Abstract
Multiple DNA double-strand break (DSB) repair pathways are active in S phase of the cell cycle; however, DSBs are primarily repaired by homologous recombination (HR) in this cell cycle phase. As the non-homologous end-joining (NHEJ) factor, Ku70/80 (Ku), is quickly recruited to DSBs in S phase, we hypothesized that an orchestrated mechanism modulates pathway choice between HR and NHEJ via displacement of the Ku heterodimer from DSBs to allow HR. Here, we provide evidence that phosphorylation at a cluster of sites in the junction of the pillar and bridge regions of Ku70 mediates the dissociation of Ku from DSBs. Mimicking phosphorylation at these sites reduces Ku's affinity for DSB ends, suggesting that phosphorylation of Ku70 induces a conformational change responsible for the dissociation of the Ku heterodimer from DNA ends. Ablating phosphorylation of Ku70 leads to the sustained retention of Ku at DSBs, resulting in a significant decrease in DNA end resection and HR, specifically in S phase. This decrease in HR is specific as these phosphorylation sites are not required for NHEJ. Our results demonstrate that the phosphorylation-mediated dissociation of Ku70/80 from DSBs frees DNA ends, allowing the initiation of HR in S phase and providing a mechanism of DSB repair pathway choice in mammalian cells.
INTRODUCTION
Genomic integrity maintenance is a fundamental function to sustain life due to the fact that DNA alterations such as mutations, chromosomal rearrangements and deletions are causative factors of disease, tumorigenesis and cell death (1). Cells encounter a large number of DNA lesions on a daily basis, jeopardizing the integrity of the genome, with DNA double strand breaks (DSBs) being the most significant. The deleterious nature of DSBs is underscored by the fact that a single unrepaired DSB can cause cell death and misrepaired DSBs can result in chromosomal mutations such as translocations and large scale deletions (2,3). To cope with DSBs, cells have evolved multiple repair pathways with the two most prominent being homologous recombination (HR) and non-homologous end-joining (NHEJ) (1,4). HR directs DSB repair by utilizing a homologous stretch of DNA to guide repair of the broken DNA strand, whereas NHEJ mediates the direct re-ligation of the broken DNA molecule.
Since there are multiple DSB repair processes, a cell must properly choose which pathway to employ for each specific DSB. A number of factors are believed to influence the selection of these pathways including direct competition for the DSB ends, cell cycle stage, specific post-translation modifications and DNA end resection (5–7). HR requires a homologous template for accurate repair; therefore, HR primarily functions in S and G2 phases because a homologous DNA template via a sister chromatid is available for repair in these cell cycle phases. NHEJ is active in all cell cycle stages as it does not require a homologous template for direct repair. However, DSB repair pathway choice is not simply mediated by limiting the availability of specific repair factors to a specific cell cycle phase as both HR and NHEJ operate in S phase, where HR is the preferred DSB pathway (8,9). Previous data suggested that direct competition likely does not tip the scale in favor of HR in S/G2 in mammalian cells as the canonical NHEJ factor, DNA-dependent protein kinase (DNA-PK), consisting of the Ku70/Ku80 heterodimer (Ku) and the DNA-PK catalytic subunit (DNA-PKcs), quickly localizes to DSBs in S phase and its initial recruitment kinetics are identical in all cell cycle phases (10–12). Furthermore, Ku has an extremely high affinity (binding constant of 2 × 109 M−1) for DNA ends and is highly abundant (∼500,000 Ku molecules/cell) in human cells. Hence, it is unlikely that competition for DNA ends is responsible for DSB repair pathway choice in mammalian cells (13–16).
The initiation of the HR pathway is dependent on 5′ to 3′ resection of the DSB ends. It is believed that once DNA end resection has initiated, NHEJ can no longer repair the DSB, indicating an important role of end resection for DSB repair pathway choice (17–19). DNA end resection is a multi-step process mediated by a number of factors including the Mre11/Rad50/Nbs1 (MRN) complex, CtIP and Exonuclease 1 (Exo1). Cell cycle-regulated factors may directly control DNA end resection as it occurs faster in S phase than other cell cycle stages, and CtIP-dependent resection is upregulated by S phase-dependent protein kinases (20–22). Furthermore, BRCA1-CtIP and 53BP1-RIF1 circuits compete to influence the initiation of DNA end resection with BRCA1-CtIP promoting the removal of 53BP1-RIF1 from DSBs in S phase, allowing the initiation of DNA end resection and the onset of HR (23,24). As DNA ends must be free for DNA end resection to occur and DNA-PK localizes to DSBs in S phase, for HR to initiate there must be a mechanism that mediates the dissociation of DNA-PK from DSBs specifically in S phase. In budding yeast, the Mre11/Rad50/Xrs2 (MRX) complex displaces yKu from DSB ends to drive pathway choice to HR (25). However, in contrast to the results obtained in yeast, our group previously showed that Ku inhibits both the Mre11/Rad50 complex and Exo1-mediated DNA end processing in vitro and that Mre11 does not displace Ku from DSBs in vitro or in vivo (26). A Ku ortholog from the Mycobacterium tuberculosis Ku (Mt-Ku) overexpressed in human cells resulted in the inhibition of DNA end resection, further suggesting that DNA ends must be free for initiation of HR (11).
These studies have led us to elucidate the mechanism mediating the displacement of Ku from DSBs to allow initiation of DNA end resection and HR. Here, we show that Ku70 is phosphorylated at the junction of the pillar and bridge regions of the molecule and that this post-translational modification is required for its dissociation from DSB ends. Blocking phosphorylation of the identified sites induced a significant decrease in DNA end resection and HR. Collectively, we have established that phosphorylation of Ku70 plays a role in DSB repair pathway choice as it mediates dissociation of the Ku heterodimer from DSBs to free the DNA ends, allowing the initiation of DNA end resection and HR in S phase.
MATERIALS AND METHODS
Cell culture and reagents
Ku70−/− mouse embryo fibroblasts (MEFs), termed DC-1(27), HCT116 Ku70+/− (a kind gift from Dr Eric A. Hendrickson) and U2-OS cells were cultured in Hyclone MEM alpha modification (GE Life Sciences) medium supplemented with 1x Pen-strep and 10% FBS and FCS (1:1 mixture). Ku70−/− MEFs complemented with human wild-type Ku70 and mutant Ku70 were maintained in Hyclone MEM media supplemented with 150 μg/ml hygromycin. For thymidine double block experiments, cells were maintained in DMEM medium (D6429, Sigma). All cells were grown as monolayers at 37°C in 5% CO2.
Irradiation of cells
Gamma radiation experiments were conducted at UT Southwestern Medical Center, Dallas using a 137Cs source irradiator.
Ku mutagenesis
Mutant Ku70 was generated by PCR directed mutagenesis using complementary oligonucleotides; 8A, 5′-GACCCGGACCTTTAATGCAGCTGCAGGCGGTTTGCTTC-3′, 5′-GGTTTGCTTCTGCCTGCCGATGCCAAGAGGTCTCAGA-3′, 5′-AATGAACCAGTGAAAGCCAAGGCCCGGGCCTTTAATGCAGCTGC-3′, 5A, 5′-GACCCGGACCTTTAATGCAGCTGCAGGCGGTTTGCTTC-3′, 5′-GGTTTGCTTCTGCCTGCCGATGCCAAGAGGTCTCAGA-3′, 5D, 5′-GACCCGGACCTTTAATGACGATGACGGCGGTTTGCTTC-3′, 5′-GGTTTGCTTCTGCCTGACGATGACAAGAGGTCTCAGA-3′ and subcloned into the pCDNA3 mammalian expression vector. The mutant Ku cDNAs were stably expressed in Ku70−/− DC1 or DC1-DR5 cell lines. For recombinant protein purification, each mutant Ku70 cDNA was subcloned into the pFastBac™ vector and recombinant baculoviruses were generated via the Bac-to-Bac baculovirus expression system (Invitrogen).
