Abstract
Poly(ADP-ribose) polymerase-2 (PARP-2) is one of three human PARP enzymes that are potently activated during the cellular DNA damage response (DDR). DDR-PARPs detect DNA strand breaks, leading to a dramatic increase in their catalytic production of the posttranslational modification poly(ADP-ribose) (PAR) to facilitate repair. There are limited biochemical and structural insights into the functional domains of PARP-2, which has restricted our understanding of how PARP-2 is specialized toward specific repair pathways. PARP-2 has a modular architecture composed of a C-terminal catalytic domain (CAT), a central Trp-Gly-Arg (WGR) domain and an N-terminal region (NTR). Although the NTR is generally considered the key DNA-binding domain of PARP-2, we report here that all three domains of PARP-2 collectively contribute to interaction with DNA damage. Biophysical, structural and biochemical analyses indicate that the NTR is natively disordered, and is only required for activation on specific types of DNA damage. Interestingly, the NTR is not essential for PARP-2 localization to sites of DNA damage. Rather, the WGR and CAT domains function together to recruit PARP-2 to sites of DNA breaks. Our study differentiates the functions of PARP-2 domains from those of PARP-1, the other major DDR-PARP, and highlights the specialization of the multi-domain architectures of DDR-PARPs.
INTRODUCTION
The poly(ADP-ribose) polymerase (PARP) family of enzymes is composed of 17 members that regulate multiple aspects of cell biology (1,2). PARP-1, PARP-2 and PARP-3 detect DNA damage and couple their interaction with DNA to the catalytic production of poly(ADP-ribose) (PAR)—a posttranslational modification and signaling molecule that plays a key role in coordinating the cellular DNA damage response (DDR). Under conditions of genotoxic stress, there is a burst in production of PAR, which participates in the repair process by recruiting cellular repair factors to sites of DNA damage and by altering the chromatin architecture to expedite repair (3,4). The DDR-PARPs (PARP-1, PARP-2 and PARP-3) are associated with multiple pathways of DNA repair, including base excision repair (5–7), homologous recombination (4,8) and non-homologous end-joining (9,10). The importance of the DDR-PARPs to normal cell function is highlighted by the embryonic lethality of the PARP-1/PARP-2 double-knockout mouse (11).
DDR-PARPs have specialized domain architectures that allow their catalytic activities to be regulated through interaction with DNA. The catalytic (CAT) domains of DDR-PARPs are composed of an ADP-ribosyl transferase (ART) fold that is conserved among all PARP family members, and a regulatory helical subdomain (HD) that is unique to the DDR-PARPs (12). DDR-PARPs also have in common a Trp-Gly-Arg (WGR) domain that is essential for DNA damage-dependent catalytic activity (12). The WGR domain contacts both DNA and the HD and thus plays a central role in the communication network that connects DNA damage detection to the regulation of DDR-PARP catalytic activity (12–14).
Despite the overall conservation of the WGR and CAT domains, the DDR-PARPs are likely to be specialized for participation in specific repair pathways (4). Indeed, we have previously shown that PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA strand breaks, suggesting that their activities are stimulated in response to specific DNA repair intermediates at particular stages of repair (12). Furthermore, the domain architecture outside of the WGR and CAT differs substantially among the DDR-PARPs. PARP-1 has four additional domains that are not found in PARP-2 and PARP-3: three zinc-binding domains (Zn1, Zn2 and Zn3) and a BRCA C-terminus (BRCT) domain (Figure 1A). The Zn1 and Zn3 domains are strictly required for PARP-1 DNA damage-dependent activity and participate in a communication network between DNA damage recognition and the CAT domain (13,14). PARP-2 and PARP-3 only have short N-terminal regions (NTRs) that extend from the WGR and CAT domains. The NTR of human PARP-2 contains a nucleolar localization sequence (NoLS; residues 4 to 7) and a putative nuclear localization signal (NLS) (Figure 1A), and is furthermore reported to participate in protein–protein interactions (15). The role of the NTR in DNA damage-dependent PARP-2 activation is not clearly defined.
Figure 1.
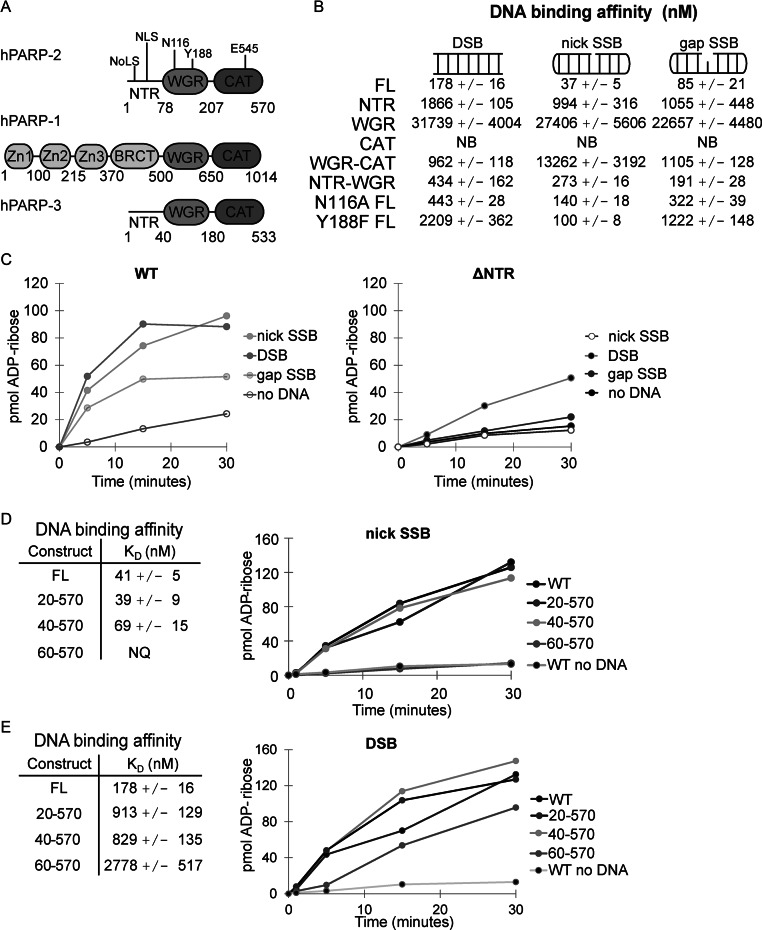
PARP-2 NTR contributes to SSB recognition. (A) Schematic of PARP-1, PARP-2 and PARP-3 domains. Specific regions of PARP-2 are noted: NoLS (nucleolar localization signal, residues 4–7), NLS (bipartite nuclear localization signal, including K21, R22 and K36, K37), key WGR residues N116 (contact to CAT) and Y188 (contact to DNA) and a key catalytic active site residue E545 (catalytic residue). (B) The DNA-binding affinities of PARP-2 WT (1–570) and mutant constructs (NTR 1–78; WGR 71–207; CAT 216–570; WGR-CAT 71–570; NTR-WGR 1–207; N116A FL 1–570; Y188F FL 1–570) were measured by fluorescence polarization using various fluorescently labeled DNA break structures (5 nM). The KD value indicated represents the average derived from three independent experiments with associated standard deviation (SD). Proteins that showed no apparent binding are labeled NB for no binding (see also Supplemental Figures S3–S5). (C) PARP-2 DNA-dependent activity was measured using a colorimetric assay. FL PARP-2 and ΔNTR (WGR-CAT) (60 nM) were incubated with various DNA templates (480 nM). Activity data shown are representative of three independent experiments performed. (D) and (E) Left: KD of PARP-2 N-terminal truncations on a nick SSB (D) or DSB (E) template measured by fluorescence polarization. The KD indicated is the average of three independent experiments with associated SD. Right: colorimetric assay showing the activity of PARP-2 N-terminal truncation constructs (60 nM) in the presence of a nick SSB (D) or DSB (E) DNA template (480 nM). Activity data shown are representative of the three independent experiments performed.
