Abstract
During eukaryotic translation initiation, 43S ribosomal complex scans mRNA leader unless an AUG codon in an appropriate context is found. Establishing the stable codon–anticodon base-pairing traps the ribosome on the initiator codon and triggers structural rearrangements, which lead to Pi release from the eIF2-bound GTP. It is generally accepted that AUG recognition by the scanning 43S complex sets the final point in the process of start codon selection, while latter stages do not contribute to this process. Here we use translation reconstitution approach and kinetic toe-printing assay to show that after the 48S complex is formed on an AUG codon, in case GTP hydrolysis is impaired, the ribosomal subunit is capable to resume scanning and slides downstream to the next AUG. In contrast to leaky scanning, this sliding is not limited to AUGs in poor nucleotide contexts and occurs after a relatively long pause at the recognized AUG. Thus, recognition of an AUG per se does not inevitably lead to this codon being selected for initiation of protein synthesis. Instead, it is eIF5-induced GTP hydrolysis and Pi release that irreversibly trap the 48S complex, and this complex is further stabilized by eIF5B and 60S joining.
INTRODUCTION
Start codon selection during eukaryotic translation initiation is a complicated process that requires a concerted action of the ribosome, Met-tRNAi, and a number of specialized proteins termed eukaryotic initiation factors (eIFs) (1). The canonical initiation pathway starts with a 43S preinitiation complex formed by the small ribosomal subunit with associated factors eIF1, eIF1A, eIF2, eIF3 and eIF5. eIF2 is loaded onto the 40S subunit in the form of a ternary complex with GTP and Met-tRNAi. After mRNA binding, 43S starts to migrate in the 5′ to 3′ direction in search of an initiation codon (usually AUG) in the appropriate nucleotide context. This process is known as ‘scanning’. It has been proved for cap-dependent mRNAs with both short and simple leaders and rather long and highly structured 5′ untranslated regions (2–4).
Once AUG is reached, the perfect codon–anticodon interaction is established, which means that the recognition has occurred. This process is controlled by a small protein eIF1 bound to the P-site (for review, see (1,5,6) and references therein). eIF1 impedes accommodation of the Met-tRNAi within the P-site until the perfect codon–anticodon duplex is formed. At this moment the scanning is ceased, and the ribosome is trapped on the selected AUG codon. The AUG recognition induces conformational rearrangements of the complex that lead to eIF1 displacement and eIF2·GDP release, further followed by eIF5B-assisted joining of the 60S ribosome subunit and dissociation of the remaining initiation factors. The resulting 80S ribosome with the correctly positioned Met-tRNAi in the P-site is competent for the second tRNA binding and starting the synthesis of a polypeptide.
Hydrolysis of the eIF2-bound GTP and subsequent Pi release are believed to be the key steps in the process of AUG selection (7–9). The GTPase activity of eIF2 requires specific GTPase activating protein (GAP), eIF5 (10–14). eIF5 is a core constituent of a multifactor complex (MFC) that also includes eIF1, eIF3, the ternary complex, and is thought to be an important intermediate of translation initiation complex assembly (15–17). As the MFC component, in most cases eIF5 must be present in the 43S ribosomal complex from the very moment of its formation, and it essentially contributes to AUG selection during scanning (1). It was also shown that eIF5-stimulated GTP hydrolysis can occur in eIF2 even before the ribosome encounters AUG (7,9). Thus, GTP is normally already hydrolyzed when the 43S arrives at AUG, although the reaction becomes irreversible only after Pi dissociates from the complex. While being crucial for the hydrolysis, eIF5 is required neither for 48S complex assembly nor for correct start codon recognition, at least on mRNAs with a single AUG (13,18,19).
In yeast, mutations in the TIF5 gene encoding eIF5 have been long known to affect start codon selection (8). More specifically, certain mutations in TIF5 produce a Sui− phenotype (i.e. increased initiation at UUG codon) by upregulation of the eIF5 GAP activity and premature Pi release (8,20). On the other hand, mutations of eIF5 that do not affect the GTPase reaction may also impair a proper AUG selection, including recognition of uAUG codons in GCN4 mRNA, thus conferring a Gcn− or a Gcd− phenotype (21,22). Studies in a reconstituted yeast translation initiation system (19) provided a mechanistic rationale for these effects by uncovering a complex pattern of conformational rearrangements within the 43S complex upon AUG recognition. These alterations involve changes in intermolecular contacts between eIF5, eIF2β, eIF1A and eIF1, and finally couple the AUG recognition to eIF1 dissociation, Pi release and stabilization of the 48S complex in a PIN state (20,23–28).
Recently, effects of eIF5 concentration changes on a stringency of AUG recognition were also reported in mammalian systems (29–31). In living cells, overexpression of eIF5 resulted in a more frequent recognition of poor-context AUGs or non-AUG start codons and also shifted the initiation in favor of uAUG codons in case of the human eIF5 mRNA itself (29). A pronounced stimulation of 48S complex formation at non-optimal start codons was also observed in a reconstituted mammalian translation initiation system (31). The ability of eIF5 to affect start codon selection is therefore an evolutionary conserved feature of this eukaryotic factor.
To conclude, although the presence of eIF5 in the 43S complex is not required for mRNA binding or scanning nor for AUG recognition, it becomes absolutely necessary to accomplish productive initiation once the complex reaches and recognizes an AUG. If the factor does not come in time, or for any reason its activity is lowered, a delay in eIF2-bound GTP hydrolysis should occur. In this work, we reconstructed this situation and found that such a delay leads to the redistribution of initiation complexes in favor to downstream AUG codons. The redistribution occurred as a result of the resumed movement of the 43S complex trapped onto 5′ proximal AUG codon along the mRNA after a pronounced pause. To distinguish this post-AUG-recognition process from the conventional scanning (and also from the ‘resumed scanning’ that takes place in some cases after translation of uORFs), we called the newly discovered process ‘sliding’. We argue that the phenomenon of 43S sliding contributes to the eIF5 impact on start codon selection.
MATERIALS AND METHODS
Plasmid constructs and in vitro transcription
A plasmid coded for the EMCV IRES-containing transcript (nts 377–1155 of the EMCV RNA) was the same as in our previous studies (32,33). To prepare ΔE012, a polymerase chain reaction (PCR) product was obtained with this plasmid and primers CAGAATTCGTGGTTTTCCTTTG and GTTTTCCCAGTCACGAC, treated with EcoRI and PstI and ligated into the same vector digested by the same enzymes. All ΔE012 derivatives were prepared by PCR mutagenesis of the initiation region; their partial sequences are shown in Supplementary Figure S1. The construct for 5uAUGs-Fluc mRNA was prepared by insertion of a duplex formed by ATGATGATGATGGCATG and CCATCATCATCATCATG oligos into the NcoI site of pFluc plasmid (34). The plasmid 1uAUG-Fluc encoded Fluc mRNA with one uAUG was described before (35). To obtain the ΔE12UMBRA construct, a fragment of the umbravirus ORF3/4 region was obtained by PCR with an umbravirus cDNA (kindly gifted by M.Taliansky) and primers CCATCAACCATGGAACAATCTTCGCAAGTGGCAAAAGC and AGAGTGCGCGCACTTATTGGCAGCGGGTTTG; this fragment was ligated into BalI and BsePI sites of the ΔE12. A series of firefly luciferase encoding constructs with 5′ UTRs of natural human mRNAs (CDK4, CFTR, MDM2, PNRC2 and EIF2D) were prepared by insertion of the corresponding cDNA fragments to the pFluc plasmid. Sequences of their 5′ proximal regions are shown in Supplementary Figure S2. The constructs encoding ATF4 and UCP2 leaders were described before (36). Prior to in vitro transcription, the ΔE012 derivatives and the 5uAUGs-Fluc plasmid were linearized by HindIII or Bpu14I, respectively. For synthesis of the polyadenylated 1uAUGs-Fluc mRNA and the mRNAs with natural 5′ UTRs, a 50T-tailed PCR product was used as described previously (34). PCR templates for synthesis of mRNAs (CA)-Fluc and (CA)uORF-Fluc were obtained using the same strategy directly from the pFluc plasmid with the corresponding T7 promoter-containing forward primers (see Supplementary Figure S2). For the transcription, RiboMAX kit (Promega) was used. The resulting transcripts were precipitated with 2M LiCl. The EMCV IRES containing mRNA was uncapped, whereas for all other transcripts Vaccinia Capping System (NEB) was used to obtain 100% capped products.