Ku heterodimer purification
Human DNA-PKcs and wild-type Ku70/80 proteins were purified as previously described (28). To purify Ku heterodimers with mutant Ku70, Sf9 cells were infected with Ku70 mutant and wild-type Ku80 baculoviruses and harvested 60 h post infection. The His6-tagged mutant Ku70 and His6-tagged wild-type Ku80 heterodimers were purified via Ni-NTA agarose (Qiagen), Superdex200 16/60 and Heparin agarose column chromatography (GE Life Sciences), as previously described (26,28).
In vitro kinase assay
In vitro DNA-PKcs kinase assays were performed as previously described (10,28). Each kinase reaction contained 25 mM Tris–HCl, pH 7.9, 25 mM MgCl2, 1.5 mM DTT, 50 mM KCl, 10% glycerol, 10 nM fork 28 DNA, 0.16 μM [γ-32P] ATP (3000 Ci/mmol), 8 nM DNA-PKcs and 20 nM Ku70/80 in a 10 μl final reaction volume. The kinase reaction mixture was incubated at 30°C for 30 min and terminated by the addition of SDS-PAGE sample buffer. The phosphorylated proteins were resolved via 7% SDS-PAGE and detected by PhosphoImager analysis (GE Life Sciences).
Pull down assay
Forked dsDNA was produced as a long single strand 5′-CGCGCCCAGCTTTCCCAGCTAATAAACTAAAAACTCCTAAGG-3′ and a short single strand 5′-CCTTAGGAGTTTTTAGTTTATTGGGCGCG-3′ (Invitrogen) as previously described (26). Purified DNA-PKcs, Ku and biotin-labeled forked 22-mer DNA were incubated in kinase reaction buffer at 30°C for 1 h in the presence or absence of ATP (26). After incubation, the in vitro reaction mixtures were incubated at 4°C for 4 h with streptavidin-agarose in binding buffer containing 50 mM Tris-HCl pH 8.0, 0.5% EDTA and 150 mM KCl (24). The streptavidin-agarose was spun down via centrifugation at 2500 rpm for 3 min. The supernatant, containing proteins that dissociated from the forked DNA, and the pellet, containing proteins still bound to the forked DNA, were resolved via 8% SDS-PAGE and transferred to a nitrocellulose membrane. Western blot analysis was performed using the antibodies indicated in the figures.
DNA end resection assay
In vitro DNA end resection assays were performed in two steps. The first step included binding of Ku70/80 to the forked DNA followed by incubation with DNA-PKcs in the presence of ATP or ATPγS to direct or block in vitro phosphorylation of Ku, respectively. The phosphorylation reaction included 20 nM forked 28mer dsDNA, 30 nM DNA-PKcs, 22 nM Ku, 1 μM ATP or 1 μM ATPγS in a buffer containing 20 mM HEPES, pH 8.0, 50 mM KCl, 5 mM MgCl2 and 1 mM DTT. In the second step, the freeing of the DNA ends was assessed via exonuclease 1 (Exo1)-mediated DNA end resection. The Exo1 protein (10 nM) was added to the reaction mixture and incubated for 30 min to allow DNA end digestion. The assay was terminated by adding formamide dye (95% formamide, 0.05% bromophenol blue, 0.05% Xylene Cyanol FF, 18 mM EDTA and 0.025% SDS) to each sample. The samples were boiled at 95°C for 5 min and allowed to cool on ice for 5 min. The samples were separated by 16% Urea-PAGE, dried and analyzed via PhosphoImager.
Live cell imaging and laser micro-irradiation
Live cell imaging combined with laser micro-irradiation was carried out as previously described (11,29). Fluorescence was monitored via an Axiovert 200 M microscope (Carl Zeiss, Inc.), with a Plan-Apochromat 63X/NA 1.40 oil immersion objective (Carl Zeiss, Inc.). A 365-nm pulsed nitrogen laser (Spectra-Physics) was directly coupled to the epifluorescence path of the microscope and used to generate DSBs in a defined area of the nucleus. For quantitative analyses, the same amount of DNA damage was generated under standardized micro-irradiation conditions (minimal laser output of 75% for 5 pulses) in each experiment. Time-lapse images were taken with an AxioCamHRm camera. The fluorescence intensities of micro-irradiated and non-irradiated areas within the cell nucleus were determined using the AxioVision Software, version 4.8 (Carl Zeiss, Inc.). Each data point is the average of 10 independent measurements.
Local damage induction and fluorescence recovery after photobleaching (FRAP) measurements
Cells (250,000–500,000) were seeded on 40 mm diameter glass cover slips (Menzel GmbH & Co KG, Braunschweig, Germany) 1 or 2 days before the experiment. Samples were mounted on a Focht Chamber System 2 (Bioptechs Inc., Butler, USA). A Leica (Leica Microsystems GmbH, Wetzlar, Germany) DM LA microscope with HCX APO UVI (337 nm irradiation) and PL Fluotar 100x (30) (NA 1.3 oil immersion lenses and CTR MIC control) unit was used. A 337 nm nitrogen laser (VSL-337ND-S; 20 Hz, 5 ns pulses) attenuated to around 6 μW at cell plane was used for local damage induction and DSB generation (31,32). A 100 mW 473 nm diode laser type DPL 473-OEM (Rapp Opto Electronics, Hamburg, Germany) was coupled to the microscope by a modified Leica AS LMD module and dichroic mirror Q480 LP (Chroma Technology Corporation, Bellows Falls, USA) for photobleaching. Both lasers were controlled by the Leica Laser Microdissection System LMD version 4.4.0.0. Fluorescence was excited with the monochromator Polychrome V (TILL Photonics GmbH, Graefelfing, Germany). Image acquisition was conducted with a sCMOS camera type Zyla (Andor Technology, Belfast, Ireland) and the corresponding AndorIQ software.
Image analysis of FRAP experiments
Image analysis was performed with the software ImageJ (http://rsweb.nih.gov/ij/). Cell motion during acquisition was compensated for via the StackReg plugin (Philippe Thevenaz, Lausanne, Switzerland). The measurements were double normalized to the pre-bleach intensity and to signal loss during image acquisition.
Cell synchronization
For thymidine double block, MEFs were plated in DMEM medium (D6429, Sigma) to reach 30% confluency one day before thymidine treatment. The protocol was optimized after determining cell cycle characteristics of the MEFs utilizing BrdU incorporation (not shown). Thymidine (T9250, Sigma) was added to a final concentration of 2 mM for the first block and the cells were incubated for 8 h. Cells were then washed three times with 1x PBS and released in normal DMEM medium for 4 h. Thymidine (2 mM) was added again to the medium and cells were treated for an additional 12 h for full synchronization to the G1/S border. Cells were then released in normal medium without thymidine for 2.5 h to achieve maximum population in the mid- to late-S phase. Synchrony at this point was ∼73–76% - S phase, 14–16% - G1 phase and 10 –11% - G2 phase. The synchronized cells were then used to perform various assays.
Survival assay with synchronous cells
For survival assays with synchronous cells, the cells were released in normal DMEM media after the second block for a period of 2.5 h and then irradiated with different doses of IR. Different cell numbers were plated in triplicates and colonies were allowed to form for 7 days. The colonies were then stained with 5% crystal violet stain in 100% methanol and counted manually using a light microscope.
Survival assay with mitomycin C (MMC)
Cells (100,000) were plated one day before mitomycin C MMC (M4287, Sigma) treatment. Different concentrations of MMC were added to the cells 1 h before plating. Different cell numbers were plated in triplicates and allowed to form colonies for 7 days. Once colonies were formed, they were stained and counted.
