Abstract
Group II introns are large RNA enzymes that can excise as lariats, circles or in a linear form through branching, circularization or hydrolysis, respectively. Branching is by far the main and most studied splicing pathway while circularization was mostly overlooked. We previously showed that removal of the branch point A residue from Ll.LtrB, the group II intron from Lactococcus lactis, exclusively leads to circularization. However, the majority of the released intron circles harbored an additional C residue of unknown origin at the splice junction. Here, we exploited the Ll.LtrB-ΔA mutant to study the circularization pathway of bacterial group II introns in vivo. We demonstrated that the non-encoded C residue, present at the intron circle splice junction, corresponds to the first nt of exon 2. Intron circularization intermediates, harboring the first 2 or 3 nts of exon 2, were found to accumulate showing that branch point removal leads to 3′ splice site misrecognition. Traces of properly ligated exons were also detected functionally confirming that a small proportion of Ll.LtrB-ΔA circularizes accurately. Overall, our data provide the first detailed molecular analysis of the group II intron circularization pathway and suggests that circularization is a conserved splicing pathway in bacteria.
INTRODUCTION
Group II introns are retromobile genetic elements found in bacteria and bacterial-derived organelles, such as plant and fungal mitochondria, and plant chloroplasts. However, no functional copies were discovered in the nuclear genome of eukaryotes, whereas only few examples are present in archaea (1,2). Bacterial group II introns have a wide phylogenetic distribution although only one quarter of all sequenced bacterial genomes harbor one to a few copies (2). They are mostly found in non-coding sequences and within other mobile genetic elements while organellar group II introns generally interrupt housekeeping genes (3).
On an evolutionary standpoint, group II introns are considered as important retromobile elements that significantly shaped the origin and evolution of contemporary eukaryotic genomes. They are the proposed ancestors of the telomerase enzyme, the spliceosome machinery as well as the highly abundant spliceosomal introns and non-LTR retroelements (3,4). Group II introns can also be disseminated within and between bacterial species by conjugation. They can invade the chromosome or resident plasmids of their new hosts following their conjugative transfer using either the retrohoming or retrotransposition pathway (5–8).
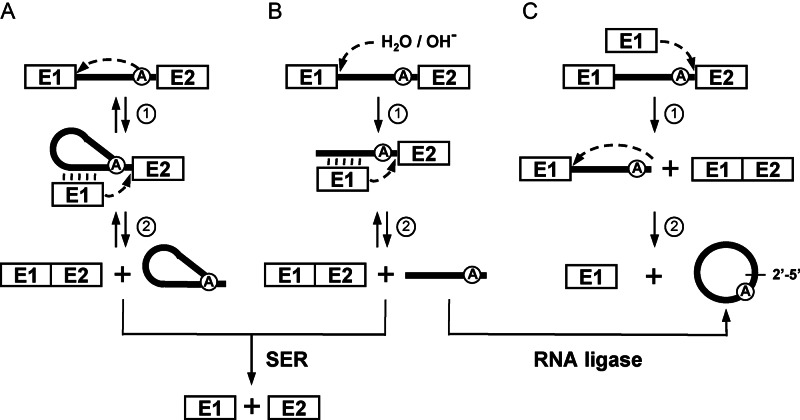
Group II introns are also large RNA enzymes that self-splice from their primary mRNA transcripts both in vivo and in vitro. Even though, they absolutely require the assistance of maturases to adopt their catalytically active tridimensional conformation in vivo, they can self-splice without protein assistance under high salt conditions in vitro (2). Group II introns can excise while concurrently ligating their flanking exons through three different splicing pathways: branching (Figure 1A), hydrolysis (Figure 1B) and circularization (Figure 1C). These splicing pathways involve two consecutive transesterification reactions (Figure 1, steps 1 and 2) and release the intron as either branched lasso-like structures called lariats, in linear forms or as closed circles, respectively (9). Branching is definitively the most studied pathway and considered as the main group II intron splicing pathway. Hydrolysis and circularization are regarded as secondary splicing pathways and are not as well characterized as branching (9,10). Although a circularization pathway for group II introns was first proposed in 2001 (Figure 1C) (11) and that the first group II intron circles were detected by electron microscopy 35 years ago (12,13) this splicing pathway was not subjected to detailed molecular analyses. Following the initial description of the branching (14–16) and the hydrolysis pathways (17,18), circularization of group II introns was mostly overlooked. Nevertheless, since that time, group II intron RNA circles from various systems were described both in vivo (19–23) and in vitro (11,24). We recently demonstrated that a significant proportion of the model group II intron, Ll.LtrB, from the Gram-positive bacterium Lactococcus lactis, excises as circles in vivo (19). Interestingly, we detected intron circles for all the Ll.LtrB mutants studied and demonstrated that both branching and circularization take place concurrently in vivo. In addition, we found that the Ll.LtrB-ΔA mutant, lacking the branch point A residue, does not produce lariats and exclusively excises as circles. However, upon circularization, a majority of the Ll.LtrB-ΔA circles were found to harbor an additional non-encoded C residue of unknown origin at the splice junction (19).
Figure 1.
Group II intron splicing pathways. (A) Branching pathway. Following transcription of the interrupted gene, the 2′-OH residue of the branch-point nucleotide (A) initiates the first nucleophilic attack at the exon 1-intron junction (step 1). This transesterification reaction connects the 5′ end of the intron to the branch point and releases exon 1 that remains associated to the intron through base pairing interactions (EBS-IBS interactions) (vertical lines). The liberated 3′-OH at the end of exon 1 then initiates a second nucleophilic attack at the intron–exon 2 junction (step 2), ligating the two exons and releasing the intron as a lariat. (B) Hydrolytic pathway. A hydroxyl ion or a water molecule initiates the first nucleophilic attack at the exon 1-intron junction (step 1). The second nucleophilic attack at the intron–exon 2 junction is initiated by the liberated 3′-OH at the end of exon 1 (step 2) which ligates the two exons and releases a linear intron. (C) Circularization pathway. The first nucleophilic attack takes place at the intron-exon 2 junction and is initiated by the 3′-OH of a free exon 1 (step 1) generating ligated exons and a circularization intermediate where the linear intron is still attached to exon 1. Next, the 2′-OH of the last intron residue is thought to initiate the second nucleophilic reaction at the exon 1-intron junction (step 2) resulting in intron circularization and the release of free exon 1. An additional potential source of free exon 1 is the spliced exon reopening (SER) reaction, where both excised lariats and linear introns can recognize and hydrolyze ligated exons at the splice junction. An alternative pathway for intron circle formation is the potential circularization of an excised linear intron by a host-encoded RNA ligase.