To better understand the function of PARP-2 in DNA repair, we have performed a focused structure and functional analysis of PARP-2 domains. Previous work identified the NTR of PARP-2 as a DNA-binding domain (16), but there has been no quantitative assessment of NTR-binding affinity and its contribution to PARP-2 interaction with DNA relative to the other domains. Sequence analysis of plant PARP-2 homologs (A. thaliana and Z. mais) predicted that the NTR has a helical SAF-A/B, Acinus, and PIAS (SAP) domain fold (17), but this structural prediction has not been experimentally validated and it is not clear that human PARP-2 shares this proposed feature (see sequence alignment in Supplementary Figure S1). Until this point, it has not been experimental determined the structure of the NTR region of PARP-2 nor its requirements in DNA damage activation. Here, we report that the NTR of human PARP-2 is natively unstructured and its DNA-binding activity is particularly important for PARP-2 activation on DNA single strand breaks (SSB). Surprisingly, the NTR is not required for PARP-2 recruitment to cellular sites of DNA damage. Rather, the WGR and CAT domains are jointly responsible for PARP-2 localization to nuclear DNA damage. Collectively, our study provides new insights into the specific domain functions and requirements of human PARP-2 and highlights the non-redundant features of the DDR-PARPs.
MATERIALS AND METHODS
Gene cloning and mutagenesis
The cloning of the DNA construct coding for human PARP-2 isoform 2 has been previously described (12). Dr. G. Cingolani provided the DNA construct coding for murine importin α1 ΔIBB (IMPα1ΔIBB; residues 70–529). The amino acid sequence for the inserted SV40T NLS was PPKKKRKVEDPG. Gene mutations, insertions and truncations were performed using the QuikChange Protocol (Stratagene) and verified by automated sequencing (Sidney Kimmel Cancer Center) or Genescript.
Protein expression and purification
PARP-2 proteins were expressed in E. coli and purified as described previously (12). The E. coli culture media was supplemented with 10 mM benzamide for all proteins containing the CAT domain of PARP-2. IMPα1ΔIBB was expressed in E. coli under similar conditions, and the cell pellets of IMPα1ΔIBB were mixed with cell pellets of NTR PARP-2. The complex was purified using a Ni2+-charged chelating column (GE Healthcare), and the eluted complex was passed over a heparin column following the PARP-2 purification protocol to remove DNA contamination (12). IMPα1ΔIBB passed through the heparin column, whereas the NTR eluted from the heparin column during the salt gradient. IMPα1ΔIBB from the flow through and the PARP-2 NTR fractions were then pooled together, concentrated, and the complex purified over an S200 Sephacryl size exclusion column in 20 mM Hepes pH 8.0, 150 mM NaCl, 0.1mM TCEP. PARP-1 Zn1 was purified as previously described (18).
Crystallization, data collection and structure determination
Crystals were grown by sitting drop vapor diffusion at room temperature in 0.6–0.7 M sodium citrate, 0.1 M sodium citrate buffer pH 5.6 and 7–10 mM DTT, and the protein complex at 15 mg/ml. Crystals were cryoprotected in reservoir solution supplemented with 25% glycerol (or ethylene glycol) and then flash-cooled in liquid nitrogen. Diffraction data were collected at beamline X29 at the National Synchrotron Light Source (NSLS, Brookhaven National Laboratory). The data were processed using Xia2 and XDS (19,20). The initial set of phases came from rigid body refinement of mouse importin α1 ΔIBB (PDB code 1Y2A, with the bound NLS removed from the structure). The human PARP-2 NTR NLS was built manually in COOT (21). Iterative rounds of refinement were performed using PHENIX (22) and REFMAC in the CCP4 Suite of programs (23–25).
PARP colorimetric activity assay
PARP colorimetric assays were performed essentially as described (12,26). Three or more independent experiments were conducted, and the results of a representative experiment are shown.
Circular dichroism spectroscopy
A JASCO J-810 spectropolarimeter was used to record the circular dichroism (CD) spectra. Wavelength scans were performed in a quartz cuvette of 1 mm path length as described previously (26). The buffer used was 5 mM Na/K phosphate pH 7.5 with 50 mM Na2SO4 and 0.1 mM TCEP. PARP-2 NTR (10 μM) was incubated with DNA (2 μM) for 30 min at room temperature prior to performing CD analysis. In Figure 2D, NTR or WGR (10 μM) was incubated 15 min at the indicated temperature before performing CD scans. The final spectrum represents the average of three scans. A spectrum representative of three independent experiments is shown in each case.
Figure 2.
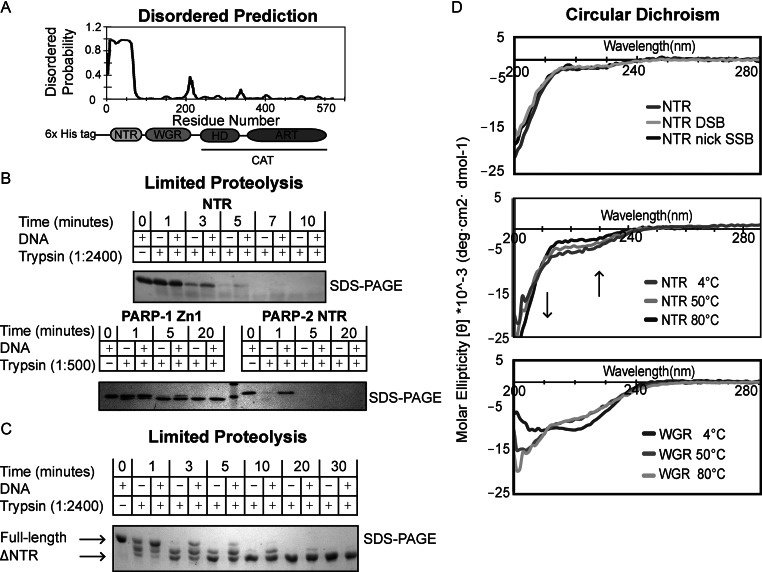
The N-terminal region (NTR) of PARP-2 is natively disordered. (A) Predicted disordered regions of human PARP-2 as determined by the bioinformatics tool DISOPRED. The NTR of PARP-2 has a high probability of being disordered. (B) Time course of human PARP-2 limited proteolysis resolved on 18% SDS-PAGE. Top: SDS-PAGE analysis of 1:2400 trypsin digest of PARP-2 NTR (2 μg) in the presence and absence of DSB (2 μM) for the indicated time points. Bottom: SDS-PAGE analysis of 1:500 trypsin digest of PARP-1 Zn1 and PARP-2 NTR (2 μg) in the presence and absence of DSB (2 μM). (C) Time course of human full-length (FL) PARP-2 limited trypsin proteolysis (1:2400) resolved on 7.5% SDS-PAGE in the presence and absence of DSB (2 μM). FL PARP-2 limited proteolysis analysis on SDS-PAGE showing a truncated, proteolytic resistant PARP-2 after 5 min (D) Top: CD analysis of PARP-2 NTR data collected at 4°C using 10 μM protein in the absence or presence of activating DNA (DSB and nick SSB) (10 μM). Middle: PARP-2 NTR displays identical CD signal at 4, 50 and 80°C. Arrows indicate regions of temperature-dependent changes in CD signal expected for intrinsically disordered proteins. Bottom: temperature-dependent CD analysis of PARP-2 WGR (10 μM) showing that WGR undergoes a structural transition from 4 to 50 to 80°C. All scans were performed in triplicates and averaged to generate the curve shown. The curves are representative of three independent experiments.
