ABSTRACT
Triple-negative breast cancer (TNBC) is a highly aggressive and recurrent type of breast carcinoma that is associated with poor patient prognosis. Because of the limited efficacy of current treatments, new therapeutic strategies need to be developed. The CXCR4-CXCL12 chemokine signaling axis guides cell migration in physiological and pathological processes, including breast cancer metastasis. Although targeted therapies to inhibit the CXCR4-CXCL12 axis are under clinical experimentation, still no effective therapeutic approaches have been established to block CXCR4 in TNBC. To unravel the role of the CXCR4-CXCL12 axis in the formation of TNBC early metastases, we used the zebrafish xenograft model. Importantly, we demonstrate that cross-communication between the zebrafish and human ligands and receptors takes place and human tumor cells expressing CXCR4 initiate early metastatic events by sensing zebrafish cognate ligands at the metastatic site. Taking advantage of the conserved intercommunication between human tumor cells and the zebrafish host, we blocked TNBC early metastatic events by chemical and genetic inhibition of CXCR4 signaling. We used IT1t, a potent CXCR4 antagonist, and show for the first time its promising anti-tumor effects. In conclusion, we confirm the validity of the zebrafish as a xenotransplantation model and propose a pharmacological approach to target CXCR4 in TNBC.
KEY WORDS: CXCR4, CXCL12, IT1t, Triple-negative breast cancer, Metastases, Inter-species crosstalk, Xenograft, Zebrafish
Summary: CXCR4-expressing human tumor cells respond to zebrafish cognate ligands and initiate metastatic events in a zebrafish xenograft model. The CXCR4 antagonist IT1t has promising tumor inhibitory effects.
INTRODUCTION
CXCR4 is a chemokine receptor, first described in the early 1990s (Baggiolini et al., 1997; Herzog et al., 1993; Jazin et al., 1993; Nomura et al., 1993) and identified as a co-receptor for human immunodeficiency virus (HIV) entry (Feng et al., 1996). It is a seven-transmembrane G-protein-coupled receptor with a major role in physiological processes such as hematopoiesis (Nagasawa et al., 1996; Rosu-Myles et al., 2000), leukocyte trafficking (Day and Link, 2012; Sallusto and Baggiolini, 2008; Sallusto et al., 2000), cell migration and organogenesis during ontogeny (Bussmann and Raz, 2015), as well as pathological conditions like HIV pathogenesis (Vicenzi et al., 2013), WHIM syndrome (warts, hypogammaglobulinemia, infections and myelokathexis syndrome) (Gulino, 2003) and cancer (Balkwill, 2004; Chatterjee et al., 2014). Its cognate ligand is the homeostatic cytokine CXCL12 (Bleul et al., 1996; Oberlin et al., 1996) [also known as stromal cell-derived factor 1 (SDF-1)]. However, recent reports indicate that ubiquitin and macrophage migration inhibitory factor (MIF) can also bind to and signal through CXCR4 (Saini et al., 2010; Bernhagen et al., 2007; Saini et al., 2011; Pawig et al., 2015). Upon CXCL12 binding, CXCR4 triggers cell migration, proliferation and transcriptional control of downstream targets via G-protein-dependent or -independent mechanisms (Pawig et al., 2015). Dissociation of Gα and Gβγ subunits leads to Ca2+ release and activation of the PI3K/AKT and MAPK signaling pathways (Teicher and Fricker, 2010). CXCR4 dimerization occurs after ligand binding; subsequently, phosphorylation by JAK kinases takes place, followed by STAT signaling initiation in a G-protein-independent mechanism (Mellado et al., 2001; Vila-Coro et al., 1999). Moreover, initiation of β-arrestin signaling can take place, resulting in G-protein coupled receptor (GPCR) signaling blockade (Lefkowitz, 1998; Shukla et al., 2011) or ERK1/2 activation (Luttrell et al., 1999). CXCR4 activity is regulated by mechanisms of desensitization, through phosphorylation of the C-terminus and internalization, which is followed by degradation or recycling to the plasma membrane (Busillo and Benovic, 2007). Moreover, CXCL12 binds also to CXCR7 (Balabanian et al., 2005). However, differently from other chemokine receptors, CXCR7 does not signal through G proteins and acts as a ligand scavenger in a β-arrestin-mediated pathway (Rajagopal et al., 2010). Interestingly, a key role of the CXCL12-CXCR4-CXCR7 axis in collective tissue migration has been studied in zebrafish embryos. In the migration of the lateral line primordium, Cxcl12 scavenging by Cxcr7 leads to the formation of a self-generated gradient and cell migration after Cxcr4 activation, along tissues where Cxcl12 is homogeneously distributed (Boldajipour et al., 2008; Dona et al., 2013; Venkiteswaran et al., 2013).
A link between CXCR4 and cancer, in particular metastatic breast cancer, has been reported (Muller et al., 2001). CXCR4-expressing tumor cells preferentially colonize distant organs that secrete high levels of CXCL12, such as brain, lungs, lymph nodes, liver and bone marrow (Janowski, 2009). Among highly aggressive malignancies of the breast, triple-negative breast cancer (TNBC) is often associated with relapse and poor patient prognosis (Palma et al., 2015; Podo et al., 2010). Conventional hormone-based therapies are not applicable owing to the absence of expression of the estrogen and progesterone receptors, and Her2 gene amplification (Anders and Carey, 2009). Accordingly, surgery and chemotherapy are the main form of medical intervention and no targeted therapies are currently available (Wahba and El-Hadaad, 2015). Therefore, a better understanding of the biology of this aggressive breast carcinoma and the development of new therapies to reduce the high mortality rate are urgently needed. The bicyclam AMD3100, also known as plerixafor, is a CXCR4 antagonist and has been introduced in clinical trials to treat different tumor types, mainly leukemia and lymphomas (Ramsey and McAlpine, 2013). However, it has been reported to cause cardiotoxicity (Hendrix et al., 2004). AMD3100 also functions as an agonist for CXCR7 (Kalatskaya et al., 2009), which has been linked to breast cancer cell proliferation (Salazar et al., 2014). In addition, an anti-CXCR4 antibody is currently in clinical trials (Kuhne et al., 2013; Vela et al., 2015). More CXCR4 antagonists have been developed and in vitro as well as animal models are required to further explore clinical applications in patients.
Zebrafish is increasingly being used as an animal model for translational research in oncology (Amatruda et al., 2002; Barriuso et al., 2015; Ghotra et al., 2015). In particular, transparent zebrafish embryos allow following the behavior of fluorescent tumor cells in a living organism. Human cancer cells engrafted in the blood circulation of 2-day-old transgenic embryos, with fluorescently traceable endothelial (Lawson and Weinstein, 2002) and immune cells (Ellett et al., 2011; Renshaw et al., 2006), have been described to induce angiogenesis and form micrometastases in concert with immune cell interaction (He et al., 2012). Tumor angiogenesis and colonization of secondary tissues can be visualized in a short time period (2-6 days) in the small and fast-developing larvae. Although numerous discoveries have been made using zebrafish embryos as a xenotransplantation model, lack of knowledge about the communication between human and zebrafish cells has questioned its validity and partially limited its use.
Here, we report that the CXCR4-CXCL12 axis acts across zebrafish and humans and drives the formation of tumor micrometastases of human TNBC cells in zebrafish. Cell treatment with IT1t, a potent CXCR4 antagonist, and genetic silencing of CXCR4 effectively inhibited early metastatic events in vivo. Therefore, using zebrafish as a xenotransplantation model, we propose a potential treatment to impair CXCR4 signaling and reduce the metastatic onset of TNBC.
RESULTS
TNBC cells display high CXCR4 expression levels and increased metastatic behavior in a zebrafish xenotransplantation model
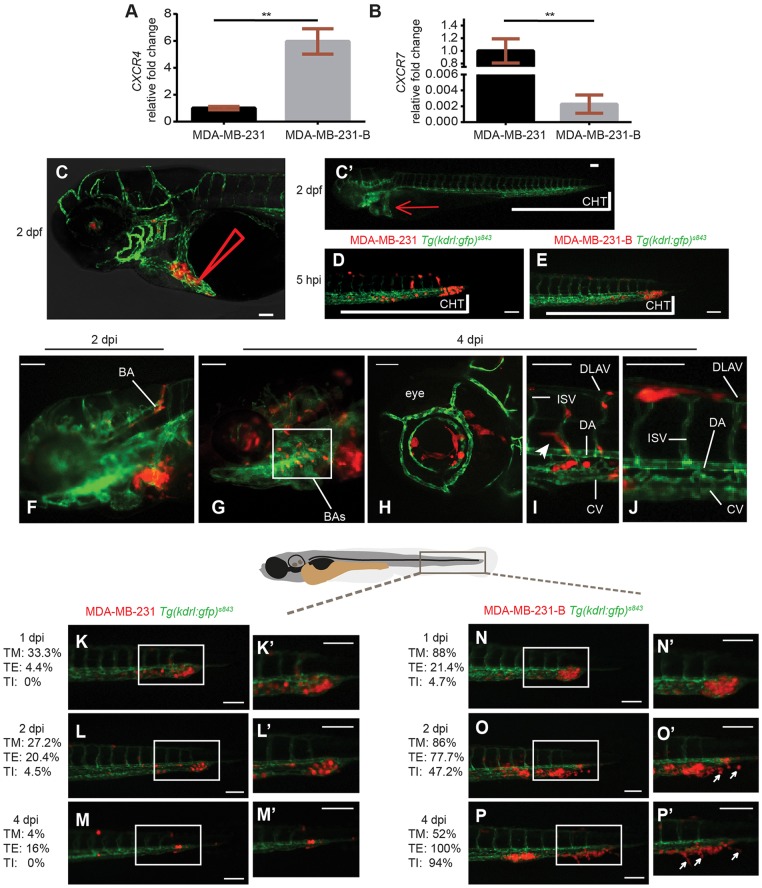
We first characterized the expression profile of CXCR4 and CXCR7, both chemokine receptors for CXCL12, in the TNBC line MDA-MB-231-B. Because this cell line derives from bone metastases of MDA-MB-231, after repeated engraftments into a murine host (Wetterwald et al., 2002), we used the parental line as a reference. We found that, compared to the original TNBC line, derived from human pleural metastases, the bone clone expressed higher CXCR4 and lower CXCR7 mRNA levels (Fig. 1A,B). Moreover, when compared in vitro, MDA-MB-231-B displayed a higher proliferation rate than the parental line (data not shown). To determine whether TNBC cells with increased CXCR4 expression displayed a different behavior, we engrafted both MDA-MB-231 and MDA-MB-231-B in zebrafish. As previously reported (He et al., 2012), tumor cells were inoculated in the blood circulation of 2-days post-fertilization (dpf) embryos via the duct of Cuvier, a vein plexus that opens into the heart (Fig. 1C,C′). Fluorescent tumor cells derived from both cell lines entered the blood vessels and, at 5 hours post-injection (hpi), they were mainly found in the tail and trunk vessels of the Tg(kdrl:EGFP)s843 zebrafish reporter line with green fluorescent vasculature (Fig. 1D,E). Injected embryos were examined by microscopy and embryos with 25-50 tumor cells hematogenously disseminating into the dorsal aorta (DA), caudal vein (CV) and vessel branches of the caudal hematopoietic tissue (CHT), in the region between the urogenital opening and the end of the tail, were selected for the experiment. Tumor cells spread through the embryo via blood circulation of the head, trunk and tail. Intravascular and perivascular cancer cells were found in the basilar artery (BA), branchial arches (BAs) and optic vessels in the head region (Fig. 1F-H), and in intersegmental vessels (ISVs), dorsal longitudinal anastomotic vessels (DLAVs) and the DA and CV in both the trunk and tail areas (Fig. 1I,J). Moreover, tumor cells were often positioned near vessel branching points (Fig. 1I), as to follow a path in a similar fashion to nascent lymphatic vessels, known to express cxcr4a/b receptors (Cha et al., 2012). Interestingly, cxcl12a and cxcl12b are expressed at these sites in developing zebrafish embryos (Cha et al., 2012; Fujita et al., 2011; Hess and Boehm, 2012). Highly aggressive cancer cells, adhering to the intravascular endothelium, initiated early metastatic events in the tail, sustaining tumor progression until 4-days post-implantation (dpi). In our model, in which tumor cells are inoculated directly into the blood circulation to study the formation of experimental micrometastases, bypassing initial modifications in a primary tumor mass, early metastatic events coincided with tumor foci formation and expansion, tumor extravasation, with adherence to the extravascular endothelium, and invasion. In line with previous work from our group, the tail fin region, in proximity of the CHT, a temporary site of hematopoiesis analogous to the fetal liver in mammalian development, was a preferential early metastatic site (He et al., 2012).