Toe-printing of ribosomal complexes in rabbit reticulocyte lysate
To assemble ribosomal complexes, we utilized commercially available nuclease treated RRL (Promega, L4960). We followed our previously published protocol (32) with some modifications. The reaction was initiated in a total volume of 9 μl containing 7 μl of RRL, 0.05 μl of RiboLock RNase inhibitor (Thermo Scientific) and 2 μl of either GTP·Mg/GMPPNP·Mg (for final concentration of 2 mM) or water solution of cycloheximide (for final concentration of 1 mg/ml). In negative control, Mg(OAc)2 concentration was elevated to 15 mM. The mixture was incubated for 5 min at 30ºC, then 1 μl of mRNA solution (0.5 pmol/μl) was added, and the mixture was incubated for an additional 10 min (or other time periods, as indicated) at 30ºC. After that, 10 μl of RT Mix were added directly into the tube at 30ºC [RT Mix was prepared during the incubation and contained 1 μl 20x Buffer (400 mM Tris-HCl pH 7.5, 1.2 M KCl, 5 mM spermidine-HCl, 20 mM DTT), 2 μl of dNTP mix (5 mM each), 1 μl of [32P]-labeled oligonucleotides (5 pmol/μl, GTAGAGCAGAGCATTTTGGG for EMCV mRNA derivatives or TGCAGTTGCTCTCCAGCG for the luciferase encoding transcripts), 1 μl of 0.5 M Mg(OAc)2, 1 μl of AMV Reverse Transcriptase (Promega) and 4 μl of water]. The mixture was incubated for 20 min at 30ºC. For kinetic toe-printing, the protocol was slightly modified. In this case, we utilized a single tube with the reaction mixture and multiple tubes with RT Mix aliquots (10 μl per tube) that were held in ice during the whole time duration. At times indicated, 10 μl aliquots of the reaction mixture were sequentially taken and placed into the tubes with the RT Mix and kept in ice. This stopped the reaction of the complex formation. After that, all the tubes were placed at 30ºC for an additional 20 min. The resulting cDNAs were then purified by thorough phenol/chloroform extraction, precipitated with ethanol and analyzed on 6% sequencing gel along with a sequence ladder obtained from the corresponding plasmid with the same primer and Sequenase 2.0 DNA sequencing Kit (USB/Affymetrix). Radioactive bands in the dried gels were visualized using the Typhoon FLA 9500 Phosphorimager (GE Healthcare Life Sciences). Relative amounts of radioactivity in the bands were determined with ImageQuant software (GE Healthcare Life Sciences).
Purification of translation initiation components
eIF2, eIF3, eIF4F, eIF5B, 40S and 60S were purified from HeLa cell extract, eIF1, eIF1A, eIF4A, eIF4B and eIF5 were expressed in Escherichia coli as described (18,32,37–39). The sequence encoding the recombinant eIF5 in pET21a-eIF5 plasmid was corrected by introducing an additional trinucleotide CCA, to restore a naturally occurring heptaproline sequence (aa 179–185) that had been disturbed in the original plasmid. eIF5 obtained from the resulting plasmid had the same activity as the original one with only six prolines. Purified tRNAfMet, a kind gift from V. Makhno and Y. Semenkov, was used as initiator tRNA. For aminoacylation, recombinant MetRSase was used as described (37).
Assembly and analysis of translation initiation complexes
48S translation initiation complexes were assembled and analyzed by toe-printing assay as described earlier (38,39). Briefly, 48S complexes were assembled by incubating 0.5 pmol of mRNA for 10 min at 30°C in a 20-μl reaction volume that contained the reconstitution buffer (20 mM Tris-HCl, pH 7.5; 110 mM KOAc; 1 mM Mg(OAc)2; 0.25 mM spermidine-HCl; 1 mM DTT), 0.4 mM GTP·Mg or GMPPNP·Mg and 1 mM ATP-Mg(OAc)2, 10 pmol of Met-tRNAiMet, 2.5 pmol of 40S ribosomal subunits and combination of factors (eIF1 (10 pmol), eIF1A (10 pmol), eIF2 or aIF2 (5 pmol), eIF3 (5 pmol), eIF4A (10 pmol), eIF4B (5 pmol), eIF4F (2 pmol), eIF5 (5 pmol) and eIF5B (5 pmol)), as described in the text. For toe-printing, the same [32P]-labeled oligonucleotides as above were used. Primer extension analysis was performed essentially as described (32).
Cell-free systems and in vitro translation
Translation in RRL in the presence of [35S]-Met was performed according to manufacturer's instruction (Promega). Krebs-2 cells S30 extract was prepared as described previously (40). Translation experiments were performed in a total volume of 10 μl, which contained 5 μl of the S30 extract, translation buffer (20 mM Hepes-KOH pH 7.6, 1 mM DTT, 0.5 mM spermidine-HCl, 0.8 mM Mg(OAc)2, 8 mM creatine phosphate, 1 mM ATP, 0.2 mM GTP, 120 mM KOAc and 25 μM of each amino acid), 2 u of RiboLock RNase inhibitor (Thermo Scientific), 0.25 pmol capped Fluc mRNA and 2 μl of eIF5 protein dilutions or an appropriate buffer, for 1 hour (41). The luciferase activities were measured using the Dual Luciferase Assay kit (Promega).
RESULTS
43S complexes assembled in the presence of GMPPNP tend to localize at distal AUG codons
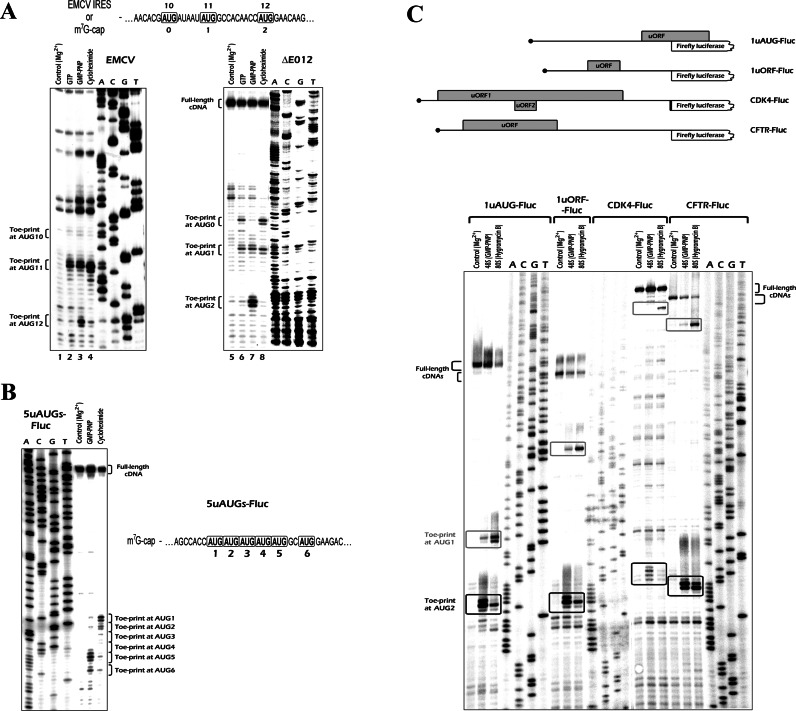
We previously reported an unusual pattern of 48S initiation complexes distribution between three closely spaced AUG codons of the encephalomyocarditis virus (EMCV) mRNA when translation had been blocked by non-hydrolysable GTP analog GMPPNP (32). At the same time, the pattern of cycloheximide-arrested 80S complexes correlated with the physiological usage of these AUG codons (42). To check whether this phenomenon is limited to the mRNA containing EMCV internal ribosome entry site (IRES) or rather is a common feature of mRNAs with two or more closely located AUG codons, we prepared a series of artificial mRNA constructs. The first was an EMCV derivative with the IRES removed. The construct ΔE012 has a 31 nt long leader followed by the EMCV initiation region that contained AUG10, AUG11 and AUG12 of the original EMCV mRNA. In the ΔE012 construct, these codons were the first, second and third AUGs from the 5′ end, respectively; however, hereafter we will call them ‘AUG0’, ‘AUG1’ and ‘AUG2’ for consistency (Figure 1A, top; for the nucleotide sequences, see Supplementary Figure S1). The in vitro transcribed and m7G-capped mRNA was incubated in rabbit reticulocyte lysate (RRL) in the presence of translation inhibitors, and then toe-printing analysis of ribosomal complexes was performed (32). For comparison, the experiment with the original EMCV IRES containing mRNA was repeated under the same conditions (Figure 1A).