Fluorescence polarization DNA-binding assay
Fluorescence polarization (FP) assays were performed as described previously (12,27) using a 28 base pair (bp) duplex DNA (DSB) carrying a 5'P on one end and a 5′ fluorescein derivative (6-FAM) on the other end, or with a dumbbell DNA carrying a nick (nick SSB) or a one nucleotide gap at its center and an internal 6-FAM (SSB gap) (Supplementary Figure S2). The KD represents the average of three independent experiments and the associated SD.
Limited proteolysis
Trypsin was added at a ratio of 1:2400 (w/w) to 2.4 μg of NTR (Figure 2B, top) or FL PARP-2 (Figure 2C). Trypsin was added at a ratio of 1:500 (w/w) to 2.4 μg of PARP-1 Zn1 and PARP-2 NTR (Figure 2B, bottom). Reactions were incubated at room temperature for the indicated time points. Reactions were quenched by the addition of SDS-PAGE loading dye and then boiled for 5 min at 95°C. Proteolytic products were resolved on 18% (NTR) or 7.5% (FL PARP-2) SDS-PAGE and the gels were treated with Imperial Stain (Thermo scientific) for visualization or used for western blot analysis. A representative image of three independent experiments is shown.
Transient transfections and live cell microscopy
For all cell-based experiments, 1.5 × 105 cells were plated and transfected 24 h later with 1 μg DNA and 3 μl Fugene (Promega) in serum free-media according to manufacturer's recommendations. Serum free media was incubated on cells for 1 h before the addition of growth media. HEK293 cell images (Figure 3) were acquired using an Olympus BX-61 microscope with an ORCA-ER (Hamamatsu, Bridgewater, NJ, USA) cooled charge-coupled device camera controlled by Slidebook version 4.0 (Intelligent Imaging Innovations, Denver, CO, USA). The displayed cells are representative of a population of cells observed in three independent experiments.
Figure 3.
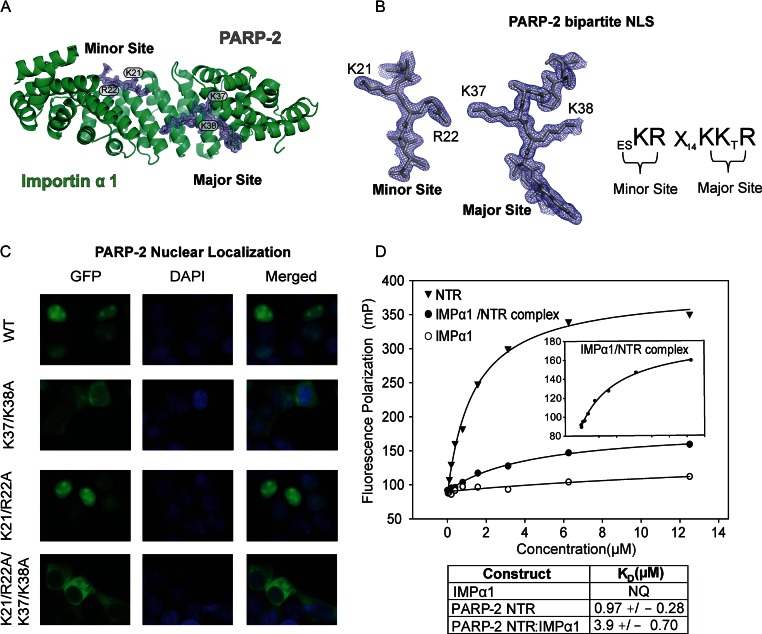
IMPα1ΔIBB:PARP-2 bipartite NLS interaction. (A) Crystal structure of human PARP-2 NLS bound to IMPα1ΔIBB. Cartoon overview of IMPα1ΔIBB (ribbons) and PARP-2 bipartite NLS (sticks). The final σ-weighted 2FO–FC electron density map contoured at 1.5σ is overlayed. (B) Left: electron density of the PARP-2 NLS minor and major binding sites. Key PARP-2 residues are shown. Right: schematic illustration of the key PARP-2 NLS residues that mediate the interaction, where X represents any amino acid. (C) Live-cell imaging of HEK 293 cells expressing GFP-PARP-2, and mutations and truncations thereof. PARP-2 requires a bipartite NLS for maintenance of strong nuclear localization. A representative image of a population of cells is shown from three independent experiments for each GFP, DAPI and the merged DAPI/GFP image. (D) Fluorescence polarization experiment showing DNA binding of human PARP-2 NTR, IMPα1ΔIBB and the IMPα1ΔIBB/NTR complex using a fluorescently labeled 5'P nick SSB template (5 nM). Inset is a zoomed-in image showing IMPα1ΔIBB/NTR DNA-binding activity. The reported KD values are averages derived from three independent experiments with associated SD. Representative binding curves are shown.
Live cell imaging microscopy and laser irradiation
Live cell microscopy and laser irradiation was performed essentially as described (28). Briefly, transfected HeLa cells were sensitized with BrdU (1 μmol/l) for 24 h, and then treated with Hoeschst stain (10 μg/ml) just prior to the start of the experiment. Using a Zeiss LSM-510 Meta Confocal laser scanning microscope, a 405-nm diode laser set to 100% power was used to locally irradiate a defined nuclear region for 1 s. Images were recorded by excitation with a 488-nm argon laser set to 50% power and focused through a 40x oil-immersion lens. Image processing and quantification were performed using Zeiss LSM Image Browser. Images are representative of observations made in three or more independent experiments. For quantification of fluorescence intensities, a non-irradiated region was selected to establish a background and to correct for the overall loss of fluorescence during the imaging time course. The irradiated region was compared to the non-irradiated region to provide the level of increase in GFP signal. Quantification of the kinetics of recruitment to laser-irradiated regions was performed using ≥4 cells allowing a calculation of average fold increase in intensity with associated SD.