Here, we took advantage of the Ll.LtrB-ΔA mutant, that exclusively splices as circles, to address at the molecular level the circularization pathway of bacterial group II introns in vivo. Using various mutants of Ll.LtrB-ΔA we demonstrated that the non-encoded C residue, present at the splice junction of released intron circles, corresponds to the first nt of exon 2. We also discovered that removal of the branch point A residue from Ll.LtrB leads to accumulation of precursor mRNAs and intron cirularization intermediates in L. lactis total RNA extracts. The Ll.LtrB-ΔA circularization intermediates were also found to harbor short CA and CAU tails at the 3′ end of the intron, corresponding to the first nts of exon 2, demonstrating that the 3′ splice site is mostly misrecognized during the first transesterification reaction of the circularization pathway. On the other hand, a functional splicing assay confirmed the presence of accurately ligated exons consistent with the detection of small amounts of perfect intron circles not harboring an extra C at the splice junction. Taken together, our data indicate that Ll.LtrB-ΔA and Ll.LtrB-WT share the same circularization pathway in vivo. Removal of the branch point A residue from Ll.LtrB prevents branching and significantly tilts the balance of the first transesterification reaction of the circularization pathway towards misrecognition of the 3′ splice site, ultimately leading to the addition of the first nt of exon 2 (C) at the splice junction of released intron circles. Overall, this work provides the first detailed molecular analysis of the group II intron circularization pathway.
MATERIALS AND METHODS
Bacterial strains and plasmids
The Escherichia coli DH10β strain was used for both cloning and plasmid amplification, and was grown in LB broth at 37°C with shaking. The Lactococcus lactis strains NZ9800ΔltrB::tet (NZ9800ΔltrB) (TetR) (25) and LMO231 (FusR) were grown in M17 broth supplemented with 0.5% glucose (GM17) at 30°C without shaking. The following antibiotic concentrations were used: chloramphenicol (Cam), 10 μg/ml; spectinomycin (Spc), 300 μg/ml; teracycline (Tet), 3 μg/ml; fusidic acid (Fus), 25 μg/ml.
Some plasmids were previously described: pDL-P232-Ll.LtrB-WT (WT), pDL-P232-Ll.LtrB-ΔLtrA (ΔLtrA) (amino acids 40 to 572 in LtrA were replaced by RT residues (26)), pDL-P232-Ll.LtrB-LtrAMat− (LtrAMat−) (amino acids SC463 were mutated to LA (26)), pLE-P232-LtrA (7), pDL-P232-Ll.LtrB-ΔA (ΔA) (19). The following kits were used for site directed mutagenesis: QuickChange Multi Site-Directed Mutagenesis, Stratagene (pDL-P232-Ll.LtrB-WT-C/U-E1, pDL-P232-Ll.LtrB-ΔA-C/U-E1, Ll.LtrB-LtrAMat−-C/U-E1) and Q5 Site-Directed Mutagenesis, New England Biolabs (pDL-P232-Ll.LtrB-WT-C/U-E2, pDL-P232-Ll.LtrB-ΔA-C/U-E2, Ll.LtrB-LtrAMat−-C/U-E2, pDL-P232-LtrB-ΔLl.LtrB, pDL-P232-LtrB-ΔLl.LtrB-ΔE2, pDL-P232-LtrB-ΔLl.LtrB-ΔHis) (primers in Supplementary Table S1).
Nucleic acid preparations, RT-PCR, poisoned primer extension, Northern blots and conjugation assays
Total RNA and nucleic acids were isolated from NZ9800ΔltrB cells harboring different intron variants as previously described (7). RT-PCR (7), conjugation (7), poisoned primer extensions (27) and Northern blots (27) were performed as previously described (primers in Supplementary Table S1).
Amplification of 3′ and 5′ intron ends
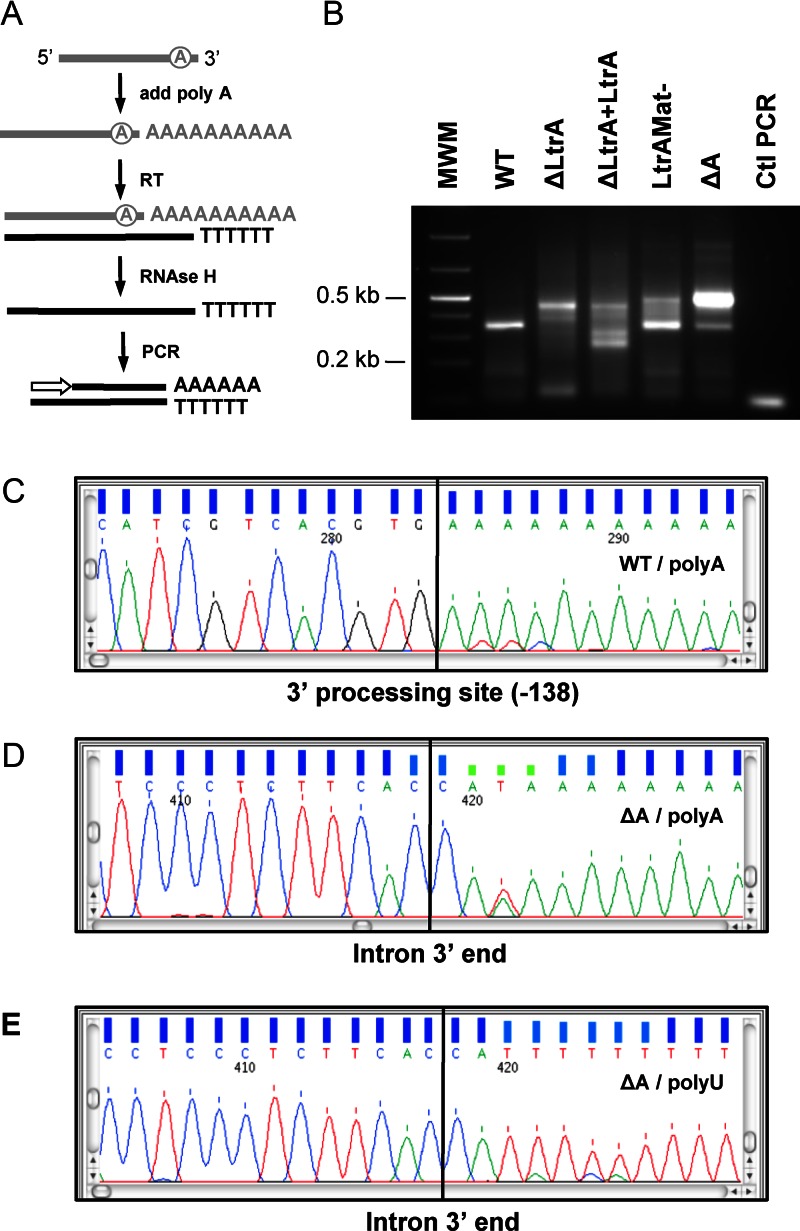
Total RNA extracts (1 μg) of L. lactis expressing various Ll.LtrB constructs were analyzed. To identify intron 3′ ends (Figure 5A), 5U of E. coli Poly(A) Polymerase (New England Biolabs) was used to extend a polyA tail (10 min at 37°C). Using an oligo dT and the SSIIRT (Invitrogen) a cDNA was synthesized at 42°C for 50 min. Following removal of the RNA strand of the RNA/DNA duplex by the RNase H (New England Biolabs) (20 min at 37°C) the single strand cDNA was amplified by PCR with the Phusion High-Fidelity DNA polymerase (New England Biolabs) using an intron specific primer (Supplementary Table S1) and the oligo dT. For Ll.LtrB-ΔA a second amplification of the 3′ end was performed where a polyU instead of a polyA tail was added using 2U of Poly(U) Polymerase (New England Biolabs) (10 min at 37°C). In that case, an oligo dA was used for cDNA synthesis. The isolated PCR bands (Figure 5C–E) were sequenced toward the 5′ end using intron specific primers (Supplementary Table S1). Amplification of free 5′ intron ends (Supplementary Figure S1A) started by cDNA synthesis using the SSIIRT (Invitrogen) and an intron specific primer (Supplementary Table S1) at 42°C for 50 min. Following removal of the RNA strand of the RNA/DNA duplex by the RNase H (New England Biolabs) (20 min at 37°C) the single strand cDNA was extended with a polyA tail with 10U of Terminal Transferase (New England Biolabs) (30 min at 37°C). The single strand cDNA was amplified by PCR with the Phusion High-Fidelity DNA polymerase (New England Biolabs) using an intron specific primer (Supplementary Table S1) and an oligo dT. The series of G residues present between the intron 5′ end and the polyA tail was most probably added by the intrinsic terminal transferase activity of the RT enzyme when it reached the 5′ end of the RNA during cDNA synthesis (28). The isolated PCR bands (Supplementary Figure S1C–E) were sequenced toward the 3′ end using intron specific primers (Supplementary Table S1).
