980; Lachance et al., 1995; Morais et al., 2005). These yeasts can easily be vectored onto the ripe grape by the host insects. However, fermentative yeast species are detectable already in early stages of ripening when the berries are still sound (intact). In addition, an important fermentative yeast, Saccharomyces does not appear to be regularly associated with the Drosophila flies in the nature. In a recent study, S. cerevisiae was not detected in 296 flies captured in vineyards, grape waste (marc) piles and wineries of two Australian wine-growing regions during grape harvest (Lam and Howell, 2015). Moreover, it is not the fruit volatiles but the yeasts, that attract Drosophila flies (Becher et al., 2012). Hanseniaspora has been found to produce aromas that are attractive to D. melanogaster (Palanca et al., 2013). So, at least certain berries have to be colonized by yeasts before the flies come, otherwise they would not come.
Vineyard soil is a potential source of the grape yeasts because the berries which fall to the ground during ripening and at harvest harbor large populations of yeasts. The work of Cordero-Bueso et al. (2011) describing different yeast populations in musts produced from grapes of wineyards in which different soil management methods were used indicates, that the condition of the soil can have an impact on the yeast communities of the grape. However, the soil is a rather unfavorable habitat for yeast overwintering because the soil microorganisms decompose the organic materials (for a review, see Treseder and Lennon, 2015) including the berries and their yeast colonists. Parle and Di Menna (1966) found very few fermentative yeasts in summer samples of vineyard soil and only Kloeckera (Hanseniaspora) apiculata in winter samples.
The goal of this study was to investigate an alternative possibility, the survival of grape yeasts in berries left-behind on the vine at harvest. These berries turn dry during winter and become mummified. To investigate whether the yeasts colonizing the ripe grape in autumn can survive the winter in these berries, mummified grapes were collected in the Tokaj wine-growing region in March. The peculiarity of this region is the extensive botrytisation leading to noble rotting of the grape on the vine. Noble rot is a benevolent Botrytis-generated process associated with dehydration (drastic increase of sugar content) and intense colonization of the ruptured berries by complex microbial consortia (Antunovics et al., 2003; Magyar and Bene, 2004). The collected grape mummies were rehydrated and used for recovering viable yeasts. The taxonomic examination of the recovered yeasts identified high numbers of basidiomyceteous and ascomyceteous yeast species, demonstrating, that the grape mummified on the vine also may contribute to the maintenance of the continuity of the vineyard yeast microflora over consecutive years.
Materials and methods
Sample collection and yeast isolation
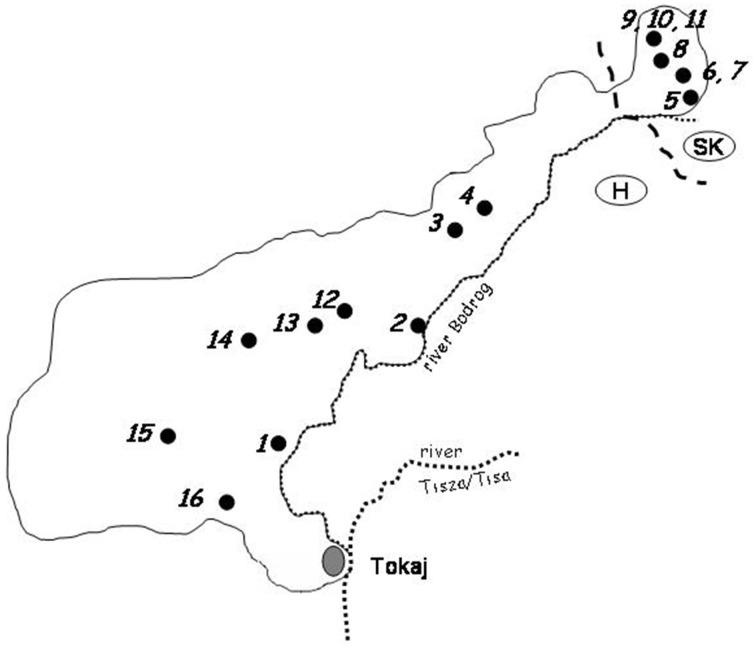
Three bunches of mummified grapes were collected from vines in each of the16 vineyards selected for the investigation (Figure 1). Five berries were picked aseptically from each bunch and placed in a sterile test-tube. As the berries were completely desiccated, 2.5 ml of YEL (1% yeast extract, 2% glucose) was added to the test-tube to rehydrate them. After 1 h of incubation at room temperature, the soaked berries were macerated with a sterile spatula and homogenized with intense vortexing. 10-μl aliquots of the homogenized sample were spread onto YEA (YEL supplemented with 2% agar) plates. The rest of the sample was incubated at room temperature overnight. Then 10-μl aliquots were plated on YEA again and 0.5-ml volumes were transferred into test-tubes containing 4.5 ml of enrichment medium (0.68% yeast nitrogen base, 1.1% raffinose, and 9% ethanol). This medium selectively supports the growth of the yeasts which tolerate high ethanol concentrations and utilize raffinose as a carbon source (Sampaio and Goncalves, 2008). The tube was incubated at 10°C. After 4 weeks of incubation, 10-μl aliquots of the enrichment culture were plated on YEA (when the cell number was high, the aliquots were diluted before plating). After 7 days of incubation at room temperature, colonies (max 150) were randomly isolated from the plates for each grape sample in order to obtain representative collections of pure isolates. The isolates were stored at 5°C on YEA plates and reinoculated onto fresh plate every month.
Figure 1.
Geographic locations of vineyards in which samples were collected. 1. Szegi; 2. Sárazsadány; 3. Hercegkút; 4. Hercegkút; 5. Viničky; 6. Bara; 7. Bara; 8. Černochov; 9. Malá Tŕňa; 10. Malá Tŕňa; 11. Malá Tŕňa; 12. Tolcsva; 13. Tolcsva; 14. Erdöbénye; 15. Abaujszántó; 16. Mád.
Phenotypic characterization of isolates
Colony morphology (color, surface ornamentation, production of pigmented zone in the medium) on YEA plates was examined and recorded for each isolate. All isolates were tested for the ability to assimilate 14 compounds as carbon-sources and lysine as a nitrogen source by replica-plating of 5-day old YEA cultures onto assimilation test plates (0.68% DIFCO yeast nitrogen base and 2% agar) supplemented with the carbon sources and onto SMA-lysine plates (2% glucose, 2% agar, 0.5% lysine and vitamins). The carbon sources tested were: cellobiose, ethanol, galactose, glucose, inulin, maltose, melesitose, melibiose, raffinose, rhamnose, ribose, saccharose, trehalose, and xylose. Growth was evaluated after 7 days of incubation at room temperature.
Molecular taxonomy
For taxonomic identification of the isolates, the D1/D2 domains of their large subunit ribosomal RNA genes and the ITS regions were amplified and sequenced with the primer pairs NL1-NL2 and ITS1-ITS4 as described earlier (Sipiczki, 2003). To assess their taxonomic positions, the resultant sequences were used to identify similar sequences in the GenBank database with the MEGABLAST-querying service of NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). As the GenBank entries are not checked for the correctness of their taxonomic assignment by the depositors the D1/D2 sequences of the isolates were then aligned with the D1/D2 sequences of the type strains of the species whose sequences were found highly similar in the GenBank search. For this, the sequences of the type strains were downloaded from the CBS database (http://www.cbs.knaw.nl/Collections/). The exact sequence similarity with the type strain sequences (number of identical nucleotides) was determined by pairwise Blast alignment using the bl2seq algorithm available at NCBI.
Osmotolerance
Dense suspensions of cells (~107 cells ml−1) were prepared from early stationary phase cultures of the isolates cultivated in YEL at room temperature for 2 days. 5-μl aliquots of the suspensions were dropped on YEA plates supplemented with 2, 20, 30, 40, and 50% of a 1:1 mixture of glucose and fructose. Yeast growth was evaluated after 5 days of incubation at 25°C by comparing the thickness of the spots.
Interaction tests
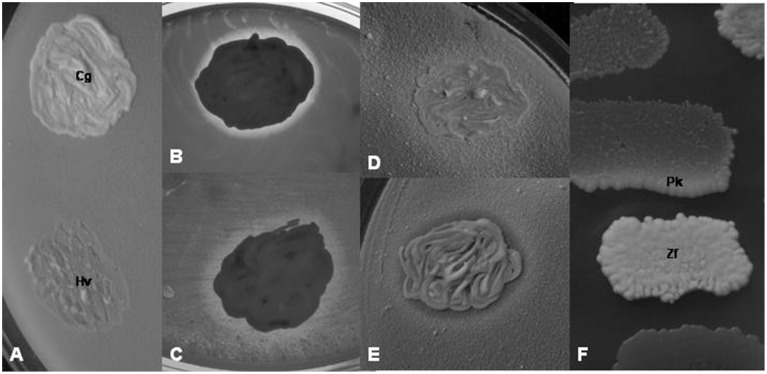
(a) Interactions among yeasts. For testing the yeast isolates for interactions with other yeast isolates, 5-day old cultures grown on YEA plates were used. Dense suspensions (~108 cells ml−1) were prepared from each isolate in 2 ml of sterile water, and YEA plates were flooded with the suspensions to obtain homogeneous lawns of cells. The rests of the suspensions were pored off. After drying the surface of the plates, loopful amounts of other isolates were smeared on the plates to form spots of ~5 mm in diameter (Figure 2). The plates were then incubated at 25°C for 7 days and examined at regular time intervals for the growth intensity of the spots and the lawn around the spots.
Figure 2.
Yeast-yeast interactions. (A) L. thermotolerans 8/2z-3 (lawn) shows no interaction with Ca. glabrata 14/1z-1 (Cg) but inhibits the growth of H. vineae 11/1z-4 (Hv). (B) Clear inhibition zone in the Ca. glabrata 14/1z-1 lawn around the W. anomalus 15/z-7 colony. (C) Turbid inhibition zone in the H. vineae 11/1z-4 lawn around the W. anomalus 15/z-7 colony. (D) Synergistic effect of the P. scaptomyzae 12/z-4 colony on the T. delbrueckii 1/1/z-10 lawn. (E) Dual effect: inhibition zone and crossfeeding of the Kl. dobzhanskii 9/z-6 lawn by the P. scaptomyzae 12z-4 colony. (F) Crossfeeding of the melibiose-minus P. kluyveri 11/2-104 colony (Pk) by the melibiose-positive Zt. florentina 7z-11 colony (Zf) on the medium containing melibiose as carbon source. (B) and (C) were photographed with transmitted light.
(b) Interactions between yeast isolates and B. cinerea. The effect of the yeast isolates on fungal growth was examined on YEA and PDA (Potato Dextrose Agar, Scharlab S.L.) plates flooded with suspensions of Botrytis conidia. The suspension of conidia was obtained by washing the surface of 2-week old B. cinerea 980 (Sipiczki, 2006) cultures grown on PDA at room temperature with sterile water. After removing the rest of the suspension and drying the surface of the plates, yeast isolates were inoculated onto the lawn of conidia as described above. The plates were incubated at 20°C for 2 weeks and examined at regular time intervals.