RESULTS
PARP-2 domains collectively bind and activate on DNA damage
We previously observed that the NTR of PARP-2 was not strictly required for DNA binding and catalytic activation using a DSB; however, deletion of the NTR resulted in a reduction in DSB-binding affinity (12) and thus suggested an NTR contribution to PARP-2 interaction with DNA. Here, we have directly and quantitatively investigated the DNA-binding activity of the NTR, and further explored the importance of the NTR for PARP-2 activation and binding to different DNA structures. The DNA-binding activity of the NTR was assessed with an FP assay using three fluorescently labeled DNA oligonucleotides representing a DSB (duplex DNA), a nick SSB (DNA dumbbell with a centrally placed break in one strand) and a gap SSB (DNA dumbbell with a centrally placed one-nucleotide gap in one strand) (Supplementary Figure S2). NTR affinities for these DNAs were compared to the affinities measured for full-length (FL) PARP-2 and deletion or mutant variants of PARP-2 (Figure 1B and see also Supplementary Figures S3–S5). The binding analysis indicated that the NTR alone has bona fide DNA-binding activity, interacting with each of the DNAs with comparable albeit weak affinity (1866, 994 and 1055 nM). In comparison, FL PARP-2 bound to the same DNAs with ∼10- to ∼26-fold higher affinity (178, 37 and 88 nM), indicating that the NTR alone does not account for the entire binding affinity of PARP-2 and that additional regions must contribute to overall binding affinity. Indeed the ΔNTR construct, comprised of the WGR and CAT domains, exhibited binding affinity for a DSB comparable to that of the NTR (ΔNTR: 962 nM, NTR: 1866 nM), consistent with regions other than the NTR contributing to DNA binding. In contrast, there was weak interaction for ΔNTR binding to a nick SSB (∼13 μM), indicating a specific requirement for the NTR to mediate PARP-2 interaction with this SSB structure. Interestingly, ΔNTR was able to bind to a SSB with a single nucleotide gap at the break site (gap SSB), suggesting that the NTR plays a specific role in allowing access to DNA ends.
The NTR is a bone fide DNA-binding domain and is the only domain of PARP-2 that binds to DNA appreciably on its own; however, both the WGR and CAT domains are required to achieve the full binding affinity of PARP-2 (Figure 1B). Even though the WGR interaction with the models of DNA damage tested is weak (∼20–30 μM) (Figure 1B), it is expected to form important DNA contacts with the 5′ end of DNA breaks based on conservation with PARP-1 and our previous analysis (12). Indeed the mutation Y188F, which targets a WGR contact with the 5′ end of DNA, reduces overall binding affinity of FL PARP-2 (Figure 1B), consistent with WGR contribution to the overall DNA-binding affinity of PARP-2. In contrast, CAT is not expected to contact DNA and indeed showed no DNA-binding activity on its own (Figure 1B). Therefore, it is particularly interesting that the WGR and CAT domains coupled together in one polypeptide bound to DNA DSB and gap SSB with appreciable affinity (Figure 1B). The WGR and CAT form key interdomain contacts during activation, and disruption of these contacts with the mutation N116A shuts down FL PARP-2 activation in response to DNA damage (12). We found that the N116A mutation of FL PARP-2 causes a reduction in DNA-binding affinity, indicating that the cross-talk between the WGR and CAT domains contributes not only to the allosteric activation mechanism, but also to the strength of PARP-2 interaction with DNA (Figure 1B). Furthermore, the CAT deletion of PARP-2, comprised of the NTR and WGR domain, exhibited a consistent decrease in binding affinity on all models of DNA damage (Figure 1B). Together, the DNA-binding measurements indicate that each of the domains of PARP-2 make contributions to overall binding affinity, with both protein–DNA and protein–protein contacts contributing to their collaboration.
We next tested the contribution of the NTR to PARP-2 catalytic activation (Figure 1C and Supplementary Figure S6) using the same DNA structures used to measure binding affinity (Figure 1B). Consistent with the binding analysis, ΔNTR showed no apparent activation in the presence of a nick SSB, but maintained a modest level of activation on a DSB (Figure 1C, right and Supplementary Figure S6B). A gap SSB was not able to stimulate the activity of ΔNTR, even though there was measurable interaction in the DNA-binding analysis. It is notable that FL PARP-2 is also less stimulated by a gap SSB relative to a nick SSB and a DSB (Figure 1C, left and Supplementary Figure S6A). These results highlight that PARP-2 activation is sensitive to the local structure of a DNA break, and this specialization likely plays a role in the timing of PARP-2 activation in DNA repair pathways. Next, we determined the minimal region of the NTR required for activation by a nick SSB and for binding to a nick SSB using successive N-terminal deletions. Catalytic stimulation in the presence of a nick SSB did not require the first 39 amino acids of PARP-2 (Figure 1D). Deletions greater than the first 39 amino acids of the NTR fully compromised activation on a nick SSB, but only had a modest effect on DSB (Figure 1E). Together, these results define the NTR of PARP-2 as an independent DNA-binding module that synergizes with the WGR and CAT domains to mediate PARP-2 interaction with DNA, and the NTR contribution to DNA binding is particularly important on SSB DNA damage intermediates.
The NTR of PARP-2 is natively disordered
Given the important contributions of the PARP-2 NTR to DNA binding and activation, we sought to better understanding its structure and function. A prediction of ordered versus disordered regions of the human PARP-2 polypeptide generated using DISOPRED (29,30) indicated that the NTR of PARP-2 has a high probability of being natively disordered (Figure 2A). In contrast, the WGR and CAT domain regions exhibited a low probability of disorder, consistent with the expected conserved structure of the PARP-2 WGR and the reported crystal structure of the human PARP-2 catalytic domain (31). Outside of the NTR, the only other regions exhibiting an elevated chance of disorder were located in linker regions connecting PARP-2 domains and subdomains (e.g. residues 208 to 217 connecting the WGR and CAT domains and residues 340 to 350 connecting the HD and ART subdomains of the CAT). Natively disordered protein regions play important functions, such as DNA binding (32,33), in a variety of proteins. We experimentally tested the prediction that the NTR of PARP-2 is natively disordered using limited proteolysis, CD spectroscopy, thermal denaturation and analytical gel filtration chromatography.
High susceptibility to proteolytic cleavage is a characteristic of natively disordered proteins, since their lack of secondary structure increases protease access to the polypeptide. A time course of proteolysis of the isolated NTR was performed using very limited catalytic amounts of trypsin (trypsin: substrate mass ratio of 1:2,400) to probe protease sensitivity (Figure 2B, top). SDS-PAGE analysis of the reaction time points revealed a rapid digestion of the NTR within 3 min. The time course of NTR proteolytic digestion was approximately the same in the presence and absence of DNA, with a slight protection observable at the 3- and 5-min time points in the presence of DNA (Figure 2B). Using 1:500 trypsin, the NTR of PARP-2 was digested in approximately one minute. In contrast, the core of the isolated DNA-binding Zn1 domain of PARP-1 was resistant to proteolysis under the same conditions for greater than 20 min (Figure 2B, bottom). The Zn1 domain is comparable in size to the NTR and has a similar distribution of Arg and Lys residues that are targeted by trypsin (Zn1 – 18, NTR – 21; Supplementary Figure S7A), and thus serves as a control and highlights the expected differences in protease sensitivity of a well-folded domain versus a natively disordered region.