Figure 5.
Identification of Ll.LtrB 3′ ends in vivo. Free intron 3′ ends were amplified by RT-PCR from L. lactis total RNA extracts. (A) Intron 3′ ends were identified by first extending the intron RNA with a polyA tail followed by the synthesis of a cDNA with an oligo dT. The RNA strand was removed by an RNAse H treatment and the single strand DNA amplified by PCR. (B) The PCR reactions were ran on a 2% agarose gel and the chromatogram of some of the sequenced bands are shown (C–D). The same procedure was repeated for the Ll.LtrB-ΔA construct but extending a polyU instead of a polyA tail at the 3′ end of the intron (E).
RESULTS
The first nucleotide of exon 2 is incorporated at the splice junction of Ll.LtrB-ΔA circles
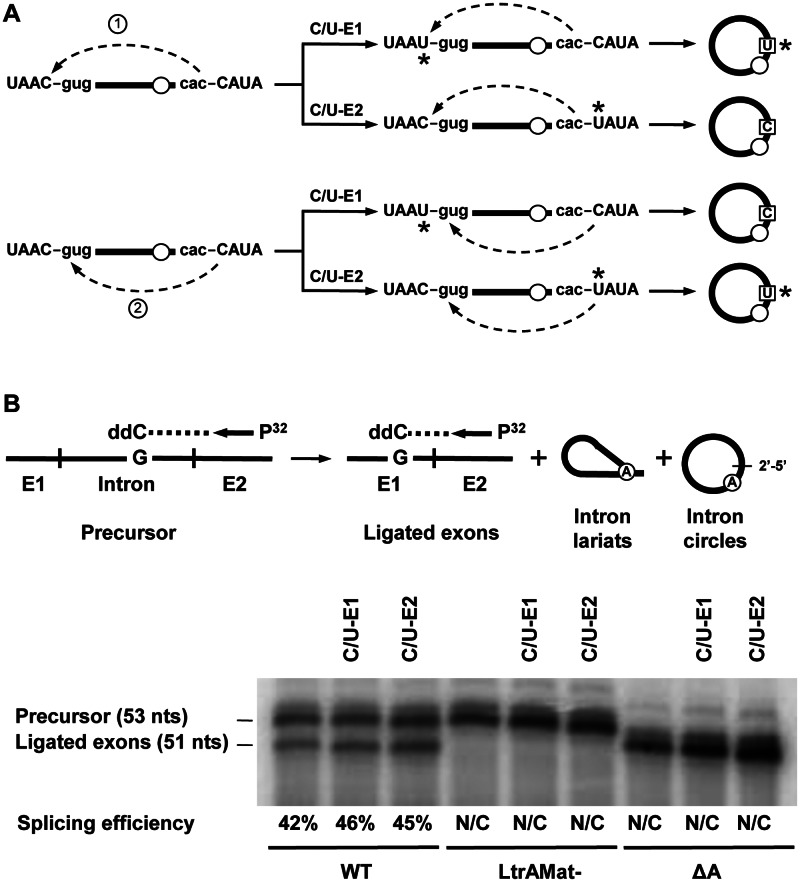
We previously demonstrated that the great majority of the released RNA circles of Ll.LtrB-ΔA and Ll.LtrB-LtrAMat− in vivo harbor an additional non-encoded C residue at the spliced junction (19). The origin of this additional residue was unclear since both nucleotides flanking Ll.LtrB are C residues (Figure 2A, left). The second nucleophilic attack of the proposed circularization pathway (Figure 1C, step 2) could thus be initiated by the last nt of the intron, misrecognizing the 5′ splice site and leading to the inclusion of the last nt of exon 1 (Figure 2A, scenario 1). Alternatively, the first nt of exon 2 may accurately identify the 5′ splice site leading to its inclusion at the intron circle splice junction (Figure 2A, scenario 2).
Figure 2.
Splicing efficiency of various Ll.LtrB constructs. (A) Schematic of the two potential transesterification reactions leading to the incorporation of an additional C residue at the Ll.LtrB-ΔA circle splice junction are depicted on the left. The second transesterification reaction of the circularization pathway can be initiated by the 2′OH of either the last nt of the intron (scenario 1), leading to the incorporation of the last residue of exon 1, or the first nt of exon 2 (scenario 2) leading to its own incorporation. The potential circularization products generated for scenarios 1 and 2 are illustrated on the right. The C to U replacements in exon 1 (C/U-E1) and exon 2 (C/U-E2) are represented by asterisks in both the precursors and the released intron circles. Exon and intron sequences are depicted in upper and lower case, respectively. The absence of the branch point A residue is illustrated by an empty circle. (B) Ll.LtrB splicing efficiency assessed by poisoned primer extension. This assay monitors splicing efficiency by comparing the relative abundance of precursor and ligated exons from total RNA extracts. A P32-labeled primer (Supplementary Table S1) complementary to exon 2 was extended from both the precusor and the ligated exons in the presence of a high concentration of ddCTP (top). Since the sequence of the two RNAs are different after the exon 2 junction the first G residue encountered is at a different distance from the primer generating differently sized bands for the precursor (53 nts) and the ligated exons (51 nts). The precursor band for the three Ll.LtrB-ΔA variants is 52 nts instead of 53 because the primer extension reaction extends past the intron branch point which is missing in these three introns. Splicing efficiency is calculated as the relative intensity of the ligated exons and precursor bands (ligated exons / precursor + ligated exons). N/C: not calculated.
To address the origin of the non-encoded C residue at the intron circle splice junction we generated the Ll.LtrB-ΔA-C/U-E1 and Ll.LtrB-ΔA-C/U-E2 mutants where the two C residues flanking the intron were independently replaced by Us (Figure 2A, right). The same two mutants were also generated for Ll.LtrB-LtrAMat− and Ll.LtrB-WT. The C residues were specifically replaced by Us in order to minimize their effect on splice site recognition and splicing efficiency. The U residues allowed for G:U instead of G:C base pair interactions at both 5′ (EBS1:IBS1) and 3′ splice sites (δ:δ′).