Results
Yeast isolation, characterization, and taxonomic identification
Yeast colonies were obtained with both isolation methods (with and without enrichment), even from those enrichment cultures which did not become turbid (no yeast growth). 3879 isolated colonies were examined for morphology, tested for the utilization of 15 compounds as sole carbon or nitrogen sources and then clustered into groups on the basis of their phenotypes. From representatives of the phenotypic groups, D1/D2 domain regions of the rDNA arrays were amplified and sequenced. Based on the similarity of the sequences to those of the type strains of known species, all but one group could be assigned to known species. 13 basidiomyceteous and 23 ascomyceteous species were identified in this way among the isolates.
Three groups of basidiomyceteous isolates could not be assigned to single species because their D1/D2 sequences were equally similar to the D1/D2 sequences of more than one type strain. In one of these groups, the similarity search found three identical type-strain sequences: Cryptococcus magnus, Filobasidium elegans, and Filobasidium floriforme. These species are indistinguishable when their D1/D2 sequences are compared but can be separated when certain physiological traits are also examined (Fell et al., 2000; Fonseca et al., 2011; Kwon-Chung, 2011). The assimilation tests of the isolates assigned this group to Cr. magnus because they grew on galactose (difference from F. elegans) and inulin (difference from both F. elegans and F. florioformae). A different group of isolates differed by one nucleotide from both Sporobolomyces coprosmae and Sp. oryzicola, a pair of very closely related sibling species which cannot be distinguished by D1/D2 sequencing (Scorzetti et al., 2002). Unfortunately, they cannot be differentiated by their growth reactions on the carbon compounds commonly tested in taxonomical studies, either (Hamamoto et al., 2011). Therefore, the ITS region also was sequenced from one of the isolates. Its sequence differed by 2 substitutions from Sp. coprosmae and 3 substitutions from Sp. oryzicola. Additional genes should be sequenced to reinforce the somewhat closer relationship to the former species. The third group showed 100% identity with the Curvibasidium pallidicorallinum type strain and differed from the Rhodotorula nothofagi type strain only by one substitution. The latter difference was due to an unambiguous nucleotide in the type strain sequence, so the somewhat higher similarity to Cu. pallidicorallinum was not relevant. The sugar assimilation tests indicated conspecificity with Cu. pallidicorallinum: the isolates grew on maltose, trehalose, and inulin, which are not assimilated by R. notophagi (Sampaio, 2011a,b).
The group represented by the isolate 6–13 in Table 1 could not be assigned to any species because its D1/D2 domain was very different from all type-strain sequences. Its closest relative type strain was Cr. keelungensis CBS 10876T, from which it differed by 24 substitutions (4%). Interestingly, it showed 99 to 100% identity to D1/D2 sequences of taxonomically uncharacterized yeast strains isolated from tree leaves, floral nectar (Alvarez-Perez and Herrera, 2013), and Iberian Pyrite Belt (Gadanho et al., 2006), indicating conspecificity with a hitherto non-described species which seems to live in diverse habitats.
Table 1.
D1/D2 sequence differences of selected representatives of the phenotypic groups of isolates from type strains of the most similar species.
| Isolate | Most similar type/reference strain | Sequence difference (number of substitutions/indels) | |||
|---|---|---|---|---|---|
| Identification number | Location of sample collection | D1/D2 accession number | Taxonomic name | D1/D2 accession number | |
| ASCOMYCOTA, PEZIZOMYCOTINA | |||||
| 10–59 | Malá Tŕňa | KU254559 | Aureobasidium subglaciale CBS 123387T | FJ150913 | 0 |
| 1/2–11 | Szegi | Kabatiella microsticta CBS 342.66 Columnosphaeria fagi CBS 171.93 |
FJ150952 AY016359 |
0 0 | |
| 6/1–5 | Bara | KU254558 | Kabatiella microsticta CBS 342.66 Columnosphaeria fagi CBS 171.93 |
FJ150952 AY016359 |
0 0 |
| 13/1–4 | Tolcsva | Kabatiella microsticta CBS 342.66 Columnosphaeria fagi CBS 171.93 |
FJ150952 AY016359 |
0 0 | |
| ASCOMYCOTA, SACCHAROMYCOTINA | |||||
| 14/1z-1 | Erdöbénye | KT933331 | Candida glabrata CBS 138T | AF399771 | 1 |
| 11/1–54 | Malá Tŕňa | Candida oleophila CBS 2219T | U45793 | 0 | |
| 11/1-55 | Malá Tŕňa | KT122406 | Candida oleophila CBS 2219T | U45793 | 0 |
| 7-9 | Bara | Kregervanrija fluxuum CBS 639T | U70247 | 3z | |
| 1/1z-2 | Szegi | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 2z-22 | Sárazsadány | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 4–3 | Hercegkút | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 4z-5 | Hercegkút | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 5/1z-3 | Vinièky | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 5/2–6 | Viničky | Hanseniaspora osmophila CBS 313T | U84228 | 1z | |
| 5/2z-6 | Viničky | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 5/2z-11 | Viničky | Hanseniaspora osmophila CBS 313T | U84228 | 1z | |
| 7/2z-1 | Bara | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 8–3 | Černochov | KT933332 | Hanseniaspora osmophila CBS 313T | U84228 | 0 |
| 8z-4 | Černochov | KT175536 | Hanseniaspora osmophila CBS 313T | U84228 | 0 |
| 9z-3 | Malá Tŕňa | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 11/2z-2 | Malá Tŕňa | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 12/2z-3 | Tolcsva | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 13/2–90 | Tolcsva | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 13/2z-5 | Tolcsva | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 15z-4 | Abaújszántó | Hanseniaspora osmophila CBS 313T | U84228 | 0 | |
| 1–3 | Szegi | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 1z-2 | Szegi | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 1/1–32 | Szegi | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 2–4 | Sárazsadány | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 4/1–6 | Hercegkút | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 7–3 | Bara | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 7/2–17 | Bara | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 8/2-28 | Černochov | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 9/1–3 | Malá Tŕňa | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 9/1–66 | Malá Tŕňa | KT156710 | Hanseniaspora uvarum CBS 314T | U84229 | 0 |
| 11/1–10 | Malá Tŕňa | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 13/2–4 | Tolcsva | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 14/1–7 | Erdöbénye | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 15/1–10 | Abaújszántó | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 15/2–1 | Abaújszántó | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 16–5 | Mád | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 16/2–2 | Mád | Hanseniaspora uvarum CBS 314T | U84229 | 0 | |
| 1/1–5 | Szegi | Hanseniaspora vineae CBS 2171T | U84224x | 3 | |
| 11/1z-4 | Malá Tŕňa | KT933333 | Hanseniaspora vineae CBS 2171T | U84224x | 1 |
| 12/1z-2 | Tolcsva | Hanseniaspora vineae CBS 2171T | U84224x | 4 | |
| 12/1z-5 | Tolcsva | Hanseniaspora vineae CBS 2171T | U84224x | 4 | |
| 5z-9 | Viničky | Kluyveromyces dobzhanskii CBS 2104T | U69575 | 0 | |
| 5z-17 | Viničky | Kluyveromyces dobzhanskii CBS 2104T | U69575 | 0 | |
| 7/1z-5 | Bara | Kluyveromyces dobzhanskii CBS 2104T | U69575 | 0 | |
| 9/z-1 | Malá Tàòa | Kluyveromyces dobzhanskii CBS 2104T | U69575 | 0 | |
| 9z-6 | Malá Tŕňa | KT122408 | Kluyveromyces dobzhanskii CBS 2104T | U69575 | 0 |
| 10z-4 | Malá Tŕňa | Kluyveromyces dobzhanskii CBS 2104T | U69575 | 0 | |
| 10/1z-1 | Malá Tŕňa | Kluyveromyces dobzhanskii CBS 2104T | U69575 | 0 | |
| 2/2z-8 | Sárazsadány | Lachancea thermotolerans CBS 6340T | U69581 | 0 | |
| 5/1z-7 | Viničky | Lachancea thermotolerans CBS 6340T | U69581 | 0 | |
| 8z-1 | Černochov | KT175534 | Lachancea thermotolerans CBS 6340T | U69581 | 0 |
| 8/2z-3 | Černochov | KT933334 | Lachancea thermotolerans CBS 6340T | U69581 | 0 |
| 9/1-15 | Malá Tŕňa | Lachancea thermotolerans CBS 6340T | U69581 | 0 | |
| 9/1z-4 | Malá Tŕňa | Lachancea thermotolerans CBS 6340T | U69581 | 0 | |
| 10/2z-4 | Malá Tŕňa | Lachancea thermotolerans CBS 6340T | U69581 | 0 | |
| 11–27 | Malá Tŕňa | Lachancea thermotolerans CBS 6340T | U69581 | 0 | |
| 11z-1 | Malá Tŕňa | Lachancea thermotolerans CBS 6340T | U69581 | 0 | |
| 11/2–112 | Malá Tŕňa | Lachancea thermotolerans CBS 6340T | U69581 | 0 | |
| 14/1z-2 | Erdöbénye | Lachancea thermotolerans CBS 6340T | U69581 | 0 | |
| 3z-1 | Hercegkút | Metschnikowia sp. 11-1090 clone d4 | KM350710 | 11 | |
| 5z-6 | Viničky | KT933337 | Pichia fermentans CBS 187T | U75726 | 2 |
| 5z-10 | Viničky | Pichia fermentans CBS 187T | U75726 | 2y | |
| 5z-12 | Viničky | Pichia fermentans CBS 187T | U75726 | 1 | |
| 11/2–104 | Malá Tŕňa | KT156709 | Pichia kluyveri CBS 188T | U75727x | 1 |
| 4/1–34 | Hercegkút | KT933335 | Pichia membranifaciens CBS 2763T | DQ198963 | 3 |
| 15/1z-9 | Abaújszántó | Pichia membranifaciens CBS 2763T | DQ198963 | 3 | |
| 12z-4 | Tolcsva | KT933336 | Pichia scaptomyzae CBS 8167T | AB045136 | 0 |
| 12z-14 | Tolcsva | Pichia scaptomyzae CBS 8167T | AB045136 | 0 | |
| 3/1z-5 | Hercegkút | Saccharomyces cerevisiae CBS 1171NT | U44806 | 0 | |
| 14/z-1 | Erdöbénye | KT933338 | Saccharomyces cerevisiae CBS 1171NT | U44806 | 0 |
| 3z-28 | Hercegkút | Saccharomyces paradoxus CBS 432NT | U68555 | 2 | |
| 3/2z-5 | Hercegkút | Saccharomyces paradoxus CBS 432NT | U68555 | 2 | |