We next tested the proteolytic sensitivity of FL PARP-2 to assess if the presence of other PARP-2 domains might influence NTR structure and thus protease sensitivity. Limited trypsin digestion of human PARP-2 in the absence or presence of DNA rapidly resulted in a stable decrease in apparent molecular weight, and the resulting fragment of PARP-2 persisted for greater than 30 min (Figure 2C). The observed decrease in molecular weight is consistent with the loss of the NTR, leaving a PARP-2 fragment with a mass approximating that of the WGR and CAT domains. Western blot analysis of the digestion reactions confirmed the loss of the N-terminus of PARP-2 (α-His), and the presence of the C-terminal catalytic domain (α-PARP), consistent with the interpretation that the stable, protease-resistant fragment consists of the WGR and CAT domains of PARP-2 (Supplementary Figure S7B). Notably, the decrease in α-His signal in the context of FL PARP-2 occurred over the same time frame as the isolated NTR digestion (Supplementary Figure S7B and Figure 2B). Together, the proteolysis results indicate that NTR digestion proceeds much more quickly than expected for a structured region, consistent with a natively disordered NTR of PARP-2. The NTR is slightly protected from proteolysis in the presence of DNA, consistent with its DNA-binding activity; however, the level of protection does not suggest that the NTR forms a secondary structure upon binding DNA.
To more directly assess the secondary structure of the PARP-2 NTR, we used far-UV CD spectroscopy. The CD spectrum of a natively disordered protein is characterized by a prominent negative molar ellipticity signal at the wavelength where random coil polypeptides absorb (∼200 nm), and a signal close to zero at the wavelength where α-helical polypeptides absorb (∼220 nm) (34). Indeed, the NTR of PARP-2 exhibited a CD spectrum indicative of an unstructured polypeptide (Figure 2D, top). Furthermore, the spectrum was largely unchanged in the presence of a DSB and nick SSB DNA structure, thus supporting that the NTR does not adopt a secondary structure upon binding to DNA. We also measured the CD spectrum at elevated temperatures, with the expectation that secondary structure will unfold as temperature is increased and thus substantially change the CD spectrum toward a random coil, whereas an intrinsically disordered protein will only show slight variations in the CD spectrum at elevated temperatures (34–36). As expected, the NTR of PARP-2 showed only slight and predictable CD spectrum variations as the temperature was increased from 4 to 50 to 80°C (Figure 2D, middle). In contrast, CD spectra measured at the same temperatures changed considerably for the PARP-2 WGR, which is expected to have both α-helix and β-strand content based on homology with PARP-1 and PARP-3 WGR structures (Figure 2D, bottom). Notably, the spectrum of the heat-denatured WGR domain was nearly identical to that of the NTR. As an additional test of thermal denaturation, we used differential scanning fluorimetry (DSF) in which a small-molecule probe emits a fluorescent signal upon binding non-specifically to hydrophobic regions that are exposed as a protein unfolds. Using DSF, the NTR of PARP-2 did not exhibit an increase in fluorescence, consistent with a natively disordered protein that does not contain a hydrophobic interior (Supplementary Figure S7C). In contrast, DSF of the PARP-2 WGR domain exhibited an increase in fluorescence signal upon thermal denaturation that is consistent with a folded domain (Supplementary Figure S7C). Lastly, analytical gel filtration analysis of the NTR indicated a greatly extended conformation based on its elution profile relative to globular protein controls (Supplementary Figure S7D), consistent with a lack of structure. In total, our analysis indicates that the NTR of PARP-2 is intrinsically disordered.
Human PARP-2 NTR contains a bipartite NLS
Natively disordered proteins are inherently difficult to study using high-resolution structural techniques like X-ray crystallography (37); however, as part of our structural analysis of the NTR of PARP-2 we took advantage of the stabilizing effects of the high-affinity interaction of the PARP-2 NTR with importin α1 (IMPα1) (38), a karyopherin that mediates nuclear import (39). IMPα1, lacking the N-terminal autoinhibitory importin β binding domain (IBB) (termed IMPα1ΔIBB), has been crystallized in complex with NLS peptides and well-folded NLS cargos, revealing the structural basis for NLS recognition by IMPα1, as well as providing useful insights into the structure of the cargo itself (39–41). We purified the NTR of PARP-2 bound to IMPα1ΔIBB and obtained crystals of the complex that diffracted to 1.9 Å. The structure of the complex was determined by molecular placement using as a search model IMPα1ΔIBB from PDB code 1Y2A (the peptide in 1Y2A was removed) (40). Initial weighted electron density maps clearly indicated regions of ordered peptide bound to IMPα1ΔIBB, and the density was unambiguously modeled with PARP-2 residues (Figure 3A and B). The atomic model of the complex was refined against data to 1.9 Å with an R/RFree of 0.17/0.19 (Table 1). The final model contains residues 19 to 23 and 33 to 40 of the PARP-2 NTR. The remaining residues of the NTR are presumed to be disordered, since washed crystals that were resolved on SDS-PAGE indicated that the entire ∼12 kDa NTR was still intact in the crystals (Supplementary Figure S8B). The crystallographic analysis is therefore consistent with the biophysical analysis above that indicates a natively disordered structure for the NTR.
Table 1. Crystallographic data and refinement statistics for the PARP-2 NLS:IMPα1ΔIBB complex.
| Data collectiona | |
| PARP-2 NTR bound to Importin α | |
| Space group | P212121 |
| Unit cell dimensions | a = 78.6 Å b = 90.0 Å, c = 100.0 Å |
| α = β = γ = 90.0 ° | |
| 1 molecule/asymmetric unit | |
| Wavelength (Å) | 1.08 |
| Resolution range (Å) | 45.0–1.9 (1.97–1.90) |
| Completeness (%) | 99.7 (99.4) |
| Average redundancy | 6.8 (6.7) |
| Mean (I/σI)b | 20.7 (1.9) |
| Rmerge (%)b | 5.9 (105.0) |
| Rpim (%)b | 2.5 (43.6) |
| Mean I CC(1/2)b | 0.999 (0.600) |
| Model refinementa | |
| Resolution range (Å) | 44.9–1.9 (1.97–1.90) |
| Number of reflections | 56617 (5556) |
| Rcrystc | 0.165 (0.266) |
| Rfreec | 0.190 (0.323) |
| Number of atoms/average B-factor (Å2) | 7177/47.3 |
| Importin α | 3697/42.8 |
| PARP-2 | 137/47.9 |
| solvent | 324/49.0 |
| Phi/Psi, most favored (%) | 98 |
| R.m.s.d. bond angles (°) | 1.27 |
| R.m.s.d. bond lengths (Å) | 0.010 |
aValues in parentheses refer to data in the highest resolution shell.
bAs calculated in SCALA (Winn et al., 2011): Rmerge = ∑hkl∑j|Ij – 〈I〉| / ∑hkl∑j Ij. 〈I〉 is the mean intensity of j observations of reflection hkl and its symmetry equivalents; Rpim takes into account measurement redundancy when calculating Rmerge; Mean I CC(1/2) is the correlation between mean intensities calculated for two randomly chosen half-sets of the data.
cRcryst = ∑hkl|Fobs – kFcalc|/ ∑hkl|Fobs|. Rfree = Rcryst for 5% of reflections excluded from crystallographic refinement.