Fig. 1.
CXCR4 expression levels correlate with metastatic potential in a zebrafish xenotransplantation model. The bone clone (MDA-MB-231-B) expressed higher levels of CXCR4 mRNA (A) and lower levels of CXCR7 mRNA (B), compared to the parental cell line MDA-MB-231, originated from metastatic triple-negative breast cancer (TNBC) [unpaired t-test, (A) **P=0.0016, (B) **P=0.0019]. Upon engraftment into the duct of Cuvier (C,C′ red arrow) of 2-dpf zebrafish embryos (C′), MDA-MB-231 circulated in the vascular system (D), in a comparable manner to MDA-MB-231-B (E). Arrowhead in C represents the site of injection. CHT, caudal hematopoietic tissue. (F-J) Tumor cells disseminated throughout the embryo, in the head (F-H), the eye (H), the trunk and the tail (I,J), and extended filopodia at vessel branching points (I, arrowhead). BA, basilar artery; BAs, branchial arches; CV, caudal vein; DA, dorsal aorta; DLAV, dorsal longitudinal anastomotic vessel; ISV, intersegmental vessel. (K-P′) Over time, a weaker phenotype was detectable for the MDA-MB-231 cell line (K-M,K′-M′), whereas evident secondary tumor mass formation, extravasation and tail fin invasion persisted when MDA-MB-231-B cells were implanted (N-P,N′-P′). Arrows in O′ and P′ indicate invasive cancer cells that are not in contact with the endothelium and are found in the tail fin tissue, after extravasation. Images were acquired using a Leica TCS SPE confocal microscope with an HC PL FLUOTAR 10× DRY objective (0.30 N.A.) in panel C and with an HC APO 20× DRY objective (0.7 N.A.) in panel H. All other images were acquired using a Leica MZ16FA fluorescent microscope coupled to a DFC420C camera. Scale bars: 50 µm. Phenotype assessment was carried out at 1, 2 and 4 dpi, evaluating the ability of both cell lines to form a secondary tumor mass, to extravasate and to invade the surrounding tail fin. Images are representative of embryos injected with MDA-MB-231 and number of individuals was n=51 (5 hpi) (D), 45 (1 dpi) (K), 44 (2 dpi) (L) and 25 (4 dpi) (M) or with MDA-MB-231-B and number of individuals was n=44 (5 hpi) (E), 42 (1 dpi) (N), 36 (2 dpi) (O) and 34 (4 dpi) (P). Percentages relative to tumor mass (TM), tumor extravasation (TE) and tumor invasion (TI) are reported for each stage, for both MDA-MB-231 and MDA-MB-231-B cell lines.
After reaching the vascular plexus that infiltrates the CHT, MDA-MB-231 and MDA-MB-231-B displayed divergent phenotypes. From 1 until 4 dpi, the parental line MDA-MB-231 showed a weakened behavior over time (Fig. 1K-M,K′-M′), whereas the bone clone induced increasingly aggressive phenotypes (Fig. 1N-P,N′-P′). The formation of a secondary tumor mass began at 1 dpi and was observed in 88% of the embryos engrafted with MDA-MB-231-B (n>40) (Fig. 1N,N′). In the MDA-MB-231 group, secondary tumors could be observed in 33.3% of the embryos (n>40) (Fig. 1K,K′). MDA-MB-231-B cells, with higher CXCR4 expression, were found to progressively extravasate (from 21.4% at 1 dpi to 100% at 4 dpi), as well as to increasingly invade the surrounding tissue of the tail (from 4.7% at 1 dpi to 94% at 4 dpi) (Fig. 1N-P,N′-P′). Invasive cells were distinguished from extravasating cells once they were no longer in contact with the external wall of the endothelium and localized in the surrounding tail fin tissue (Fig. 1O′,P′). On the other hand, the MDA-MB-231 line, with lower CXCR4 mRNA levels, displayed maximum extravasation at 2 dpi (20.4%), with a reduction at 4 dpi (16%). Invasive events were detected at 2 dpi (4.5%), whereas no invading cells were found in the tail fin at 4 dpi (Fig. 1M,M′). In conclusion, formation of a compact tumor structure, cancer cell extravasation, and invasion of the CHT and tail fin tissues increased over time in MDA-MB-231-B and decreased in MDA-MB-231. Taken together, our data show that the TNBC cell line MDA-MB-231-B displays high CXCR4 expression and enhanced metastatic behavior in the zebrafish embryo.
The CXCR4-CXCL12 signaling axis is cross-activated in zebrafish and humans
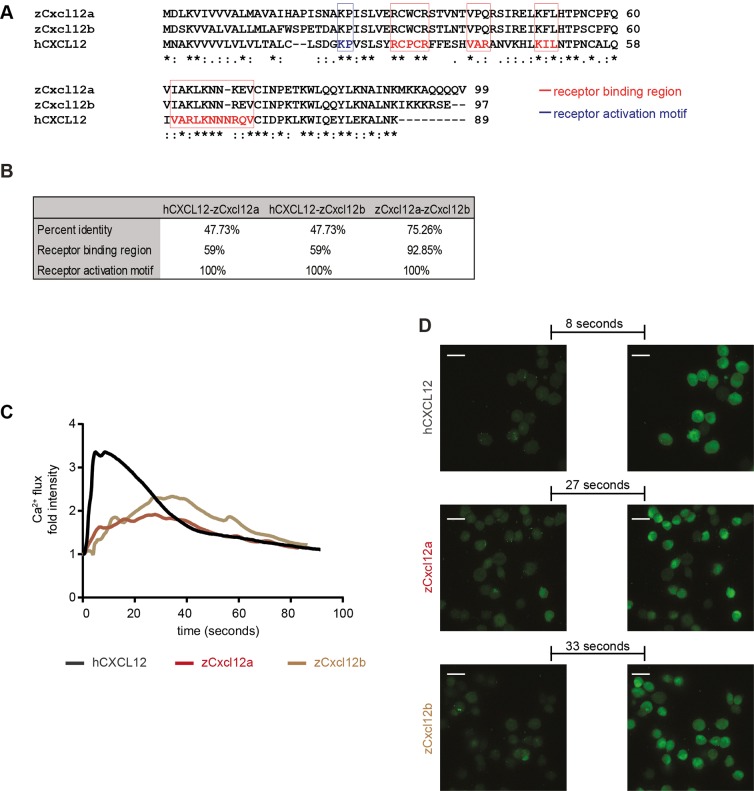
Cancer cells expressing CXCR4 form distant metastases in secondary organs that produce high levels of CXCL12, in human specimens and murine models (Muller et al., 2001). Our initial findings showed that TNBC cells initiating early metastatic events in the zebrafish xenotransplantation model express high levels of CXCR4. To establish whether CXCR4 sustained tumor metastatic properties in a Cxcl12-dependent manner, we first assessed whether the CXCR4-CXCL12 axis acts across zebrafish and human. Two cxcl12 genes, cxcl12a and cxcl12b, are found in zebrafish, following duplication events during teleost evolution. In a multiple alignment (Fig. 2A), human CXCL12 (α-isoform) shared 47.73% identical residues with both zebrafish Cxcl12a and Cxcl12b, whereas 75.26 was the percentage of identity between zebrafish Cxcl12 paralogs. Pair-wise sequence alignment showed 59% identity on residues involved in receptor binding, when the human ligand was compared to each zebrafish homolog, and 92.85% between Cxcl12a and Cxcl12b. Full identity in the motif involved in receptor activation was found (Fig. 2B). Therefore, considering the level of conservation, we verify that the CXCR4 signaling on human tumor cells is activated by both zebrafish Cxcl12 ligands. For this purpose, Ca2+ flux was measured. Whereas human CXCL12 (100 nM) failed to induce Ca2+ mobilization from intracellular storage into the cytoplasm in the parental line MDA-MB-231 (Fig. S1), time-lapse microscopy revealed that calcium sensor fluorescent signal intensity increased when MDA-MB-231-B cells were stimulated with either the human or the zebrafish ligands. In the bone clone, human CXCL12 elicited a response that increased and decreased rapidly. Zebrafish Cxcl12a and Cxcl12b triggered a slower but still significant response, in a non-synchronized fashion. In addition, the fluorescent signal gradually faded and failed to extinguish at once (Fig. 2C,D and Movies 1-3). Hence, we show that zebrafish Cxcl12 ligands trigger CXCR4 signal activation in human TNBC cells.
Fig. 2.
Zebrafish Cxcl12 ligands activate CXCR4 signaling in human cancer cells. Human CXCL12 was aligned to the zebrafish homologs using ClustalW (A). Amino acid residues were conserved in the receptor binding region and activation motif of CXCL12/Cxcl12 chemokines (A,B). Asterisks (*) represent fully conserved residues; colons (:) and periods (.) indicate positions at which residues share strong or weak similarity, respectively. (C,D) Human CXCL12-α and zebrafish ligands Cxcl12a and Cxcl12b induced calcium flux in MDA-MB-231-B cells, as detected by increased fluorescence intensity. The human ligand initiated an immediate response that extinguished rapidly (31 s to register half fluorescence intensity after the highest response), whereas the zebrafish ligands triggered a slower and prolonged signal induction (>55 s for zCxcl12a and >52 s for zCxcl12b to register half fluorescence intensity). (C) The fold intensity increase is calculated by normalization on fluorescence intensity correspondent to signal before response activation. (D) Frames show intensity before signaling activation was triggered by each ligand and at the highest peak of response. The time length to reach the strongest response is indicated.