| 5z-7 | Viničky | Saccharomyces paradoxus CBS 432NT | U68555 | 2 | |
| 7/2z-2 | Bara | Saccharomyces paradoxus CBS 432NT | U68555 | 2 | |
| 7/2z-3 | Bara | Saccharomyces paradoxus CBS 432NT | U68555 | 2 | |
| 10z-2 | Malá Tŕňa | KT122407 | Saccharomyces paradoxus CBS 432NT | U68555 | 2 |
| 4/2z-11 | Hercegkút | KT933339 | Saccharomyces uvarum CBS 395T | AJ279065 | 0 |
| 1/1z-1 | Szegi | Torulaspora delbrueckii CBS 1146T | U72156 | 0 | |
| 1/1z-10 | Szegi | KT933340 | Torulaspora delbrueckii CBS 1146T | U72156 | 0 |
| 15z-7 | Abaújszántó | KT933341 | Wickerhamomyces anomalus CBS 5759T | U74592 | 0 |
| 8z-2 | Černochov | KT175535 | Zygoascus meyerae CBS 7521T | AY447014 | 0 |
| 4–24 | Hercegkút | KT933342 | Zygosaccharomyces bailii CBS 680T | U72161 | 0 |
| 8–29 | Černochov | Zygosaccharomyces bailii CBS 680T | U72161 | 0 | |
| 2z-30 | Sárazsadány | Zygotorulaspora florentina CBS 746T | U72165 | 0 | |
| 3/2–1 | Hercegkút | Zygotorulaspora florentina CBS 746T | U72165 | 0 | |
| 7z-11 | Bara | KU254556 | Zygotorulaspora florentina CBS 746T | U72165 | 0 |
| 9/2z-9 | Malá Tŕňa | Zygotorulaspora florentina CBS 746T | U72165 | 0 | |
| BASIDIOMYCOTA, AGARIMYCOTINA | |||||
| 13/1–34 | Tolcsva | KT933343 | Bulleromyces albus CBS 500T | AF416643 | 0 |
| 6/2–10 | Bara | KT933344 | Cryptococcus carnescens CBS 973T | AB035054 | 0 |
| 7/2–10 | Bara | Cryptococcus flavescens CBS 942T | AB035042 | 0 | |
| 10/2–3 | Malá Tŕňa | Cryptococcus flavescens CBS 942T | AB035042 | 0 | |
| 8–30 | Černochov | KT933345 | Cryptococcus flavescens CBS 942T | AB035042 | 0 |
| 12/2–18 | Tolcsva | Cryptococcus flavescens CBS 942T | AB035042 | 0 | |
| 13/1–29 | Tolcsva | Cryptococcus flavescens CBS 942T | AB035042 | 0 | |
| 6–13 | Bara | KT001494 | Cryptococcus keelungensis CBS 10876T | EF621562 | 24 |
| 6–21 | Bara | KT933346 | Cryptococcus magnus CBS 140T Filobasidium elegans CBS 7640 Filobasidium floriforme CBS 6241 |
AF181851 AF181548 AF075498 |
0 0 0 |
| 7/1–40 | Bara | KT933352 | Cryptococcus magnus CBS 140T Filobasidium elegans CBS 7640 Filobasidium floriforme CBS 6241 |
AF181851 AF181548 AF075498 |
0 0 0 |
| 7/1–55 | Bara | Cryptococcus magnus CBS 140T Filobasidium elegans CBS 7640 Filobasidium floriforme CBS 6241 |
AF181851 AF181548 AF075498 |
1 1 1 | |
| 10/2–10 | Malá Tŕňa | KT933347 | Cryptococcus stepposus CBS 10265T | DQ222456 | 0 |
| 11/2–10 | Malá Tŕňa | KT933348 | Cryptococcus victoriae CBS 8685T | AF363647 | 2 |
| 12/2–50 | Tolcsva | Cryptococcus victoriae CBS 8685T | AF363647 | 2 | |
| 13/2–49 | Tolcsva | KU254557 | Cryptococcus carnescens CBS 973T Cryptococcus victoriae CBS 8685T |
AB035054 AF363647 |
8 8 |
| 13/1–37 | Tolcsva | KT933349 | Cryptococcus wieringae CBS 1937T | AF181541 | 0 |
| 7/1–56 | Bara | KT933353 | Holtermanniella festucosa VKM Y-2930T | AY462119 | 6 |
| BASIDIOMYCOTA, PUCCINIOMYCOTINA | |||||
| 8/1–14 | Černochov | KT933350 | Curvibasidium cygneicollum CBS 4551T | AF189928 | 0 |
| 10–10 | Malá Tŕňa | Curvibasidium cygneicollum CBS 4551T | AF189928 | 0 | |
| 10–20 | Malá Tŕňa | Curvibasidium cygneicollum CBS 4551T | AF189928 | 0 | |
| 15–23 | Abaújszántó | Curvibasidium cygneicollum CBS 4551T | AF189928 | 0 | |
| 15–25 | Abaújszántó | Curvibasidium cygneicollum CBS 4551T | AF189928 | 1 | |
| 6–23 | Bara | Curvibasidium pallidicorallinum CBS 9091T Rhodotorula nothofagi CBS 8166T |
AF444736 AF189950w |
0 1 | |
| 7/1–1 | Bara | KT156708 | Curvibasidium pallidicorallinum CBS 9091T Rhodotorula nothofagi CBS 8166T |
AF444736 AF189950w |
0 1 |
| 7/1–2 | Bara | Curvibasidium pallidicorallinum CBS 9091T Rhodotorula nothofagi CBS 8166T |
AF444736 AF189950w |
0 1 | |
| 7/1–50 | Bara | Curvibasidium pallidicorallinum CBS 9091T Rhodotorula nothofagi CBS 8166T |
AF444736 AF189950w |
0 1 | |
| 9–25 | Malá Tŕňa | KT933351 | Curvibasidium pallidicorallinum CBS 9091T Rhodotorula nothofagi CBS 8166T |
AF444736 AF189950w |
1 2 |
| 9–50 | Malá Tŕňa | Curvibasidium pallidicorallinum CBS 9091T Rhodotorula nothofagi CBS 8166T |
AF444736 AF189950w |
0 1 | |
| 11–2 | Malá Tŕňa | Curvibasidium pallidicorallinum CBS 9091T Rhodotorula nothofagi CBS 8166T |
AF444736 AF189950w |
0 1 | |
| 13/1z-1 | Tolcsva | Curvibasidium pallidicorallinum CBS 9091T Rhodotorula nothofagi CBS 8166T |
AF444736 AF189950w |
0 1 | |
| 13/1–16 | Tolcsva | KT933354 | Rhodotorula graminis CBS 2826T | AF070431 | 0 |
| 6/2–4 | Bara | KT933355 | Sporobolomyces coprosmae CBS 7899T Sporobolomyces oryzicola CBS 7228T |
AF189980 AF189990 |
1 1 |
, Type strain;
, neotype strain;
, contains one ambiguous nucleotide: N (A, G, C or T);
, isolate contains two ambiguous nucleotides: S (G or C) and Y (C or T);
, isolate contains one ambiguous nucleotide: Y (C or T);
, contains one ambiguous nucleotide: Y (C or T);
z, isolate contains two ambiguous nucleotides: Y (C or T) and R (A or G).
Almost all samples harbored cells, that produced colonies showing the morphology characteristic of the ascomycetous yeast-like fungus Aureobasidium pullulans. These rapidly growing colonies contained yeast-like cells and septate hyphae producing lateral blastospores. Since A. pullulans plays a very marginal role in wine-making and is quite ubiquitous on the phylloplane, only a few isolates of this morphology were characterized taxonomically in this study. Unexpectedly, their D1/D2 sequences showed closer relationship to A. glaciale, Kabatiella microsticta, and Columnosphaeria fagi than to A. pullulans. The examples shown in Table 1 have D1/D2 sequences identical with those of these species and differ from that of the A. pullulans type strain by 17 and 4 substitutions, respectively.
The isolates producing pigmented halos in the medium around their colonies appeared to belong to the pulcherrima clade of Metschnikowia (Lachance, 2011). However, their exact taxonomic position could not be determined because their D1/D2 sequences were diverse and different from the corresponding sequences of all type strains of the clade. For example, the GenBank database sequences most similar to the example shown in Table 1 were KM350710 and KM275362 which were cloned from the rDNA arrays of the M. sinensis and M. andauensis type strains CBS 10359T and CBS 10357T, respectively.
Tables S1, S2 show the occurrence and relative abundance of species in the 48 samples. The most frequently encountered species belonged to the genera Metschnikowia, Hanseniaspora, Cryptococcus before enrichment and Kluyveromyces, Lachancea, and Pichia after enrichment. Basidiomyceteous yeasts were detected in 69% of the samples before enrichment and only in 4% of the samples after enrichment (Table S2), indicating that the enrichment conditions were lethal to this group. The yeasts most frequently occurring in the enrichment cultures were strains of the ascomycetous genera Metschnikowia (in 33%), Lachancea thermotolerance (in 31%), and Hanseniaspora osmophila (in 31%; Table S1). Among these species only L. thermotolerance can utilize raffinose as carbon source. The other two species must have gained energy from alternative sources released from the dehydrated berry tissues and could overgrow the other yeasts due to their better ability to tolerate the high alcohol concentration and the low incubation temperature. Saccharomyces strains were found only in enriched cultures and only in 6 vineyards: S. paradoxus in 4, S. cerevisiae in 2, S. uvarum in 1 vineyard.
Lysine utilization was included in the phenotypic characterization of the isolates because one of the most frequently used methods in wine microbiology to identify Saccharomyces versus non-Saccharomyces yeasts is plating on lysine agar. Saccharomyces does not grow on lysine as a sole nitrogen source, therefore only non-Saccharomyces yeasts will grow on these plates (Angelo and Siebert, 1987). Consistent with this, none of the three Saccharomyces species identified in this study could utilize lysine as a nitrogen source. However, certain Hanseniaspora, Candida glabrata, Zygoascus meyerae, Zygosaccharomyces bailii, Kl. Dobzhanskii, and L. thermotolerans isolates identified by sequencing were also lysine minus.
Osmotolerance
The results are shown in Table 2. Surprisingly, only 7 isolates grew at 50% sugar, but neither the Saccharomyces species nor Zs. bailii were among them. The most osmotolerant species was Wickerhamomyces anomalus.
Table 2.
Osmotolerance of representative isolates of ascomyceteous yeast species.
| Species | Isolate | Growth on media supplemented with 1:1 fructose:glucose | |||
|---|---|---|---|---|---|
| 2% | 30% | 40% | 50% | ||
| Candida glabrata | 14/1z-1 | +++ | +++ | ++ | + |
| Candida oleophila | 11/1–55 | +++ | +++ | ++ | + |
| Hanseniaspora osmophila | 8-3 | +++ | +++ | ++ | + |
| Hanseniaspora uvarum | 9/1-66 | +++ | +++ | + | (+) |
| Hanseniaspora vineae | 11/1z-4 | +++ | +++ | +(+) | (+) |
| Kluyveromyces dobzhanskii | 9z-6 | +++ | +++ | ++(+) | – |
| Lachancea thermotolerans | 8/2z-3 | +++ | +++ | ++(+) | + |
| Metschnikowia sp. | 11/1-3 | +++ | +++ | ++ | + |
| Pichia fermentans | 5z-6 | +++ | +++ | – | – |
| Pichia kluyveri | 11/2–104 | +++ | +++ | + | – |
| Pichia membranifaciens | 4/1-34 | +++ | +++ | ++(+) | – |
| Pichia scaptomyzae | 12z-4 | +++ | +++ | + | – |
| Saccharomyces cerevisiae | 14z-1 | +++ | +++ | ++ | – |
| Saccharomyces paradoxus | 10z-2 | +++ | +++ | ++ | – |
| Saccharomyces uvarum | 4/2z-11 | +++ | +++ | ++ | – |
| Torulaspora delbrueckei | 1/1z-10 | +++ | +++ | ++ | + |
| Wickerhamomyces anomalus | 15z-7 | +++ | +++ | ++(+) | ++ |
| Zygoascus meyerae | 8z-2 | +++ | +++ | + | – |
| Zygosaccharomyces bailii | 4–24 | +++ | +++ | ++ | – |
| Zygotorulaspora florentina | 7z-11 | +++ | +++ | ++ | – |
+, growth; (+), weak growth; –, no growth.