Splicing efficiency of the intron mutants was assessed by poisoned primer extension (27). A P32-labeled primer complementary to exon 2 was extended from both the precursor and ligated exons in the presence of a high concentration of ddCTP (Figure 2B, top). Since the sequence of the two RNAs are different after the exon 2 junction the first G residue encountered is at a different distance from the primer generating differently sized bands for the precursor (53 nts) and ligated exons (51 nts) (Figure 2B, top). The splicing efficiency was found to be the same for the two C/U mutants compared to Ll.LtrB-WT (Figure 2B, bottom). This shows that substituting either the last nt of exon 1 or the first nt of exon 2 from C to U does not affect Ll.LtrB-WT splicing efficiency. For the Ll.LtrB-ΔA and Ll.LtrB-LtrAMat− variants, that do not splice efficiently, only the precursor band was detected for all constructs showing that the C/U mutations on both sides of the intron do not influence their splicing phenotype.
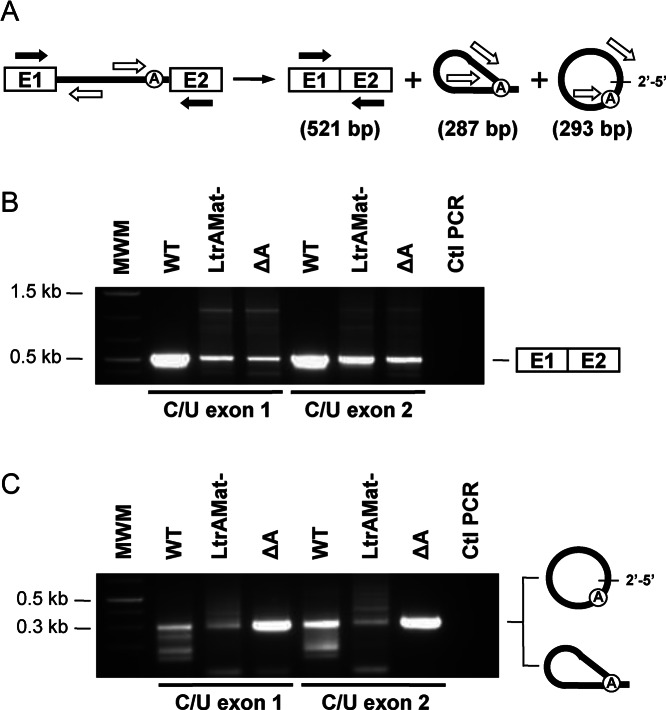
Splicing accuracy of the C/U mutants was next assessed and compared to their wild-type intron counterparts by RT-PCR of ligated exons and released intron junctions (Figure 3) (19). Direct sequencing of the ligated exons bands (Figure 3B, 521 bp) confirmed accurate exon joining for the six C/U intron mutants. On the other hand, the RT-PCR bands of released intron junctions (Figure 3C, 287 or 293 bp) were isolated, cloned and the sequence of independent clones was obtained. Similarly to Ll.LtrB-WT (19) we detected intron lariats, intron circles and alternatively circularized introns for Ll.LtrB-WT-C/U-E1 and Ll.LtrB-WT-C/U-E2. As previously described (19), the great majority of intron circles did not harbor additional nt at the splice junction regardless if the intron was flanked by a C or a U residue on its 5′ or 3′ end. This confirms that the C/U mutations flanking Ll.LtrB-WT do not affect 5′ and 3′ splice site recognition. In contrast, both C/U mutations significantly affect the splicing accuracy of Ll.LtrB-LtrAMat− leading to almost exclusively alternatively spliced circles and preventing further analysis. Similarly to Ll.LtrB-ΔA (19), all Ll.LtrB-ΔA-C/U-E1 circles harbored an extra C at the splice junction (Figure 2A, scenario 2). However, the splice junction of the Ll.LtrB-ΔA-C/U-E2 circles exclusively harbored an additional U residue instead of a C (Figure 2A, scenario 2). Interestingly, the limited number of Ll.LtrB-WT-C/U-E1 and Ll.LtrB-WT-C/U-E2 circles, harboring respectively a non-encoded C or U residue, also supported scenario 2 (Figure 2A, scenario 2). The identification of an additional U at the splice junction of the Ll.LtrB-ΔA-C/U-E2 and some Ll.LtrB-WT-C/U-E2 circles demonstrates that the non-encoded nt is accurately read by the RT enzyme during RT-PCR and validate the identity of the C residue at the junction of Ll.LtrB-ΔA-C/U-E1 and Ll.LtrB-ΔA circles.
Figure 3.
RT-PCR amplification of ligated exons and excised intron junctions. (A) Schematic of the Ll.LtrB splicing pathway. Position of the primers (Supplementary Table S1) used to amplify ligated exons (black arrows, 521 bp) and the intron splice junction (open arrows, 287 or 293 bp) is depicted. RT-PCR amplifications (B) (ligated exons) (C) (intron splice junction) were performed on total RNA extracts from L. lactis (NZ9800ΔltrB) harboring different Ll.LtrB constructs expressed under the control of the P23 constitutive promoter. RT-PCR amplicons of ligated exons (B, 521 bp) were excised and directly sequenced while the intron splice junction bands (C, 287 or 293 bp) were phosphorylated, cloned in pBS (SmaI) and sequenced.
Overall, these results demonstrate that the non-encoded C residue present at the splice junction of Ll.LtrB-ΔA circles correspond to the first nt of exon 2 (Figure 2A, scenario 2). The additional C residue is incorporated at the intron circle splice junction during the second step of the circularzation pathway through accurate recognition of the 5′ splice site by the 2′OH residue of the first nt of exon 2 (Figure 2A, scenario 2). Our data also suggest that the 3′ splice site is mostly misrecognized during the first transesterification reaction of the circularization pathway (Figure 1C, step 1) leaving at least one extra nt at the intron 3′ end.
Accumulation of Ll.LtrB-ΔA circularization intermediates in L. lactis
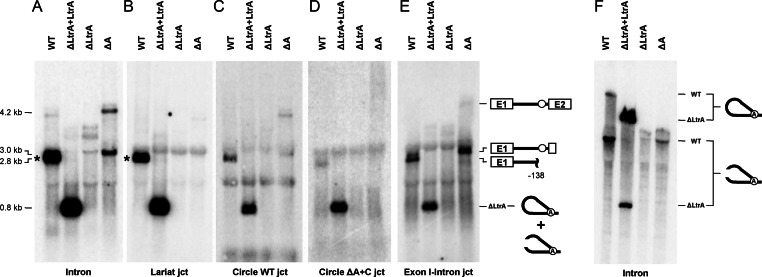
We next compared splicing intermediates of Ll.LtrB-ΔA with controls by Northern blots using a series of probes specific to either the intron (Intron), the intron lariat junction (Lariat jct), the intron circle junction (Circle WT jct), the Ll.LtrB-ΔA circle junction (Circle ΔA+C jct) and the 5′ splice site (Exon 1-Intron jct) (Figure 4). The four junction probes were designed to recognize the same number of nts on both sides of their respective junctions (Supplementary Table S1). The intron (Figure 4A) and 5′ splice site (Figure 4E) probes revealed that the full-length precursor mRNA of Ll.LtrB-ΔA (∼4.2 kb) accumulates when compared to Ll.LtrB-WT and Ll.LtrB-ΔLtrA + LtrA that both splice very efficiently (27). A more intense Ll.LtrB-ΔA band (∼3.0 kb) was also detected by the same two probes (Figure 4A and E). According to its size and the fact that it is specifically revealed by the 5′ splice site junction probe, this band most likely corresponds to the splicing intermediate of the circularization pathway consisting of exon 1 attached to full-length Ll.LtrB-ΔA (Figure 1C). We were unable to detect Ll.LtrB-ΔA circles from L. lactis total RNA extracts even if we used a probe specific to the circular junction of Ll.LtrB-ΔA (Figure 4D). In addition, following migration through a denaturing polyacrylamide gel, no retarded bands corresponding to intron circles, in the size range of the control intron lariats, were revealed with the general intron probe (Figure 4F).