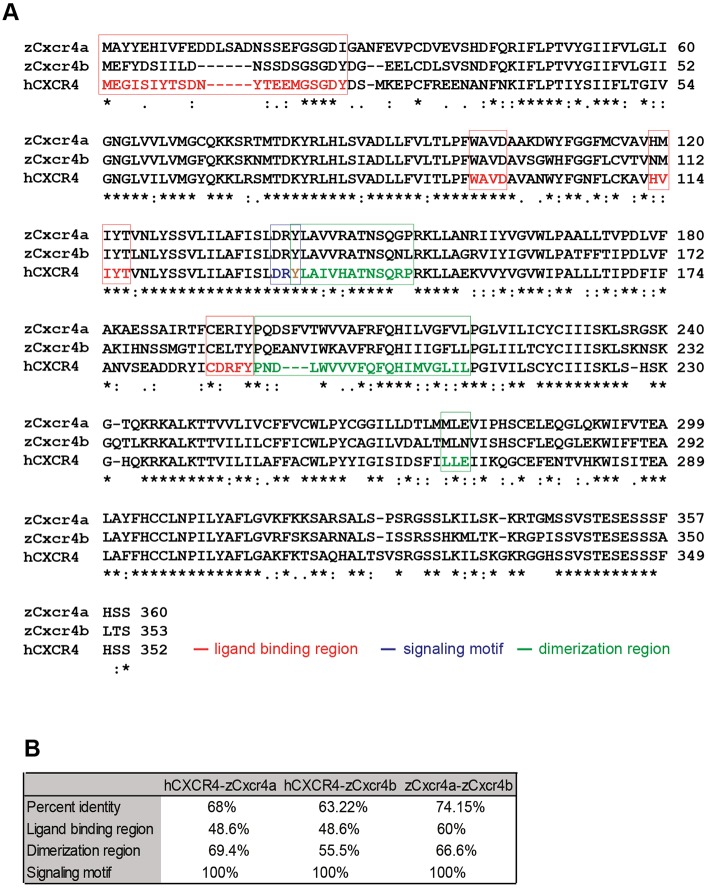
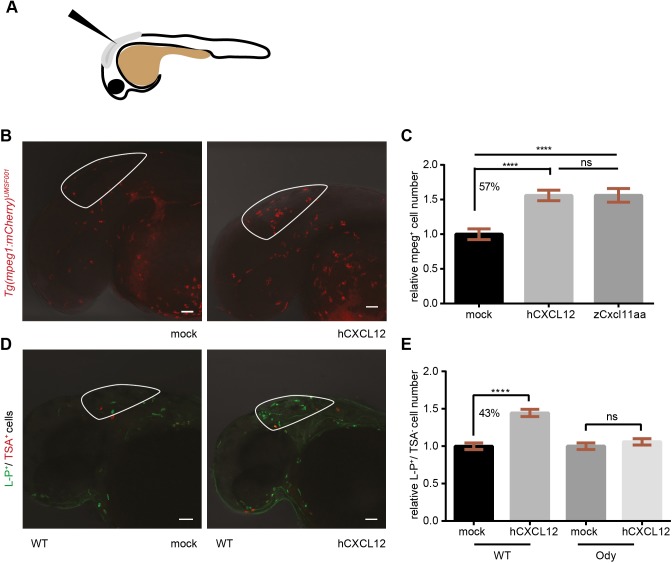
Next, we investigated whether human CXCL12 activates zebrafish Cxcr4. As for the Cxcl12 ligands, two Cxcr4 receptors have been described in zebrafish, Cxcr4a and Cxcr4b. ClustalW (Goujon et al., 2010; Larkin et al., 2007) alignments of the human CXCR4 with the zebrafish Cxcr4a and Cxcr4b (Fig. 3A) showed a percentage of identity equal to 68 and 63.22 on whole sequence, respectively (Fig. 3B). When ligand-binding regions were considered, the pair-wise identity reached 48.6% (CXCR4-Cxcr4a and CXCR4-Cxcr4b), whereas, in the dimerization regions, 69.4% (CXCR4-Cxcr4a) and 55.5% (CXCR4-Cxcr4b) of the residues were identical. Moreover, the signaling motif was 100% conserved (Fig. 3B). In addition, the zebrafish Cxcr4 paralogs displayed 74.15%, 60%, 66.6% and 100% identity at the whole-sequence level, ligand-binding and receptor-dimerization regions, and signaling motif, respectively (Fig. 3B). Besides partial redundancy, Cxcl12 and Cxcr4 zebrafish paralogs seemed to play distinct functions and to have a different spatial expression during embryo development, as reviewed (Bussmann and Raz, 2015). In particular, cxcr4b is found to be expressed by myeloid cells (Walters et al., 2010). To verify whether inter-species crosstalk exists between human ligands and zebrafish receptors, human recombinant CXCL12 (0.4 mg/ml) was injected in the hindbrain ventricle (HBV) of 30-32 hours post-fertilization (hpf) Tg(mpeg1:mCherry)UMSF001 embryos, where macrophages are fluorescently labeled (Fig. 4A). A 57% increase in the number of cells that migrated to the site of injection was observed compared to the mock-injected group, and in a similar fashion to the zebrafish chemokine Cxcl11aa, previously shown to be a chemoattractant for this class of phagocytes (Torraca et al., 2015) (Fig. 4B,C). Furthermore, macrophage motility towards the human CXCL12 (α-isoform) was found to be Cxcr4-dependent. Macrophages did not respond to the human CXCL12 in the ody mutant line, with a non-functional Cxcr4b receptor. Injection of the human ligand in the HBV led to a 43% increase in macrophage number compared to the water-injected group, in the wild-type (wt) siblings, whereas no differences in mean cell number was detected when CXCL12- and mock-injected groups were compared in the ody mutants (Fig. 4D,E). Moreover, we excluded a possible Cxcr4b-dependent alteration of basal motility and total macrophage number in ody mutants compared to wild-type siblings (Fig. S2). In conclusion, zebrafish is a valuable in vivo model to study human cancers, particularly focusing on the interaction between cancer and host stromal cells, because human attractants trigger zebrafish cell migration and zebrafish ligands are sensed by human cells.
Fig. 3.
CXCR4 alignment shows similarity between human and zebrafish proteins. Human CXCR4 was aligned to the zebrafish homologs using ClustalW (A). Asterisks (*) represent fully conserved residues, colons (:) and periods (.) indicate positions at which residues share strong or weak similarity, respectively. The tyrosine (Y) in brown belongs to both the signaling motif and dimerization region. Amino acid residues were conserved in the ligand-binding region, and dimerization and signaling motifs in the CXCR4/Cxcr4 receptors (B).
Fig. 4.
Human CXCL12 triggers zebrafish macrophage migration in a Cxcr4-dependent manner. (A) Scheme of a 30- to 32-hpf embryo and injection site are shown. (B,C) Zebrafish macrophages were found to be responsive to human CXCL12 (0.4 mg/ml) 3 h after injection into the hindbrain ventricle (HBV) of 30- to 32-hpf embryos. No increase in macrophage number occurred when a mock solution (water) was inoculated. Zebrafish Cxcl11aa (1.5 mg/ml) was used as a positive control (C). Data in C are pooled observations from two independent experiments (n=55 in mock; n=48 in hCXCL12; n=57 in zCxcl11aa). (D,E) Macrophages were recruited by human CXCL12 in a Cxcr4-dependent manner: a higher number (43%) of L-P+/TSA− cells was found in the HBV compared to the mock-injected group in wild-type (wt) siblings (D,E), whereas no differences were detected in the cxcr4b−/− (ody) mutants (E). In B, mCherry-expressing macrophages are recruited by hCXCL12 as in D, where L-P staining combined to TSA detection is used to distinguish macrophages (L-P+/TSA−) from neutrophils (L-P+/TSA+). ****P<0.0001, ns P>0.05 one-way ANOVA, Bonferroni post-hoc test. Data in E are pooled observations from five independent experiments (n=171 in mock/wt; n=180 in hCXCL12/wt; n=139 in mock/ody; n=160 in hCXCL12/ody).
Zebrafish Cxcl12-sensing by human CXCR4 receptor sustains TNBC cancer burden in zebrafish larvae
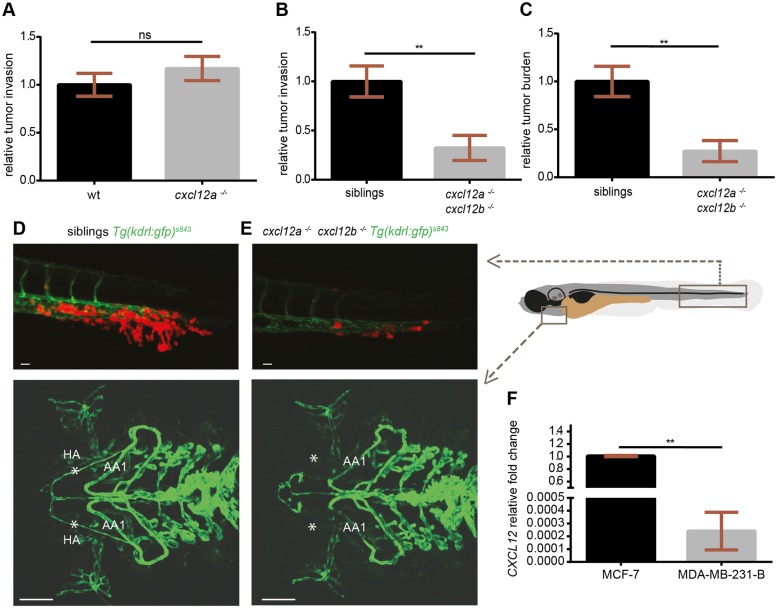
The tumor microenvironment plays a crucial role in the establishment of a favorable niche for the onset of cancer metastasis, and the CXCR4-CXCL12 axis, among other signaling pathways, guides the communication between tumor and microenvironment. We showed that crosstalk between human and zebrafish receptors and ligands (CXCR4/Cxcl12 and Cxcr4/CXCL12) occurs in vitro and in the zebrafish embryo model. Hence, we investigated whether CXCR4-expressing TNBC cells initiated early metastatic events in vivo after sensing zebrafish Cxcl12 ligands. MDA-MB-231-B cells were engrafted in the blood circulation of zebrafish embryos carrying null mutations in cxcl12a or cxcl12b. MDA-MB-231-B showed reduced localization in the head (BA and BAs) and trunk (ISVs) in larvae with a non-functional Cxcl12a (Fig. S3). However, tumor invasion developed similarly in cxcl12a−/−/Tg(kdrl:EGFP)s843 larvae and wt siblings at 4 dpi (Fig. 5A). The same effect was observed in the cxcl12b−/−/Tg(kdrl:EGFP)s843 mutants and wt siblings at 2 and 4 dpi (Fig. S4A). Like tumor invasion, also the overall micrometastasis burden in the tail fin was the same in wt, cxcl12a−/−/Tg(kdrl:EGFP)s843 and cxcl12b−/−/Tg(kdrl:EGFP)s843 at 2 dpi (Fig. S4B,C). However, the response of tumor cells in the Ca2+ assay to both zebrafish Cxcl12a and Cxcl12b supports the hypothesis that human tumor cells sense the Cxcl12a ligand in a cxcl12b mutant and the Cxcl12b ligand in a cxcl12a mutant. Therefore, in this scenario, tumor invasion and tumor burden could still occur in each single-ligand mutant line. Hence, the xenogeneic implantation was performed in the cxcl12a−/−/cxc12b−/−/Tg(kdrl:EGFP)s843 double-mutant embryos. For this purpose, the cxcl12a−/−/cxc12b+/−/Tg(kdrl:EGFP)s843 family was in-crossed and tumor engraftments were performed in the siblings of the F1 generation (experimental groups were blinded). Tumor burden and tumor invasion were significantly decreased in the double mutants compared to cxcl12a−/−/cxc12b+/−/Tg(kdrl:EGFP)s843 and cxcl12a−/−/cxc12b+/+/Tg(kdrl:EGFP)s843 siblings at 4 dpi (Fig. 5B,C and D,E, top panels). Mutant larvae for both ligands were distinguished from the siblings by screening for abnormal formation of the hypobranchial arteries at 6 dpf (Fig. 5D,E, bottom panels). Therefore, we suggest that the CXCR4-CXCL12 axis functions in a paracrine fashion across species and is responsible for driving the formation of TNBC micrometastases in zebrafish. Importantly, a potential role of the human CXCL12 autocrine loop in driving the formation of TNBC micrometastases in vivo is unlikely. CXCL12 expression levels were undetectable in the parental line MDA-MB-231 (data not shown) and significantly lower in MDA-MB-231-B compared to the Luminal A (ER+, PR+/−, Her2−) MCF-7 breast cancer cell line (Fig. 5F). Accordingly, a very low expression of CXCL12 in MDA-MB-231-B argued against the generation of a potential autocrine loop to activate the receptor CXCR4. Taken together, the onset of early metastatic events in this experimental system is enhanced by the CXCR4-CXCL12 axis in a zebrafish xenotransplantation model in which human tumor cells respond to zebrafish ligands.