Antagonistic and Synergistic interactions among Yeast isolates
Negative interaction (growth inhibition) was detected as an inhibition zone in the sensitive lawn around the colony of the antagonist (Figures 2B,C) or as the inhibition of the growth of the sensitive colony on the lawn of the antagonistic isolate (Figure 2A). Two types of inhibition zones could be distinguished: clear (Figure 2B) and turbid (Figure 2C) zones. The positive, growth intensifying effect was noticed as a halo of stronger growth of the lawn (lawn thickening) around the colony of the isolate which had such effect (Figure 2D). In a few combinations of isolates simultaneous antagonistic and synergistic effects could be seen (Figure 2E). The results of the interaction tests carried out with isolates representing all ascomycetous yeasts are shown in Table 3.
Table 3.
Yeast antagonism.
| Lawn | Inoculated on the lawn | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Isolate | 14/1z-1 | 11/1–55 | 8-3 | 9/1–66 | 11/1z-4 | 9z-6 | 8/2z-3 | 11/1–3 | 4/1–34 | 12z-4 | 5z-6 | 11/2–104 | 10z-2 | 14z-1 | 4/2z-11 | 1/1z-10 | 15z-7 | 8z-2 | 4–24 | 7z-11 |
| Candida glabrata | 14/1z-1 | – | – | –* | –* | –* | 1 | 1 | 0.5 | – | – | – | – | – | – | – | – | 2 | – | – | – |
| Candida oleophila | 11/1–55 | – | – | – | – | –* | 0.5 | – | –* | – | – | – | – | – | – | – | – | 1* | – | – | – |
| Hanseniaspora osmophila | 8–3 | ± | 2T | – | – | ± | – | –* | –* | ± | –*+ | –* | –* | –* | –* | –* | ± | –*+ | – | –* | –* |
| Hanseniaspora uvarum | 9/1–66 | – | – | +* | – | –* | – | –* | –* | –* | –* | –* | –* | –* | –* | –* | –* | –* | – | –* | –* |
| Hanseniaspora vineae | 11/1z-4 | – | – | –* | –* | – | – | –* | –* | –*+ | –*+ | –* | –* | –* | –* | –* | 1t* | 3t* | – | –* | –* |
| Kluyveromyces dobzhanskii | 9z-6 | – | – | – | – | – | – | – | 2t* | 2T | 1t+ | – | 0.5 | 1 | 1T | 0.5 | – | 1* | – | 0.5 | 1.5 |
| Lachancea thermotolerans | 8/2z-3 | – | – | –* | –* | –* | 1T | – | –* | 1t+ | ± | –* | –* | –* | – | – | – | 0.5* | – | – | – |
| Metschnikowia sp. | 11/1–3 | – | – | – | – | – | 2T | 1T | – | 1T | – | – | – | 0.5 | – | 0.5 | 1T | 1T | 0.5T | – | – |
| Pichia fermentans | 5z-6 | – | – | –* | – | – | 0.5T | – | – | ± | ± | – | – | – | – | – | – | –* | – | – | – |
| Pichia kluyveri | 11/2–104 | – | – | – | – | – | – | – | –* | 1T | – | – | – | – | – | – | – | –* | – | – | – |
| Pichia membranifaciens | 4/1–34 | – | – | ± | ± | ± | 2T | 3T | – | – | – | – | – | – | – | – | – | 2t* | – | – | – |
| Pichia scaptomyzae | 12z-4 | – | – | –*+ | –*+ | –*+ | 1T | – | – | –* | – | – | – | – | – | – | – | 0.5t* | – | – | – |
| Saccharomyces cerevisiae | 14z-1 | – | – | –* | –* | –* | 0.5T | 0.5 | –* | – | – | –* | –* | – | – | – | – | 2.5t* | – | – | – |
| Saccharomyces paradoxus | 10z-2 | – | – | –* | –* | –* | 2 | – | 1* | – | – | –* | – | – | – | – | – | 2* | – | – | – |
| Saccharomyces uvarum | 4/2z-11 | – | – | – | –* | –* | 1 | – | 1.5* | – | ± | –* | – | – | – | – | – | 1* | – | – | – |
| Torulaspora delbrueckei | 1/1z-10 | – | – | –* | –* | –* | 0.5 | – | –* | – | ± | –* | – | – | – | – | – | 2* | – | – | – |
| Wickerhamomyces anomalus | 15z-7 | – | – | – | – | – | – | 0.5 | 2T | 3T | – | – | – | 1 | 2 | 0.2 | 1 | – | 1T | – | – |
| Zygoascus meyerae | 8z-2 | – | – | – | – | – | 0.5T | – | –* | – | –*+ | – | – | – | – | – | – | 1t* | – | – | – |
| Zygosaccharomyces bailii | 4–24 | – | – | – | – | – | 1 | 0.5T | –* | –* | – | –* | –* | – | – | – | – | 1t* | – | – | – |
| Zygotorulaspora florentina | 7z–11 | – | – | – | – | – | 2T | – | – | – | – | – | – | – | – | – | – | –* | – | – | – |
Numerals, width of the inhibition zone in mm; –, no interaction is detected; +, crossfeeding (stronger lawn growth around the yeast colony);
, colony does not grow or grow poorly on the background lawn;
, the inhibition zone is turbid.
When the isolates were tested for the assimilation of sugars as carbon sources (see above), an interesting mode of growth stimulation was noticed. On the plates supplemented with saccharose or melibiose, certain isolates able to utilize these disaccharides stimulated the growth of certain other isolates which were unable to assimilate them. An example is shown in Figure 2F. The print of the colony of the melibiose-negative isolate 11/2-104 (Pichia kluyveri) replica-plated on the melibiose medium grew along its edge facing the colony of the melibiose-positive isolate 7z–11 (Zygotorulaspora florentina). Apparently, the positive isolate supplied the negative isolate with utilizable carbon sources (crossfeeding).
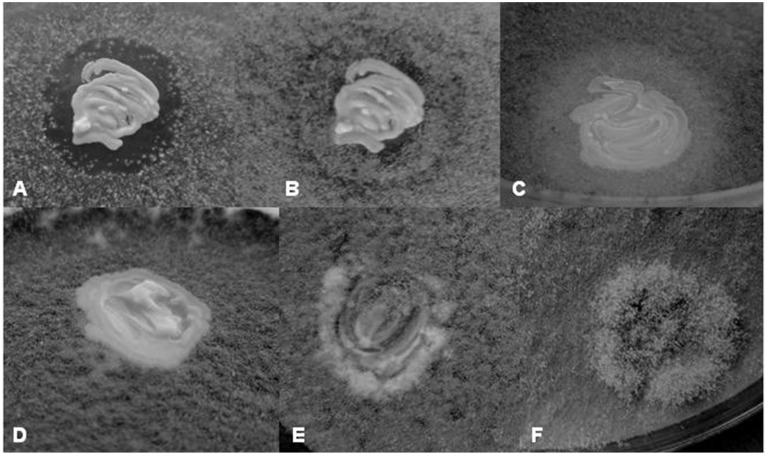
The effect of yeast isolates on the growth of Botrytis
The isolates selected for yeast-yeast interactions tests were also tested for effects on the growth of B. cinerea. Four types of interactions with the fungus were observed: (1) Clear inhibition zone around the yeast colony (Figure 3A), (2) turbid inhibition zone around the yeast colony (Figure 3C), (3) contact inhibition at the edge of the yeast colony (Figure 3D), and (4) overgrowth of the yeast colony by the Botrytis mycelium (Figures 3E,F). The clear zone can be interpreted as total inhibition of the growth of the fungus, whereas the turbid zone can be attributed to weaker inhibition or to competition for nutrients. The former morphology was observed only around Metschnikowia, P. fermentans, Zygosaccharomyces, and Zygotorulaspora isolates. Both types of zones and the contact inhibition (11 isolates) were visible only for 4 to 5 days, then the mycelium gradually grew into the zones (Figure 3B) and on the yeast colonies, indicating, that either the inhibitory conditions were changing with time or the hyphae invading the zones and the colonies were fed by cytoplasmic transport from the mycelium growing outside of the inhibition zone (e.g., Sipiczki, 2006). Representative results are shown in Table 4. Surprisingly, conspecificity did not always correlate with the intensity of the antagonistic activity. For species showing intraspecific diversity (H. osmophila, H. vineae, Kl. dobzhanskii, L. thermotolerance, Metschnikowia sp., P. membranifaciens, S. cerevisiae, S. paradoxus, Zs. bailii, Zt. florentina) more than one strain is listed in the table. Notably, more isolates produced zones on YEA than on PDA. The difference might be attributed to the much more vigorous growth of Botrytis on PDA.
Figure 3.
Yeast-Botrytis cinerea interactions. (A) Inhibition of the growth of B. cinerea around the Metschnikowia sp. 11/1–3 colony on YEA after 5 days of incubation. (B) Growth of the Botrytis mycelium into the inhibition zone after 11 days of incubation. Note that the contact with the yeast colony halts the growth of the mycelium. (C) Reduced mycelial growth around the P. kluyveri 11/2–104 colony on YEA after 5 days of incubation. (D) Contact inhibition: the Botrytis mycelium stops growing at the contact with the P. fermentans 5z-6 colony. (E) Gradual invasion of the H. osmophila 5/1z-3 colony by the Botrytis mycelium on YEA after 5 days of incubation. (F) Growth of the Botrytis mycelium on the L. thermotolerans 2/2z-8 colony. Note that the mycelium is thicker on the yeast colony.
Table 4.