Figure 4.
Detection of Ll.LtrB splicing intermediates in vivo. Northern blot hybridizations were performed with various P32-labeled probes (Supplementary Table S1). L. lactis RNA extracts were run on denaturing agarose (A–E) or acrylamide gels (F). A general intron probe (A and F, Intron) was used to detect all splicing intermediates and intron forms while the other probes were designed to detect specific junctions recognizing the same number of nts on each side of the junction (B–E). Position of precursor mRNA, intron lariats, processed intron lariats and splicing intermediates is denoted. Position of the full-length released intron lariat (A and B) (2491 nts) is indicated by asterisks.
Overall these data show accumulation of full-length precursor mRNA and splicing intermediates of Ll.LtrB-ΔA suggesting that both steps of the circularization pathway are not very efficient in vivo. They also suggest that the released intron RNA circles identified by RT-PCR (Figure 3C) (19) are present in low amounts not detectable by Northern blots.
Circularization intermediates of Ll.LtrB-ΔA harbor short tails at the 3′ end
To further characterize the extremities of the Ll.LtrB-ΔA circularization intermediate detected by Northern blot we performed RT-PCR assays on L. lactis total RNA extracts. The intron 3′ ends were analyzed by first extending the intron RNA with a polyA tail followed by the synthesis of a cDNA with an oligo dT. The RNA strand from the RNA/DNA duplex was removed by RNAse H treatment followed by PCR amplification of the cDNA (Figure 5A). The specific band amplified for Ll.LtrB-WT (Figure 5B) corresponds to a processing site located within the ltrA gene at position -138 from the intron 3′ end (Figure 5C). It is consistent with one of the bands detected by Northern blot (∼2.8 kb) (Figure 4E, WT) and a processing site previously detected in the same area of Ll.LtrB (26). The same processing site was confirmed for Ll.LtrB-ΔA and Ll.LtrB-LtrAMat− by sequencing the corresponding bands. The processing site is absent for Ll.LtrB-ΔLtrA and Ll.LtrB-ΔLtrA+LtrA where the LtrA coding gene is missing or expressed in trans, respectively. The higher and more intense band for Ll.LtrB-ΔA revealed the presence of at least an additional C at the 3′ end of the intron (Figure 5D). In order to confirm the exact nature of the 3′ end of Ll.LtrB-ΔA, the same experiment was repeated but extending a polyU instead of a polyA tail at the 3′ end of the intron (Figure 5A). A similar PCR band was obtained and its sequence (Figure 5E), in conjunction with the previous sequence (Figure 5D), demonstrates that Ll.LtrB-ΔA harbors either two (CA) or three extra nts (CAU) at its 3′ end corresponding to the first residues of exon 2. The intensity of the overlapping peaks at position 421 (Figure 5D) indicates that the Ll.LtrB-ΔA splicing intermediates harboring either a CA or CAU tail are in comparable amounts in vivo. Sequences of the corresponding bands for Ll.LtrB-ΔLtrA, Ll.LtrB-ΔLtrA+LtrA and Ll.LtrB-LtrAMat− revealed the presence of similar tails at the 3′ end of these intron variants (Figure 5B). Using a similar RT-PCR approach, free 5′ intron end was only detected for Ll.LtrB-WT and Ll.LtrB-ΔLtrA+LtrA (Supplementary Figure S1). However, the amplified band for Ll.LtrB-ΔA was higher, more diffuse and significantly less intense (Supplementary Figure S1B). Sequence of this band was shown to harbor the last 22 nts of exon 1 and to be heterogeneous beyond that point showing that precise 5′ ends of Ll.LtrB-ΔA are not present in vivo (Supplementary Figure S1E). Thus, no trace of perfect linear intron was detected since Ll.LtrB-ΔA harbors either the first 2 or 3 nts of exon 2 at the 3′ end and at least 22 nts of exon 1 at the 5′ end.
These results demonstrate the presence, in similar amounts, of two major circularization intermediates consisting of exon 1 attached to full-length Ll.LtrB-ΔA with the two (CA) or three (CAU) first nts of exon 2 at their 3′ end. They also corroborate the accumulation of these circularization intermediates in vivo (Figure 4) and confirm that the 3′ splice site is mostly misrecognized by exon 1 during the first transesterification reaction of the Ll.LtrB-ΔA circularization pathway (Figure 1C).
Detection of Ll.LtrB-ΔA accurate circularization in vivo using a functional splicing assay
Analysis of the 3′ end of Ll.LtrB-ΔA indicated that the 3′ splice site is mostly misrecognized leading to the accumulation of circularization intermediates haboring either a CA or a CAU tail (Figure 5). Misrecognition of the 3′ splice site by exon 1 during the first transesterification reaction should also lead to inaccurate exon ligation (Figure 1C, step 1). The absence of 2 nts (CA) at the exon splice junction changes the reading frame and creates a premature stop codon early in exon 2 producing a truncated relaxase (LtrB-ΔE2). The absence of 3 nts (CAU) at the splice junction leads to the deletion of a conserved catalytic Histidine residue (LtrB-ΔHis) (31). On the other hand, we detected by RT-PCR the presence of properly ligated exons (Figure 3) (19) as well as low levels of perfect intron circles, not harboring an extra C at the splice junction (Figure 3) (19) suggesting that both Ll.LtrB-ΔA splice sites are sometimes properly recognized during circularization.
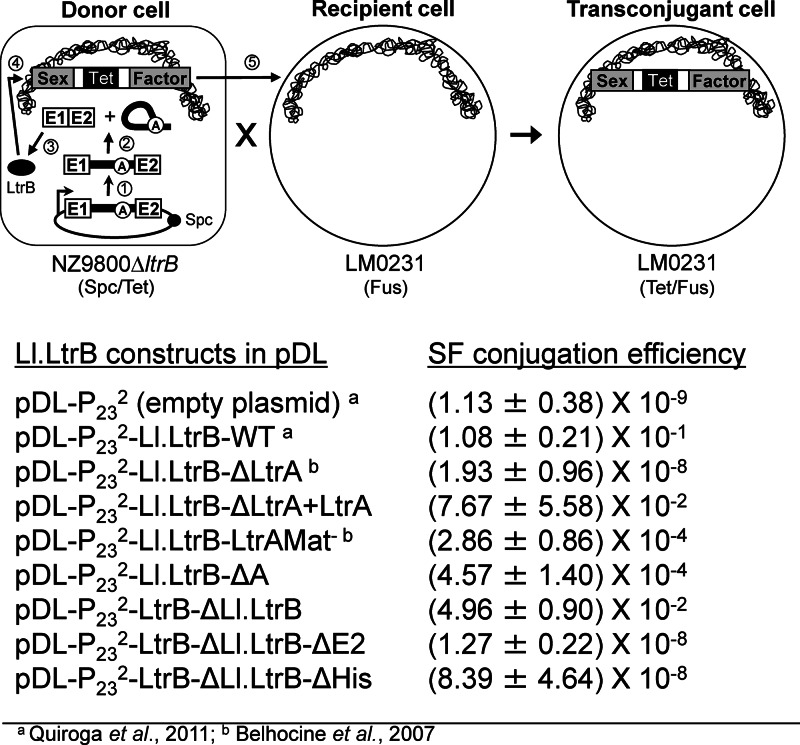
To functionally address the accurate circularization of Ll.LtrB-ΔA in vivo we used a very sensitive splicing/conjugation assay where Ll.LtrB splicing efficiency is monitored by the transfer rate of the chromosomal sex factor (SF) between strains of L. lactis (Figure 6, top) (7,29). Ll.LtrB interrupts the gene coding for a relaxase enzyme (LtrB) that initiates SF transfer by conjugation. The transfer rate of the SF between L. lactis strains was shown to be directly proportional to Ll.LtrB splicing efficiency from the relaxase transcript (7,30).