The crystal structure of PARP-2 bound to IMPα1ΔIBB provided detailed insight into the nature of the IMPα1ΔIBB /PARP-2 interaction (Figure 3A). The PARP-2 NTR was clearly modeled as a classical bipartite NLS. In a classical bipartite NLS, there may be as many as five points of contact between the NLS and IMPα1, in both the major and the minor binding pockets (42). These points of contact are referred to as P1–P5, in the major site, and P1’-P5’, in the minor site. Human PARP-2 engages the major pocket primarily with residues K37 and K38 (P2 and P3) and engages the minor binding pocket primarily with residues K21 and R22 (P1’ and P2’) consistent with the manner in which importin alpha would bind to cargo presenting a bipartite NLS (Figure 3A and B). A bipartite NLS was predicted for human PARP-2 based on sequence analysis (15); however, cell localization data and mutagenesis indicated a monopartite NLS for murine PARP-2 (38). Consistent with sequence analysis and the crystal structure, we used live-cell localization experiments in HEK 293 cells to demonstrate that human PARP-2 utilizes a bipartite NLS for efficient nuclear localization (Figure 3C). Individual mutations of critical residues in the major (K37A, K38A) or minor sites (K21A, R22A) reduced, but did not abolish nuclear localization, ruling out that PARP-2 utilizes a monopartite NLS. In contrast, disrupting both binding sites led to efficient exclusion from the nucleus and enrichment in the cytoplasm (Figure 3C).
We next tested whether the IMPα1ΔIBB/NTR interaction influenced the DNA-binding activity of the NTR, since IMPα1 binding sometimes overlaps with DNA-binding regions and can thereby influence the activity of certain DNA-binding proteins (43,44). The complex of NTR with IMPα1ΔIBB was still able to measurably bind DNA, although with approximately 4-fold lower affinity compared to the NTR alone (3.9 μM versus 0.99 μM, Figure 3D and see also Supplementary Figure S8). The reduction in apparent affinity most likely indicates that IMPα1 binding introduces a degree of steric constraint on NTR. IMPα1 contacts with K37 and K38, located near the minimal NTR region supporting DNA-binding activity, are likely to be strong contributors to these steric constraints. Furthermore, we note that the overall decrease in polarization signal observed for the IMPα1/NTR complex likely reflects that these steric restraints prevent the binding of multiple NTR molecules onto DNA. Overall, these results indicate that the natively disordered NTR of human PARP-2 contains a bipartite NLS that is required for efficient transport into the nucleus.
The NTR of PARP-2 is not sufficient or required for localization to sites of DNA damage
With multiple domains contributing to PARP-2 interaction with DNA damage in vitro (Figure 1B), we were interested in establishing the domain requirements for the reported localization of PARP-2 to sites DNA damage (45). GFP was fused to human PARP-2 and PARP-2 mutant and deletion variants, and the localization kinetics of GFP-PARP-2 constructs to sites of laser-induced DNA damage were monitored using live-cell microscopy of HeLa cells. In constructs that deleted the NLS of PARP-2, a substitute NLS from SV40T antigen was inserted in order to preserve PARP-2 ability to localize to the nucleus (15,46,47).
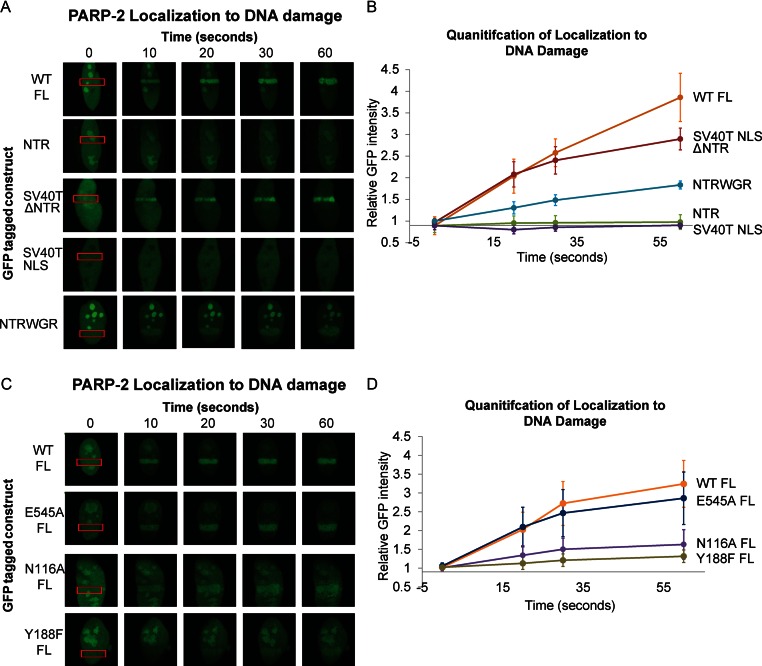
As expected and reported by others (15,38,45), wild-type (WT) FL PARP-2 localized to the nucleus and to nucleolar regions, and was rapidly recruited within seconds to sites of laser-induced DNA damage (Figure 4A–D). Also as expected, deletion of the PARP-2 NTR (ΔNTR) and replacement with the SV40T NLS (SV40T NLS ΔNTR) maintained PARP-2 in the nucleus, but did not lead to enrichment in the nucleolus. Interestingly, the NTR deletion construct efficiently localized to sites of DNA damage, with kinetics similar to that observed for FL WT PARP-2 (Figure 4A and B). This result was surprising given that the NTR of PARP-2 interacts with a variety of models for DNA damage (albeit at modest binding affinities; Figure 1B), and has frequently been assumed to fulfill the same damage recognition function as the PARP-1 NTR (zinc finger domains; Figure 1A). Instead, we found that the isolated NTR of PARP-2 was not enriched at sites of DNA damage, despite being abundantly produced in the nucleus and nucleolar regions (Figure 4A and B).
Figure 4.
PARP-2 recruitment to cellular sites of DNA damage. (A) and (C) Live-cell imaging of recruitment of GFP-PARP-2, truncation constructs (A) or FL mutants (C) to sites of laser-induced DNA damage. The region of laser irradiation is indicated with a red box. The images were captured at the various time points indicated. The images shown are representative the results obtained from at least three independent experiments. SV40T NLS is an N-terminal peptide tethered to PARP-2 truncations that lack the canonical NLS of PARP-2. (B) and (D) Quantitation of relative GFP intensity within the laser path relative to background (a non-irradiated area of the nucleus). Relative GFP signal averaged for ≥4 cells. Error bars represent the SD.