Figure 1.
Toe-printing analysis of ribosomal complexes assembled in RRL on mRNAs with two or more AUG codons. (A) mRNA containing the EMCV IRES (left) and the m7G-capped mRNA ΔE012 (right) were incubated in RRL with 15 mM Mg(OAc)2 (lanes 1 and 5), 2 mM GTP·Mg/GMPPNP·Mg (lanes 2/3 and 6/7) or 1 mg/ml cycloheximide (lanes 4 and 8). Nucleotide sequences of the initiation region is shown on the top. Positions of the toe-prints are indicated. Sequencing lanes obtained with the same primer and the corresponding cDNA are shown on the right. (B) Ribosomal complexes assembled on the 5uAUG-Fluc mRNA in the presence of GMPPNP or cycloheximide. (C) Toe-printing analysis of the 48S and 80S complexes assembled in RRL on Fluc encoding mRNAs with artificial or natural uORF-containing 5′ UTRs. The 5′ proximal regions of the transcripts are schematically shown at the top.
In the case of the cap-dependent mRNA, 80S ribosomes arrested by cycloheximide were detected exclusively at the two 5′ proximal codons (Figure 1A, line 8), in accordance with the scanning model (3). Most of them recognized the first AUG (AUG0), but a significant portion reached the second one (AUG1) presumably via leaky scanning due to a rather short leader (3). In the case of the EMCV IRES, the cycloheximide-arrested ribosomes were detected almost exclusively at the AUG11, as expected (Figure 1A, line 4) (42). However, in the presence of GMPPNP·Mg, the 43S complexes demonstrated a clear preference for the 5′ distal AUGs, with very strong toe-print signals corresponding to the furthermost start codons (Figure 1A, lanes 3 and 7). The effect was not an artifact of magnesium sequestration (or some other consequences of the guanine nucleotide addition) since the adding GTP·Mg to the same concentration did not lead to the aberrant ribosome distribution (Figure 1A, lanes 2 and 6).
Since in both above cases the initiation regions were identical, one could not exclude that it was some feature in a local nucleotide context of the three AUGs, and not their order, that biased the 43S distribution. In other words, AUG12 (and the corresponding AUG2 in the case of ΔE012) might be preferential for 48S formation in the presence of GMPPNP due to some peculiarities of its environment and not because it was the most distal start codon. We therefore prepared several additional constructs on the basis of the ΔE012 that differed in their initiation regions (Supplementary Figures S1 and S3). First of all, to eliminate a likely impact of leaky scanning through AUG0, we removed this codon by replacing it with UAG. Then, we inserted AUG together with its adjacent 9 nts corresponding to the context of AUG1 or AUG2 (2 codons upstream and 1 codon downstream to the AUG) either between the original AUG1 and AUG2 or after AUG2. We also modified the initiation region by shortening or lengthening the spacer between AUG1 and AUG2. In all experimental settings, we observed strong signals corresponding to pre-initiation complexes formed at the last AUG present within the initiation region (Supplementary Figure S3). This contradicted the predictions of the canonical scanning model.
To avoid any impact of EMCV sequences, we repeated the experiment with another mRNA that had a leader derived from an irrelevant artificial vector sequence followed by 6 consecutive AUG codons and firefly luciferase (Fluc) coding region (Figure 1B). As expected, in the presence of cycloheximide, we observed one major toe-print corresponding to 80S ribosomes formed at the first AUG of the mRNA and a much less prominent signal at the second AUG. In contrast, in GMPPNP-supplemented lysate we detected toe-print bands that corresponded to 48S complexes located at the two most distal (fifth and sixth) AUG codons.
Then we turned to more physiologically relevant situations and performed the same experiments with luciferase encoding transcripts having uORFs in their 5′ UTRs (Figure 1C and Supplementary Figure S2). We used two artificial (plasmid-derived) sequences and two natural leaders of the human CDK4 and CFTR mRNAs. To produce 80S complexes, we utilized hygromycin B instead of the cycloheximide. This allowed us to avoid toe-print smearing caused by an incomplete elongation arrest, and to improve the signal intensity (32). In all four cases, a similar and clear difference between 48S and 80S complex distribution patterns was observed (Figure 1C). The 80S complexes were preferentially localized at 5′ proximal AUG codons, in accordance with the scanning model. However, a major portion of the 48S complexes were detected at the 5′ distal AUG codons, in full compliance with the observations made previously. In the case of the CFTR-Fluc mRNA, a significant portion of the 80S was formed at the second AUG presumably due to leaky scanning through the first initiation codon located in a weaker nucleotide context, caaAUGc (see Supplementary Figure S2).
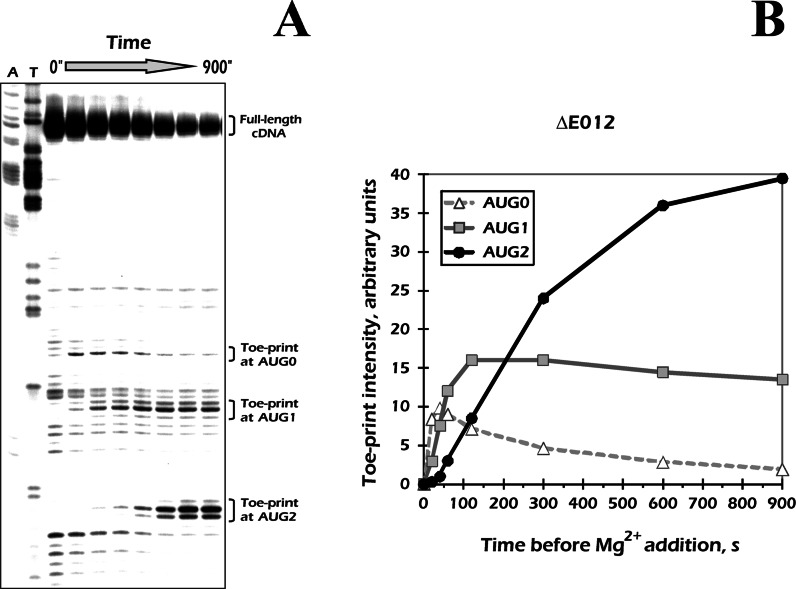
Delayed redistribution of the GMPPNP-arrested 43S complexes toward distal AUG codons revealed by kinetic toe-printing
In all of the above experiments, we incubated mRNA in RRL for 10 min before fixation of the complexes by elevating a Mg2+ concentration. To address the phenomenon in greater detail, we performed time-course analysis of the reaction. We tracked the formation of the complexes on ΔE012 mRNA from 0 to 15 min, terminating the reaction at different time points by adding 15 mM Mg(OAc)2. At the beginning of incubation, intensive toe-print bands corresponding to 48S complex formed at the AUG0 were observed, while the complex disappeared at later times. From the second time point (≈40 s), 48S complexes also began to form at the AUG1, while at the last AUG codon (AUG2) they accumulated pronouncedly only as late as at ≈300 s of incubation (Figure 2A).
Figure 2.
Time course of 48S formation at three AUG codons of the ΔE012 mRNA assayed by kinetic toe-printing technique. (A) Sequencing gel with results of the kinetic toe-printing assay. The ΔE012 mRNA was incubated in RRL in the presence of GMPPNP·Mg for different times, then the reaction was stopped by addition of high concentration of Mg(OAc)2. Positions of the toe-prints are indicated. (B) Quantification of toe-print signals corresponding to the 48S complexes at the three AUG codons. The values were normalized to the overall signal densities in the corresponding lanes.