Figure 6.
Ll.LtrB splicing/conjugation assay and SF conjugation rates. The splicing efficiency of Ll.LtrB is monitored by the conjugation rate of the SF between two L. lactis strains. The donor strain harbors the chromosomal SF with a defective relaxase gene (NZ9800ΔltrB::tet) while the recipient strain lacks the SF (LM0231). The Ll.LtrB intron, along with portions of its exons, was replaced in the chromosome of NZ9800 by a tetracycline resistance marker (tet), which prevents expression of the relaxase (LtrB). The LtrB deficient strain is complemented by providing the interrupted ltrB gene from a plasmid. Following transcription of the ltrB gene (step 1) Ll.LtrB splices and the flanking exons are ligated (step 2). If the exons are precisely ligated the mature mRNA will be properly translated leading to expression of the relaxase enzyme (LtrB, black oval) (step 3). LtrB will then recognize the SF origin of transfer (oriT) (step 4) and initiate SF transfer from a donor to a recipient cell by conjugation (step 5). Transfer efficiency of conjugative elements between L. lactis strains was previously shown to be directly proportional to Ll.LtrB splicing from the relaxase transcript (7,30). In variations of the assay, the pLE-P232-LtrA plasmid was cotransformed with pDL-P232-Ll.LtrB-ΔLtrA to provide LtrA in trans (pDL-P232-Ll.LtrB-ΔLtrA+LtrA) or the uninterrupted relaxase gene enzyme was expressed (pDL-P232-LtrB-ΔLl.LtrB, pDL-P232-LtrB-ΔLl.LtrB-ΔE2, pDL-P232-LtrB-ΔLl.LtrB-ΔHis).
The level of SF conjugation efficiency supported by Ll.LtrB-ΔA was found to be similar to Ll.LtrB-LtrAMat− (10−4) three logs lower than Ll.LtrB-WT (10−1) but significantly higher than background (10−9) (Figure 6, bottom). In contrast, the mutant relaxase genes, not interrupted by the intron (LtrB-ΔLl.LtrB-ΔE2, LtrB-ΔLl.LtrB-ΔHis), only promote SF conjugation efficiency in the 10−8 range (Figure 6, bottom). This suggests that SF conjugation efficiency is supported by the presence of low levels of accurately ligated exons and not by the improperly ligated exons complementary to the two major circularization intermediates.
Taken together, our functional splicing assay confirms the presence of low levels of accurately ligated exons in vivo and suggests that accurate recognition of the 3′ splice site by exon 1 leads to the appropriate recognition of the 5′ splice site by the last nt of the intron and the precise excision of Ll.LtrB-ΔA cirles not harboring additional non-encoded nts at the splice junction.
DISCUSSION
Even though a pathway for the excision of group II intron circles was initially proposed many years ago (11), this splicing mechanism was mostly ignored and not examined in details at the molecular level. Nevertehless, we recently demonstrated that, concurrently with branching, a significant proportion of the Ll.LtrB group II intron from the Gram-positive bacterium L. lactis uses the circularization pathway in vivo (19). However, since the RT enzyme is not as processive moving through the 2′-5′ bond at the branch point of lariats during RT-PCR, the relative abundance between intron lariats and circles cannot be precisely determined (19). In this study, we took advantage of the Ll.LtrB-ΔA mutant that does not produce lariats and exclusively excises as circles to address experimentally the circularization pathway of bacterial group II introns.
Upon circularization, a majority of the Ll.LtrB-ΔA circles were found to harbor an additional non-encoded C residue of unknown origin at the intron splice junction while some circular junctions were perfectly joined (19). RT-PCR assays using Ll.LtrB-ΔA mutants where the flanking C residues were independently replaced by Us demonstrated that the non-encoded C residue present at the splice junction of circular introns originates exclusively from the first nt of exon 2 accurately recognizing the 5′ splice site (Figure 2A, scenario 2). This result strongly suggested that the 3′ splice site of Ll.LtrB-ΔA is mostly misrecognized during the first transesterification reaction of the circularization pathway (Figure 1C) leaving at least the first nt of exon 2 attached to the 3′ end of the intron. In accordance, we showed that the 3′ end of the Ll.LtrB-ΔA circularization intermediates harbor the first two (CA) or three (CAU) nts of exon 2 (Figure 5). These two main circularization intermediates, consisting of full length intron still attached to exon 1, were found to be in approximately the same amount in L. lactis total RNA extracts (Figure 5E) and to accumulate in vivo (Figure 4). As expected, free 3′ intron ends were not detected since we previously demonstrated that only a small proportion of Ll.LtrB-ΔA circles do not harbor an extra C at the circularization junction (19). On the other hand, misrecognition of the 3′ splice site suggested the generation of improperly ligated exons missing either 2 (CA) or 3 (CAU) nts at the splice junction and producing respectively a truncated (LtrB-ΔE2) or catalytically inactive (LtrB-ΔHis) relaxase enzyme. Using a functional splicing assay we showed that the ltrB gene interrupted by Ll.LtrB-ΔA supports SF conjugation efficiency significantly higher than background (10−4 vs 10−9) (Figure 6). We concluded that the SF conjugation efficiency is supported by trace amounts of accurately ligated exons produced during accurate circularization of Ll.LtrB-ΔA since both LtrB-ΔLl.LtrB-ΔE2 (10−8) and LtrB-ΔLl.LtrB-ΔHis (10−8) mutated proteins, that should be generated through 3′ splice site misrecognition, cannot support SF conjugation in the 10−4 range (Figure 6).