Fig. 5.
CXCR4-expressing TNBC cells fail to initiate metastatic events in cxcl12a- and cxcl12b-null zebrafish mutants. (A) No differences in tumor cell invasion were found in the medusa (cxcl12a−/−) mutants, compared to wild-type (wt) siblings. (B,C) Breast cancer cells failed to form micrometastases in 4-dpi zebrafish embryos deficient for both cxcl12a and cxcl12b ligands, whereas tumor invasion (B) and tumor burden (C) occurred in the cxcl12a−/−/cxcl12b+/+ and cxcl12a−/−/cxcl12b+/− siblings. Unpaired t-test: (A) ns, P>0.05 (wt: n=73; medusa: n=66), (B) **P=0.0012 and (C) **P=0.0033 (n=11 in each group). Graphs in A-C are cumulative of two independent experiments. (D,E) Top panels: MDA-MB-231-B breast cancer cells were highly invasive in the siblings, whereas few tumor cells remained in the metastatic region in zebrafish that were mutant for both ligands. Images were acquired using a Leica MZ16FA fluorescent microscope coupled to a DFC420C camera. Bottom panels: vessel connections are compared to distinguish siblings and double mutants: the hypobranchial arteries (HA), indicated by asterisks, failed to connect to the mandibular arch (AA1) in the cxcl12a−/−/cxcl12b−/− larvae. Images were acquired using a Leica TCS SPE confocal microscope with an HC PL FLUOTAR 10× DRY objective (0.30 N.A.). Scale bars: 50 µm. (F) CXCL12 expression is lower in the MDA-MB-231-B compared to MCF-7 breast cancer cell line. Unpaired t-test, with Welch's correction. **P=0.006. qPCR was performed on two biological replicates.
The CXCR4 antagonist IT1t reduces the formation of TNBC early metastases in vivo
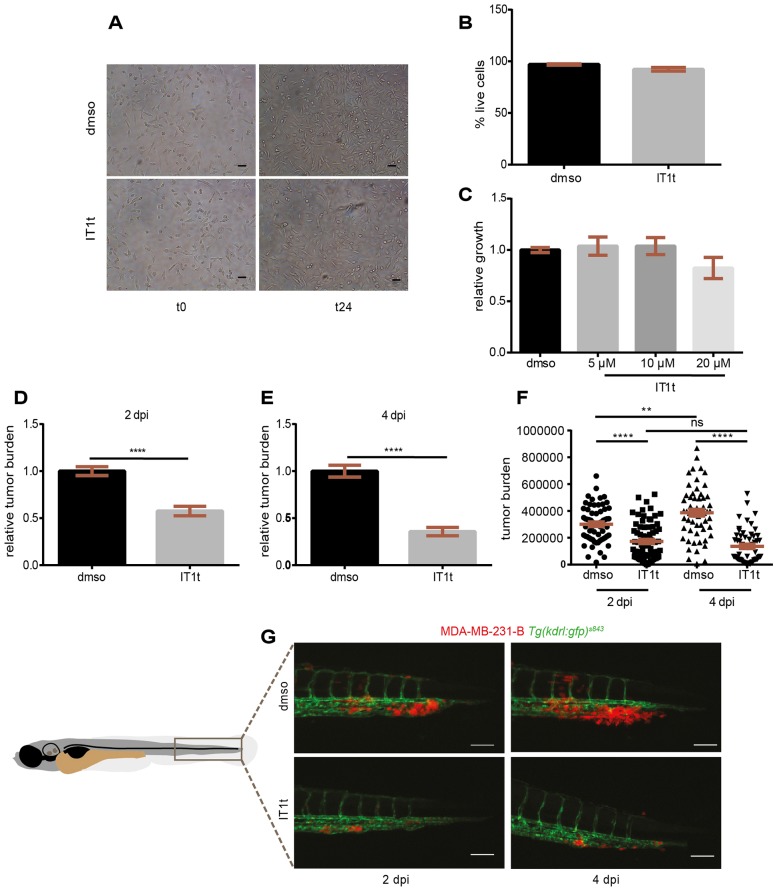
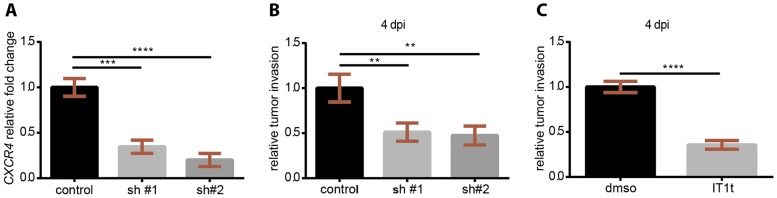
The use of CXCR4 antagonists as a therapeutic targeted approach to inhibit tumor spreading and the formation of metastases has been introduced in clinical trials for different cancer types. Despite the fact that CXCR4 is highly involved in the establishment of secondary neoplasias, there are no approved FDA drugs to block CXCR4 in TNBC. In the zebrafish xenograft model in which CXCR4-Cxcl12 inter-species communication supports TNBC early metastasis onset, we test whether the isothiourea derivative IT1t, a recently described CXCR4 antagonist (Thoma et al., 2008), displays anti-neoplastic functions. This small molecule is an orthosteric competitor of the CXCL12 N-terminal signaling peptide and it impairs signaling activation by interfering with the docking of the ligand domain to the receptor (Wu et al., 2010). MDA-MB-231-B cells were treated in vitro for 24 h and subsequently engrafted in zebrafish embryos. Cells proliferated in treated (20 μM) and untreated conditions (Fig. 6A). Cell survival was not significantly changed: the percentage of live cells was found to be comparable in both groups (97% DMSO, 92% IT1t) (Fig. 6B). To monitor cell viability, a WST-1 (tetrazolium salt) proliferation assay was performed. After a 24 h incubation period with increasing concentrations of IT1t (5, 10 and 20 µM), cancer cell metabolic activity was not changed when compared to vehicle control (Fig. 6C). After pre-treatment (20 µM), engraftment of cells in the blood circulation of 2-dpf zebrafish embryos was performed and tumor burden assessed at the metastatic site at 2 and 4 dpi (Fig. 6D-G). CXCR4 chemical inhibition affected tumor burden, with a 39.5% and 60% reduction at 2 and 4 dpi, respectively (Fig. 6D,E). An increase in tumor burden was found from 2 to 4 dpi for MDA-231-B pre-treated with DMSO, whereas no difference was detected in the IT1t group (Fig. 6F). At 2 dpi, TNBC cells associated to form a secondary mass inside the CV and invaded the tail fin, forming micrometastases at 4 dpi (Fig. 6G, top panel). Blocking CXCR4 in vitro impaired tumor mass formation in vivo: few cells remained in the CV at 2 dpi and consequently minor invasive events occurred at 4 dpi (Fig. 6G, bottom panel). To phenocopy the suppressive effect of CXCR4 pharmacological inhibition on tumor aggressiveness, we used RNA interference. Stable CXCR4 knockdown was achieved via lentiviral transduction of two independent CXCR4 short hairpin RNAs (shRNAs). We confirmed CXCR4 silencing on a gene expression level, via quantitative PCR (qPCR). Notably, CXCR4 mRNA levels were decreased in sh#1 and sh#2 compared to scrambled control shRNA, showing a knockdown efficiency of 66% and 80%, respectively (Fig. 7A). Subsequently, xenograft experiments were performed. Tumor cell invasion at the metastatic site was effectively reduced upon CXCR4 silencing (Fig. 7B), similar to the antagonist IT1t (Fig. 7C). Therefore, using chemical and genetic approaches, we demonstrate that CXCR4 signaling inhibition reduces the formation of TNBC early metastases in vivo and describe IT1t as a potential therapeutic for metastatic TNBC.
Fig. 6.
The CXCR4 antagonist IT1t reduces metastatic tumor burden in vivo. IT1t (20 µM) was applied into the cell medium for 24 h prior to engraftment in zebrafish embryos to antagonize CXCR4 receptor activation (A). The percentage of live cells after treatment was not significantly different than in the control condition (three independent experiments) (B). The metabolic activity, readout of cell growth, was not significantly affected when increasing concentrations of IT1t were used (C). Pre-treatment in vitro caused a reduction in tumor burden at the secondary site in vivo, both at 2 dpi (D) and 4 dpi (E). Cancer cell burden increased over time from 2 to 4 dpi in the control group, whereas it remained at comparable levels upon treatment (F). Data set in F is obtained by using the same data points as shown in D and E. Number of larvae is n=64 (DMSO) and n=75 (IT1t) at 2 dpi and n=59 (DMSO) and n=56 (IT1t) at 4 dpi. (G) Effect of CXCR4 inhibition on tumor burden over time is shown. Scale bars: 50 µm. Data are mean±s.e.m. from two independent experiments. Statistical analysis: two-tailed, unpaired t-test and ANOVA with Bonferroni post-hoc test for datasets with two or more groups respectively. ****P<0.0001; **P=0.005; ns, P>0.05.
Fig. 7.