It is worth noting, that each particular mRNA molecule in our system was able to acquire only a single 43S complex onto its initiation region, since the distances between the AUG codons were too short to accommodate more than one ribosome. Thus, the disappearance of signals at the AUG0 could not be explained by a declined number of reverse transcriptase molecules that reached this codon. We concluded that after the transcripts were added into the system, they quickly acquired the ribosomes and formed 48S complexes at their 5′ proximal AUGs, AUG0. The amount of these complexes peaked as early as at 20–40 s (Figure 2B), indicating a high rate of ribosomal scanning through the preceded 31-nt leader. This correlates with the previously estimated speed of scanning ribosomes (4). Although the distance between AUG0 and the following AUG1 was as small as 6 nt, the maximal amount of 48S formation at the latter codon was reached only at 120 s. The most striking results were obtained with the AUG2 which was separated from AUG0 only by 16 nt but continued to acquire the complexes gradually until the very end of the incubation. Interestingly, the 48S at the AUG2 started to form at much earlier times when the intermediate AUG codon (AUG1) was deleted, leaving only AUG0 and AUG2 in the mRNA ΔE02 (Supplementary Figure S4).
The slow ribosome accumulation at the 5′ distal AUGs was even more evident when we compared the kinetics of the 48S complexes formation on the mRNA ΔE12 (with only two AUGs) with those on an equimolar mixture of two single-AUG mRNAs that had start codons in the same context, ΔE1 and ΔE2 (Supplementary Figure S5). In both cases, the complex accumulation kinetics were much faster than those at the ΔE012 mRNA, but a profound delay in 48S formation on the second AUG was observed at the ΔE12 mRNA as compared to ΔE2 mRNA.
This slow 43S movement along the mRNA could be explained by an ability of the 40S subunit to resume scanning after the initial AUG recognition; this is the phenomenon that we decided to term ‘sliding’. Sliding presupposes the movement of the 43S complex along the mRNA to a 5′ distal AUG codon after a pronounced pause at the 5′ proximal AUG.
An alternative explanation could be related to the accumulation of AUG0-less and AUG1-less transcripts due to mRNA degradation from the 5′ end in the course of the incubation. This could result in 48S formation at the AUG2 of such 5′ truncated transcripts by conventional scanning. However, the 5′ truncated mRNAs obviously do not possess m7G-cap and consequently should not be able to form initiation complexes efficiently. This idea is supported by a control experiment with uncapped transcripts (Supplementary Figure S6). One more argument against any impact of the mRNA degradation is that in that case we should have observed the same effects on both 48S and 80S formation, which was not the case (Figure 1).
Another possible explanation could be a hypothetical depletion of some factor(s) that affect start codon recognition due to recruiting the factor(s) into the ‘dead-end’ GMPPNP-arrested 48S complexes. For example, increasing the mRNA concentration in RRL was shown to deplete eIF2, causing a preferential initiation from downstream AUG codons (43). In this case, the reduced mRNA concentration should correct the dysfunction, as it was shown earlier (43). However, we did not observe any difference when we decreased the ratio of mRNA to lysate 5-fold (Supplementary Figure S7). Moreover, when we tested the eIF2 requirements for the proper start codon selection in a reconstituted translation initiation system (Supplementary Figure S8, see below), we found the opposite correlation. The more eIF2 we added to the ΔE012 mRNA, the stronger signal at the last AUG codon as compared to that at the AUG0 and AUG1 we observed. Thus, the most probable explanation of the described phenomenon was the sliding of the 43S complex to the downstream AUG codons under conditions when hydrolysis of the eIF2-bound GTP analog was impossible.
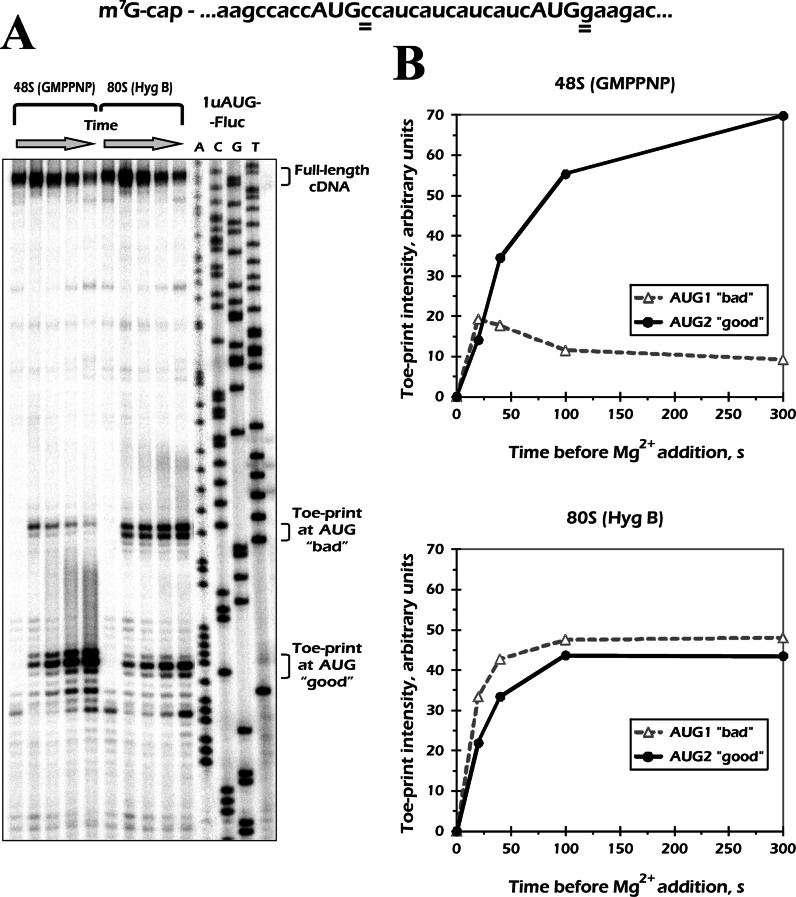
43S sliding is conceptually distinguished from leaky scanning
A well-known phenomenon that leads to AUG skipping during translation initiation is leaky scanning (3). It usually operates at AUG codons placed in a poor nucleotide context (with pyrimidines in either −3 or +4 positions, or both). Kinetics of leaky scanning is largely unknown, therefore one could suggest that sliding is just an ‘extreme’ particular case of the former. To discriminate between the two phenomena, we performed an additional time course experiment with an mRNA having two AUG codons, the first of which was in a nucleotide context of intermediate strength (with A in −3 but C in +4 position). On this 1uAUG-Fluc mRNA, the kinetics of 80S assembly at both AUG codons was quite comparable. The complexes formed rapidly, with a small delay in the case of the 5′ distal one (Figure 3). This delay most probably reflected the time needed for the 43S skipping AUG1 to arrive to AUG2 and thus illustrates the process of leaky scanning. The final ratio of the toe-print intensities quickly reached equilibrium (close to 1:1 in this case, Figure 3B, bottom). In contrast, the 48S complex formation occurred with quite distinct kinetics for the two codons. Here, the ribosomes accumulated much slower at AUG2 than at AUG1, whereas the ratio of toe-print intensities finally reached ≈1:7 in favor of the distal AUG codon (Figure 3B, top). The distinct 48S and 80S accumulation kinetics at AUG2 clearly shows the difference between the two processes, namely leaky scanning and 43S sliding.
Figure 3.
Time course of 48S and 80S formation at the 1uAUG-Fluc mRNA reveals a difference in kinetics of leaky scanning and ribosomal sliding. The 1uAUG-Fluc initiation region is shown on the top with +4 nucleotide positions underlined. (A) Results of the kinetic toe-printing assay. The 1uAUG-Fluc mRNA was incubated in RRL in the presence of GMPPNP·Mg or hygromycin B to allow the 48S or 80S complexes to be formed, respectively. The reaction was stopped by addition of high concentration of Mg(OAc)2 at different time points. Positions of the toe-prints are indicated. (B) Quantification of toe-print signals corresponding to the 48S and 80S complexes at the two AUG codons. The values were normalized to the overall signal densities in the corresponding lanes.