Antagonistic effect of representative isolates of ascomyceteous yeast species on B. cinerea.
| Yeast | Botrytis cinerea | ||||||
|---|---|---|---|---|---|---|---|
| Species | Isolate | Is inhibited around the yeast colony (inhibition zone in mm) | Grows onto the yeast colony | ||||
| 5 day | 7 day | 11 day | |||||
| YEA | PDA | YEA | PDA | YEA | PDA | ||
| Candida glabrata | 14/1z-1 | – | – | – | – | (+) | (+) |
| Candida oleophila | 11/1–55 | 3T | – | – | – | (+) | – |
| Hanseniaspora osmophila | 4–3 | 3T | – | + | + | + | + |
| 8–3 | – | – | + | + | + | + | |
| Hanseniaspora uvarum | 9/1–66 | 5T | – | + | + | + | + |
| Hanseniaspora vineae | 1/1–5 | – | – | – | – | (+) | (+) |
| 11/1z-4 | 3T | – | + | – | + | + | |
| 12/1z-2 | 3T | – | + | – | + | – | |
| Kluyveromyces dobzhanskii | 5z-9 | 1T | – | – | – | + | (+) |
| 9z-6 | – | – | – | – | + | – | |
| Lachancea thermotolerans | 2/2z-8 | – | – | + | + | + | + |
| 8/2z-3 | 4T | – | + | + | + | + | |
| 14/1z-2 | 2T | – | + | + | + | + | |
| Metschnikowia sp. | 11/1–3 | 4 | 1 | – | – | – | – |
| 15–8 | 3 | – | – | – | – | – | |
| Pichia fermentans | 5z-6 | 4 | 4 | – | – | (+) | – |
| Pichia kluyveri | 11/2–104 | 5T | 5T | – | – | (+) | – |
| Pichia membranifaciens | 4/1–34 | 2T | – | (+) | – | + | (+) |
| 15/1z-9 | 2T | – | – | – | + | (+) | |
| Pichia scaptomyzae | 12z-4 | – | – | (+) | – | + | (+) |
| Saccharomyces cerevisiae | 3/1z-5 | – | – | – | – | + | + |
| 14z-1 | – | – | – | – | (+) | (+) | |
| Saccharomyces paradoxus | 3z-28 | – | – | – | – | (+) | – |
| 7/2z-2 | 3T | – | – | – | + | + | |
| 10z-2 | 3T | 3T | – | – | (+) | – | |
| Saccharomyces uvarum | 4/2z-11 | 2T | 2T | – | – | + | + |
| Torulaspora delbrueckei | 1/1z-10 | – | – | – | – | (+) | – |
| Wickerhamomyces anomalus | 15z-7 | – | – | – | – | – | (+) |
| Zygoascus meyerae | 8z-2 | 4T | 4T | – | – | + | – |
| Zygosaccharomyces bailii | 4–24 | 3 | 3 | – | – | (+) | (+) |
| 8–29 | 2 | 2 | – | – | (+) | – | |
| Zygotorulaspora florentina | 2z-30 | – | – | – | – | – | – |
| 3/2–1 | – | – | + | (+) | + | + | |
| 7z-11 | 3 | 1 | – | – | (+) | – | |
| 9/2z-9 | 3 | 1 | – | – | (+) | (+) | |
YEA and PDA are media (see Materials and Methods for description); numerals, width of inhibition zone in mm; +, growth; (+), weak growth; –, no growth;
, inhibition zone is turbid.
Discussion
Recovery of viable yeasts
When the grape is harvested, it is unavoidable, that berries fall to the ground and bunches remain on the vines. The former fall prey to rotting soil fungi and bacteria, which can also decompose the associated yeasts. The bunches on the vines are isolated from the destructive soil microorganisms and will be dehydrated (mummified) by frost and wind on the vines. The results of this study demonstrate, that fermentative yeasts relevant for wine making can tide over winter in these mummies.
This study was focused on the surviving yeasts, therefore the yeast communities of the overwintering grapes were examined by the conventional agar-plate method to obtain viable yeasts able to form colonies on a laboratory medium. The culture-independent strategies (e.g., metagenomics methods, DGGE) based on the analysis of DNA extracted directly from the yeast-containing substrates can identify yeast DNA but cannot distinguish between the DNA sequences of dead and live organisms (Mills et al., 2002; Prakitchaiwattana et al., 2004; Bokulich et al., 2014; Setati et al., 2015).
As certain yeasts of wine-making relevance are usually very sparse on grape berries if present at all (e.g., Mortimer and Polsinelli, 1999), an enrichment step was also included in the procedure. After plating out small aliquots on the agar plates, larger volumes of the samples were added to the enrichment medium restrictive for most non-Saccharomyces yeasts. By plating out aliquots before and after enrichment, large number of colonies were randomly isolated from 48 mummified grape bunches collected in 16 locations covering the entire Tokaj wine-growing region, (shared by Hungary and Slovakia; Figure 1) at the end of winter, shortly before pruning.
Taxonomy of isolates
To broaden the spectrum of the phenotypic traits applicable to clustering of the isolates, the colonies were tested for the utilization of 14 compounds as carbon sources and lysine as a nitrogen source. The taxonomic affiliation of the phenotypic groups was then determined by sequencing the chromosomal segments coding for the D1/D2 domains of the LSU (large subunit) rRNA from randomly chosen isolates of the clusters.
Among the isolates, more ascomycetous species and fewer basidiomycetous species were identified but neither group was represented in all samples. All basidiomyceteous species are known phylloplane yeasts (for a review, see Fonseca and Inacio, 2006) belonging to Agaricomycotina or to Pucciniomycotina and have been detected on grape as well (e.g., Yanagida et al., 1992; Sabate et al., 2002; Raspor et al., 2006; Li et al., 2010; Cadez et al., 2010; Bourret et al., 2013; Lederer et al., 2013; Brysch-Herzberg and Seidel, 2015; Nemcová et al., 2015; Setati et al., 2015). The basidiomyceteous yeasts were not considered further in this study because of their marginal significance in winemaking.
The ascomycetevous yeasts most frequently detected in the mummified samples were Metschnikowia strains producing pigmented colonies and Hanseniaspora strains producing apiculate cells. The Hanseniaspora isolates were assigned to three species, H. osmophila, H. uvarum, and H. vineae, with H. uvarum being more frequent in samples directly spread on agar plates and H. osmophila being more frequent in the enriched cultures. The apiculate yeasts usually predominate the early phase of fermentation and produce compounds, that enrich the aroma profile of the wine (e.g., Zironi et al., 1993; Romano et al., 2003; Moreira et al., 2011). Here, H. osmophila was found in the enrichment cultures even if undetected in the non-enriched samples, indicating, that H. osmophila can better tolerate high alcohol concentrations than the other two species of the genus. The abundance of the Hanseniaspora cells in the samples demonstrates that these yeasts cope well with the harsh microclimatic conditions during overwintering.
Pulcherrimin-producing Metschnikowia strains are common on ripe grapes. They are usually assigned to M. pulcherrima (C. pulcherrima) or less frequently to M. fructicola in the oenological literature. However, the pigmented colonies isolated in this study differed from the type strains of all known pulcherrimin-producing species (pulcherrima clade) in their D1/D2 sequences and usually contained several ambiguous nucleotides. D1/D2 differences between grape-borne Metschnikowia strains and the type strains of the related species were already noticed in a previous study of Tokaj grape yeasts (Sipiczki, 2006). Recently, Brysch-Herzberg and Seidel (2015) encountered a similar problem with Metschikowia yeasts isolated from ripe wine grapes in Germany. These difficulties indicate, that the deficiency of the rDNA homogenization process recently discovered in M. fructicola and M. andauensis (Sipiczki et al., 2013) might characterize all pigmented Metschnikowia strains and obscure the species boundaries in the pulcherrima clade. As these species cannot be clearly separated by physiological tests either (Lachance, 2011), the taxonomic assignment of the Metschnikowia yeasts described in the oenological literature should be treated with prudence. Nevertheless, the isolates examined in this work undoubtedly belong to the pulcherrima clade which harbors the pigmented species of the genus (Lachance, 2011). Strains of the clade are usually present in the must during the early phase of fermentation. Their contribution to the quality of the wine is beyond doubt but not yet fully explored (e.g., Gil et al., 1996; Sadineni Naresh et al., 2012; Jolly et al., 2014; Contreras et al., 2015).
The other ascomyceteous yeast species were less abundant than Metschnikowia and Hanseniaspora in the sampled mummified bunches. Except for a few bunches, ascomyceteous non-Metschnikowia and non-Hanseniaspora yeasts were found only in the enrichment cultures. Interestingly, even species which are unable to utilize raffinose (Ca. glabrata. Ca. oleophila, P. kluyvei, P. membranifaciens, T. delbruckei, Za. meyerae, Zs. bailii) were enriched. In their case, the enriching factor could have been the high alcohol concentration of the medium lethal to Metschnikowia, most Hanseniaspora species, and the basidiomycetes.
The principal wine yeasts, S. cerevisiae and S. uvarum, are rarely isolated from grapes by conventional direct agar plating procedures, and there is an ongoing debate about their natural origin in wine fermentation (e.g., Fleet et al., 2002). S. cerevisiae was occasionally isolated from mature, overripe, and damaged grapes, but usually enrichment steps had to be applied, that elicit the recovery of minority species which would not be detected by direct plating (e.g., Mortimer and Polsinelli, 1999; Mercado et al., 2007; Sampaio and Goncalves, 2008; Peter et al., 2011). Consistent with these earlier observations, no Saccharomyces was found in this study among the colonies when the samples were plated directly on the agar medium. Upon enrichment, S. paradoxus, S. cerevisiae, and S. uvarum could be recovered from certain cultures. Remarkably, S. paradoxus was more frequent than the other Saccharomyces species. This finding is consistent with the reports on large distribution of S. paradoxus in certain grape-growing areas (Redzepovic et al., 2002) but inconsistent with the microbiological analyses which detected only S. cerevisiae and S. uvarum in fermenting Tokaj wines (Sipiczki et al., 2001; Naumov et al., 2002; Antunovics et al., 2003, 2005). Another interesting finding is the presence of S. uvarum in one of the overwintering populations because this yeast has not been reported yet from grape samples. These results unanimously prove, that S. cerevisiae, S. paradoxus and S. uvarum can participate in the colonization of grape berries and can also be transmitted in mummified grape berries over consecutive vegetation periods. Nevertheless, their rather sporadic occurrence indicates, that either they are not regular components of the colonizing yeast communities or they have poor winter tolerance, a property assumed to depend on the sporulation efficiency (Sipiczki, 2010). Further, experiments could reveal to what extent their survival in the mummified grape can contribute to the maintenance of the continuity of the Saccharomyces populations in vineyards. It is worth noting that the inability to utilize lysine as a nitrogen source was not an exclusive trait of Saccharomyces isolates in this study. Many isolates assigned to 6 other ascomyceteous and 2 basidiomyceteous species could not utilize lysine either. Thus, the widely used method of differentiation of Saccharomyces (lys−) and non-Saccharomyces yeasts (lys+) on the basis of lysine utilization (Angelo and Siebert, 1987) can lead to false results when not combined with other tests.
Surprisingly, no strains of Ca. zemplinina were found among the isolates although this osmotolerant and psychrotolerant species (Sipiczki, 2003) is quite regularly encountered on ripe grape (e.g., Li et al., 2010; Brežná et al., 2010; Cadez et al., 2010; Sun et al., 2014; Brysch-Herzberg and Seidel, 2015; Setati et al., 2015) and is the third major wine yeast in the Tokaj region (Csoma and Sipiczki, 2008). As the locations of sample collection covered the entire Tokaj region, the lack of Ca. zemplinina among the viable yeasts can be attributed to its inability to survive in the overwintering grapes rather than to its absence on the ripe grapes.
Certain grape samples yielded rapidly extending colonies of yeast-like cells fringed by wide hyphal halos. Their morphology suggested conspecificity with A. pullulans, a widespread phylloplane fungus (e.g., Grube et al., 2011) belonging to Pezizomycotina. Surprisingly, the D1/D2 sequences of the isolates indicated closer genetic affinity with A. subglaciale, Ka. Microsticta, and Co. fagi than with A. pullulans. A. subglaciale was described from subglacial ice (Zalar et al., 2008; Gostinčar et al., 2014) and has not yet been detected in the wine-related environment. As for the isolates showing 100% D1/D2 sequence identity with Ka. microsticta and Co. fagi, it is worth mentioning, that Ka. microsticta ITS sequences were recently amplified from grape must in South Africa (Setati et al., 2015). However, the similarity to database Ka. microsticta ITS sequences does not prove conspecificity with Ka. microsticta because no Co. fagi sequences are available in the databases. In addition, Setati et al. (2015) did not culture the strains from the samples. Thus, this is the first report on the isolation of A. subglaciale and Ka. microsticta/Co. fagi from grape. As these species are closely related and can easily be confused with A. pullulans (Zalar et al., 2008), earlier reports on the occurrence of A. pullulans on grapes should be taken with caution if not supported with adequate taxonomic analyses.