CXCR4 genetic impairment via RNA interference recapitulates chemical treatment effects on early metastatic events. (A) CXCR4 stable knockdown efficiency obtained via shRNA was 66% and 80% for the shRNA #1 (sh#1) and sh#2, respectively. A reduced tumor cell invasion was observed in cell lines carrying one of the CXCR4 targeting shRNAs (B) as well as upon pre-treatment before engraftment with the CXCR4 antagonist (C). (A,B) One-way ANOVA with Bonferroni post-hoc test: ****P<0.0001, ***P=0.0002, **P<0.01. (C) Un-paired t-test: ****P<0.0001.
DISCUSSION
Metastatic TNBC is a major challenge for biopharmaceutical and clinical research because tumor relapse and cell spreading represent the main cause of death for patients. The development of targeted therapies, in combination with conventional chemotherapy, is an important approach to prolong patient lifetime. Although steps forward in elucidating cancer dissemination have been made, generally the pathogenesis of metastases is not fully understood. Monitoring single tumor cells while crossing the blood vessel boundaries in vivo is an optimal scenario to unravel early metastatic events. For this purpose, zebrafish is an advantageous model. The transparency of the embryos and the use of reporter lines make the zebrafish an excellent host to study human tumor cell growth and invasion at early stages (Movie 4). In the last decade the zebrafish xenotransplantation model has been used to study human tumor progression (Konantz et al., 2012) and to discover potential treatments (van der Ent et al., 2014a,b; Veinotte et al., 2014; Zoni et al., 2015). However, the translational validity of a zebrafish xenotransplantation approach has been questioned. Concerns have emerged on possible inter-species crosstalk and lack of species-specific environmental cues. Here, we show that the cross communication between human tumor cells and zebrafish ligands is maintained, because zebrafish Cxcl12 activates human CXCR4 signaling in vitro and supports the formation of TNBC early metastases in vivo. We found that TNBC cells with high CXCR4 expression levels exhibit aggressive features in zebrafish, in agreement with findings in patients and other models. In addition to CXCR4 and CXCR7 mRNA levels, other differences in gene expression between MDA-MB-231-B and MDA-MB-231 might be present and influence tumor cell behavior. Notably, we proved that CXCR4-linked tumor burden occurred in a zebrafish Cxcl12-dependent manner. The involvement of the human CXCL12 ligand is questioned owing to very low expression levels, in line with the evidence that CXCL12 expression is higher in non-metastatic breast tumors, compared to metastatic ones (Zhao et al., 2014). However, CXCR4 activation by autocrine mechanisms cannot be fully excluded and CXCL12 knockdown is required to completely rule out this possibility. Moreover, Cxcr4-expressing macrophages migrate towards human CXCL12, demonstrating that the intercommunication takes place in both directions and confirming the validity of the zebrafish embryo model to study human tumors.
Using the zebrafish xenograft model, we observed that TNBC cells make contact with the endothelium, after inoculation in the blood circulation. Then, tumor-endothelium interaction favors tumor mass formation and, consequently, tumor extravasation and invasion. Interestingly, the invasive process frequently recurs when tumor mass formation and growth are observed at earlier stages, whereas a minimized or absent invasion takes place if no tumor mass is present. This scenario is in accordance with the hypothesis, proposed by Ewing in 1929 (Ewing, 1929), that there is a link between metastasis and organization of the vascular system, stressing the mechanical nature of cancer homing to different sites in the body. At the same time, we could not observe tumor aggregates and invasion phenomena in other tissues along the trunk that are also perfused by the DA and the CV in the zebrafish larvae. The aggressive tumor phenotype occurred mainly in the CHT, a site of hematopoiesis in zebrafish larvae. This is in line with the frequent presence of TNBC metastases in the bone marrow of adult mammals (Shi et al., 2014) and correlates with Cxcl12 ligand expression in zebrafish. In the zebrafish tail region, cxcl12a is generally expressed in the CHT (Walters et al., 2010), and expression of cxcl12a and cxcl12b is normally found in the endothelium of the CV and DA, respectively (Cha et al., 2012). Perhaps both Cxcl12a and Cxcl12b work in concert in sustaining tumor adhesion to the endothelium and consequent tumor burden. Using the zebrafish model, we propose that receptor activation via ligand stimulation and not necessarily in response to a Cxcl12 gradient enhance tumor burden and subsequent invasion. This observation is in line with the ‘seed and soil’ theory proposed by Paget in 1889 (Paget, 1989): tumor cells, the ‘seed’, form a secondary mass in a growth-supportive microenvironment or ‘fertile soil’. Moreover, the preferential growth and invasion in the CHT partially explain why the bone clone and not the parental line MDA-MB-231, more commonly used in other animal models, showed aggressive features in the zebrafish embryo. In conclusion, our data are in agreement with previous theories (Fidler, 2003) that both mechanical and microenvironment-related factors contribute to tumor mass formation and cell invasion to ultimately initiate the metastatic process.
Attempts in the clinic have been made to pharmaceutically interfere with CXCR4-CXCL12 signaling. AMD3100 (plerixafor), the most commonly used drug to inhibit the receptor CXCR4, is currently in clinical trials for glioma, leukemia, Ewing sarcoma, neuroblastoma and brain tumors and is already FDA-approved for Non-Hodgkin's lymphomas and multiple myeloma. However, long-term secondary effects and mobilization of hematopoietic stem cells have been registered. Alternatively, CXCL12 targeting agents are currently in clinical trials and under investigation (Scala, 2015). IT1t is an orally available isothiourea compound that antagonizes CXCR4 activation with high specificity and potency, as shown in the Ca2+ flux assay, inhibition of X4-HIV attachment and whole-blood actin polymerization assay in rats (Thoma et al., 2008). Moreover, residues involved in IT1t binding to CXCR4 are not conserved in CXCR7 (Yoshikawa et al., 2013). We showed for the first time that CXCR4 inhibition via IT1t results in a reduction of early metastasis formation in vivo. A reduction in tumor invasion as well as tumor mass formation was observed at 4 dpi in zebrafish larvae, after 24 h pre-incubation in vitro. We propose that the treatment affects ligand sensing in vivo, therefore affecting the ability of tumor cells to survive in the blood circulation, colonize the CHT, and to make contact with the endothelium to subsequently proliferate and extravasate, initiating early metastatic events. Cancer cell proliferation can occur inside blood vessels (Hanahan and Weinberg, 2011) and extravasation events are linked to tumor-cell–endothelium interaction (Stoletov et al., 2010). Moreover, highly adherent tumor cells have been reported to have stem-cell-like features (Bansal et al., 2014). Hence, it is not to be excluded that TNBC cells that initiate early metastatic events in our model have stem-like properties and might express high levels of CXCR4.
In conclusion, the zebrafish xenotransplantation model, in which inter-species crosstalk is maintained, has provided new insight into the metastatic events associated with TNBC and into the employment of a potential compound to limit CXCR4-dependent tumor early metastases.
MATERIALS AND METHODS
Zebrafish husbandry
Zebrafish lines were maintained according to standard protocols, described in zfin.org, and handled in accordance with the Dutch animal welfare regulations and the EU Animal Protection Directive 2010/63/EU.
Zebrafish lines
In the present study, the reporter lines Tg(kdrl:EGFP)s843 (Jin et al., 2005), Tg(mpeg1:mCherry)UMSF001 (Bernut et al., 2014) and Tg(mpeg1:EGFP)gl22 (Ellett et al., 2011) were used to monitor tumor cell behavior, macrophage recruitment and motility, respectively. Mutant lines used were cxcl12at30516 (medusa) (Valentin et al., 2007), cxc12bmu100 (Bussmann et al., 2011) and cxcr4bt26035 [odysseus (ody)] (Knaut et al., 2003). cxcl12at30516/t30516 (cxcl12a−/−) and cxcr4bt26035/t26035 (cxcr4b−/−) mutants were identified, before raising, for incomplete migration of the lateral line primordium at the larval stage. Adult fins were clipped and DNA extraction for genotyping was performed. Genotype identification was carried out using a KASP assay. The following primers were used: A1 (reverse) 5′-CTGTGTTGACTGTGGAACGGCAC-3′, A2 (reverse) 5′-CTGTGTTGACTGTGGAACGGCAT-3′ and C1 (forward) 5′-AGCCAAGCCCATCAGCCTGGTA-3′ for cxcl12a; A1 (forward) 5′-GTGCTGGTGTCGCTCCACC-3′, A2 (forward) 5′-GTGCTGGTGTCGCTCCACG-3′ and C1 (reverse) 5′-AACTTGATCTCTCGGATGCTCCGTT-3′ for cxcl12b; and A1 (reverse) 5′-TGACGGTGGTCTTCAGTGCCTT-3′, A2 (reverse) 5′-TGACGGTGGTCTTCAGTGCCTA-3′ and C1 (forward) 5′-CAAGAACTCCAAGGGTCAGACTCTA-3′ for cxcr4b. KASP assay results were confirmed by sequencing, using the following primers: 5′-AGGATGCTGTTCCGTTTTAC-3′ (forward) and 5′-TGTGTGTGTCTGACTAAGCA-3′ (reverse) for cxcl12a; and 5′-AAGCCCATCAGTCTGGTGGAGAGG-3′ (forward) and 5′-GTGCCCTTTGTCTGGTGTAACCTG-3′ (reverse) for cxcl12b. Primers for cxcl12b−/− (Bussmann et al., 2011) and cxcr4b−/− (Miyasaka et al., 2007) identification were previously described and used for sequencing. For experiments, cxcl12a−/−/cxc12b+/−/Tg(kdrl:EGFP)s843 were in-crossed and double mutants were identified based on impaired connection of the hypobranchial artery branches to the first aortic arch at 6 dpf. Cxcl12a−/− siblings, wild-type or heterozygote for cxcl12bmu100, were considered as control groups.
Cell culture
The breast cancer cell line MDA-MB-231 [American Type Culture Collection (ATCC)], the derived bone clone MDA-MB-231-B and MCF-7 were cultured in DMEM complemented with 10% fetal calf serum (FCS) and grown at 37°C and 5% CO2. pLenti-tdtomato plasmid was introduced via lentiviral transduction in MDA-MB-231, whereas MDA-MB-231-B cells stably expressed dsRed fluorescent protein. Blasticidin and G418 were used to select cell clones that were tdtomato- or dsRed-positive, respectively. Cell lines were regularly tested for mycoplasma with the Universal Mycoplasma Detection Kit (30-1012k, ATCC).
Proliferation assay
MDA-231-B tumor cells were treated with increasing concentrations (5, 10 and 20 μM) of CXCR4 antagonist IT1t (239821, Calbiochem) to assess cell growth. A total of 30,000 cells were seeded in a single well of a 96-well plate. The following day, inhibitor treatment was carried on for 24 h. WST-1 tetrazolium salt (05015944001, Roche) was added into the cell medium and absorbance of the reduced by-product was measured to quantify cell viability.