RPA2/Rad51 foci kinetics
Cells were grown on coverslips one day before the experiment. On the day of experiment, EdU (C10338, kit from Molecular Probes) was added to a final concentration of 30 μM and incubated for 40 min under normal growth conditions (10). Cells were subsequently washed twice with 1x PBS followed by replacement with normal medium and then subjected to 8 Gy γ-rays. At different time points after IR, cells were washed twice with ice cold 1x PBS and then treated with freshly made CSK extraction buffer (10 mM HEPES pH 7.4, 300 mM Sucrose, 100 mM NaCl, 3 mM MgCl2 and 0.1% TritonX-100) for 10 min on ice. Cells were then washed 5–6 times with ice cold 1x PBS. Cells were fixed with 4% paraformaldehyde (in 1x PBS) at room temperature for 20 min (RT), washed 4–5 times with 1x PBS (final 2 times 3–5 min interval) and incubated in 0.5% Triton X-100 on ice for 10 min. Cells were further washed 4–5 times with 1x PBS and incubated in blocking solution (5% goat serum (Jackson Immuno Research) in 1x PBS) overnight. The blocking solution was replaced with the RPA2 (NA-19L, EMD Millipore) / Rad51(SC-8349, Santa Cruz) primary antibody diluted in 5% goat serum in 1x PBS and incubated for 2 h. Cells were then washed 3–5 times with wash buffer (5 min interval; 1% BSA in 1x PBS). EdU detection using Click-iT reaction cocktail was performed according to manufacturer's protocol (C10338, kit from Molecular Probes). After incubation of the cells with the Click-iT reaction cocktail in the dark for 1 h, cells were washed 5 times with wash buffer. Cells were incubated with the Alexa Fluor 488 (1:1000) (Molecular Probes) secondary antibody in 1% BSA, 2.5% goat serum in 1x PBS for 1 h in the dark, followed by 5 washes. After the last wash, cells were mounted in VectaShield mounting medium containing DAPI. The images were acquired using a Zeiss Axio Imager fluorescence microscope utilizing 63x oil objective lens and a LSM 510 Meta laser scanning confocal microscope with a 63 × 1.4 NA Plan-Apochromat oil immersion objective. The foci were analyzed and counted using the Imaris (Bitplane) image analysis software.
53BP1 foci kinetics
Cells were grown on coverslips one day before the experiment. On the day of the experiment, EdU was added to a final concentration of 30 μM and incubated for 40 min under normal growth conditions. After incubation, cells were washed twice with 1x PBS and replaced with normal medium. Cells were then exposed to 1Gy of γ-rays. At different time points after IR, cells were washed twice with ice cold 1x PBS and fixed with 4% paraformaldehyde (in 1x PBS) for 20 min at RT, washed 4–5 times with 1x PBS (final 2 times 3–5 min interval) and incubated in 0.5% Triton X-100 on ice for 10 min. Cells were further washed 4–5 times with 1x PBS and incubated in blocking solution (5% goat serum (Jackson Immuno Research) in 1x PBS) overnight. The blocking solution was then replaced with the 53BP1 (SC-22760, Santa Cruz) primary antibody diluted in 5% goat serum in 1x PBS and the cells were incubated for 2 h. The rest of the protocol is similar to the one described above for RPA/Rad51 foci kinetics.
HR assay
The DR-GFP vector was inserted into the Ku70−/− DC-1 cell line and the single insertion was confirmed by Southern hybridization (not shown). This cell line was complemented with either the wild-type or various Ku70 mutants and individual stable clones were isolated. Ku expression was confirmed for all the stable clones (Supplementary Figure S6). HR assays were performed by transiently transfecting 5 μg of the RFP-ISce-GR plasmid to initiate the assay. Cells undergoing HR successfully expressed GFP, which was detected by flow cytometry. The percentage of HR was calculated as GFP+ / (GFP+ + RFP+). Cells that were only RFP+ indicated the fraction of transfected cells that failed to carry out HR.
RESULTS
Phosphorylation of Ku leads to its dissociation from DNA ends in vitro
The canonical NHEJ factor DNA-PK immediately localizes to DSBs in all cell cycle phases (10–12). As HR is the primary pathway mediating the repair of DSBs in mid- to late-S phase, we postulated that a post-translational modification, in particular phosphorylation of Ku70/80 (Ku), was the active mechanism responsible for displacing DNA-PK from DSBs to allow HR to occur (5,8,9). To assess if phosphorylation affects Ku's dynamics at DSBs, Ku80 was fluorescently labeled (YFP-tag) and its dynamics monitored at laser-generated DNA damage sites utilizing live cell microscopy in the presence or absence of the broad spectrum PI3K kinase inhibitor wortmannin (10,11,33). We found that treatment with wortmannin resulted in the sustained retention of Ku at laser-induced DSBs, indicating that phosphorylation may be a plausible mechanism by which Ku is actively displaced from DSBs (Supplementary Figure S1A) (34).
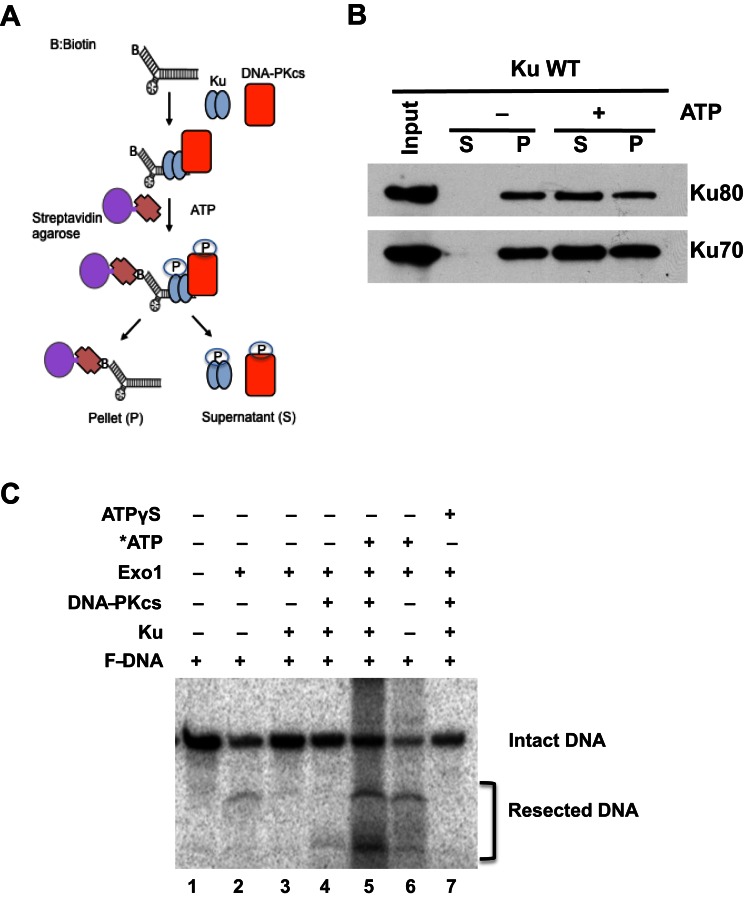
Next, an in vitro pull-down assay using biotinylated forked dsDNA and purified Ku was developed to test whether Ku phosphorylation via the kinase, DNA-PKcs, induced dissociation of Ku from biotin-labeled forked dsDNA (Figure 1A). Ku binds to DNA ends in an oriented manner and its dissociation from the forked dsDNA is very slow in the absence of activated DNA-PKcs (Figure 1B, -ATP lanes). On the other hand, DNA-PKcs activation, via the addition of ATP in the pull-down assay, results in the phosphorylation of Ku70/80 and increased dissociation of the Ku heterodimer from the forked dsDNA (Figure 1B and Supplementary Figure S2A). As phosphorylation of Ku induces its dissociation from forked dsDNA, we tested if this causes the freeing of DNA ends for processing. To assess this, we performed DNA end resection assays with exonuclease 1 (Exo1). Our biochemically defined in vitro DNA end resection assay previously showed that Ku blocks Exo1-mediated DNA end resection when the Ku70/80:DNA stoichiometry is 1:1 (Figure 1C, compare lanes 2 and 3; and Supplementary Figure S2B) (26). As expected, phosphorylation of Ku by DNA-PKcs results in the dissociation of the Ku heterodimer from the forked dsDNA and the subsequent freeing of the dsDNA ends allowed for Exo1-mediated DNA end resection (Figure 1C, compare lanes 3 and 5; Supplementary Figure S2A). Phosphorylation-mediated Ku dissociation was also confirmed by the addition of non-hydrolyzable ATP-γS, which blocked the DNA-PKcs phosphorylation of Ku, resulting in its sustained binding to the forked dsDNA and the blocking of Exo1-mediated processing (Figure 1C, compare lanes 5 and 7)(35). Finally, we tested if blocking of the DNA-PKcs kinase activity attenuated Ku's dissociation in vivo. Treatment with the DNA-PKcs inhibitor NU7441 resulted in the sustained retention of Ku at laser-generated DSBs (Supplementary Figure S1B). Taken together, the data show that phosphorylation of the Ku heterodimer leads to its dissociation from DSBs.