Osmotolerance of isolates
Since, the yeasts residing in the overwintering grape berries have to cope with unfavorable factors, such as the antagonistic effects of other microbes and the osmotic pressure increasing during the dehydration of the berries, representatives of the ascomycetous yeast isolates were tested for response to these challenges. To investigate the ability of the isolates to cope with high osmotic pressure, representatives of the identified ascomyceteous species were tested for growth on agar plates supplemented with various concentrations of sugar. To mimic the real situation, glucose, and fructose were used in 1:1 proportion for supplementation. The W. anomalus isolates surpassed all other isolates in osmotolerance. This species has been described before as halophilic (Kagiyama et al., 1988) and a frequent spoilage yeast of fruit juice concentrates (e.g., Combina et al., 2008). As most isolates did not grow or poorly grew at 50% sugar, it can be assumed that the increasing osmotic pressure may also be involved in the preservation of the yeast community in the berries.
Antagonistic and synergistic interactions
The interaction tests revealed both antagonistic (growth inhibition) and synergistic (growth promotion) interactions among the isolates. In the test method applied in this study, the lawn of the antagonistic isolate hampered the growth of the colony of the sensitive isolate inoculated on it. In the reversed situation, the colony of the antagonistic isolate elicited an inhibition zone around its colony in the lawn of the sensitive isolate. Turbid zones indicated milder antagonisms (reduction of growth in the sensitive lawn) probably attributable to the depletion of the medium of certain nutrients by the colony of the “antagonist” (competition for nutrients). Clear zones were produced when the antagonist caused total growth inhibition in the lawn of the sensitive isolate. Several mechanisms might account for total inhibition. One possibility is, that the antagonistic isolate killed the cells of the sensitive isolate by secreting a toxic agent into the medium. Numerous yeasts species have been found to have strains harboring extracellular genetic elements (killer factors) encoding secretable agents referred to as killer toxins (for a review, see Schmitt and Breinig, 2002). The clear zones in the lawn of certain non-Saccharomyces isolates around the Saccharomyces colonies might be caused by killer toxins. Testing the antagonistic isolates identified in this study for the presence of such killer factors in their cells will be the subject of a different study. Nevertheless, it is rather unlikely that the Metschnikowia isolates inhibited the growth of other yeasts by killer toxins. They all formed colonies fringed by maroon-red pigmented zones in the agar media. In a previous study (Sipiczki, 2006), the pigmented zones were found to coincide with the growth inhibition zones around the Metschnikowia colonies inoculated onto lawns of sensitive microorganisms. It turned out that the growth inhibition was due to the immobilization of the free iron in the medium by complexing the ferric ions with a compound secreted by the Metschnikowia cells (Sipiczki, 2006). The complex referred to as pulcherrimin is water-insoluble and has maroon-red color (Cook and Slater, 1956). The behavior of the Wickerhamomyces strain shown in Table 2 and Figure 2 demonstrates how complex the interactions within the yeast communities can be. It grew poorly on the lawn of most isolates, but generated clear inhibition zones in the lawn of many of them and its lawn was inhibited by the colonies of certain other isolates. This diversity of interactions is consistent with the diversity of modes by which W. anomalus (P. anomala) can antagonize other microorganisms. Its strains can produce inhibitory amounts of ethyl acetate (Fredlund et al., 2004), secrete killer toxins (Kagiyama et al., 1988), and cel-wall lytic enzymes (Jijakli and Lepoivre, 1998). Synergistic, growth-facilitating effects of certain isolates on other isolates were observed on all media used in this study and could be attributed to crossfeeding with nutrients. On media containing mellibiose [D-galactose-α(1 → 6)-D-glucose] as carbon sources, the melibiose-utilizing isolates (Zs. florentina) promoted the growth of the melibiose-minus (P. kluyveri) isolates. Most probably, the former hydrolyzed the disaccharide in excessive amount and released some of the monosaccharides utilizable by the latter into the medium. On the grapes similar synergistic interactions can take place but with different nutrients.
In the Tokaj region, grape is harvested late in the autumn after a long period of ripening during which high proportions of berries undergo noble rotting generated by the B. cinerea infection. The invasion of the berries by the hyphae of the fungus causes ruptures in the skin which are then colonized by yeasts and bacteria. It was found, that at least one type of the colonizing yeasts, the pulcherrimin-producing Metschnikowia strains can antagonize the growth of Botrytis by inhibiting the germination of its conidia and the extension of its hyphae (Sipiczki, 2006). Consistent with this observation, all Metschnikowia isolates investigated in this study showed anti-Botrytis antagonism manifested in clear inhibition zones in the mycelium around their colonies. As the inhibition zones and the pigmented halos coincided, it is likely that the growth inhibition by Metschnikowia was due to iron immobilization by a secreted compound as described above. The clear zones around P. fermentans, Zs. Bailii, and Za. florentina colonies are most probably due to different mechanisms because these yeasts do not produce pulcherrimin. The mild inhibition of the fungal growth by S. paradoxus and S. uvarum isolates is an unexpected result. As the zones around their colonies were turbid, the inhibition can be ascribed to competition for nutrients rather than to the secretion of agents having antifungal activities. Contact inhibition observed at the colonies of 11 isolates is a phenomenon which has been noticed in certain yeasts before this study but the mechanisms by which these yeasts exert their influence on the hyphae has not yet been understood (for a review, see Liu et al., 2013).
Potential contribution of the mummified grapes to the maintenance of the vineyard yeast microflora
Taken together, the findings of this study demonstrate, that the grapes mummified on the vine can serve as a safe reservoir of fermentative yeasts, including Saccharomyces, and can transmit these yeasts between consecutive years in the vineyard. It can be reasonably assumed, that these yeasts may contribute to the maintenance of a complex vineyard yeast flora of wine-making relevance over years, together with those dispersed by insects (e.g., wasps, bees, Drosophila), and birds (Stevic, 1962; Francesca et al., 2012; Stefanini et al., 2012; Lam and Howell, 2015) visiting the ripening berries. Further, studies are needed to reveal the relative significance of these sources in the maintenance of the autochtonous vineyard yeast communities because soil is a rather unfavorable environment for yeast overwintering (Parle and Di Menna, 1966), the ROS-based antimicrobial defense system kills the ingested yeasts very fast in Drosophila (Hoang et al., 2015) the persistence of yeasts in bird cloacae has been shown to be very short (Francesca et al., 2012), the social wasp Vesta crabro (“European hornet” originally native only to Europe) recently found to harbor fermentative yeasts in guts (Francesca et al., 2012) is not common (or even absent) in large areas of the globe, where wine is produced. Moreover, certain yeast species were detected in the V. crabro guts after grape maturation, suggesting, that the wasps gathered those yeasts from the grapes rather than delivered them there. Mummified grapes can also be rare when modern harvesting technology is used.
Author contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author thanks Anita Csabai-Olah and Anita Bordan-Kovacs for excellent technical assistance. The Hungarian National Research Fund, Grants No. OTKA 81792 and 101323, supported this work.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00212
References
- Alvarez-Perez S., Herrera C. M. (2013). Composition, richness and nonrandom assembly of culturable bacterial-microfungal communities in floral nectar of Mediterranean plants. FEMS Microbiol. Ecol. 83, 685–699. 10.1111/1574-6941.12027 [DOI] [PubMed] [Google Scholar]
- Angelo J., Siebert K. J. (1987). A new medium for the detection of wild strains in brewing culture yeast. J. Am. Soc. Brew. Chem. 45, 135–140. [Google Scholar]
- Antunovics Z., Csoma H., Sipiczki M. (2003). Molecular and genetic analysis of the yeast flora of botrytized Tokaj wines. Bull. l'O.I.V. 76, 380–397. [Google Scholar]
- Antunovics Z., Irinyi L., Sipiczki M. (2005).Combined application of methods to taxonomic identification of Saccharomyces strains in fermenting botrytized grape must. J. Appl. Microbiol. 98, 971–979. 10.1111/j.1365-2672.2005.02543.x [DOI] [PubMed] [Google Scholar]
- Barata A., Malfeito-Ferreira M., Loureiro V. (2012).The microbial ecology of wine grape berries. Int. J. Food Microbiol. 153, 243–259. 10.1016/j.ijfoodmicro.2011.11.025 [DOI] [PubMed] [Google Scholar]
- Becher P. G., Flick G., Rozpędowska E., Schmidt A., Hagman A., Lebreton S., et al. (2012). Yeast, not fruit volatiles mediate Drosophila melanogaster attraction, oviposition and development. Funct. Ecol. 26, 822–828. 10.1111/j.1365-2435.2012.02006.x26776734 [DOI] [Google Scholar]
- Bisson L. F., Joseph C. M. L. (2009). Yeasts, in Biology of Microorganisms on Grapes, in Must and in Wine, eds König H., Unden G., Fröhlich J. (Berlin; Heidelberg: Springer-Verlag; ), 45–60. [Google Scholar]
- Bokulich N. A., Thorngate J. H., Richardson P. M., Mills D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111, E139–E148. 10.1073/pnas.1317377110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret T. B., Grove G. G., George J., Vandemark G. J., Henick-Kling T., Glawe D. A. (2013). Diversity and molecular determination of wild yeasts in a central Washington State vineyard. N. Am. Fungi 8, 1–32. 10.2509/naf2013.008.015 [DOI] [Google Scholar]