RNA interference
CXCR4 stable knockdown in MDA-MB-231-B was obtained using shRNA containing constructs derived from Sigma MISSION library [TRCN0000004054 (or #1): 5′-CTTTGTCATCACGCTTCCC-3′ and TRCN0000004056 (or #2): 5′-GAATCACGTAAAGCTAGAA-3′]. Lentivirus virions were produced by transfecting HEK293T cells with pKLO1-puro plasmid (containing the CXCR4-targeting shRNA or a non-mammalian shRNA control), pCMV-VSV-G (envelope plasmid), pMDLg-RRE (gag and pol elements) and pRSV-REV (rev or HIV1gp6) as packaging vectors. Plasmid mix was added to cell medium together with CaCl2 and incubated for 18-20 h. Virus-containing supernatant was collected 48 and 72 h post-transfection and virus concentration was measured using Lenti-X™ p24 Rapid Titer Kit (Clontech). For transduction, MDA-MB-231-B cells were seeded in a 24-well plate (25,000 cells/well) and lentiviruses [1-3 multiplicity of infection (MOI)] added together with polybrene overnight (O.N.). Cells were cultured with complete medium containing 1 µg/ml puromycin for four passages and samples were collected for RNA isolation.
RNA isolation, cDNA synthesis and qPCR
RNA was isolated using a High Pure RNA Isolation Kit (Roche). After DNAase treatment, complementary DNA (cDNA) synthesis was performed (i-Script™ cDNA Synthesis Kit, Bio-Rad) and expression levels were measured by qPCR (iQ™ SYBR® Green Supermix, Bio-Rad). Relative fold changes of gene expression were calculated using the ΔΔCt method. CXCR4 primers (Fw: 5′-CAGCAGGTAGCAAAGTGACG-3′; Rv: 5′-GTAGATGGTGGGCAGGAAGA-3′; amplicon size: 150 bp) were kindly provided by Dr S. B. Geutskens (LUMC, Leiden, The Netherlands) and CXCL12 (Fw: 5′-CACATCTAACCTCATCTTC-3′; Rv: 5′-GACTTACTCTTCACATAGC-3′; amplicon size: 180 bp) primers were described in Costantini et al. (2013). GAPDH was used as housekeeping gene (Fw: 5′-AATCCCATCACCATGTTCCA-3′; Rv: 5′-TGGACTCCACGACGTACTCA-3′; amplicon size: 160 bp) (van der Vaart et al., 2014).
Ca2+ flux assay
MDA-MB-231-B and MDA-MB-231 cells (5×104 to 1×105) were seeded in uncoated µ-Dish35mm ibidi dishes (81156, ibidi) to adhere overnight. Adherent cells forming a 50% confluent monolayer were pre-incubated for 30 min at 37°C with 1-10 µM cell permeant calcium sensor Fluo-4, AM (F14217, Invitrogen). Cells were kept in Dulbecco's phosphate buffered saline (DPBS) until and during imaging. Ca2+ flux was measured upon stimulation with recombinant human CXCL12-α (300-28A, Peprotech) (100-300 nM), zebrafish CXCL12a (500 nM) or CXCL12b (100 nM) ligands (Boldajipour et al., 2011) by fluorescent signal imaging, using an Axiovert200 microscope (Zeiss, Germany) combined with a spinning disk unit (CSU-X1, Yokogawa, Japan) and a CCD camera (iXon 897, Andor, UK). Time-lapse imaging was performed using a 20× objective with a laser illumination at 488 nm (Crystal), at a 200 ms or 1 s time interval. Image analysis was performed using self-written algorithms in MatLab (MathWorks Inc., USA).
Inoculum preparation for engraftment and xenotransplantation
Cell suspension was prepared once cells had grown to a 70-80% confluent monolayer. After detachment with trypsin-EDTA (30-2101, ATCC®), tumor cells were washed once in complete medium and twice in DPBS (GIBCO® by Life Technologies). Centrifugation steps were performed for 5 min at 200 g (Eppendorf 5702). 2% PVP40 (polyvinylpyrrolidone-40) was used for the final cell suspension. 2-day-old zebrafish embryos, manually dechorionated and treated with 0.003% PTU (1-phenyl-2-thiourea, Sigma-Aldrich) at 24 hpf, were anesthetized using 0.02% Tricaine (MS-222) and transferred in a Petri dish with a 1.5% agarose coating layer. Cell suspension was loaded in a glass capillary, prepared using a Flaming/Brown micropipette puller (model P-97, Sutter Instrument Co.). Forceps were used to cut the end of the needle and injection tests were performed in a drop of water to set pressure and time parameters, in order to engraft 300-500 cells. Tumor cells were inoculated in the blood circulation, via the duct of Cuvier. Engrafted embryos were transferred to a new Petri dish and kept at 34°C. Embryos were checked 3- to 5-h post implantation (hpi) for correct engraftment and the ones showing tumor cells in the blood circulation were selected for experiments.
Microscopy and phenotype assessment in vivo
Tumor burden of early metastases, extravasation and tissue invasion were assessed via imaging of the tail fin region, in proximity of the CHT, at 2 and 4 dpi. In order to quantify tumor burden, single-embryo pictures were acquired. Cell aggregates inside the blood vessels, as well as extravasating and invading single cells, were included in the analysis of tumor burden and comprehended in the definition of early metastases and micrometastases. Tumor cell extravasation and invasion were also quantified separately from intravascular tumor mass by counting the number of cells per embryo and acquiring representative micrographs. A Leica MZ16FA fluorescent microscope coupled to a DFC420C camera was used. GFP and dsRed channels were overlaid in LAS AF Lite software and snapshots were analyzed in Image-Pro Analyzer 7.0 (Media Cybernetics). For each larva, tumor burden was calculated based on the number of objects multiplied by mean area and mean intensity, generated with a macro designed by H. de Bont (Toxicology, LACDR, Leiden University) and previously used to quantify tumor migration and proliferation (Ghotra et al., 2012; van der Ent et al., 2014b). In each micrograph, larvae are shown in lateral view and oriented head (left)-to-tail (right), as shown by representative cartoons.
Chemokine injection, L-P staining and in silico analysis
Human CXCL12-α (0.4 mg/ml) (300-28A, Peprotech) or zebrafish Cxcl11aa (1.5 mg/ml) (Torraca et al., 2015) chemokines, or water control (1 nl), were injected in the hindbrain ventricle (HBV) of 30-32 hpf embryos. Sample fixation was done at 3-3.5 hpi with 4% paraformaldehyde (PFA) (O.N. at 4°C or for 3 h at room temperature). Macrophages were counted as mpeg+ cells in the Tg(mpeg1:mCherry)UMSF001 line and L-plastin (L-P)+/Tyramide Signal Amplification (TSA)− cells when immunohistochemistry was performed, as described previously (Cui et al., 2011; Loynes et al., 2010). Images were acquired using a Leica TCS SPE confocal microscope with an HC PL FLUOTAR 10× DRY objective (0.30 N.A.). In each micrograph, embryos are shown in lateral view and oriented head (left)-to-tail (right) and injection site shown by schematic drawing. Human and zebrafish CXCL12/Cxcl12 and CXCR4/Cxcr4 sequences were obtained in UniProt (The UniProt Consortium, 2015) and aligned in ClustalW (Goujon et al., 2010; Larkin et al., 2007). Specific domains of the human proteins for ligand binding and receptor activation were reported in UniProt.
Statistical analysis
Unpaired, two-tailed t-test was used to compare the means of two groups and Welch's correction applied when variances were significantly different (P<0.05). For datasets of three or more groups, one-way ANOVA with Bonferroni post-hoc test was performed. Raw or normalized data are mean±s.e.m. of pooled data points from at least two independent experiments. Statistics were performed with GraphPad Prism 6.
Acknowledgements
The authors would like to thank A. H. Meijer for critical reading of the manuscript and V. Torraca, A. Groenewoud (Leiden University), M. J. Smit (VU, Amsterdam) and E. Raz (University of Münster) for scientific discussion. The authors are grateful to M. Olszewski (IIMB, Warsaw) for providing MDA-MB-231 transduced with the pLenti-tdtomato plasmid, and P. ten Dijke and Y. Drabsch for MDA-MB-231-B dsRed cells (LUMC, Leiden). CXCR4-targeting shRNAs were provided by R. Hoeben and M. Rabelink (LUMC, Leiden, NL). Zebrafish Cxcl11aa was a kind gift of V. Torraca (Leiden University). The authors thank A. F. Siekmann (Max-Plank-Institute, Münster), D. Gilmour (EMBL, Heidelberg) and H. Knaut (Max-Plank-Institute, Tübingen) for providing zebrafish mutant lines used in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
C.T., C.S. and E.B. performed experiments and data analysis. J.B. generated the zebrafish cxcl12a−/−/cxc12b+/−/Tg(kdrl:EGFP)s843 mutant line and identified cxcl12a−/−/cxc12b−/−/Tg(kdrl:EGFP)s843 mutant larvae. K.T. established the zebrafish Cxcl12 chemokine purification system and produced Cxcl12a and Cxcl12b used in Fig. 2. J.B. and T.S. gave valuable suggestions and revised the paper. C.T. and B.E.S.-J. designed experiments and wrote the paper. All authors approved the final version of the manuscript.
Funding
The present work was supported by the Netherlands Organization for Scientific Research (TOP GO Grant: 854.10.012). J.B. was supported by the STW VENI fellowship (no.12520), which is financed by the Netherlands Organization for Scientific Research (NWO).