Figure 1.
Ku phosphorylation mediates its dissociation from DNA ends. (A) Schematic diagram of the in vitro DNA-Ku pull-down assay. (B) Phosphorylation of Ku led to its displacement from dsDNA ends in vitro. Purified Ku and DNA-PKcs were incubated with biotin-labeled forked dsDNA followed by the addition of ATP to allow phosphorylation of Ku by DNA-PKcs. The dsDNA was pulled down via streptavidin agarose. Dissociation from the forked dsDNA was assessed with Ku either still bound (pellet, P) or dissociated from the dsDNA (supernatant, S). (C) In vitro DNA resection assay. Exonuclease 1 (Exo1) digests the fork DNA substrate (lane 2 and 6). In the presence of Ku or the DNA-PK complex (DNA-PKcs and Ku), Exo1 could not resect the DNA (lane 3 and 4). The addition of ATP to the forked DNA substrate in the presence of the DNA-PK complex resulted in phosphorylation of Ku, leading to the dissociation of Ku and freeing DNA ends for Exo1-mediated resection (lane 5). Non-hydrolyzable ATP (ATPγS) was used as a control for phosphorylation mediating the freeing of the dsDNA ends (lane 7).
Putative Ku70 phosphorylation sites located at the junction of the pillar and bridge regions of Ku70 affects Ku's dissociation from DSBs
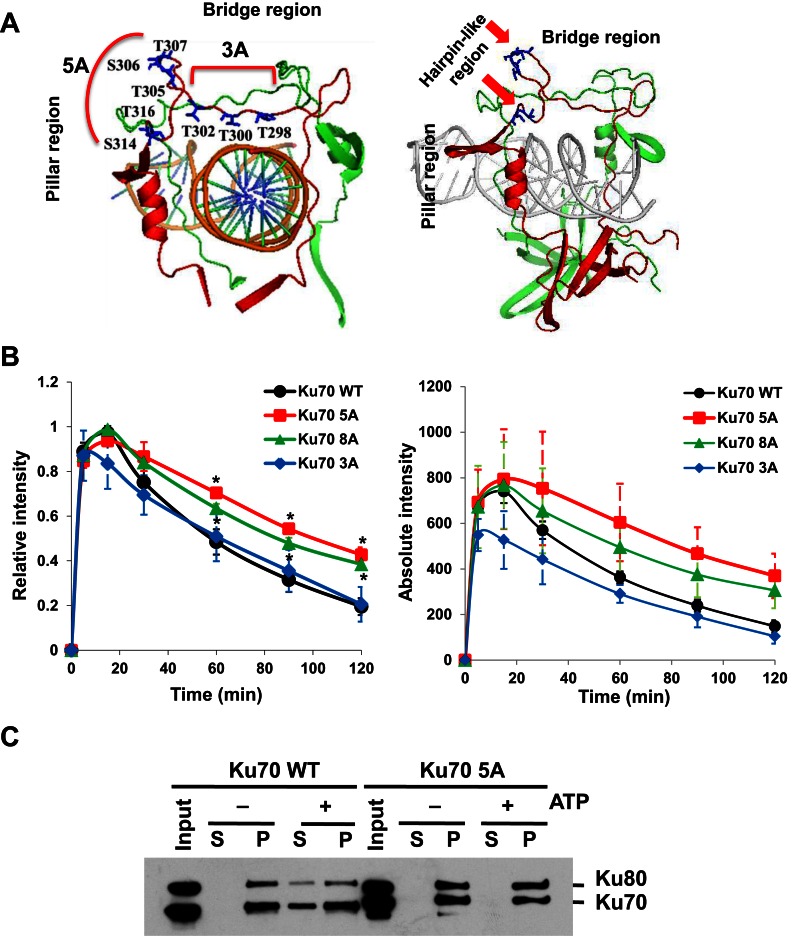
A knowledge-based approach was used to identify possible phosphorylation site(s) responsible for the dissociation of Ku70/80 from DSBs. We focused on residues in the bridge and pillar regions of Ku70 and Ku80, as they form the ring structure of Ku that is used to slide onto the DNA molecule. We postulated that a phosphorylation-induced conformational change in these regions might alter Ku's DNA affinity (14). A number of serine and threonine residues in the bridge and pillar regions of Ku70 and Ku80 were identified and mutated to alanine to ablate phosphorylation at these residues (Figure 2A and Supplementary Figure S3A). To screen which of these putative phosphorylation sites played an important role in regulating Ku's dissociation from DSBs, Ku mutants were YFP-tagged and their dynamics monitored at laser-generated DNA damage sites in asynchronous cells. Mutations in Ku80 had a minimal effect on Ku's dynamics at laser-generated DNA damage sites (Supplementary Figure S3A and B). In contrast, mutations at eight putative phosphorylation sites (T298, T300, T302, T305, S306, T307, S314 and T316; termed 8A) in Ku70 resulted in significant retention of Ku at damage sites (Figure 2A and B). To further delineate the sites important for mediating the dissociation of Ku from DSBs, the putative phosphorylation sites were split into two clusters termed 5A (T305, S306, T307, S314 and T316) and 3A (T298, T300, T302) (Figure 2A). The 5A mutant was retained at DSBs, while 3A was not. This implicates that the 5A cluster sites are required for phosphorylation-mediated dissociation of Ku from DSBs (Figure 2B). The 5A amino acids are located in hairpin-like regions of Ku70 with threonine 305 (T305), serine 306 (S306), threonine 307 (T307) sitting at the junction of the bridge and pillar of Ku70, whereas serine 314 (S314) and threonine 316 (T316) sit at the bottom of the pillar which connects to the base of Ku70 (Figure 2A). To test if these sites were required for the phosphorylation-induced dissociation of Ku from DNA ends in vitro, the Ku heterodimer was purified containing wild-type Ku80 and either wild-type Ku70 or 5A (Supplementary Figure S4A) and the in vitro pull-down assay with the forked dsDNA was repeated. We observed that the phosphorylation-induced dissociation of Ku from forked dsDNA ends was blocked in the 5A mutant protein (Figure 2C). Furthermore, phosphorylation of Ku70 in the 5A mutant protein was significantly reduced compared to wild-type Ku70 (Supplementary Figure S4B). Finally, we assessed if the phosphorylation-mediated dissociation of Ku from DSBs occurs in S phase. To test this, we analyzed the kinetics of wild-type Ku70 (WT) and 5A at laser-generated DSBs in S phase of the cell cycle. The kinetics data showed that WT quickly dissociates from laser-induced DSBs in S phase cells, whereas the 5A protein is clearly retained (Supplementary Figure S5). Taken together, the data showed that phosphorylation at the 5A cluster in Ku70 is required for Ku's dissociation from DSBs in S phase of the cell cycle.
Figure 2.
Phosphorylation of Ku70 is required for the dissociation of Ku from DSBs. (A) Structure of Ku70/80 with the DNA. Ku70 is indicated in red, while Ku80 is depicted in green. The locations of putative phosphorylation sites in Ku70 are denoted. Ku70 8A cluster sites include both 5A and 3A phosphorylation clusters. (B) Relative and absolute fluorescence intensity of YFP-tagged Ku70 and mutant proteins in Ku70−/− MEFs at DSBs after micro-irradiation. Student's t-test was performed to assess statistical significance (*P < 0.05). (C) Phosphorylation at the 5A sites is required for Ku's dissociation from DSBs. Results were similar to those indicated in 1B, but experiment was performed with purified Ku heterodimer with Ku80 with either wild-type Ku70 or 5A.