We focused therefore on the WGR and CAT domains for mediating PARP-2 recruitment to sites of DNA damage. We confirmed that the SV40T NLS does not recruit to sites of DNA damage on its own, and therefore is not likely to assist the WGR-CAT domains (ΔNTR) in their robust recruitment to sites of DNA damage (Figure 4A and B). Furthermore, a strict NTR deletion from PARP-2 without the addition of the SV40T NLS was also able to recruit to sites of DNA damage with rapid kinetics (Supplementary Figure S9). Consistent with the lack of an NLS, the cellular distribution of this protein was both nuclear and cytoplasmic. The WGR domain is expected to make key DNA contacts based on homology with the PARP-1 WGR (12,14), thus we anticipated that the WGR domain might contribute the key interactions that recruit PARP-2 to sites of DNA damage. However, the WGR domain alone was only weakly enriched at sites of DNA damage (Supplementary Figure S9), suggesting that the combination of the WGR and CAT domain are essential for efficient recruitment. Indeed, deletion of the CAT domain led to a decrease in the kinetics of PARP-2 recruitment to site of DNA damage (Figure 4A and B). These results are consistent with our in vitro binding experiments that suggest a synergistic cross-talk between the WGR and CAT domains during DNA binding (Figure 1B). Consistent with the biochemical analysis, we observed that the N116A mutant of FL PARP-2 displayed reduced recruitment to sites of DNA damage (Figure 4C and D). Furthermore, a mutation expected to directly target WGR interaction with DNA (Y188F) was also deficient in recruitment to sites of DNA damage (Figure 4C and D). Importantly, the reduced recruitment observed for N116A, Y188F and NTR-WGR (delta CAT) is not due to their catalytic deficiencies, since the recruitment kinetics of the inactive catalytic active site mutant E545A is nearly identical to the kinetics observed for WT PARP-2 (Figure 4C and D). Together, the cell localization experiments indicate an unsuspected requirement for a combination of the WGR and CAT domains to mediate PARP-2 recruitment to sites of DNA damage, and highlight the importance of interdomain contacts for PARP-2 recognition of DNA damage.
DISCUSSION
The DDR-PARPs appear to have both overlapping and specific roles in DNA repair pathways (4). In this study, we have undertaken a detailed structural and functional analysis to better understand how PARP-2 domain functions and requirements have been specialized, in particular relative to PARP-1. Interestingly, we found that the NTR of PARP-2 is a natively unstructured region of the protein that contributes to the overall binding affinity and specificity of PARP-2 for DNA. The DNA-binding contribution is particularly important for interaction with and activation on SSB DNA. We envision that the structural plasticity common to natively disordered proteins allows the NTR of PARP-2 to adapt to a range of damaged DNA structures, and perhaps can act to tether PARP-2 on DNA (Figure 5). Although the NTR is not a strictly required component of the PARP-2 activation mechanism, it likely plays an important role in the cell by contributing to the overall robust DNA-binding activity that allows PARP-2 to act as a DNA damage sensor.
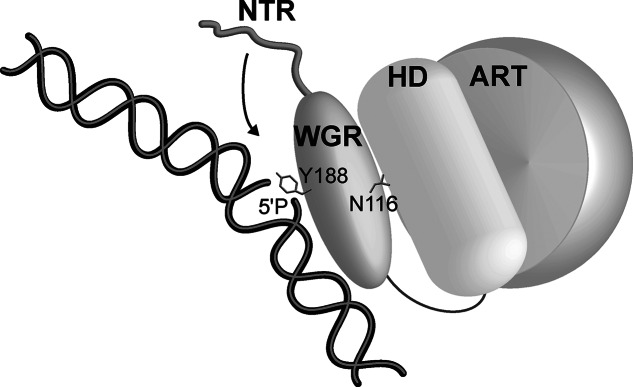
Figure 5.
Model for PARP-2 recognition of DNA damage. The WGR domain forms direct contacts with DNA damage (e.g. Y188); however, WGR domain cross-talk with the HD region of the CAT domain (e.g. N116) is required to allow the domains to synergize and effectively detect DNA damage. These same contacts are necessary for DNA damage-dependent activation. The inherent flexibility of the NTR allows it to engage DNA in an adaptable manner, dependent on the nature of the damage DNA intermediate (e.g. gapped or nicked SSB).
Earlier studies have reported on the DNA-binding affinity of the PARP-2 NTR, demonstrating that the NTR indeed binds to DNA and suggesting that the PARP-2 NTR could fulfill the function of the more elaborate NTR of PARP-1 (16), which contains a series of specialized zinc finger domains. The PARP-1 zinc fingers partake in the allosteric activation mechanism and are responsible for recruitment to sites of DNA damage. Thus, our initial expectation was that the NTR of PARP-2 would similarly serve as a DNA damage sensor. We unexpectedly found that the NTR does not localize to sites of DNA damage on its own, and furthermore that it is not required for PARP-2 localization to sites of DNA damage. Moreover, our results have confirmed and extended the understanding of the NTR as key to nuclear import and nucleolar localization. Our structural and functional analysis revealed that human PARP-2 NTR contains a bipartite NLS sequence. Key binding determinants at position K37/K38 and K21/R22 are essential to confer nuclear localization in a live-cell assay.
Most importantly, our results have demonstrated that the WGR and CAT domains of PARP-2 are necessary and sufficient for recruitment to sites of laser-induced DNA damage in live cells. Recent structural and biochemical studies have highlighted the role of the WGR domain in DNA binding and the allosteric activation mechanism of DDR-PARPs (12–14,48). Our study extends these results to show that the WGR domain of PARP-2 is involved in targeting to sites of DNA damage, in addition to playing a central role in the activation mechanism. Another surprising result from this study was that the CAT domain contributes to the DNA-binding affinity of PARP-2 and the ability of PARP-2 to localize to sites of DNA damage. Disrupting a key contact point between the WGR and the CAT domains lowered PARP-2 affinity for DNA, and comprised the speed of recruitment to sites of DNA damage. Thus, these results illustrate how the communication between PARP-2 domains is important not just for activation but also for DNA binding, and the results highlight how the multiple domains of PARP-2 work together. In summary, we have provided new structural and biochemical insights into PARP-2 that improve our understanding of the specialization of DDR-PARPs.
Supplementary Material
Acknowledgments
We would like to thank the Sidney Kimmel Cancer Center X-ray Crystallography and Molecular Interactions Facility at Thomas Jefferson University (CA56036) for assistance with circular dichroism, and the Bioimaging Shared Resource of the Sidney Kimmel Cancer Center (CA56036), particularly Dr. Covarrubias, for assistance performing live cell imaging experiments. We would also like to thank Dr. Phillip Wedegaertner and his laboratory for assistance with and use of the Olympus BX-61 microscope. We thank NSLS staff for access and assistance with data collection (Beamline X29A, Brookhaven National Laboratory).
Author contributions: A.A.R. performed all of the experiments. A.A.R. and J.M.P. designed the experiments, analysed data and wrote the manuscript. G.C. contributed to the design of Importin alpha crystallization. J.M.P. directed the research.