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.023275/-/DC1
References
- Amatruda J. F., Shepard J. L., Stern H. M. and Zon L. I. (2002). Zebrafish as a cancer model system. Cancer Cell 1, 229-231. 10.1016/S1535-6108(02)00052-1 [DOI] [PubMed] [Google Scholar]
- Anders C. K. and Carey L. A. (2009). Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin. Breast Cancer 9 Suppl. 2, S73-S81. 10.3816/CBC.2009.s.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggiolini M., Dewald B. and Moser B. (1997). Human chemokines: an update. Annu. Rev. Immunol. 15, 675-705. 10.1146/annurev.immunol.15.1.675 [DOI] [PubMed] [Google Scholar]
- Balabanian K., Lagane B., Infantino S., Chow K. Y. C., Harriague J., Moepps B., Arenzana-Seisdedos F., Thelen M. and Bachelerie F. (2005). The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 280, 35760-35766. 10.1074/jbc.M508234200 [DOI] [PubMed] [Google Scholar]
- Balkwill F. (2004). The significance of cancer cell expression of the chemokine receptor CXCR4. Semin. Cancer Biol. 14, 171-179. 10.1016/j.semcancer.2003.10.003 [DOI] [PubMed] [Google Scholar]
- Bansal N., Davis S., Tereshchenko I., Budak-Alpdogan T., Zhong H., Stein M. N., Kim I. Y., DiPaola R. S., Bertino J. R. and Sabaawy H. E. (2014). Enrichment of human prostate cancer cells with tumor initiating properties in mouse and zebrafish xenografts by differential adhesion. Prostate 74, 187-200. 10.1002/pros.22740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriuso J., Nagaraju R. and Hurlstone A. (2015). Zebrafish: a new companion for translational research in oncology. Clin. Cancer Res. 21, 969-975. 10.1158/1078-0432.CCR-14-2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhagen J., Krohn R., Lue H., Gregory J. L., Zernecke A., Koenen R. R., Dewor M., Georgiev I., Schober A., Leng L. et al. (2007). MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat. Med.USA 13, 587-596. 10.1038/nm1567 [DOI] [PubMed] [Google Scholar]
- Bernut A., Herrmann J.-L., Kissa K., Dubremetz J.-F., Gaillard J.-L., Lutfalla G. and Kremer L. (2014). Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc. Natl. Acad. Sci. USA 111, E943-E952. 10.1073/pnas.1321390111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleul C. C., Farzan M., Choe H., Parolin C., Clark-Lewis I., Sodroski J. and Springer T. A. (1996). The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382, 829-833. 10.1038/382829a0 [DOI] [PubMed] [Google Scholar]
- Boldajipour B., Mahabaleshwar H., Kardash E., Reichman-Fried M., Blaser H., Minina S., Wilson D., Xu Q. and Raz E. (2008). Control of chemokine-guided cell migration by ligand sequestration. Cell 132, 463-473. 10.1016/j.cell.2007.12.034 [DOI] [PubMed] [Google Scholar]
- Boldajipour B., Doitsidou M., Tarbashevich K., Laguri C., Yu S. R., Ries J., Dumstrei K., Thelen S., Dorries J., Messerschmidt E.-M. et al. (2011). Cxcl12 evolution - subfunctionalization of a ligand through altered interaction with the chemokine receptor. Development 138, 2909-2914. 10.1242/dev.068379 [DOI] [PubMed] [Google Scholar]
- Busillo J. M. and Benovic J. L. (2007). Regulation of CXCR4 signaling. Biochim. Biophys. Acta 1768, 952-963. 10.1016/j.bbamem.2006.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann J. and Raz E. (2015). Chemokine-guided cell migration and motility in zebrafish development. EMBO J. 34, 1309-1318. 10.15252/embj.201490105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann J., Wolfe S. A. and Siekmann A. F. (2011). Arterial-venous network formation during brain vascularization involves hemodynamic regulation of chemokine signaling. Development 138, 1717-1726. 10.1242/dev.059881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha Y. R., Fujita M., Butler M., Isogai S., Kochhan E., Siekmann A. F. and Weinstein B. M. (2012). Chemokine signaling directs trunk lymphatic network formation along the preexisting blood vasculature. Dev. Cell 22, 824-836. 10.1016/j.devcel.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Azad B. B. and Nimmagadda S. (2014). The intricate role of CXCR4 in cancer. Adv. Cancer Res. 124, 31-82. 10.1016/b978-0-12-411638-2.00002-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini S., Di Bernardo G., Cammarota M., Castello G. and Colonna G. (2013). Gene expression signature of human HepG2 cell line. Gene 518, 335-345. 10.1016/j.gene.2012.12.106 [DOI] [PubMed] [Google Scholar]
- Cui C., Benard E. L., Kanwal Z., Stockhammer O. W., van der Vaart M., Zakrzewska A., Spaink H. P. and Meijer A. H. (2011). Infectious disease modeling and innate immune function in zebrafish embryos. Zebrafish 105, 273-308. 10.1016/b978-0-12-381320-6.00012-6 [DOI] [PubMed] [Google Scholar]
- Day R. B. and Link D. C. (2012). Regulation of neutrophil trafficking from the bone marrow. Cell. Mol. Life Sciences 69, 1415-1423. 10.1007/s00018-011-0870-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dona E., Barry J. D., Valentin G., Quirin C., Khmelinskii A., Kunze A., Durdu S., Newton L. R., Fernandez-Minan A., Huber W. et al. (2013). Directional tissue migration through a self-generated chemokine gradient. Nature 503, 285-289. 10.1038/nature12635 [DOI] [PubMed] [Google Scholar]
- Ellett F., Pase L., Hayman J. W., Andrianopoulos A. and Lieschke G. J. (2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49-e56. 10.1182/blood-2010-10-314120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing J. (1929). Neoplastic Diseases, 6th edn Philadelphia: W. B. Saunders. [Google Scholar]
- Feng Y., Broder C. C., Kennedy P. E. and Berger E. A. (1996). HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272, 872-877. 10.1126/science.272.5263.872 [DOI] [PubMed] [Google Scholar]
- Fidler I. J. (2003). Timeline: the pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 3, 453-458. 10.1038/nrc1098 [DOI] [PubMed] [Google Scholar]
- Fujita M., Cha Y. R., Pham V. N., Sakurai A., Roman B. L., Gutkind J. S. and Weinstein B. M. (2011). Assembly and patterning of the vascular network of the vertebrate hindbrain. Development 138, 1705-1715. 10.1242/dev.058776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghotra V. P. S., He S., de Bont H., van der Ent W., Spaink H. P., van de Water B., Snaar-Jagalska B. E. and Danen E. H. J. (2012). Automated whole animal bio-imaging assay for human cancer dissemination. PLoS ONE 7, e31281 10.1371/journal.pone.0031281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghotra V. P. S., He S., van der Horst G., Nijhoff S., de Bont H., Lekkerkerker A., Janssen R., Jenster G., van Leenders G. J. L. H., Hoogland A. M. M. et al. (2015). SYK is a candidate kinase target for the treatment of advanced prostate cancer. Cancer Res. 75, 230-240. 10.1158/0008-5472.CAN-14-0629 [DOI] [PubMed] [Google Scholar]
- Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J. and Lopez R. (2010). A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695-W699. 10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulino A. V. (2003). WHIM syndrome: a genetic disorder of leukocyte trafficking. Curr. Opin. Allergy Clin. Immunol. 3, 443-450. 10.1097/00130832-200312000-00005 [DOI] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646-674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- He S., Lamers G. E. M., Beenakker J.-W. M., Cui C., Ghotra V. P. S., Danen E. H. J., Meijer A. H., Spaink H. P. and Snaar-Jagalska B. E. (2012). Neutrophil-mediated experimental metastasis is enhanced by VEGFR inhibition in a zebrafish xenograft model. J. Pathol. 227, 431-445. 10.1002/path.4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix C. W., Collier A. C., Lederman M. M., Schols D., Pollard R. B., Brown S., Jackson J. B., Coombs R. W., Glesby M. J., Flexner C. W. et al. (2004). Safety, pharmacokinetics, and antiviral activity of AMD3100, a selective CXCR4 receptor inhibitor, in HIV-1 infection. J. Acquir. Immune Defic. Syndr. 37, 1253-1262. 10.1097/01.qai.0000137371.80695.ef [DOI] [PubMed] [Google Scholar]
- Herzog H., Hort Y. J., Shine J. and Selbie L. A. (1993). Molecular cloning, characterization, and localization of the human homolog to the reported bovine NPY Y3 receptor: lack of NPY binding and activation. DNA Cell Biol. 12, 465-471. 10.1089/dna.1993.12.465 [DOI] [PubMed] [Google Scholar]
- Hess I. and Boehm T. (2012). Intravital imaging of thymopoiesis reveals dynamic lympho-epithelial interactions. Immunity 36, 298-309. 10.1016/j.immuni.2011.12.016 [DOI] [PubMed] [Google Scholar]
- Janowski M. (2009). Functional diversity of SDF-1 splicing variants. Cell Adh. Migr. 3, 243-249. 10.4161/cam.3.3.8260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazin E. E., Yoo H., Blomqvist A. G., Yee F., Weng G., Walker M. W., Salon J., Larhammar D. and Wahlestedt C. (1993). A proposed bovine neuropeptide Y (NPY) receptor cDNA clone, or its human homologue, confers neither NPY binding sites nor NPY responsiveness on transfected cells. Regul. Pept. 47, 247-258. 10.1016/0167-0115(93)90392-L [DOI] [PubMed] [Google Scholar]
- Jin S.-W., Beis D., Mitchell T., Chen J.-N. and Stainier D. Y. R. (2005). Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199-5209. 10.1242/dev.02087 [DOI] [PubMed] [Google Scholar]
- Kalatskaya I., Berchiche Y. A., Gravel S., Limberg B. J., Rosenbaum J. S. and Heveker N. (2009). AMD3100 is a CXCR7 ligand with allosteric agonist properties. Mol. Pharmacol. 75, 1240-1247. 10.1124/mol.108.053389 [DOI] [PubMed] [Google Scholar]
- Knaut H., Werz C., Geisler R., The Tübingen 2000 Screen Consortium Robert and Nusslein-Volhard C. (2003). A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature 421, 279-282. 10.1038/nature01338 [DOI] [PubMed] [Google Scholar]
- Konantz M., Balci T. B., Hartwig U. F., Dellaire G., Andre M. C., Berman J. N. and Lengerke C. (2012). Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann. N. Y. Acad. Sci. 1266, 124-137. 10.1111/j.1749-6632.2012.06575.x [DOI] [PubMed] [Google Scholar]
- Kuhne M. R., Mulvey T., Belanger B., Chen S., Pan C., Chong C., Cao F., Niekro W., Kempe T., Henning K. A. et al. (2013). BMS-936564/MDX-1338: a fully human anti-CXCR4 antibody induces apoptosis in vitro and shows antitumor activity in vivo in hematologic malignancies. Clin. Cancer Res. 19, 357-366. 10.1158/1078-0432.CCR-12-2333 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R. et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947-2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lawson N. D. and Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307-318. 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- Lefkowitz R. J. (1998). G protein-coupled receptors: III. New roles for receptor kinases and beta-arrestins in receptor signaling and desensitization. J. Biol. Chem. 273, 18677-18680. 10.1074/jbc.273.30.18677 [DOI] [PubMed] [Google Scholar]
- Loynes C. A., Martin J. S., Robertson A., Trushell D. M., Ingham P. W., Whyte M. K. B. and Renshaw S. A. (2010). Pivotal advance: pharmacological manipulation of inflammation resolution during spontaneously resolving tissue neutrophilia in the zebrafish. J. Leukoc. Biol. 87, 203-212. 10.1189/jlb.0409255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell L. M., Ferguson S. S. G., Daaka Y., Miller W. E., Maudsley S., Della Rocca G. J., Lin F.-T., Kawakatsu H., Owada K., Luttrell D. K. et al. (1999). Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science 283, 655-661. 10.1126/science.283.5402.655 [DOI] [PubMed] [Google Scholar]