Mimicking phosphorylation of the five putative sites lessens Ku's DNA binding affinity
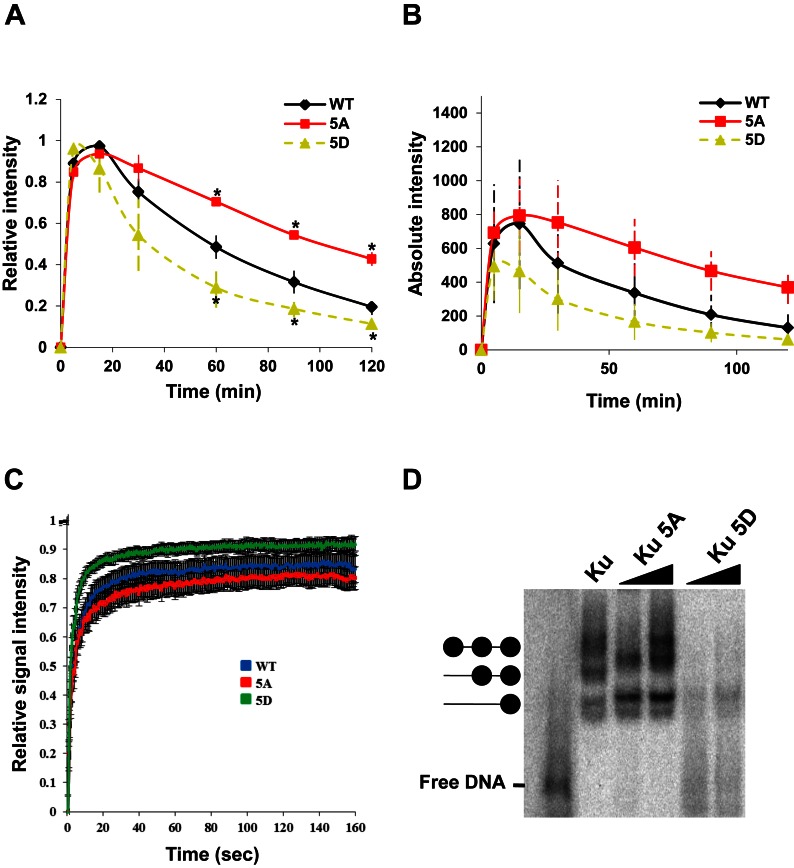
The five putative phosphorylation sites identified in our screening are all located at the hairpin-like structures in the junction of pillar and bridge regions of Ku70. We postulated that phosphorylation-induced conformational changes in these hairpin-like regions may affect the DNA binding affinity of Ku. To assess this, the dynamics of 5A were compared to Ku70 in which the sites were mutated to aspartic acid to mimic phosphorylation (5D). The kinetics showed that 5A is retained at DSBs longer than wild-type Ku70, whereas 5D clearly dissociates more quickly (Figure 3A). The recruitment of wild-type Ku70 and its mutants to DSBs, as shown by their absolute intensity values, revealed an increase in the amount of 5A protein localizing to DSBs compared to 5D, indicating that mimicking phosphorylation at these regions may affect Ku's affinity for DSBs (Figure 3B). This was supported by fluorescence recovery after photobleaching analysis with wild-type, 5A, and 5D proteins, which revealed the exchange rate of Ku to be much faster in 5D compared to 5A in asynchronously growing cells (Figure 3C) (33). The difference in recovery rates between wild-type, 5A, and 5D likely reflects a difference in the stability of the DNA-protein species, with the 5D-DNA interaction being less stable. Electrophoretic mobility shift assays with purified Ku heterodimer containing wild-type Ku80 and wild-type Ku70, 5A, or 5D indicated that Ku80/Ku70-WT and Ku80/Ku70–5A bind more strongly to dsDNA as compared to Ku80/Ku70–5D (Figure 3D and purified protein used shown in Supplementary Figure S4A). Collectively, the data illustrate that phosphorylation of Ku70 in the pillar and bridge regions reduces its affinity for dsDNA, which is responsible for Ku's dissociation from DNA ends in vivo and in vitro.
Figure 3.
Mimicking phosphorylation of the five putative sites reduced Ku's DNA binding affinity. (A) Relative and (B) Absolute fluorescence intensity of YFP-tagged Ku70 and mutant proteins in Ku70−/− MEFs at DSBs after micro-irradiation. Student's t-test was performed to assess statistical significance (*P < 0.05). (C) FRAP curves of Ku70 WT, 5A and 5D at the DSB site. Each data point is the average of 15 independent, normalized measurements. Error bars represent the standard deviation (SD). Fluorescence images were captured every 3 s up to 160 s. Pre-bleach intensity levels were normalized to 1, while post-bleach intensity levels were normalized to 0. Zero second represents the time of bleaching signal. (D) Electrophoretic mobility shift assays with purified wild-type Ku80 and wild-type Ku70, 5A or 5D with forked dsDNA.
Removal of Ku from DNA ends is essential for initiation of DNA end resection and homologous recombination
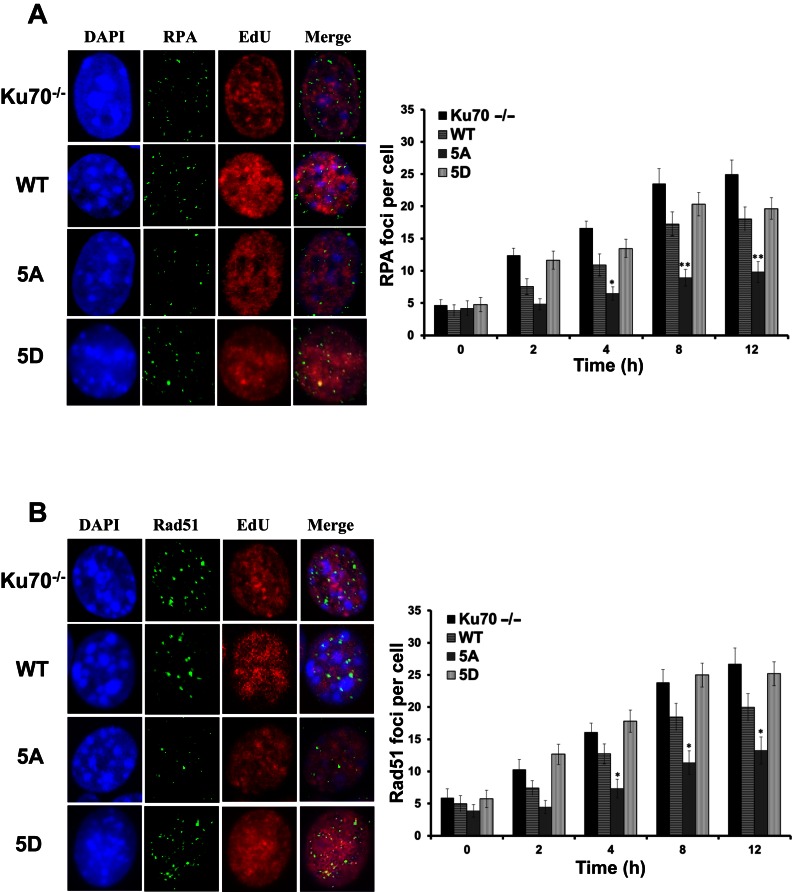
Our hypothesis that Ku is actively displaced to allow initiation of DNA end resection and HR suggests that the inability of bound Ku to be displaced from DNA ends in S phase may result in attenuation of HR. To assess if this was the case with the human Ku70 mutants, DNA end resection and Rad51 nucleofilament formation were monitored via the tracking of IR-induced RPA and Rad51 focus formation in S phase cells in Ku70−/− MEFs that were left either uncomplemented or complemented with wild-type Ku70, 5A or 5D (Supplementary Figure S6) (10,11). To ensure the detection of HR-specific DNA end resection and ongoing HR-mediated repair, differences in EdU staining were used to identify cells in mid- to late-S phase, where HR is thought to be the preferred DSB repair pathway (8,9). As previously observed, IR-induced RPA and Rad51 focus formation was increased in the absence of Ku compared to cells with wild-type Ku (Figure 4A and B) (11). Cells expressing 5A showed a marked decrease in IR-induced RPA and Rad51 foci per nucleus compared to wild-type complemented cells, indicating that blocking Ku dissociation caused a defect in the initiation of HR-mediated repair in S phase (Figure 4A and B). In contrast, cells expressing the 5D mutant showed increased RPA and Rad51 foci formation compared to 5A, and slightly higher than WT cells (Figure 4A and B). The data show that Ku must be displaced from DSB ends for initiation of HR.