Omission of eIF5 recapitulates the effect of GMPPNP in the reconstituted translation initiation system in the presence of GTP
GMPPNP closely resembles GTP in structural terms; however, its inability to be hydrolyzed might affect a ternary complex conformation, resulting in inadequate AUG codon recognition. To be able to use GTP instead of its nonhydrolyzable analog in the reaction of 48S complex formation, we took advantage of the reconstituted mammalian translation initiation system (33,38). In the absence of eIF5, the 48S complexes were formed mostly at the 5′ distal AUG codons, irrespectively of whether GTP or GMPPNP were present in the mixture (Figure 4, lanes 2–3). In the presence of eIF5 and GTP, however, we could clearly observe toe-print bands corresponding to complexes on the first and the second AUGs (Figure 4, lane 5). Such bands were much weaker when GMPPNP was used instead of GTP in the presence of eIF5 (Figure 4, lane 4). We observed even a further stabilization of the 5′ proximal complexes, especially at the first AUG, if eIF5B (or eIF5B and 60S) were included in the mixture (Figure 4, lanes 7 and 9). We conclude that it is eIF5-stimulated hydrolysis of the eIF2-bound GTP and subsequent Pi release that irreversibly stop the complexes at the 5′ proximal AUGs and that these complexes are further stabilized by eIF5B. Obviously, the effect of the latter was caused by blocking the Met-tRNAi within the ribosomal P-site, which otherwise may easily dissociate after departure of eIF2-GDP from the 43S complex (compare toe-print intensities in lanes 2 and 3). It means, eIF5B additionally fixes the ribosome on the 5′ proximal AUG codons.
Figure 4.
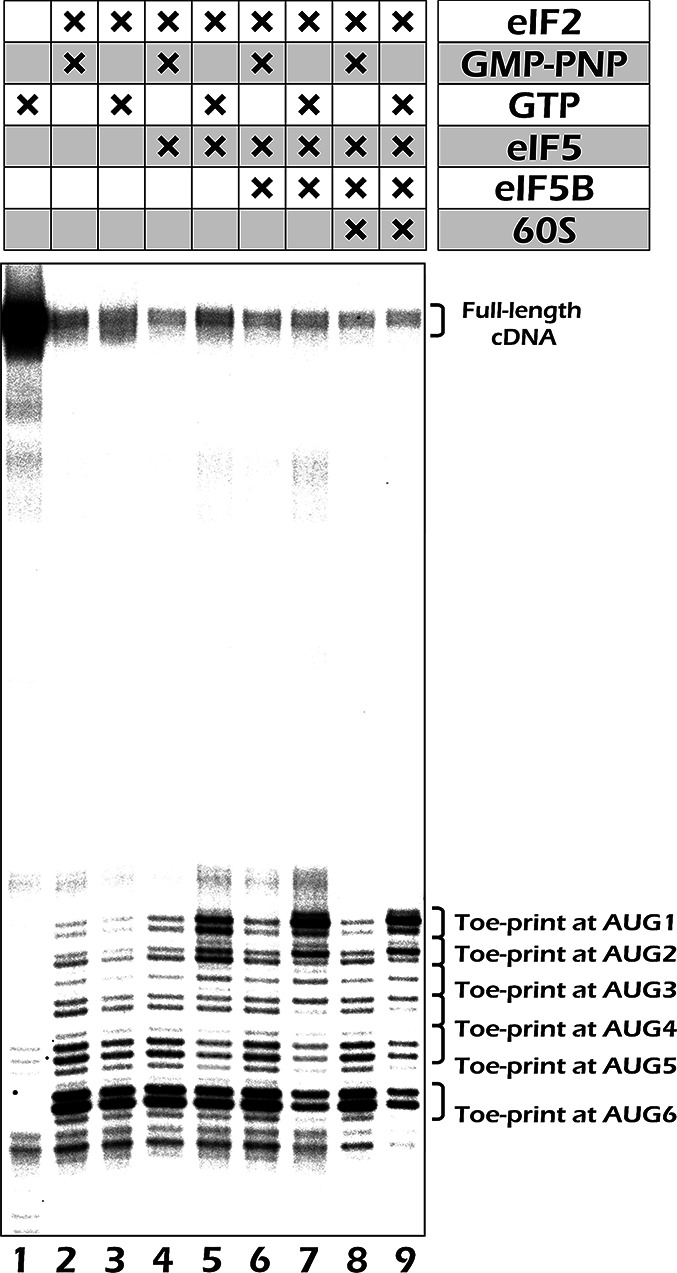
Toe-printing analysis of ribosomal complexes assembled from purified components on mRNA 5uAUG-Fluc. The reaction mixtures contained all individual components necessary for the 48S complex reconstitution (40S, Met-tRNAi, eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, eIF4F) and the additional ingredients as indicated in the header. The reaction was stopped after 10 min incubation by addition of high concentration of Mg(OAc)2.
Elevated eIF5 concentration shifts the translation initiation toward the 5′ proximal start codons in complete cytoplasmic lysates
To date, the effects of altering the eIF5 concentration on mRNA translation in mammalian systems have been reported in a very limited number of studies (29,30). In particular, it has been observed that the overproduced factor in cultured cells down-regulated the expression of mRNAs containing uORFs (29). However, dramatic changes in eIF5 concentration could induce secondary effects in living cells, so using a cell-free translation system supplemented with eIF5 would be preferential for investigating its effects on individual mRNA translation. Barth-Bause et al. recently reported differential impacts of eIF5 addition on translation products originating from two in-frame AUGs of a model mRNA in RRL (30). It was interesting, however, to systematically assess an effect of eIF5 concentration on a selection between two alternative open reading frames in a single mRNA. To investigate this, we prepared ΔE12UMBRA (Figure 5A), an artificial mRNA with the ΔE12-based initiation region, where the AUG1 and AUG2 opened alternative long overlapping ORFs derived from umbravirus mRNA (44). In nuclease-treated RRL, translation of this mRNA produced two [35S]-labeled proteins of different molecular weight (Figure 5A). Adding recombinant eIF5 increased AUG1-driven translation, while it inhibited the one initiated from the second start codon (AUG2).
Figure 5.
Effects of eIF5 concentration on mRNA translation in mammalian cell-free systems. (A) The ΔE12UMBRA mRNA (schematically represented on the top) was translated in RRL for 1 hour in the presence of [35S]-Met and increasing amount of added recombinant eIF5. The translation mixtures were resolved by SDS-PAGE (left panel), and the bands corresponding to AUG1- and AUG2-initiated products were quantified and normalized to the volumes in a mixture without exogenous eIF5 (right panel). (B) The 1uAUG-Fluc mRNA (schematically represented on the top) or the same mRNA without uAUG were translated in S30 extract of the mouse Krebs-2 ascite cells for 1 hour. An Rluc encoding mRNA was added into all probes as an internal control. The Fluc activities were normalized to Rluc ones, and the resulting volumes obtained for the 1uAUG-Fluc mRNA were divided to the volumes for the Fluc mRNA without uAUG. (C) Effects of eIF5 addition (100 ng/μl) on in vitro translation of the luciferase encoding mRNAs with artificial or natural mRNA 5′ UTRs. The mRNA leaders are schematically shown on the left. The luciferase activities in translation mixtures supplemented with eIF5 were divided by the values for the ones containing buffer.
As much as a half of mammalian mRNAs possess uAUGs in their 5′ UTRs (45,46). Thus, it was of special interest to analyze the effect of eIF5 on translation of uAUG containing mRNA. To perform such a test in an in vitro translation system, we used 1uAUG-Fluc, the luciferase encoding construct with an uAUG codon in a nucleotide context of intermediate strength (accAUGc). The uAUG was out-of-frame with the Fluc ORF (Figure 5B), and thus uORF translation should have inhibited the Fluc production. Using the luciferase as a reporter allowed us to utilize another cell-free system, the nuclease untreated S30 cytoplasmic extract from Krebs-2 mouse ascite cells, which is much closer to living cells and more appropriate for studying translation initiation (40). The same Fluc encoding mRNA without uAUG served as a control, while an mRNA with a similar simple leader encoding Renilla luciferase (Rluc) was translated in the same mixtures and used for normalization. Addition of eIF5 to the system resulted in a gradual decrease in Fluc activity in the case when the mRNA reporter possessed uAUG codon, as related to the values obtained with the uAUG-less mRNA (Figure 5B). From these data, we concluded that elevated eIF5 concentration shifted translation initiation toward the 5′ proximal AUGs and affected translation of mRNAs with alternative start codons.