- Mellado M., Rodriguez-Frade J. M., Manes S. and Martinez-A C. (2001). Chemokine signaling and functional responses: the role of receptor dimerization and TK pathway activation. Annu. Rev. Immunol. 19, 397-421. 10.1146/annurev.immunol.19.1.397 [DOI] [PubMed] [Google Scholar]
- Miyasaka N., Knaut H. and Yoshihara Y. (2007). Cxcl12/Cxcr4 chemokine signaling is required for placode assembly and sensory axon pathfinding in the zebrafish olfactory system. Development 134, 2459-2468. 10.1242/dev.001958 [DOI] [PubMed] [Google Scholar]
- Muller A., Homey B., Soto H., Ge N., Catron D., Buchanan M. E., McClanahan T., Murphy E., Yuan W., Wagner S. N. et al. (2001). Involvement of chemokine receptors in breast cancer metastasis. Nature 410, 50-56. 10.1038/35065016 [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Hirota S., Tachibana K., Takakura N., Nishikawa S.-I., Kitamura Y., Yoshida N., Kikutani H. and Kishimoto T. (1996). Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature 382, 635-638. 10.1038/382635a0 [DOI] [PubMed] [Google Scholar]
- Nomura H., Nielsen B. W. and Matsushima K. (1993). Molecular cloning of cDNAs encoding a LD78 receptor and putative leukocyte chemotactic peptide receptors. Int. Immunol. 5, 1239-1249. 10.1093/intimm/5.10.1239 [DOI] [PubMed] [Google Scholar]
- Oberlin E., Amara A., Bachelerie F., Bessia C., Virelizier J.-L., Arenzana-Seisdedos F., Schwartz O., Heard J.-M., Clark-Lewis I., Legler D. F. et al. (1996). The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1 (vol 382, pg 833, pg 1996). Nature 384, 288-288 10.1038/384288a0 [DOI] [PubMed] [Google Scholar]
- Paget S. (1989). The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 8, 98-101. [PubMed] [Google Scholar]
- Palma G., Frasci G., Chirico A., Esposito E., Siani C., Saturnino C., Arra C., Ciliberto G., Giordano A. and D'Aiuto M. (2015). Triple negative breast cancer: looking for the missing link between biology and treatments. Oncotarget 6, 26560-26574. 10.18632/oncotarget.5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawig L., Klasen C., Weber C., Bernhagen J. and Noels H. (2015). Diversity and inter-connections in the CXCR4 chemokine receptor/ligand family: molecular perspectives. Front. Immunol. 6, 429 10.3389/fimmu.2015.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podo F., Buydens L. M. C., Degani H., Hilhorst R., Klipp E., Gribbestad I. S., Van Huffel S., van Laarhoven H. W. M., Luts J., Monleon D. et al. (2010). Triple-negative breast cancer: present challenges and new perspectives. Mol. Oncol. 4, 209-229. 10.1016/j.molonc.2010.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal S., Kim J., Ahn S., Craig S., Lam C. M., Gerard N. P., Gerard C. and Lefkowitz R. J. (2010). beta-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc. Natl. Acad. Sci. USA 107, 628-632. 10.1073/pnas.0912852107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey D. M. and McAlpine S. R. (2013). Halting metastasis through CXCR4 inhibition. Bioorg. Med. Chem. Lett. 23, 20-25. 10.1016/j.bmcl.2012.10.138 [DOI] [PubMed] [Google Scholar]
- Renshaw S. A., Loynes C. A., Trushell D. M. I., Elworthy S., Ingham P. W. and Whyte M. K. B. (2006). A transgenic zebrafish model of neutrophilic inflammation. Blood 108, 3976-3978. 10.1182/blood-2006-05-024075 [DOI] [PubMed] [Google Scholar]
- Rosu-Myles M., Gallacher L., Murdoch B., Hess D. A., Keeney M., Kelvin D., Dale L., Ferguson S. S. G., Wu D., Fellows F. et al. (2000). The human hematopoietic stem cell compartment is heterogeneous for CXCR4 expression. Proc. Natl. Acad. Sci. USA 97, 14626-14631. 10.1073/pnas.97.26.14626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini V., Marchese A. and Majetschak M. (2010). CXC chemokine receptor 4 is a cell surface receptor for extracellular ubiquitin. J. Biol. Chem. 285, 15566-15576. 10.1074/jbc.M110.103408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini V., Staren D. M., Ziarek J. J., Nashaat Z. N., Campbell E. M., Volkman B. F., Marchese A. and Majetschak M. (2011). The CXC chemokine receptor 4 ligands ubiquitin and stromal cell-derived factor-1 function through distinct receptor interactions. J. Biol. Chem. 286, 33466-33477. 10.1074/jbc.M111.233742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar N., Muñoz D., Kallifatidis G., Singh R. K., Jordà M. and Lokeshwar B. L. (2014). The chemokine receptor CXCR7 interacts with EGFR to promote breast cancer cell proliferation. Mol. Cancer 13, 198 10.1186/1476-4598-13-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F. and Baggiolini M. (2008). Chemokines and leukocyte traffic. Nat. Immunol. 9, 949-952. 10.1038/ni.f.214 [DOI] [PubMed] [Google Scholar]
- Sallusto F., Mackay C. R. and Lanzavecchia A. (2000). The role of chemokine receptors in primary, effector, and memory immune responses. Annu. Rev. Immunol. 18, 593-620. 10.1146/annurev.immunol.18.1.593 [DOI] [PubMed] [Google Scholar]
- Scala S. (2015). Molecular pathways: targeting the CXCR4-CXCL12 axis-untapped potential in the tumor microenvironment. Clin. Cancer Res. 21, 4278-4285. 10.1158/1078-0432.ccr-14-0914 [DOI] [PubMed] [Google Scholar]
- Shi J., Wei Y., Xia J., Wang S., Wu J., Chen F., Huang G. and Chen J. (2014). CXCL12-CXCR4 contributes to the implication of bone marrow in cancer metastasis. Future Oncol. 10, 749-759. 10.2217/fon.13.193 [DOI] [PubMed] [Google Scholar]
- Shukla A. K., Xiao K. and Lefkowitz R. J. (2011). Emerging paradigms of beta-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem. Sci. 36, 457-469. 10.1016/j.tibs.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletov K., Kato H., Zardouzian E., Kelber J., Yang J., Shattil S. and Klemke R. (2010). Visualizing extravasation dynamics of metastatic tumor cells. J. Cell Sci. 123, 2332-2341. 10.1242/jcs.069443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher B. A. and Fricker S. P. (2010). CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. 16, 2927-2931. 10.1158/1078-0432.CCR-09-2329 [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium (2015). UniProt: a hub for protein information. Nucleic Acids Res. 43, D204-D212. 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma G., Streiff M. B., Kovarik J., Glickman F., Wagner T., Beerli C. and Zerwes H.-G. (2008). Orally bioavailable isothioureas block function of the chemokine receptor CXCR4 in vitro and in vivo. J. Med. Chem. 51, 7915-7920. 10.1021/jm801065q [DOI] [PubMed] [Google Scholar]
- Torraca V., Cui C., Boland R., Bebelman J.-P., van der Sar A. M., Smit M. J., Siderius M., Spaink H. P. and Meijer A. H. (2015). The CXCR3-CXCL11 signaling axis mediates macrophage recruitment and dissemination of mycobacterial infection. Dis. Model. Mech. 8, 253-269. 10.1242/dmm.017756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin G., Haas P. and Gilmour D. (2007). The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr. Biol. 17, 1026-1031. 10.1016/j.cub.2007.05.020 [DOI] [PubMed] [Google Scholar]
- van der Ent W., Burrello C., Teunisse A. F. A. S., Ksander B. R., van der Velden P. A., Jager M. J., Jochemsen A. G. and Snaar-Jagalska B. E. (2014a). Modeling of human uveal melanoma in zebrafish xenograft embryos. Invest. Ophthalmol. Vis. Sci. 55, 6612-6622. 10.1167/iovs.14-15202 [DOI] [PubMed] [Google Scholar]
- van der Ent W., Jochemsen A. G., Teunisse A. F. A. S., Krens S. F. G., Szuhai K., Spaink H. P., Hogendoorn P. C. W. and Snaar-Jagalska B. E. (2014b). Ewing sarcoma inhibition by disruption of EWSR1-FLI1 transcriptional activity and reactivation of p53. J. Pathol. 233, 415-424. 10.1002/path.4378 [DOI] [PubMed] [Google Scholar]
- van der Vaart M., Korbee C. J., Lamers G. E. M., Tengeler A. C., Hosseini R., Haks M. C., Ottenhoff T. H. M., Spaink H. P. and Meijer A. H. (2014). The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense. Cell Host Microbe 15, 753-767. 10.1016/j.chom.2014.05.005 [DOI] [PubMed] [Google Scholar]
- Veinotte C. J., Dellaire G. and Berman J. N. (2014). Hooking the big one: the potential of zebrafish xenotransplantation to reform cancer drug screening in the genomic era. Dis. Model. Mech. 7, 745-754. 10.1242/dmm.015784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela M., Aris M., Llorente M., Garcia-Sanz J. A. and Kremer L. (2015). Chemokine receptor-specific antibodies in cancer immunotherapy: achievements and challenges. Front. Immunol. 6, 12 10.3389/fimmu.2015.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkiteswaran G., Lewellis S. W., Wang J., Reynolds E., Nicholson C. and Knaut H. (2013). Generation and dynamics of an endogenous, self-generated signaling gradient across a migrating tissue. Cell 155, 674-687. 10.1016/j.cell.2013.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicenzi E., Lio P. and Poli G. (2013). The puzzling role of CXCR4 in human immunodeficiency virus infection. Theranostics 3, 18-25. 10.7150/thno.5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Coro A. J., Rodríguez-Frade J. M., Martín De Ana A., Moreno-Ortíz M. C., Martínez-A C. and Mellado M. (1999). The chemokine SDF-1alpha triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. FASEB J. 13, 1699-1710. [PubMed] [Google Scholar]
- Wahba H. A. and El-Hadaad H. A. (2015). Current approaches in treatment of triple-negative breast cancer. Cancer Biol. Med. 12, 106-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters K. B., Green J. M., Surfus J. C., Yoo S. K. and Huttenlocher A. (2010). Live imaging of neutrophil motility in a zebrafish model of WHIM syndrome. Blood 116, 2803-2811. 10.1182/blood-2010-03-276972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterwald A., van der Pluijm G., Que I., Sijmons B., Buijs J., Karperien M., Lowik C. W. G. M., Gautschi E., Thalmann G. N. and Cecchini M. G. (2002). Optical Imaging of cancer metastasis to bone marrow - a mouse model of minimal residual disease. Am. J. Pathol. 160, 1143-1153. 10.1016/S0002-9440(10)64934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Chien E. Y. T., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C. et al. (2010). Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066-1071. 10.1126/science.1194396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y., Oishi S., Kubo T., Tanahara N., Fujii N. and Furuya T. (2013). Optimized method of G-protein-coupled receptor homology modeling: its application to the discovery of novel CXCR7 ligands. J. Med. Chem. 56, 4236-4251. 10.1021/jm400307y [DOI] [PubMed] [Google Scholar]
- Zhao S., Chang S. L., Linderman J. J., Feng F. Y. and Luker G. D. (2014). A comprehensive analysis of CXCL12 isoforms in breast cancer. Transl. Oncol. 7, 429-438. 10.1016/j.tranon.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoni E., van der Horst G., van de Merbel A. F., Chen L., Rane J. K., Pelger R. C. M., Collins A. T., Visakorpi T., Snaar-Jagalska B. E., Maitland N. J. et al. (2015). miR-25 modulates invasiveness and dissemination of human prostate cancer cells via regulation of alpha(v)- and alpha(6)-integrin expression. Cancer Res. 75, 2326-2336. 10.1158/0008-5472.CAN-14-2155 [DOI] [PubMed] [Google Scholar]