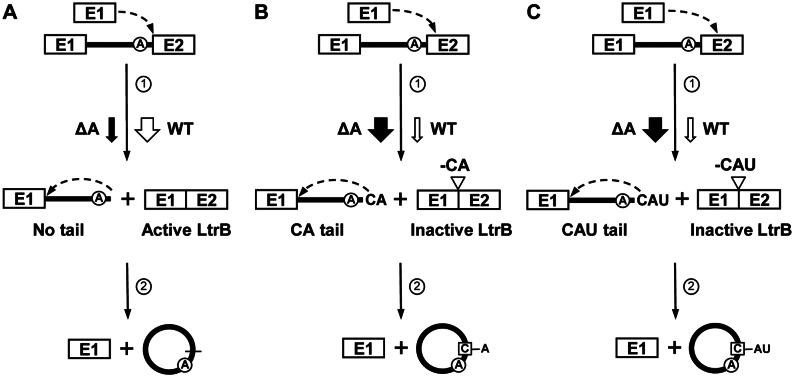
Our work suggests that the Ll.LtrB-ΔA circularization pathway (Figure 7, black arrows) is mostly initiated by misrecognition of the 3′ splice site by the 3′OH of the last nt of free exon 1 (Figure 7B, C, step 1). This first transesterification reaction concurrently leads to the production of two major circularization intermediates with short tails of two (CA) (Figure 7B) or three (CAU) (Figure 7C) nts at the 3′ end of the intron and the inaccurate joining of flanking exons leading to the production of inactive LtrB relaxase enzymes (Figure 7B and C). However, small amounts of perfect intron circles and accurately ligated exons were detected by RT-PCR and conjugation showing that the 3′ splice site is also accurately recognized albeit at a significant lower level (Figure 7A). The second transesterification reaction, that accurately recognizes the 5′ splice site, is mainly initiated by the 2′OH of the first nt of exon 2 leading to its incorporation at the intron circle splice junction (Figure 7B and C, step 2). When the intron does not harbor a tail at the 3′ end (Figure 7A), the 2′OH of the last nt of the intron precisely attack the 5′ splice site releasing perfect end-to-end intron circles (Figure 7A, step 2). Precise recognition of the 5′ splice site releases more exon 1 that can initiate another round of intron circularization (Figure 7A–C). Accumulation of full-length precursor mRNAs (Figure 4) and circularization intermediates of Ll.LtrB-ΔA (Figures 4 and 5) suggests that both transesterification reactions are not very efficient in vivo when compared to the branching pathway (Figure 1A).
Figure 7.
Ll.LtrB circularization pathway. Ll.LtrB-WT (WT) and Ll.LtrB-ΔA (ΔA) use the same circularization pathway (A–C) in vivo. (A) The first nucleophilic attack at the intron-exon 2 junction is initiated by the 3′-OH of a free exon 1 (step 1). Accurate recognition of the 3′ splice site properly ligates the flanking exons leading to the expression of an active relaxase (Active LtrB). The 2′-OH of the last intron residue initiates the second nucleophilic reaction at the exon 1-intron junction (step 2) resulting in precise intron circularization and the release of free exon 1 available to initiate another circularization reaction. (B and C) The 3′-OH of a free exon 1 misrecognizes the intron-exon 2 junction during the first nucleophilic attack (step 1) leaving tails of 2 (CA) (B) or 3 (CAU) (C) nts at the 3′ end of the intron. Misrecognition of the 3′ splice site also improperly ligates the flanking exons creating a premature stop codon just after the splice junction (-CA) (Inactive LtrB, LtrB-ΔE2) (B) or deleting an important catalytic amino acid from the LtrB relaxase enzyme (-CAU) (Inactive LtrB, LtrB-ΔHis) (C). The 2′-OH of the first nt of exon 2 then accurately initiates the second nucleophilic reaction at the exon 1-intron junction (step 2) resulting in its incorporation at the intron circularization junction and the release of free exon 1 available to initiate another circularization reaction. Ll.LtrB-WT circularization predominantly involves the release of perfect intron circles (A) while the great majority of Ll.LtrB-ΔA circles harbor the first nt of exon 2 at their splice junction (B, C). However, low level of accurate (perfect intron circles, precisely ligated exons, SF conjugation) and inaccurate (first nt of exon 2 at circle splice junction) circularization can still be detected for Ll.LtrB-ΔA and Ll.LtrB-WT respectively. Removal of the branch point A residue significantly affects 3′ splice site recognition tilting the balance from accurate (A) to inaccurate (B and C) 3′ splice site recognition ultimately leading to the addition of the first nt of exon 2 (C) at the circle splice junction (cf. WT (open arrows) and ΔA (black arrows)).
Taken together, our data also indicate that Ll.LtrB-WT uses the same circularization pathway as Ll.LtrB-ΔA in vivo. In accordance, we detected circularization intermediates with 3′ tails similar to Ll.LtrB-ΔA for Ll.LtrB-ΔLtrA+LtrA, Ll.LtrB-ΔLtrA and Ll.LtrB-LtrAMat− showing some level of 3′ splice site misrecognition despite the presence of the branch point A residue (Figure 5B). We assume that low amounts of circularization intermediates with similar tails are also present for Ll.LtrB-WT but that the occurrence of a prominent processing site at position -138 prevented their detection (Figure 5B and C). The detection of some Ll.LtrB-WT circles also harboring the first nt of exon 2 at the splice junction is another indication that Ll.LtrB-WT and Ll.LtrB-ΔA share the same circularization pathway. Even though both Ll.LtrB variants use the same overall circularization pathway the balance between accurate and misrecognition of the 3′ splice site during the first transesterification reaction was found to be considerably different. While the 3′ splice site of Ll.LtrB-WT is most of time accurately recognized, leading to precise ligation of its flanking exons, and releasing almost exclusively perfect intron circles, the 3′ splice site of Ll.LtrB-ΔA is generally misrecognized, leading to improperly ligated exons and releasing almost exclusively intron circles haboring the first nt of exon 2 at the splice junction (Figure 7, cf. black arrows (Ll.LtrB-ΔA) and open arrows (Ll.LtrB-WT)). Removal of the branch point A residue from Ll.LtrB thus not only prevents branching but also significantly tilts the balance of the first transesterification reaction of the circularization pathway towards misrecognition of the 3′ splice site.
Interestingly, variants of the RmInt1 intron from the bacterium Sinorhizobium meliloti were also found to harbor an additional non-encoded C residue at the splice junction of released intron circles (21,22). Similarly to Ll.LtrB, RmInt1 harbors a C residue as the first nt of exon 2 which was proposed to be at the origin of the additional nt at the circle splice junction following 3′ splice site misrecognition (32). This suggests that these two bacterial introns, even though not from the same class, share the same circularization pathway in vivo.
In summary, this work represents a thorough molecular characterization of the previously proposed group II intron circularization pathway (11). Our data also suggest that circularization is a conserved secondary splicing pathway for bacterial group II introns. Given the variety of known and predicted functions associated with circular RNAs from various biological systems (33) one could envisage that group II intron RNA circles also serve a specific function in bacterial cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Félix LaRoche-Johnston and Deeva Uthayakumar for providing comments on the manuscript.