Figure 4.
Blocking phosphorylation of Ku70 attenuates initiation of end resection and onset of homologous recombination. Panel of images depict (A) RPA and (B) Rad51 foci in EdU positive cells after 8 h after 8Gy gamma-ray exposure, respectively (Left panels). Immunostaining of Ku70−/− MEFs or Ku70−/− MEFs complemented with Ku70 wild-type, 5A or 5D after 8Gy of γ-rays. Cells were pre-extracted and fixed 2, 4, 8, or 12 h after IR and immunostained for RPA or Rad51. RPA and Rad51 foci were counted for each cell and averaged (Right panels). Student's t-test was performed to assess statistical significance (*P < 0.01 and **P < 0.001). Error bars denote standard error of the mean (SEM) for at least 3 independent experiments.
Phosphorylation-mediated dissociation of Ku from DNA ends is dispensable for non-homologous end joining in G1 but is required for homologous recombination in S/G2 phase
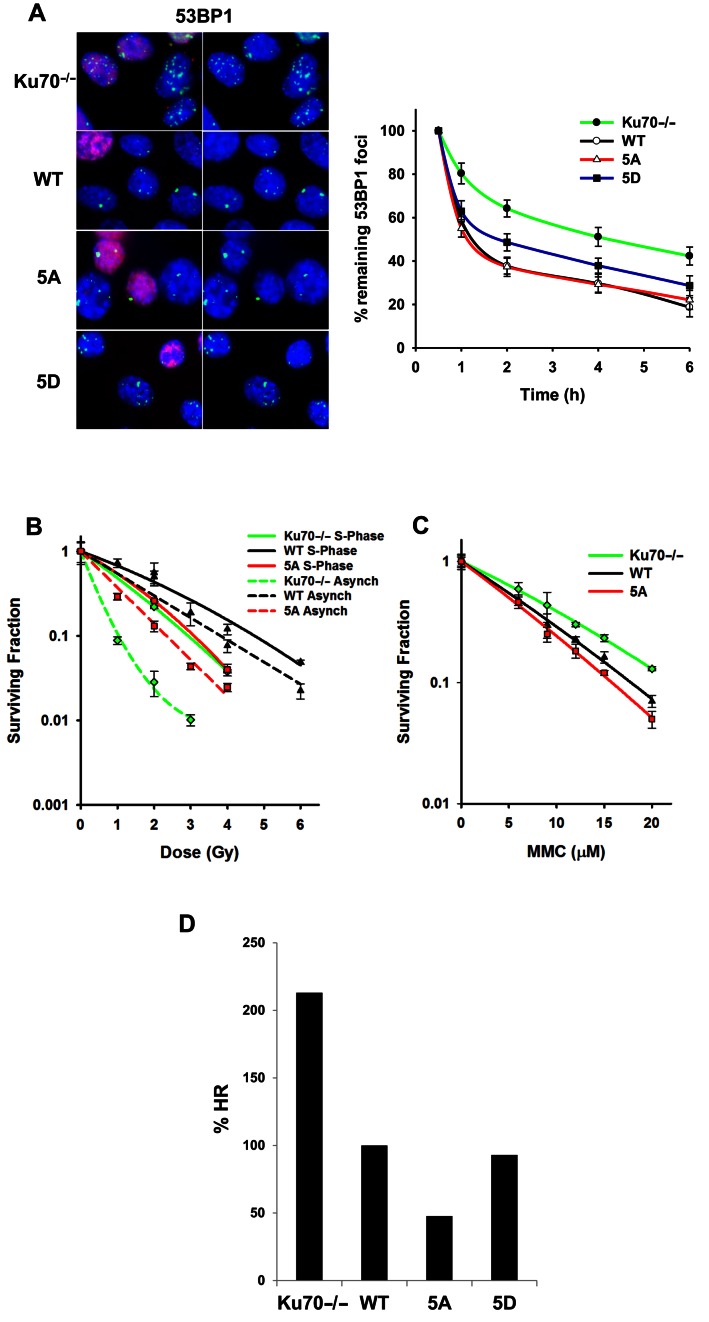
The observed decrease in HR initiation in the 5A cells may be due to a loss of overall NHEJ. This may result in blocking overall DSB repair, including HR, similarly to when phosphorylation of DNA-PKcs at the threonine 2609 cluster is blocked (36). To evaluate if blocking putative Ku phosphorylation affects NHEJ, 53BP1 focus formation in G1 cells was monitored in Ku70−/− MEFs that were left uncomplemented or complemented with wild-type Ku70 or 5A after exposure to 1 Gy of gamma rays. Non-EdU stained cells were tracked to identify G1 cells. Ku70−/− MEFs showed a defect in NHEJ as assessed by 53BP1 foci resolution, whereas 5A cells showed resolution of 53BP1 foci in a similar manner to wild-type complemented cells. (Figure 5A). In contrast, Ku70−/− MEF expressing 5D showed some degree of impairment in the resolution of 53BP1 foci in non-EdU stained cells. This finding suggests that NHEJ is attenuated in 5D complemented cells as compared to WT and 5A complemented cells (Figure 5A).
Figure 5.
Ablation of Ku70 phosphorylation results in attenuation of homologous recombination. (A) Images depicting 53BP1 foci, enumerated in EdU negative G1 phase cells after 6 h of 1Gy gamma-ray exposure (Left panel). Ku70−/− MEFs or Ku70−/− MEFs complemented with Ku70 wild-type, 5A or 5D were irradiated with 1Gy and 53BP1. Foci formation was assessed 1, 2, 4, and 6 h later (Right panel). Data were normalized to 53BP1 foci number enumerated at 30 min post IR. Remaining foci number per time point assessed was calculated and plotted. Error bars denote SEM of at least three independent experiments. (B) Colony formation assays were performed to compare radiation sensitivities of Ku70−/− MEFs or Ku70−/− MEFs complemented with Ku70 wild-type or 5A in S phase or as an asynchronous cell population. Cell lines were left cycling or were synchronized by the double thymidine block method and then released. Subsequently, cells were irradiated at the indicated doses and plated for analysis of survival and colony-forming ability. Error bars denote SD values. (C) Colony formation assays were performed to compare mitomycin C (MMC) sensitivity of Ku70−/− MEFs or Ku70−/− MEFs complemented with Ku70 wild-type or 5A. Cell lines were treated with the indicated concentrations of MMC and plated for analysis of survival and colony-forming ability. Error bars denote SD values. (D) Ku70−/− MEFs, containing one copy of the HR reporter assay vector integrated into the genome, were either uncomplemented or complemented with FLAG-tagged Ku70 wild-type, 5A or 5D. I-SceI was transiently expressed and induced a DSB at the I-SceI restriction site. Cells were analyzed by flow cytometry and the percent HR value was calculated as GFP / (RFP + GFP). The percent HR value of wild-type was normalized to 100% and the percent HR of Ku70−/− MEFs, 5A and 5D cells were calculated and plotted.