In many mammalian mRNAs, an uAUG codon opens a short reading frame that ends before the main initiation codon, thus forming a non-overlapping uORF (45,46). In some cases, the uORFs have been shown to play a very important role in translational control of the main coding region (see (1,36,47) and references therein). Still, impact of eIF5 activity on regulation of their expression is poorly studied. Thus, we prepared a set of reporter mRNA constructs based on both artificial and natural human mRNA leaders (Supplementary Figure S2) and assessed effects of elevated eIF5 concentration on their translation efficiency (Figure 5C). The addition of eIF5 to the system brought about a decrease in the luciferase expression, but to a different extent for various mRNAs. Importantly, the same eIF5 concentration produced only a minor effect on uORF-less mRNAs’ translation. It was also true for the transcripts in which the original uAUG(s) were mutated to non-AUG codons, UCP2mut-Fluc and (CA)-Fluc mRNAs. Since uORF-dependent translational control has been documented for some of the leaders we used (e.g. MDM2, CFTR, ATF4 mRNAs) (36,48–51), we propose that intracellular eIF5 level and changes in its activity play a vital but still underestimated role in regulation of their expression. However, this issue requires additional extensive studies.
DISCUSSION
It is generally accepted that during eukaryotic translation initiation the establishment of a stable codon–anticodon base paring in the 48S initiation complex is the ultimate step in the process of a start codon selection. The AUG recognition therefore should unambiguously determine the point of protein synthesis initiation, while later events (including eIF2-bound GTP hydrolysis, initiation factors dissociation and 60S subunit joining) could not contribute to AUG selection. In this model, the only way for the 43S to reach a 5′ distal start codon is with leaky scanning, which strongly requires a suboptimal nucleotide context for the 5′ proximal AUG or its location within a short distance from the 5′ terminus.
In this work, we documented a differential distribution of 48S or 80S ribosomal complexes among two or more closely spaced AUG codons of mRNA under conditions when the translation process had been arrested by either GMPPNP (subunit joining inhibitor) or cycloheximide (an elongation inhibitor). The pattern of the 80S complexes distribution (produced by elongation inhibitors, e.g. cycloheximide or hygromycin B) reflected physiological usage of initiation codons, whereas the 48S complexes formed in the presence of non-hydrolysable GTP analog (GMPPNP) tended to accumulate at 5′ distal AUGs. We showed that such aberrant GMPPNP-arrested 48S allocation is a general phenomenon for closely spaced AUGs, since the same difference was observed for both m7G-cap- and IRES-dependent mRNAs. We interpreted this observation as a consequence of the ability of 43S to skip the 5′ proximal AUG codons in the absence of GTP hydrolysis. Using kinetic toe-printing assay and a set of mRNA constructs, we demonstrated that under conditions of impaired eIF2-bound GTP hydrolysis, the 40S ribosomal subunit is able to slide down along the mRNA from already recognized AUG to a downstream initiation codon. Otherwise stated, despite the established codon–anticodon base-pairing, the ribosome may leave the already recognized AUG and resume scanning. To distinguish the original scanning and the post-recognition movement of the 43S, we called this novel process ‘sliding’. The two mechanisms proceed with a clearly different kinetics, separated by a pronounced pause at the AUG. Sliding leads to AUG skipping even after its recognition has occurred and the 48S complex therefore should have acquired a closed conformation. Notably, this resumption of ribosome movement occurs even when the AUG is placed in the optimal Kozak context (Figure 1 and Supplementary Figure S3). This is the first evidence that an AUG recognition may be not a final point in start codon selection during the translation initiation process in eukaryotes.
Based on the described observations, we propose a modified model of eukaryotic translation initiation that includes the 43S sliding as a novel phenomenon. It occurs under conditions of delayed or otherwise impaired eIF2-bound GTP hydrolysis (Figure 6). According to this model, the scanning 43S complex that faces an AUG codon can either recognize it or skip it by leaky scanning. In the former case, it can then either produce 80S and begin to elongate a polypeptide (if eIF2-bound GTP hydrolysis has occurred in time), or it can resume the movement toward the next initiator codon (if GTP hydrolysis has been delayed). Our data suggest that sliding may have a substantial contribution to the process of start selection on mRNAs with two or more initiating AUGs and may be especially important for uORFs-mediated translational control. Unlike leaky scanning, sliding seems to be not limited to AUGs in a poor nucleotide context. It could also account for start codon selection in some other cases that are hardly explained by ‘canonical’ leaky scanning, e.g. a significantly high expression level of mRNAs with multiple uAUG codons (52).
Figure 6.
A model of ribosomal sliding between two adjacent AUG codons during mammalian translation initiation. After binding to the m7G-cap of the mRNA, the 43S complex comprising the small ribosomal subunit (yellow) loaded with eIF2 (blue), Met-tRNAi (red), GTP (green) and other factors (not shown for simplicity) begins to move in a 5′ to 3′ direction in a search of a start codon. In this process (called scanning), the ribosomal subunit encounters the first AUG codon. If recognition of the AUG1 does not occur, the 43S keeps on moving to the downstream AUG (leaky scanning, the upper panel). If the recognition of the AUG1 does occur, the 43S complex stops (the second panel). If the eIF2-bound hydrolysis takes place in time, the eIF2-GDP dissociates and the large ribosomal subunit joins and elongation starts. However, in the case of a delay in the GTP hydrolysis, the 43S may resume movement and slides down to the downstream AUG codon (the lower panel). To distinguish the original scanning and the similar process after the resumption, we suggest to name this phenomenon ‘sliding’. In fact, the sliding 43S complex may differ from the scanning one in a composition of translation initiation factors (not shown).
In eukaryotes, reaction of the eIF2-bound GTP hydrolysis requires the special GAP: eIF5 (10,14). Besides providing the GAP activity, eIF5 regulates Pi release and stabilization of the closed conformation of the 43S complex after it recognizes an AUG codon (1). The additional function of eIF5 is to stabilize the binding of GDP to eIF2 after its dissociation from the 48S complex by inhibition of the GDP–GTP exchanging factor eIF2B (53).
During the whole process of translation initiation, eIF5 is thought to be present in the 43S complex, although it is not absolutely necessary neither for scanning nor for AUG recognition (13,19,54). However, it is required for selection of the appropriate start codon among two or more adjacent AUGs or near-cognate start codons (8,20–22). eIF5 forms multiple contacts with other components of the 43S complex, including eIF1, eIF1A and eIF2. And what is more important, it governs rearrangements of intermolecular contacts within the complex upon AUG recognition to promote the release of eIF1 and to stabilize PIN conformation of the 48S (reviewed in (1,5,6)).
All the evidence presented provides a mechanistic basis for sliding. In our work, we discovered this phenomenon under artificial conditions when GTP hydrolysis was completely excluded (either due to usage of the non-hydrolysable analog GMPPNP or due to the absence of eIF5 from the reaction mixture). However, we believe that in living cells sliding may occur under physiological conditions when eIF5 concentration is low or its activity is reduced. 43S complexes, that lack eIF5 or bear an inactive form of the factor, will tend to skip AUG codons due to the sliding event. This may substantially affect translation of mRNAs that have alternative initiation codons or uAUGs in their 5′ UTRs. Such an effect of eIF5 concentration on uAUG-regulated translation has been documented by Loughran et al. for living mammalian cells (29) and was confirmed in cell-free systems in this work (Figure 5). In yeast, the eIF5 mutations that reduce affinity of the factor to the 43S was shown to impair uORF-dependent regulation of GCN4 protein synthesis (21,22). The eIF5-deficient 43S complexes may be argued to skip the 5′ proximal AUG codons due to a defect in recognition and not because of the post-recognition sliding, for eIF5 is needed for appropriate factor interactions within the scanning complex (27,28). However, our experiments with GMPPNP in the presence of the same amount of eIF5 and the results of the kinetic toe-printing assay support the existence of sliding. Most importantly, in these experiments (performed both in reconstituted system and in the complete cell lysate), eIF5 was present in the same concentration as in case of GTP-charged reaction mixtures. Thus, it is the inability of GTP hydrolysis, and not the absence of eIF5, that provokes shifting of the 43S complexes to the downstream AUG codons.