FUNDING
Discovery Grant from Natural Sciences and Engineering Research Council of Canada [227826 to B.C.]; William Dawson Scholar Award from McGill University [100705 to B.C.]. The open access publication charge for this paper has been waived by Oxford University Press - NAR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ferat J.L., Michel F. Group II self-splicing introns in bacteria. Nature. 1993;364:358–361. doi: 10.1038/364358a0. [DOI] [PubMed] [Google Scholar]
- 2.Lambowitz A.M., Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimmerly S., Semper C. Evolution of group II introns. Mob. DNA. 2015;6:7. doi: 10.1186/s13100-015-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambowitz A.M., Belfort M. Mobile bacterial group II introns at the crux of eukaryotic evolution. Microbiol. Spectr. 2015;3:1. doi: 10.1128/microbiolspec.MDNA3-0050-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belhocine K., Plante I., Cousineau B. Conjugation mediates transfer of the Ll.LtrB group II intron between different bacterial species. Mol. Microbiol. 2004;51:1459–1469. doi: 10.1111/j.1365-2958.2004.03923.x. [DOI] [PubMed] [Google Scholar]
- 6.Belhocine K., Yam K.K., Cousineau B. Conjugative transfer of the Lactococcus lactis chromosomal sex factor promotes dissemination of the Ll.LtrB group II intron. J. Bacteriol. 2005;187:930–939. doi: 10.1128/JB.187.3.930-939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belhocine K., Mak A.B., Cousineau B. Trans-splicing of the Ll.LtrB group II intron in Lactococcus lactis. Nucleic Acids Res. 2007;35:2257–2268. doi: 10.1093/nar/gkl1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nisa-Martínez R., Jiménez-Zurdo J.I., Martínez-Abarca F., Muñoz-Adelantado E., Toro N. Dispersion of the RmInt1 group II intron in the Sinorhizobium meliloti genome upon acquisition by conjugative transfer. Nucleic Acids Res. 2007;35:214–222. doi: 10.1093/nar/gkl1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fedorova O., Zingler N. Group II introns: structure, folding and splicing mechanism. Biol. Chem. 2007;388:665–678. doi: 10.1515/BC.2007.090. [DOI] [PubMed] [Google Scholar]
- 10.Pyle A.M. The tertiary structure of group II introns: implications for biological function and evolution. Crit. Rev. Biochem. Mol. Biol. 2010;45:215–232. doi: 10.3109/10409231003796523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray H.L., Mikheeva S., Coljee V.W., Turczyk B.M., Donahue W.F., Bar-Shalom A., Jarrell K.A. Excision of group II introns as circles. Mol. Cell. 2001;8:201–211. doi: 10.1016/s1097-2765(01)00300-8. [DOI] [PubMed] [Google Scholar]
- 12.Arnberg A.C., Van Ommen G.J., Grivell L.A., Van Bruggen E.F., Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980;19:313–319. doi: 10.1016/0092-8674(80)90505-x. [DOI] [PubMed] [Google Scholar]
- 13.Halbreich A., Pajot P., Foucher M., Grandchamp C., Slonimski P. A pathway of cytochrome b mRNA processing in yeast mitochondria: specific splicing steps and an intron-derived circular DNA. Cell. 1980;19:321–329. doi: 10.1016/0092-8674(80)90506-1. [DOI] [PubMed] [Google Scholar]
- 14.Peebles C.L., Belcher S.M., Zhang M., Dietrich R.C., Perlman P.S. Mutation of the conserved first nucleotide of a group II intron from yeast mitochondrial DNA reduces the rate but allows accurate splicing. J. Biol. Chem. 1993;268:11929–11938. [PubMed] [Google Scholar]
- 15.Peebles C.L., Perlman P.S., Mecklenburg K.L., Petrillo M.L., Tabor J.H., Jarrell K.A., Cheng H.L. A self-splicing RNA excises an intron lariat. Cell. 1986;44:213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- 16.van der Veen R., Arnberg A.C., van der Horst G., Bonen L., Tabak H.F., Grivell L.A. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell. 1986;44:225–234. doi: 10.1016/0092-8674(86)90756-7. [DOI] [PubMed] [Google Scholar]
- 17.Podar M., Chu V.T., Pyle A.M., Perlman P.S. Group II intron splicing in vivo by first-step hydrolysis. Nature. 1998;391:915–918. doi: 10.1038/36142. [DOI] [PubMed] [Google Scholar]
- 18.Jarrell K.A., Peebles C.L., Dietrich R.C., Romiti S.L., Perlman P.S. Group II intron self-splicing. Alternative reaction conditions yield novel products. J. Biol. Chem. 1988;263:3432–3439. [PubMed] [Google Scholar]
- 19.Monat C., Quiroga C., Laroche-Johnston F., Cousineau B. The Ll.LtrB intron from Lactococcus lactis excises as circles in vivo: insights into the group II intron circularization pathway. RNA. 2015;21:1286–1293. doi: 10.1261/rna.046367.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalby S.J., Bonen L. Impact of low temperature on splicing of atypical group II introns in wheat mitochondria. Mitochondrion. 2013;13:647–655. doi: 10.1016/j.mito.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Molina-Sánchez M.D., Barrientos-Durán A., Toro N. Relevance of the branch point adenosine, coordination loop, and 3′ exon binding site for in vivo excision of the Sinorhizobium meliloti group II intron RmInt1. J. Biol. Chem. 2011;286:21154–21163. doi: 10.1074/jbc.M110.210013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molina-Sánchez M.D., Martinez-Abarca F., Toro N. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J. Biol. Chem. 2006;281:28737–28744. doi: 10.1074/jbc.M602695200. [DOI] [PubMed] [Google Scholar]
- 23.Li-Pook-Than J., Bonen L. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res. 2006;34:2782–2790. doi: 10.1093/nar/gkl328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy V., Pirakitikulr N., Zhou K.I., Chillón I., Luo J., Pyle A.M. Predicted group II intron lineages E and F comprise catalytically active ribozymes. RNA. 2013;19:1266–1278. doi: 10.1261/rna.039123.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichiyanagi K., Beauregard A., Lawrence S., Smith D., Cousineau B., Belfort M. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol. Microbiol. 2002;46:1259–1272. doi: 10.1046/j.1365-2958.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- 26.Matsuura M., Saldanha R., Ma H., Wank H., Yang J., Mohr G., Cavanagh S., Dunny G.M., Belfort M., Lambowitz A.M. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plante I., Cousineau B. Restriction for gene insertion within the Lactococcus lactis Ll.LtrB group II intron. RNA. 2006;12:1980–1992. doi: 10.1261/rna.193306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zajac P., Islam S., Hochgerner H., Lönnerberg P., Linnarsson S. Base preferences in non-templated nucleotide incorporation by MMLV-derived reverse transcriptases. PLoS One. 2013;31:e85270. doi: 10.1371/journal.pone.0085270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quiroga C., Kronstad L., Ritlop C., Filion A., Cousineau B. Contribution of base-pairing interactions between group II intron fragments during trans-splicing in vivo. RNA. 2011;17:2212–2221. doi: 10.1261/rna.028886.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein J.R., Chen Y., Manias D.A., Zhuo J., Zhou L., Peebles C.L., Dunny G.M. A conjugation-based system for genetic analysis of group II intron splicing in Lactococcus lactis. J. Bacteriol. 2004;186:1991–1998. doi: 10.1128/JB.186.7.1991-1998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pansegrau W., Schröder W., Lanka E. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J. Biol. Chem. 1994;269:2782–2789. [PubMed] [Google Scholar]
- 32.Chillón I., Molina-Sánchez M.D., Fedorova O., García-Rodríguez F.M., Martínez-Abarca F., Toro N. In vitro characterization of the splicing efficiency and fidelity of the RmInt1 group II intron as a means of controlling the dispersion of its host mobile element. RNA. 2014;20:2000–2010. doi: 10.1261/rna.047407.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.