- Brežná B., Zenišová K., Chovanová K., Chebeňová V., Kraková L., Kuchta T., et al. (2010). Evaluation of fungal and yeast diversity in Slovakian wine-related microbial communities. Antonie Van Leeuwenhoek 98, 519–529. 10.1007/s10482-010-9469-6 [DOI] [PubMed] [Google Scholar]
- Brysch-Herzberg M., Seidel M. (2015). Yeast diversity on grapes in two German wine growing regions. Int. J. Food Microbiol. 214, 137–144. 10.1016/j.ijfoodmicro.2015.07.034 [DOI] [PubMed] [Google Scholar]
- Cadez N., Zupan J., Raspor P. (2010). The effect of fungicides on yeast communities associated with grape berries. FEMS Yeast Res. 10, 619–630. 10.1111/j.1567-1364.2010.00635.x [DOI] [PubMed] [Google Scholar]
- Chavan P., Mane S., Kulkarni G., Shaikh S., Ghormade V., Nerkar D. P., et al. (2009). Natural yeast flora of different varieties of grapes used for winemaking in India. Food Microbiol. 26, 801–808. 10.1016/j.fm.2009.05.005 [DOI] [PubMed] [Google Scholar]
- Ciani M., Mannazzu I., Marinangeli P., Clementi F., Martini A. (2004). Contribution of winery-resident Saccharomyces cerevisiae strains to spontaneous grape must fermentation. Antonie Van Leeuwenhoek 85, 159–164. 10.1023/B:ANTO.0000020284.05802.d7 [DOI] [PubMed] [Google Scholar]
- Combina M., Daguerre C., Massera A., Mercado L., Sturm M. E., Ganga A., et al. (2008). Yeast identification in grape juice concentrates from Argentina. Lett. Appl. Microbiol. 46, 192–197. 10.1111/j.1472-765X.2007.02291.x [DOI] [PubMed] [Google Scholar]
- Combina M., Mercado L., Borgo P., Elia A., Jofré V., Ganga A., et al. (2005). Yeasts associated to Malbec grape berries from Mendoza, Argentina. Int. J. Food Microbiol. 98, 1055–1061. 10.1111/j.1365-2672.2005.02540.x [DOI] [PubMed] [Google Scholar]
- Contreras A., Curtin C., Varela C. (2015). Yeast population dynamics reveal a potential ‘collaboration’ between Metschnikowia pulcherrima and Saccharomyces uvarum for the production of reduced alcohol wines during Shiraz fermentation. Appl. Microbiol. Biotechnol. 99, 1885–1895. 10.1007/s00253-014-6193-6 [DOI] [PubMed] [Google Scholar]
- Cook A. H., Slater C. A. (1956). The structure of pulcherrimin. J. Chem. Soc. 1956, 4133–4135. 10.1039/jr9560004133 [DOI] [Google Scholar]
- Cordero-Bueso G., Arroyo T., Serrano A., Tello J., Aporta I., Vélez M. D., et al. (2011). Influence of the farming system and vine variety on yeast communities associated with grape berries. Int. J. Food Microbiol. 145, 132–139. 10.1016/j.ijfoodmicro.2010.11.040 [DOI] [PubMed] [Google Scholar]
- Csoma H., Sipiczki M. (2008). Taxonomic reclassification of Candida stellata strains reveals frequent occurrence of Candida zemplinina in wine fermentation. FEMS Yeast Res. 8, 328–336. 10.1111/j.1567-1364.2007.00339.x [DOI] [PubMed] [Google Scholar]
- Di Maio S., Polizzotto G., Di Gangi E., Foresta G., Genna G., Verzera A., et al. (2012). Biodiversity of indigenous Saccharomyces populations from old wineries of South-Eastern Sicily (Italy): preservation and economic potential. PLoS ONE 7:e30428. 10.1371/journal.pone.0030428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., Cooper D. M., Phaff H. J., Knapp E. P., Carson H. L. (1956). Differential attraction of species of Drosophila to different species of yeasts. Ecology 37, 544–550. 10.2307/1930178 [DOI] [Google Scholar]
- Egli C. M., Edinger W. D., Mitrakul C. M., Henick-Kling T. (1998). Dynamics of indigenous and inoculated yeast populations and their effect on the sensory character of Riesling and Chardonnay wines. J. Appl. Microbiol. 85, 779–789. 10.1046/j.1365-2672.1998.00521.x [DOI] [PubMed] [Google Scholar]
- Fell J. W., Boekhout T., Fonseca A., Scorzetti G., Statzell-Tallman A. (2000). Biodiversity and systematics of basidiomycetous yeasts as determined by large subunit rDNA D1/D2/domain sequence analysis. Int. J. Syst. Bacteriol. 50, 1351–1371. 10.1099/00207713-50-3-1351 [DOI] [PubMed] [Google Scholar]
- Fleet G. H., Prakitchaiwattana C., Beh A. L., Heard G. (2002). The yeast ecology of wine grapes, in Biodiversity and Biotechnology of Wine Yeast, ed Ciani M. (Kerala: Research Signpost; ), 1–17. [Google Scholar]
- Fonseca A., Boekhout T., Fell J. W. (2011). Cryptococcus Vuillemin (1901), in The Yeasts - A Taxonomic Study, eds Kurtzman C. P., Fell J. W., Boekhout T. (London: Elsevier; ), 1661–1737. [Google Scholar]
- Fonseca A., Inacio J. (2006). Phylloplane yeasts, in Biodiversity and Ecophysiology of Yeasts, eds Rosa C., Peter G. (Berlin; Heidelberg: Springer-Verlag; ), 263–301. [Google Scholar]
- Francesca N., Canale D. E., Settanni L., Moschetti G. (2012). Dissemination of wine-related yeasts by migratory birds. Envir. Microbiol. Rep. 4, 105–112. 10.1111/j.1758-2229.2011.00310.x [DOI] [PubMed] [Google Scholar]
- Fredlund E., Druvefors U. A., Olstorpe M. N., Passoth V., Schnürer J. (2004). Influence of ethyl acetate production and ploidy level on the anti-mould activity of Pichia anomala. FEMS Microbiol. Lett. 238, 133–137. 10.1016/j.femsle.2004.07.027 [DOI] [PubMed] [Google Scholar]
- Gadanho M., Libkind D., Sampaio J. P. (2006). Yeast diversity in the extreme acidic environments of the Iberian Pyrite Belt. Microb. Ecol. 52, 552–563. 10.1007/s00248-006-9027-y [DOI] [PubMed] [Google Scholar]
- Gil J. V., Mateo J. J., Jimenez M., Pastor A., Huerta T. (1996). Aroma compounds in wine as influenced by apiculate yeasts. J. Food Sci. 61, 1247–1249. 10.1111/j.1365-2621.1996.tb10971.x [DOI] [Google Scholar]
- Gilbert D. G. (1980). Dispersal of yeasts and bacteria by Drosophila in temperate rain forest. Oecologia 46, 135–137. 10.1007/BF00346979 [DOI] [PubMed] [Google Scholar]
- Goddard M. R., Anfang N., Tang R., Gardner R. C., Jun C. (2010). A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ. Microbiol. 12, 63–73. 10.1111/j.1462-2920.2009.02035.x [DOI] [PubMed] [Google Scholar]
- Gostinčar C., Ohm R. O., Kogej T., Sonjak S., Turk M., Zajc J., et al. (2014). Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics 15:549. 10.1186/1471-2164-15-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube M., Schmid F., Berg G. (2011). Black fungi and associated bacterial communities in the phyllosphere of grapevine. Fungal. Biol. 115, 978–986. 10.1016/j.funbio.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Gutierrez A. R., Santamaria P., Epifanio S., Garijo P., Lopez R. (1999). Ecology of spontaneous fermentation in one winery during 5 consecutive years. Letts. Appl. Microbiol. 29, 411–415. 10.1046/j.1472-765X.1999.00657.x [DOI] [Google Scholar]
- Hamamoto M., Boekhout T., Nakase T. (2011). Sporobolomyces Kluyver and van Niel (1924), in The Yeasts - A Taxonomic Study, eds Kurtzman C. P., Fell J. W., Boekhout T. (London: Elsevier; ), 1929–1990. [Google Scholar]
- Hoang D., Kopp A., Chandler J. A. (2015). Interactions between Drosophila and its natural yeast symbionts - Is Saccharomyces cerevisiae a good model for studying the fly-yeast relationship? PeerJ 3:e1116. 10.7717/peerj.1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jijakli M. H., Lepoivre P. (1998). Characterization of an exo-b-1,3- glucanase produced by Pichia anomala strain K, antagonist of Botrytis cinerea on apples. Phytopathology 88, 335–343. 10.1094/PHYTO.1998.88.4.335 [DOI] [PubMed] [Google Scholar]
- Jolly N. P., Varela C., Pretorius I. S. (2014). Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215–237. 10.1111/1567-1364.12111 [DOI] [PubMed] [Google Scholar]
- Kagiyama S., Aiba T., Kadowaki K., Mogi K. (1988). New killer toxins of halophilic Hansenula anomala. Agric. Biol. Chem. 52, 1–7. 10.1271/bbb1961.52.1 [DOI] [Google Scholar]
- Kwon-Chung K. J. (2011). Filobasidium Olive (1968), in The Yeasts - A Taxonomic Study, eds Kurtzman C. P., Fell J. W., Boekhout T. (London: Elsevier; ), 1457–1465. [Google Scholar]
- Lachance M. A. (2011). Metschnikowia Kamienski (1899), in The Yeasts - A Taxonomic Study, eds Kurtzman C. P., Fell J. W., Boekhout T. (London: Elsevier; ), 575–619. [Google Scholar]
- Lachance M. A., Gilbert D. G., Starmer W. T. (1995). Yeast communities associated with Drosophila species and related flies in an eastern oak-pine forest: a comparison with western communities. J. Ind. Microbiol. 14, 484–494. 10.1007/BF01573963 [DOI] [PubMed] [Google Scholar]
- Lam S. S. T. H., Howell K. S. (2015). Drosophila-associated yeast species in vineyard ecosystems. FEMS Microbiol. Lett. 362:fnv170. 10.1093/femsle/fnv170 [DOI] [PubMed] [Google Scholar]
- Lederer M. A., Nielsen D. S., Toldam-Andersen T. B., Herrmann J. V., Arneborg N. (2013). Yeast species associated with different wine grape varieties in Denmark. Acta Agric. Scand. B Soil Plant Sci. 63, 89–96. 10.1080/09064710.2012.723738 [DOI] [Google Scholar]
- Li S. S., Cheng C., Li Z., Chen J. Y., Yan B., Han B. Z., et al. (2010). Yeast species associated with wine grapes in China. Int. J. Food Microbiol. 138, 85–90. 10.1016/j.ijfoodmicro.2010.01.009 [DOI] [PubMed] [Google Scholar]
- Liu J., Sui Y., Wisniewski M., Droby S., Liu Y. (2013). Review: utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. Int. J. Food Microbiol. 167, 153–160. 10.1016/j.ijfoodmicro.2013.09.004 [DOI] [PubMed] [Google Scholar]
- Loureiro V., Malfeito-Ferreira M. (2003). Spoilage yeasts in the wine industry. Int. J. Fodd Microbiol. 86, 23–50. 10.1016/S0168-1605(03)00246-0 [DOI] [PubMed] [Google Scholar]
- Magyar I., Bene Z. (2004). Characterization of yeast and mould biota of botrytized grapes in Tokaj wine region in the years 2000 and 2001. Acta Alimentaria 3, 259–267. 10.1556/AAlim.33.2004.3.6 [DOI] [Google Scholar]
- Martini A. (1993). The origin and domestication of the wine yeast Saccharomyes cerevisiae. J. Wine Res. 4, 165–176. 10.1080/09571269308717966 [DOI] [Google Scholar]
- Mercado L., Dalcero A., Masuelli R., Combina M. (2007). Diversity of Saccharomyces strains on grapes and winery surfaces: analysis of their contribution to fermentative flora of Malbec wine from Mendoza (Argentina) during two consecutive years. Food Microbiol. 24, 403–412. 10.1016/j.fm.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Mills D. A., Eric J., Eric A., Cocolin L. (2002). Yeast diversity and persistence in Botrytis-affected wine fermentation. Appl. Environ. Microbiol. 68, 4884–4893. 10.1128/AEM.68.10.4884-4893.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais P. B., Lachance M.-A., Rosa C. A. (2005). Saturnispora hagleri sp. nov., a yeast species isolated from Drosophila flies in Atlantic rainforest in Brazil. Int. J. System. Evol. Microbiol. 55, 1725–1727. 10.1099/ijs.0.63673-0 [DOI] [PubMed] [Google Scholar]