Footnotes
Present address: John M. Pascal, Department of Biochemistry and Molecular Medicine, Université de Montréal, Montréal, QC H3C 3J7, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM087282]; American Cancer Society [RSG0918301]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 2.Vyas S., Chesarone-Cataldo M., Todorova T., Huang Y.-H., Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013;4:2240. doi: 10.1038/ncomms3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Žaja R., Mikoč A., Barkauskaite E., Ahel I. Molecular insights into poly(ADP-ribose) recognition and processing. Biomolecules. 2012;3:1–17. doi: 10.3390/biom3010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Vos M., Schreiber V., Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem. Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Kutuzov M.M., Khodyreva S.N., Amé J.C., Ilina E.S., Sukhanova M. V., Schreiber V., Lavrik O.I. Interaction of PARP-2 with DNA structures mimicking DNA repair intermediates and consequences on activity of base excision repair proteins. Biochimie. 2013;95:1208–1215. doi: 10.1016/j.biochi.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Kutuzov M.M., Khodyreva S.N., Ilina E.S., Sukhanova M. V., Amé J.-C., Lavrik O.I. Interaction of PARP-2 with AP site containing DNA. Biochimie. 2015 doi: 10.1016/j.biochi.2015.02.010. doi:10.1016/j.biochi.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Huber A., Bai P., Murcia J.M.D., Murcia G.D. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair. 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Beck C., Boehler C., Guirouilh Barbat J., Bonnet M.E., Illuzzi G., Ronde P., Gauthier L.R., Magroun N., Rajendran A., Lopez B.S., et al. PARP3 affects the relative contribution of homologous recombination and nonhomologous end-joining pathways. Nucleic Acids Res. 2014;42:5616–5632. doi: 10.1093/nar/gku174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rulten S.L., Fisher A.E.O., Robert I., Zuma M.C., Rouleau M., Ju L., Poirier G., Reina-San-Martin B., Caldecott K.W. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Boehler C., Gauthier L.R., Mortusewicz O., Biard D.S., Saliou J.-M., Bresson A., Sanglier-Cianferani S., Smith S., Schreiber V., Boussin F., et al. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ménissier de Murcia J., Ricoul M., Tartier L., Niedergang C., Huber A., Dantzer F., Schreiber V., Amé J.C., Dierich A., LeMeur M., et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langelier M.-F., Riccio A.A., Pascal J.M. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014;42:7762–7775. doi: 10.1093/nar/gku474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langelier M.F., Pascal J.M. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol. 2013;23:134–143. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langelier M.-F., Planck J.L., Roy S., Pascal J.M. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meder V.S., Boeglin M., de Murcia G., Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J. Cell Sci. 2005;118:211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- 16.Amé J.C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Höger T., Ménissier-de Murcia J., De Murcia G. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 17.Aravind L., Koonin E. V. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 18.Langelier M.F., Planck J.L., Roy S., Pascal J.M. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 2011;286:10690–10701. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winter G. Xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 2010;43:186–190. [Google Scholar]
- 21.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams P.D., Afonine P. V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langelier M.F., Servent K.M., Rogers E.E., Pascal J.M. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J. Biol. Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 27.Langelier M.F., Ruhl D.D., Planck J.L., Kraus W.L., Pascal J.M. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 2010;285:18877–18887. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steffen J.D., Tholey R.M., Langelier M.F., Planck J.L., Schiewer M.J., Lal S., Bildzukewicz N.A., Yeo C.J., Knudsen K.E., Brody J.R., et al. Targeting PARP-1 allosteric regulation offers therapeutic potential against cancer. Cancer Res. 2014;74:31–37. doi: 10.1158/0008-5472.CAN-13-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchan D.W.A., Minneci F., Nugent T.C.O., Bryson K., Jones D.T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 31.Karlberg T., Hammarström M., Schütz P., Svensson L., Schüler H. Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888. Biochemistry. 2010;49:1056–1058. doi: 10.1021/bi902079y. [DOI] [PubMed] [Google Scholar]
- 32.Ward J.J., Sodhi J.S., McGuffin L.J., Buxton B.F., Jones D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 34.Chemes L.B., Alonso L.G., Noval M.G., de Prat-Gay G. Circular dichroism techniques for the analysis of intrinsically disordered proteins and domains. Methods Mol. Biol. (Clifton, NJ) 2012;895:387–404. doi: 10.1007/978-1-61779-927-3_22. [DOI] [PubMed] [Google Scholar]
- 35.Kjaergaard M., Brander S., Poulsen F.M. Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J. Biomol. NMR. 2011;49:139–149. doi: 10.1007/s10858-011-9472-x. [DOI] [PubMed] [Google Scholar]
- 36.Gast K., Zirwer D., Damaschun G. Are there temperature-dependent structural transitions in the ‘intrinsically unstructured’ protein prothymosin alpha? Eur. Biophys. J. 2003;31:586–594. doi: 10.1007/s00249-002-0254-y. [DOI] [PubMed] [Google Scholar]
- 37.Linding R., Jensen L.J., Diella F., Bork P., Gibson T.J., Russell R.B. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Haenni S.S., Hassa P.O., Altmeyer M., Fey M., Imhof R., Hottiger M.O. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int. J. Biochem. Cell Biol. 2008;40:2274–2283. doi: 10.1016/j.biocel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Pumroy R.A., Ke S., Hart D.J., Zachariae U., Cingolani G. Molecular determinants for nuclear import of Influenza A PB2 by importin α isoforms 3 and 7. Structure. 2015;23:374–384. doi: 10.1016/j.str.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen M.H., Ben-Efraim I., Mitrousis G., Walker-Kopp N., Sims P.J., Cingolani G. Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin α. J. Biol. Chem. 2005;280:10599–10606. doi: 10.1074/jbc.M413194200. [DOI] [PubMed] [Google Scholar]
- 41.Lott K., Bhardwaj A., Sims P.J., Cingolani G. A minimal Nuclear Localization Signal (NLS) in human phospholipid scramblase 4 that binds only the minor NLS-binding site of importin α1. J. Biol. Chem. 2011;286:28160–28169. doi: 10.1074/jbc.M111.228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lokareddy R.K., Hapsari R.A., van Rheenen M., Pumroy R.A., Bhardwaj A., Steen A., Veenhoff L.M., Cingolani G. Distinctive properties of the nuclear localization signals of inner nuclear membrane proteins Heh1 and Heh2. Structure (London, England) 2015;23:1305–1316. doi: 10.1016/j.str.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaCasse E.C., Lefebvre Y. a. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeiri M., Horie K., Takahashi D., Watanabe M., Horie R., Simizu S., Umezawa K. Involvement of DNA binding domain in the cellular stability and importin affinity of NF-κB component RelB. Org. Biomol. Chem. 2012;10:3053. doi: 10.1039/c2ob07104e. [DOI] [PubMed] [Google Scholar]
- 45.Mortusewicz O., Amé J.C., Schreiber V., Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McConnell B. V, Koto K., Gutierrez-Hartmann A. Nuclear and cytoplasmic LIMK1 enhances human breast cancer progression. Mol. Cancer. 2011;10:75. doi: 10.1186/1476-4598-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith S., Lange T. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- 48.Pascal J.M., Ellenberger T. The rise and fall of poly(ADP-ribose): an enzymatic perspective. DNA Repair. 2015;32:10–16. doi: 10.1016/j.dnarep.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.