To assess if the phosphorylation-dependent dissociation of Ku was required for homologous recombination, clonogenic survival assays were performed following S phase synchronization. The assay was performed via the thymidine double block method in Ku70−/− MEFs which were either left uncomplemented or complemented with wild-type Ku70 or 5A cells (Figure 5B). Clonogenic survival assays with asynchronous cells revealed that 5A cells are more radiosensitive than WT complemented Ku70−/− MEFs, but less radiosensitive than the uncomplemented Ku70−/− MEFs (Figure 5B). To assess if the radiosensitivity of the 5A complemented cells was due to impaired HR, clonogenic survival assays were performed in cells synchronized in S phase. Clonogenic survival data with S phase synchronized cells showed a marked decrease in radiosensitivity in the Ku70−/− MEFs. In contrast, minimal changes in radiosensitivity were noted in the 5A cells, indicating that the radiosensitivity observed in the asynchronous 5A cells is due to a HR defect in a population of S phase cells (Figure 5B). To confirm that the repair defect in 5A cells was S phase specific, DSB repair was evaluated with a neutral Comet assay in cells synchronized in S phase (Supplementary Figure S7A). The data showed that DSB repair in S phase was attenuated in the 5A cells compared to complemented wild-type and Ku70−/− cells (Supplementary Figure S7B and S7C). Next, it was examined if blocking phosphorylation of Ku70 results in a sensitivity to DNA cross-links, a type of DNA damage that is repaired by the HR pathway. Clonogenic survival assays with the cross-linking agent MMC revealed that 5A cells were more susceptible to DNA cross-links as compared to wild-type and Ku70−/− cells (Figure 5C). Finally, the efficiency of homologous recombination was assessed in vivo via an HR reporter assay. 5A complemented cells showed a marked decrease (47.7%) in HR-mediated DSB repair when compared to the complemented wild-type (normalized to 100%) and Ku70−/− cells (213%) (Figure 5D). As expected, 5D cells showed HR activity similar to WT, and increased activity when compared to 5A cells (Figure 5D). Collectively, our data clearly indicate that the phosphorylation of Ku70 is required for Ku's dissociation from DSBs to free DNA ends for HR-mediated DSB repair.
DISCUSSION
Cells have access to multiple mechanisms to repair potentially lethal DSBs. Nevertheless, the selection of one pathway over another is still unclear. Since each DSB repair pathway has known initiating factors, competition between these proteins is believed to be responsible for DSB repair pathway choice. However, we previously showed that the NHEJ factors Ku70/80 and DNA-PKcs quickly localize to DSBs in S phase, where HR is the preferred DSB repair pathway (10,11). These data suggest that competition between NHEJ and HR factors is likely not responsible for choice of the DSB pathway in mammalian cells. Hence, we postulated that DNA ends must be freed from Ku in S phase to allow HR to occur. The need for freeing DSB ends for initiation of DNA end resection was supported by one of our previous studies. We showed that an ortholog of human Ku from Mycobacterium tuberculosis Ku (Mt-Ku) is recruited to laser-generated DSB ends similarly to human Ku, while Mt-Ku is retained at DSBs resulting in inhibition of DNA end resection and HR (11). Furthermore, we used a biochemically defined in vitro DNA end resection assay with a forked dsDNA to show that Ku cannot be displaced from DNA ends by either the endo- or exonuclease activity of Mre11 or the Mre11/Rad50 complex in vitro, which is in contrast with a previous report (26). Therefore, we sought to identify the mechanism mediating the dissociation of Ku from DSB ends in S phase.
Structurally, the Ku protein forms an expansive base and a very narrow bridge, producing a toroidal structure that allows Ku to slide onto the ends of DSBs (14,37). Furthermore, Ku has oriented DNA binding and along with its unique toroidal dsDNA binding region results in tight and sustained binding to DNA ends limiting the ability for Ku to simply be pushed off of a DSB (38,39). Since the unique physical properties of Ku help to maintain its sustained and firm binding to DNA ends, we investigated whether these physical properties could be reversed by post-translational modifications. Ku undergoes significant structural changes upon interaction with DNA-PKcs, although the preformed ring structure is stable even in absence of DNA binding (14). We report that Ku70 is phosphorylated, and this phosphorylation results in displacement of the Ku heterodimer from DNA ends in S phase. The phosphorylation sites lay in the hairpin-like loop regions of the bridge and pillar regions of Ku70. We believe that phosphorylation at these residues results in the unzipping of the hairpin-like loop regions of the bridge-pillar junction, causing the enlargement of the dsDNA binding channel and less stringent DNA affinity (Supplementary Figure S8). This will ultimately lead to Ku being easily slid off of the dsDNA ends. In vivo dynamic studies with point mutants ablating phosphorylation of Ku indicated sustained retention at DSBs compared to wild-type, supporting our hypothesis. Phospho-mimetic Ku mutants showed very low DNA binding affinity in in vivo FRAP analysis and in vitro gel shift assays, further supporting the notion that phosphorylation of Ku70 leads to less stringent DNA binding. Our data indicate for the first time that the high DNA end binding affinity of Ku can be altered by phosphorylation.
Previous studies have implicated that ubiquitylation of Ku may play a role in the displacement of the Ku heterodimer from DNA ends. In Xenopus egg extracts, xKu80 is polyubiquitylated via K48-linkage by Skp1-Cul1-Fbxl12, resulting in Ku's dissociation from dsDNA and subsequent degradation by the proteasome (40,41). Furthermore, the E3 ubiquitin ligase RNF8 has also been implicated in mediating the dissociation of Ku from DNA ends as depletion of RNF8 resulted in the prolonged retention of Ku80 at laser-generated DSBs (42). A recent study showed that neddylation promotes ubiquitylation and release of Ku from DSBs (43). The ubiquitination/neddylation-mediated Ku dissociation was hypothesized to be responsible for mediating Ku's dissociation near or following completion of NHEJ-mediated repair and/or required for the dissolution of the entire NHEJ machinery after a repair event (44). Our studies showed that treatment with the neddylation inhibitor MLN4924 induced an insignificant increase in Ku retention at laser generated DSBs in S phase cells (Supplementary Figure S9). This result suggests that neddylation may only play a minimal role in the dissociation of Ku from DSBs in S phase (compare Figure 2B and Supplementary Figure S9). Our findings collectively indicate that phosphorylation-dependent dissociation of Ku from DSBs is a unique mechanism for quickly and actively mediating the dissociation of the DNA-PK complex from DSBs to allow DNA end processing in S phase. This mechanism for Ku dissociation from DNA ends is distinct from that regulated by neddylation/ubiquitylation as we believe it is S phase specific and occurs before NHEJ is completed. However, we also believe that future studies could focus on understanding if these post-translational modification events are either separate entities or coordinated to modulate NHEJ.
DNA end resection is required for HR-mediated repair. It is believed that DNA end resection plays a role in the selection of the DSB pathway because once the DSB has been resected, the cell has committed to HR (17). Recent studies have implicated the 53BP1/PTIP/RIF1 and BRCA1/CtIP circuits to regulate DNA end resection in a cell cycle specific manner (23,24,45,46). We hypothesize that Ku must be actively displaced to allow initiation of DNA end resection and that this is upstream of the competition between the two circuits. This is supported by our RPA2/Rad51 foci and survival assays data, which show that the biological consequence of blocking phosphorylation of Ku70, and thus abrogating Ku dissociation in S phase, results in attenuation of DNA end resection and defective HR-mediated repair. In contrast, phospho-mimetic mutations at these same Ku70 (5D) sites result in reduced DNA binding affinity for the Ku heterodimer and defects in NHEJ, as indicated by an increase in radiosensitivity in 5D cells (Supplementary Figure S10). Taken together our results suggest that phosphorylation of these five sites in the pillar and bridge regions of Ku70 is a key mechanism driving the choice of the DNA repair pathway in S phase of the cell cycle.
We believe DSB repair pathway choice is likely orchestrated by multiple regulatory mechanisms including chromatin-associated factors, cell cycle dependent regulations, and the coordinated interplay between the NHEJ and HR repair machineries. We propose the following model. First, the DNA-PK complex quickly localizes to DSBs in all cell cycle phases. In S phase, when HR is the preferred DSB repair pathway, Ku is phosphorylated by DNA-PKcs in the pillar and bridge regions of Ku70. This phosphorylation induces a conformational change leading to a less stringent DNA binding and promoting the release of Ku from DSB ends. Ku displacement is either mediated by a reduced affinity for dsDNA and/or assisted by the MRN/CtIP proteins. This frees the DSB ends and allows for the initiation of DNA end resection, ultimately resulting in HR-mediated DSB repair in S phase. It should be noted that it remains to be elucidated how these regulatory mechanisms coordinate and work in conjunction for selecting DSB repair pathways for optimum maintenance of genomic stability in a cell. A better understanding of the regulatory mechanisms governing DSB repair pathway choice in specific phases of the cell cycle could potentially lead to the development of targeted therapies for tumor cells in S/G2 phases after radiotherapy, reducing the tumor burden.