- Moreira N., Pina C., Mendes F., Couto J. A., Hogg T., Vasconcelos I. (2011). Volatile compounds contribution of Hanseniaspora guilliermondii and Hanseniaspora uvarum during red wine vinifications. Food Control 22, 662–667. 10.1016/j.foodcont.2010.07.025 [DOI] [Google Scholar]
- Mortimer R., Polsinelli M. (1999). On the origins of wine yeast. Res. Microbiol. 150, 199–204. 10.1016/S0923-2508(99)80036-9 [DOI] [PubMed] [Google Scholar]
- Naumov G. I., Naumova E. S., Antunovics Z., Sipiczki M. (2002). Saccharomyces bayanus var. uvarum in Tokaj wine-making of Slovakia and Hungary. Appl. Microbiol. Biotechnol. 59, 727–730. 10.1007/s00253-002-1077-6 [DOI] [PubMed] [Google Scholar]
- Nemcová K., Breierová E., Vadkertiová R., Molnárová J. (2015). The diversity of yeasts associated with grapes and musts of the Strekov winegrowing region, Slovakia. Folia Microbiol. 60, 103–109. 10.1007/s12223-014-0347-x [DOI] [PubMed] [Google Scholar]
- Palanca L., Gaskett A. C., Günther C. S., Newcomb R. D., Goddard M. R. (2013). Quantifying variation in the ability of yeasts to attract Drosophila melanogaster. PLoS ONE 8:e75332. 10.1371/journal.pone.0075332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parle J. N., Di Menna M. E. (1966). The source of yeasts in New Zealand wines. N.Z. J. Agric. Res. 9, 98–107. 10.1080/00288233.1966.10418122 [DOI] [Google Scholar]
- Peter G., Dlauchy D., Szucs E., Tornai-Lehoczki J. (2011). Enrichment in methanol containing broth – a simple method for the isolation of Saccharomyces from grapes. Acta Alimentaria 40, 376–384. 10.1556/AAlim.40.2011.3.8 [DOI] [Google Scholar]
- Phaff H. J., Knapp E. P. (1956). The taxonomy of yeasts found in exudates of certain trees and other natural breeding sites of some species of Drosophila. Antonie Van Leeuwenhoek 22, 117–130. 10.1007/BF02538319 [DOI] [PubMed] [Google Scholar]
- Prakitchaiwattana C. J., Fleet G. H., Heard G. M. (2004). Application and evaluation of denaturing gradient gel electrophoresis to analyse the yeast ecology of wine grapes. FEMS Yeast Res. 4, 865–877. 10.1016/j.femsyr.2004.05.004 [DOI] [PubMed] [Google Scholar]
- Raspor P., Milek D. M., Polanc J., Mozina S. S., Cadez N. (2006). Yeasts isolated from three varieties of grapes cultivated in different locations of the Dolenjska vine-growing region, Slovenia. Int. J. Food Microbiol. 109, 97–102. 10.1016/j.ijfoodmicro.2006.01.017 [DOI] [PubMed] [Google Scholar]
- Redzepovic S., Orlic S., Sikora S., Majdak A., Pretorius I. S. (2002). Identification and characterization of Saccharomyces cerevisiae and Saccharomyces paradoxus strains isolated from Croatian vineyards. Lett. Appl. Microbiol. 35, 305–310. 10.1046/j.1472-765X.2002.01181.x [DOI] [PubMed] [Google Scholar]
- Renouf V., Claisse O., Lonvaud-Funel A. (2007). Inventory and monitoring of wine microbial consortia. Appl. Microbiol. Biotechnol. 75, 149–164. 10.1007/s00253-006-0798-3 [DOI] [PubMed] [Google Scholar]
- Romano P., Fiore C., Paraggio M., Caruso M., Capece A. (2003). Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 86, 169–180. 10.1016/S0168-1605(03)00290-3 [DOI] [PubMed] [Google Scholar]
- Rosini G. (1984). Assessment of dominance of added yeast with fermentation and origin of Saccharomyces cerevisiae in wine making. J. Gen. Appl. Microbiol. 30, 249–256. 10.2323/jgam.30.249 [DOI] [Google Scholar]
- Sabate J., Cano J., Esteve-Zarzoso B., Guillamón J. M. (2002). Isolation and identification of yeasts associated with vineyard and winery by RFLP analysis of ribosomal genes and mitochondrial DNA. Microbiol. Res. 157, 267–274. 10.1078/0944-5013-00163 [DOI] [PubMed] [Google Scholar]
- Sadineni Naresh V., Kondapalli N., Obulam V. (2012). Effect of co-fermentation with Saccharomyces cerevisiae and Torulaspora delbrueckii or Metschnikowia pulcherrima on the aroma and sensory properties of mango wine. Ann. Microbiol. 62, 1353–1360. 10.1007/s13213-011-0383-6 [DOI] [Google Scholar]
- Sampaio J. P. (2011a). Curvibasidium Sampaio and Golubev (2004), in The Yeasts - A Taxonomic Study, eds Kurtzman C. P., Fell J. W., Boekhout T. (London: Elsevier; ), 1413–1418. [Google Scholar]
- Sampaio J. P. (2011b). Rhodotorula Harrison (1928), in The Yeasts - A Taxonomic Study, eds Kurtzman C. P., Fell J. W., Boekhout T. (London: Elsevier; ), 1873–1927. [Google Scholar]
- Sampaio J. P., Goncalves P. (2008). Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl. Environ. Microbiol. 74, 2144–2152. 10.1128/AEM.02396-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamarıa P., Garijo P., Lopez R., Tenorio C., Rosa Gutierrez A. (2005). Analysis of yeast population during spontaneous alcoholic fermentation: effect of the age of the cellar and the practice of inoculation. Int. J. Food Microbiol. 103, 49–56. 10.1016/j.ijfoodmicro.2004.11.024 [DOI] [PubMed] [Google Scholar]
- Schmitt M. J., Breinig F. (2002). The viral killer system in yeast: from molecular biology to application. FEMS Microbiol. Rev. 748, 1–20. 10.1111/j.1574-6976.2002.tb00614.x [DOI] [PubMed] [Google Scholar]
- Schuller D., Alves H., Dequin S., Casal M. (2005). Ecological survey of Saccharomyces cerevisiae strains from vineyards in the Vinho Verde region of Portugal. FEMS Microbiol. Ecol. 51, 167–177. 10.1016/j.femsec.2004.08.003 [DOI] [PubMed] [Google Scholar]
- Scorzetti G., Fell J. W., Fonseca A., Statzell-Tallman A. (2002). Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2, 495–517. 10.1111/j.1567-1364.2002.tb00117.x [DOI] [PubMed] [Google Scholar]
- Setati M. E., Jacobson D., Andong U.-C., Bauer F. (2012). The vineyard yeast microbiome, a mixed model microbial map. PLoS ONE 7:e52609. 10.1371/journal.pone.0052609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setati M. E., Jacobson D., Bauer F.F. (2015). Sequence-based analysis of the Vitis vinifera L. cv Cabernet Sauvignon grape must mycobiome in three South African vineyards employing distinct agronomic systems. Front. Microbiol. 6:1358. 10.3389/fmicb.2015.01358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. (2003). Candida zemplinina sp. nov., an osmotolerant and psychrotolerant yeast that ferments sweet botrytised wines. Int. J. Syst. Evol. Microbiol. 53, 2079–2083. 10.1099/ijs.0.02649-0 [DOI] [PubMed] [Google Scholar]
- Sipiczki M. (2006). Metschnikowia strains isolated from botrytised grapes antagonize fungal and bacterial growth by iron depletion. Appl. Environ. Microbiol. 72, 6716–6724. 10.1128/AEM.01275-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. (2010). Diversity, variability and fast adaptive evolution of the wine yeast (Saccharomyces cerevisiae) genome. Ann. Microbiol. 61, 85–93. 10.1007/s13213-010-0086-4 [DOI] [Google Scholar]
- Sipiczki M., Pfliegler W. P., Holb I. J. (2013). Metschnikowia species share a pool of diverse rRNA genes differing in regions that determine hairpin-loop structures and evolve by reticulation. PLoS ONE 8:e67384. 10.1371/journal.pone.0067384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M., Romano P., Lipani G., Miklos I., Antunovics Z. (2001). Analysis of yeasts derived from natural fermentation in a Tokaj winery. Antonie Van Leeuwenhoek 79, 97–105. 10.1023/A:1010249408975 [DOI] [PubMed] [Google Scholar]
- Starmer W. T., Heed W. B., Miranda M., Miller M. W., Phaff H. J. (1976). The ecology of yeast flora associated with cactiphilic Drosophila and their host plants in the Sonoran desert. Microb. Ecol. 3, 11–30. 10.1007/BF02011450 [DOI] [PubMed] [Google Scholar]
- Stefanini I., Dapporto L., Legras J.-L., Antonio Calabretta A., Di Paola M., De Filippo C., et al. (2012). Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 13398–13403. 10.1073/pnas.1208362109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevic B. (1962). The importance of bees (Apis sp) and wasps (Vespa sp) as carrier of yeasts for the microflora of grapes and the quality of wines. Archiv Poljoprivredre Nauke Beograd 15, 80–91. [Google Scholar]
- Sun Y., Guo J., Liu F., Liu Y. (2014). Identification of indigenous yeast flora isolated from the five winegrape varieties harvested in Xiangning, China. Antonie Van Leeuwenhoek 105, 533–540. 10.1007/s10482-013-0105-0 [DOI] [PubMed] [Google Scholar]
- Taylor M. W., Tsai P., Anfang N., Ross H. A., Goddard M. R. (2014). Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 16, 2848–2858. 10.1111/1462-2920.12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treseder K. K., Lennon J. T. (2015). Fungal traits that drive ecosystem dynamics on land. Microbiol. Mol. Biol. Rev. 79, 243–262. 10.1128/MMBR.00001-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero E., Cambon B., Schuller D., Casal M., Dequin S. (2007). Biodiversity of Saccharomyces yeast strains from grape berries of wine producing areas using starter commercial yeasts. FEMS Yeast Res. 7, 317–329. 10.1111/j.1567-1364.2006.00161.x [DOI] [PubMed] [Google Scholar]
- Vaughan-Martini A., Martini A. (1995). Facts, myths and legends on the prime industrial microorganisms. J. Indus. Microbiol. 14, 514–522. 10.1007/BF01573967 [DOI] [PubMed] [Google Scholar]
- Yanagida F., Ichinose F., Shinohara T., Goto S. (1992). Distribution of wild yeasts in the white grape varieties at Central Japan. J. Gen. Appl. Microbiol. 38, 505–509. 10.2323/jgam.38.501 [DOI] [Google Scholar]
- Zahavi T., Droby S., Cohen L., Weiss B., Ben-Arie R. (2002). Characterisation of the yeast flora on the surface of grape berries in Israel. Vitis 41, 203–308. [Google Scholar]
- Zalar P., Gostinčar C., de Hoog G.S., Uršiè V., Sudhadham M., Gunde-Cimerman N. (2008). Redefinition of Aureobasidium pullulans and its varieties. Stud. Mycol. 61, 21–38. 10.3114/sim.2008.61.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zironi R., Romano P., Suzzi G., Battistutta F., Comi G. (1993). Volatile metabolites produced in wine by mixed and sequential cultures of Hanseniaspora guilliermondii or Kloeckera apiculata and Saccharomyces cerevisiae. Biotechnol. Lett. 15, 235–238. 10.1007/BF0012831117440687 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.