Supplementary Material
Acknowledgments
The authors thank Dr Benjamin Chen for assistance and comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [CA092584, CA162804 and CA134991]; Cancer Prevention Research Institute of Texas [RP110465-P1 to D.J.C.]. Funding for open access charge: National Institutes of Health (NIH) [CA092584, CA162804 and CA134991]; Cancer Prevention Research Institute of Texas [RP110465-P1].
Conflict of interest statement. None declared.
REFERENCES
- 1.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schipler A., Iliakis G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013;41:7589–7605. doi: 10.1093/nar/gkt556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 4.Goodarzi A.A., Jeggo P.A. The repair and signaling responses to DNA double-strand breaks. Adv. Genet. 2013;82:1–45. doi: 10.1016/B978-0-12-407676-1.00001-9. [DOI] [PubMed] [Google Scholar]
- 5.Thompson L.H. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat. Res. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Ann. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 7.Chapman J.R., Taylor M.R., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 8.Hartlerode A., Odate S., Shim I., Brown J., Scully R. Cell cycle-dependent induction of homologous recombination by a tightly regulated I-SceI fusion protein. PloS One. 2011;6:e16501. doi: 10.1371/journal.pone.0016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karanam K., Kafri R., Loewer A., Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell. 2012;47:320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis A.J., Chi L., So S., Lee K.J., Mori E., Fattah K., Yang J., Chen D.J. BRCA1 modulates the autophosphorylation status of DNA-PKcs in S phase of the cell cycle. Nucleic Acids Res. 2015;42:11487–11501. doi: 10.1093/nar/gku824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao Z., Davis A.J., Fattah K.R., So S., Sun J., Lee K.J., Harrison L., Yang J., Chen D.J. Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair (Amst) 2012;11:310–316. doi: 10.1016/j.dnarep.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britton S., Coates J., Jackson S.P. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 2013;202:579–595. doi: 10.1083/jcb.201303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mimori T., Hardin J.A., Steitz J.A. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J. Biol. Chem. 1986;261:2274–2278. [PubMed] [Google Scholar]
- 14.Walker J.R., Corpina R.A., Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 15.Blier P., Griffith A., Craft J., Hardin J. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J. Biol. Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- 16.Downs J.A., Jackson S.P. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 17.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niu H., Raynard S., Sung P. Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev. 2009;23:1481–1486. doi: 10.1101/gad.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernstein K.A., Shor E., Sunjevaric I., Fumasoni M., Burgess R.C., Foiani M., Branzei D., Rothstein R. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J. 2009;28:915–925. doi: 10.1038/emboj.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zierhut C., Diffley J.F. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huertas P., Cortes-Ledesma F., Sartori A.A., Aguilera A., Jackson S.P. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata A., Conrad S., Birraux J., Geuting V., Barton O., Ismail A., Kakarougkas A., Meek K., Taucher-Scholz G., Lobrich M., et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman J.R., Barral P., Vannier J.B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D., Boulton S.J. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Escribano-Diaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkac J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Mimitou E.P., Symington L.S. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun J., Lee K.J., Davis A.J., Chen D.J. Human Ku70/80 protein blocks exonuclease 1-mediated DNA resection in the presence of human Mre11 or Mre11/Rad50 protein complex. J. Biol. Chem. 2012;287:4936–4945. doi: 10.1074/jbc.M111.306167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G.C., Ouyang H., Li X., Nagasawa H., Little J.B., Chen D.J., Ling C.C., Fuks Z., Cordon-Cardo C. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]
- 28.Davis A.J., Lee K.J., Chen D.J. The amino-terminal region of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is required for its DNA double-strand break-mediated activation. J. Biol. Chem. 2013;288:7037–7046. doi: 10.1074/jbc.M112.434498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.So S., Davis A.J., Chen D.J. Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J. Cell Biol. 2009;187:977–990. doi: 10.1083/jcb.200906064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y.G., Saidi A., Frappart P.O., Min W., Barrucand C., Dumon-Jones V., Michelon J., Herceg Z., Wang Z.Q. Conditional deletion of Nbs1 in murine cells reveals its role in branching repair pathways of DNA double-strand breaks. EMBO J. 2006;25:5527–5538. doi: 10.1038/sj.emboj.7601411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobias F., Lob D., Lengert N., Durante M., Drossel B., Taucher-Scholz G., Jakob B. Spatiotemporal dynamics of early DNA damage response proteins on complex DNA lesions. PloS One. 2013;8:e57953. doi: 10.1371/journal.pone.0057953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Splinter J., Jakob B., Lang M., Yano K., Engelhardt J., Hell S.W., Chen D.J., Durante M., Taucher-Scholz G. Biological dose estimation of UVA laser microirradiation utilizing charged particle-induced protein foci. Mutagenesis. 2010;25:289–297. doi: 10.1093/mutage/geq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uematsu N., Weterings E., Yano K., Morotomi-Yano K., Jakob B., Taucher-Scholz G., Mari P.O., van Gent D.C., Chen B.P., Chen D.J. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell. Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mari P.O., Florea B.I., Persengiev S.P., Verkaik N.S., Bruggenwirth H.T., Modesti M., Giglia-Mari G., Bezstarosti K., Demmers J.A., Luider T.M., et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan D., Ye R., Veillette C., Lees-Miller S. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry. 1999;38:1819–1828. doi: 10.1021/bi982584b. [DOI] [PubMed] [Google Scholar]
- 36.Zhang S., Yajima H., Huynh H., Zheng J., Callen E., Chen H.T., Wong N., Bunting S., Lin Y.F., Li M., et al. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J. Cell Biol. 2011;193:295–305. doi: 10.1083/jcb.201009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones J.M., Gellert M., Yang W. A Ku bridge over broken DNA. Structure. 2001;9:881–884. doi: 10.1016/s0969-2126(01)00658-x. [DOI] [PubMed] [Google Scholar]
- 38.Hu S., Cucinotta F.A. Modelling the way Ku binds DNA. Radiat. Prot. Dosimetry. 2011;143:196–201. doi: 10.1093/rpd/ncq519. [DOI] [PubMed] [Google Scholar]
- 39.Krishna S.S., Aravind L. The bridge-region of the Ku superfamily is an atypical zinc ribbon domain. J. Struct. Biol. 2010;172:294–299. doi: 10.1016/j.jsb.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postow L., Funabiki H. An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle. 2013;12:587–595. doi: 10.4161/cc.23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postow L., Ghenoiu C., Woo E.M., Krutchinsky A.N., Chait B.T., Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J. Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng L., Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat. Struct. Mol. Biol. 2012;19:201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown J.S., Lukashchuk N., Sczaniecka-Clift M., Britton S., le Sage C., Calsou P., Beli P., Galanty Y., Jackson S.P. Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites. Cell Rep. 2015;11:704–714. doi: 10.1016/j.celrep.2015.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meir M., Galanty Y., Kashani L., Blank M., Khosravi R., Fernandez-Avila M.J., Cruz-Garcia A., Star A., Shochot L., Thomas Y., et al. The COP9 signalosome is vital for timely repair of DNA double-strand breaks. Nucleic Acids Res. 2015;43:4517–4530. doi: 10.1093/nar/gkv270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callen E., Di Virgilio M., Kruhlak M.J., Nieto-Soler M., Wong N., Chen H.T., Faryabi R.B., Polato F., Santos M., Starnes L.M., et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J., Aroumougame A., Lobrich M., Li Y., Chen D., Chen J., Gong Z. PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev. 2014;28:2693–2698. doi: 10.1101/gad.252478.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.