In this respect, it is interesting to note that the eIF5 mRNA is highly differentially expressed in human tissues (55) and regulated by a tumor suppressor microRNA-107 (56). This suggests a quite variable eIF5 concentration in different human cells under various conditions. The affinity of eIF5 to other components of the 43S may also be regulated by post-translational modifications of its partners or the factor itself (see below). Finally, the presence of eIF5 in the 43S may be abolished by competitors like eIF5-mimic proteins BZW2/5MP1 and BZW1/5MP2 (57,58).
Even more intriguing is a possibility of the temporal regulation of the eIF5 GAP activity in the cell. It is well established that in yeast the eIF5 mutations that accelerate the GAP activity strongly influence start codon selection (8,20). A similar effect may be supposed to be caused by modification(s) of the protein. Indeed, regulation of eIF5 GAP activity by phosphorylation was recently documented upon glucose depletion in yeast, and this modification affected start codon selection (59). This is not the only example, as eIF5 is known to be phosphorylated by CK2 under a variety of conditions in human, plant and yeast cells (59–64). In some cases significant effects of this phosphorylation on various aspect of cell physiology were shown (17,65).
Finally, another notable peculiarity is an additional effect of eIF5B and 60S on AUG selection observed in the reconstituted translation initiation system (Figure 4). When this manuscript was already in preparation, Pisareva and Pisareva reported similar effects of eIF5B on 48S complex formation at non-optimal start codons (e.g. CUG or poor-context AUG) (31). Influence of eIF5B on stringency of start codon selection was also shown in a mammalian cell-free system (30) and has been especially well documented in yeast (66–69). In our experiments, eIF5B shifted the translation initiation toward a 5′ proximal start codon. eIF5B obviously stabilizes Met-tRNAi within the ribosomal P-site after departure of eIF2-GDP from the 43S complex and thus additionally promotes ribosome clamping on the particular codon (70). The final fixation occurs when the large subunit joins the 43S complex. The contribution of the 60S joining step in AUG selection is supported by a finding that mutation in the 60S ribosomal protein L33A (eL33) impairing this stage affected translation regulation of the GCN4 mRNA (68). It should be mentioned that eIF5B level is tightly regulated in cells under various physiological conditions and at particular developmental stages (71).
Another question is a composition of the sliding 43S complex (as compared to the canonical scanning 43S). Recent structural and biochemical data (5,6,72) provide evidence for eIF1 displacement from its original position as a direct consequence of codon–anticodon duplex formation. Thus, in accordance with earlier data (7,25,73), eIF1 may dissociate upon AUG recognition and can do this even without eIF5 present in the complex. Although the dissociation may not necessarily occur in some mammalian systems (9), nevertheless, it should be kept in mind that the composition of the 48S complex just after the AUG recognition is most probably distinct from the original one. However, we believe that eIF1 can re-associate during the pause that precedes sliding, and this may provoke the sliding event. Some indirect data from eIF1 overexpression experiments suggest that eIF1 might be able to re-bind to the 43S after the recognition has occurred (23,74). It should be noted, that we did not observe any additional toe-prints at downstream non-AUG codons under conditions which favored sliding. This speaks in favour of eIF1 presence in the sliding complex. Finally, our data also argue that initiation factors driving scanning (e.g. eIF4s) most probably do not dissociate upon the AUG recognition.
In summary, we present a novel mode of start codon selection that is based on the kinetics of eIF2-bound GTP hydrolysis after the AUG recognition and is directly linked to activities of the subunit joining factors – eIF5 and eIF5B. The described phenomenon of ribosomal sliding contributes to our knowledge regarding the control of gene expression at the level of translation.
Supplementary Material
Acknowledgments
We are grateful to M.Taliansky (The James Hutton Institute, UK) for the original umbravirus construct, L.Shagam for the plasmid with the corrected eIF5 sequence, A.Anisimova for the (CA)-Fluc and (CA)uORF-Fluc mRNA preparations, M.Orlova (The Research Institute of McGill University Health, Canada) and A.Sharapkova (Lomonosov Moscow State University) for critical reading of the manuscript and helpful suggestions. We thank the Moscow State University Development Program PNR5 for providing access to the Typhoon FLA 9500 Phosphorimager.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Russian Foundation for Basic Research [RFBR13-04-01121а to S.E.D.]. The in vitro translation experiments were supported by the Russian Science Foundation [RSF 14-14-00127] to D.E.A.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 2.Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA. Cell. 1978;15:1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- 3.Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vassilenko K.S., Alekhina O.M., Dmitriev S.E., Shatsky I.N., Spirin A.S. Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res. 2011;39:5555–5567. doi: 10.1093/nar/gkr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain T., Llacer J.L., Fernandez I.S., Munoz A., Martin-Marcos P., Savva C.G., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014;159:597–607. doi: 10.1016/j.cell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llacer J.L., Hussain T., Marler L., Aitken C.E., Thakur A., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V. Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex. Mol. Cell. 2015;59:399–412. doi: 10.1016/j.molcel.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Algire M.A., Maag D., Lorsch J.R. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Huang H.K., Yoon H., Hannig E.M., Donahue T.F. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unbehaun A., Borukhov S.I., Hellen C.U., Pestova T.V. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakrabarti A., Maitra U. Function of eukaryotic initiation factor 5 in the formation of an 80 S ribosomal polypeptide chain initiation complex. J. Biol. Chem. 1991;266:14039–14045. [PubMed] [Google Scholar]
- 11.Paulin F.E., Campbell L.E., O'Brien K., Loughlin J., Proud C.G. Eukaryotic translation initiation factor 5 (eIF5) acts as a classical GTPase-activator protein. Curr. Biol. 2001;11:55–59. doi: 10.1016/s0960-9822(00)00025-7. [DOI] [PubMed] [Google Scholar]
- 12.Das S., Ghosh R., Maitra U. Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J. Biol. Chem. 2001;276:6720–6726. doi: 10.1074/jbc.M008863200. [DOI] [PubMed] [Google Scholar]
- 13.Pestova T.V., Lomakin I.B., Lee J.H., Choi S.K., Dever T.E., Hellen C.U. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 14.Trachsel H., Staehelin T. Binding and release of eukaryotic initiation factor eIF-2 and GTP during protein synthesis initiation. Proc. Natl. Acad. Sci. U.S.A. 1978;75:204–208. doi: 10.1073/pnas.75.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asano K., Clayton J., Shalev A., Hinnebusch A.G. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokabe M., Fraser C.S., Hershey J.W. The human translation initiation multi-factor complex promotes methionyl-tRNAi binding to the 40S ribosomal subunit. Nucleic Acids Res. 2012;40:905–913. doi: 10.1093/nar/gkr772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis M.D., Person M.D., Browning K.S. Phosphorylation of plant translation initiation factors by CK2 enhances the in vitro interaction of multifactor complex components. J. Biol. Chem. 2009;284:20615–20628. doi: 10.1074/jbc.M109.007658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestova T.V., Borukhov S.I., Hellen C.U. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 19.Algire M.A., Maag D., Savio P., Acker M.G., Tarun S.Z., Jr, Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–397. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saini A.K., Nanda J.S., Martin-Marcos P., Dong J., Zhang F., Bhardwaj M., Lorsch J.R., Hinnebusch A.G. Eukaryotic translation initiation factor eIF5 promotes the accuracy of start codon recognition by regulating Pi release and conformational transitions of the preinitiation complex. Nucleic Acids Res. 2014;42:9623–9640. doi: 10.1093/nar/gku653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen K.H., Szamecz B., Valasek L., Jivotovskaya A., Shin B.S., Hinnebusch A.G. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh C.R., Curtis C., Yamamoto Y., Hall N.S., Kruse D.S., He H., Hannig E.M., Asano K. Eukaryotic translation initiation factor 5 is critical for integrity of the scanning preinitiation complex and accurate control of GCN4 translation. Mol. Cell. Biol. 2005;25:5480–5491. doi: 10.1128/MCB.25.13.5480-5491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valasek L., Nielsen K.H., Zhang F., Fekete C.A., Hinnebusch A.G. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol. 2004;24:9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maag D., Algire M.A., Lorsch J.R. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J. Mol. Biol. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- 25.Nanda J.S., Cheung Y.N., Takacs J.E., Martin-Marcos P., Saini A.K., Hinnebusch A.G., Lorsch J.R. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 2009;394:268–285. doi: 10.1016/j.jmb.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nanda J.S., Saini A.K., Munoz A.M., Hinnebusch A.G., Lorsch J.R. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J. Biol. Chem. 2013;288:5316–5329. doi: 10.1074/jbc.M112.440693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luna R.E., Arthanari H., Hiraishi H., Nanda J., Martin-Marcos P., Markus M.A., Akabayov B., Milbradt A.G., Luna L.E., Seo H.C., et al. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2beta. Cell Rep. 2012;1:689–702. doi: 10.1016/j.celrep.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luna R.E., Arthanari H., Hiraishi H., Akabayov B., Tang L., Cox C., Markus M.A., Luna L.E., Ikeda Y., Watanabe R., et al. The interaction between eukaryotic initiation factor 1A and eIF5 retains eIF1 within scanning preinitiation complexes. Biochemistry. 2013;52:9510–9518. doi: 10.1021/bi4009775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loughran G., Sachs M.S., Atkins J.F., Ivanov I.P. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 2012;40:2898–2906. doi: 10.1093/nar/gkr1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barth-Baus D., Bhasker C.R., Zoll W.L., Merrick W.C. Influence of translation factor activities on start site selection in six different mRNAs. Translation. 2013;1:e24419. doi: 10.4161/trla.24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pisareva V.P., Pisarev A.V. eIF5 and eIF5B together stimulate 48S initiation complex formation during ribosomal scanning. Nucleic Acids Res. 2014;42:12052–12069. doi: 10.1093/nar/gku877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dmitriev S.E., Pisarev A.V., Rubtsova M.P., Dunaevsky Y.E., Shatsky I.N. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 2003;533:99–104. doi: 10.1016/s0014-5793(02)03776-6. [DOI] [PubMed] [Google Scholar]
- 33.Pestova T.V., Hellen C.U., Shatsky I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dmitriev S.E., Andreev D.E., Terenin I.M., Olovnikov I.A., Prassolov V.S., Merrick W.C., Shatsky I.N. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol. Cell. Biol. 2007;27:4685–4697. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dmitriev S.E., Stolboushkina E.A., Terenin I.M., Andreev D.E., Garber M.B., Shatsky I.N. Archaeal translation initiation factor aIF2 can substitute for eukaryotic eIF2 in ribosomal scanning during mammalian 48S complex formation. J. Mol. Biol. 2011;413:106–114. doi: 10.1016/j.jmb.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 36.Andreev D.E., O'Connor P.B., Fahey C., Kenny E.M., Terenin I.M., Dmitriev S.E., Cormican P., Morris D.W., Shatsky I.N., Baranov P.V. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreev D.E., Terenin I.M., Dunaevsky Y.E., Dmitriev S.E., Shatsky I.N. A leaderless mRNA can bind to mammalian 80S ribosomes and direct polypeptide synthesis in the absence of translation initiation factors. Mol. Cell. Biol. 2006;26:3164–3169. doi: 10.1128/MCB.26.8.3164-3169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dmitriev S.E., Terenin I.M., Dunaevsky Y.E., Merrick W.C., Shatsky I.N. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol. Cell. Biol. 2003;23:8925–8933. doi: 10.1128/MCB.23.24.8925-8933.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terenin I.M., Dmitriev S.E., Andreev D.E., Royall E., Belsham G.J., Roberts L.O., Shatsky I.N. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol. Cell. Biol. 2005;25:7879–7888. doi: 10.1128/MCB.25.17.7879-7888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dmitriev S.E., Andreev D.E., Adyanova Z.V., Terenin I.M., Shatsky I.N. Efficient cap-dependent translation of mammalian mRNAs with long and highly structured 5′-untranslated regions in vitro and in vivo. Mol. Biol. (Mosk.) 2009;43:108–113. [PubMed] [Google Scholar]
- 41.Andreev D.E., Dmitriev S.E., Terenin I.M., Prassolov V.S., Merrick W.C., Shatsky I.N. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009;37:6135–6147. doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaminski A., Howell M.T., Jackson R.J. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dasso M.C., Milburn S.C., Hershey J.W., Jackson R.J. Selection of the 5′-proximal translation initiation site is influenced by mRNA and eIF-2 concentrations. Eur. J. Biochem. 1990;187:361–371. doi: 10.1111/j.1432-1033.1990.tb15313.x. [DOI] [PubMed] [Google Scholar]
- 44.Taliansky M.E., Robinson D.J., Murant A.F. Complete nucleotide sequence and organization of the RNA genome of groundnut rosette umbravirus. J. Gen. Virol. 1996;77:2335–2345. doi: 10.1099/0022-1317-77-9-2335. [DOI] [PubMed] [Google Scholar]
- 45.Kochetov A.V., Sarai A., Rogozin I.B., Shumny V.K., Kolchanov N.A. The role of alternative translation start sites in the generation of human protein diversity. Mol. Genet. Genomics. 2005;273:491–496. doi: 10.1007/s00438-005-1152-7. [DOI] [PubMed] [Google Scholar]
- 46.Iacono M., Mignone F., Pesole G. uAUG and uORFs in human and rodent 5′ untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 47.Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdisp. Rev. RNA. 2014;5:765–778. doi: 10.1002/wrna.1245. [DOI] [PubMed] [Google Scholar]
- 48.Jin X., Turcott E., Englehardt S., Mize G.J., Morris D.R. The two upstream open reading frames of oncogene mdm2 have different translational regulatory properties. J. Biol. Chem. 2003;278:25716–25721. doi: 10.1074/jbc.M300316200. [DOI] [PubMed] [Google Scholar]
- 49.Lukowski S.W., Rothnagel J.A., Trezise A.E. CFTR mRNA expression is regulated by an upstream open reading frame and RNA secondary structure in its 5′ untranslated region. Hum. Mol. Genet. 2015;24:899–912. doi: 10.1093/hmg/ddu501. [DOI] [PubMed] [Google Scholar]
- 50.Lu P.D., Harding H.P., Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vattem K.M., Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X.Q., Rothnagel J.A. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 2004;32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jennings M.D., Pavitt G.D. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature. 2010;465:378–381. doi: 10.1038/nature09003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jivotovskaya A.V., Valasek L., Hinnebusch A.G., Nielsen K.H. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Si K., Das K., Maitra U. Characterization of multiple mRNAs that encode mammalian translation initiation factor 5 (eIF-5) J. Biol. Chem. 1996;271:16934–16938. doi: 10.1074/jbc.271.28.16934. [DOI] [PubMed] [Google Scholar]
- 56.Song N., Ma X., Li H., Zhang Y., Wang X., Zhou P., Zhang X. microRNA-107 functions as a candidate tumor suppressor gene in renal clear cell carcinoma involving multiple genes. Urol. Oncol. 2015;33:205.e1–205.e11. doi: 10.1016/j.urolonc.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Singh C.R., Watanabe R., Zhou D., Jennings M.D., Fukao A., Lee B., Ikeda Y., Chiorini J.A., Campbell S.G., Ashe M.P., et al. Mechanisms of translational regulation by a human eIF5-mimic protein. Nucleic Acids Res. 2011;39:8314–8328. doi: 10.1093/nar/gkr339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hiraishi H., Oatman J., Haller S.L., Blunk L., McGivern B., Morris J., Papadopoulos E., Gutierrez W., Gordon M., Bokhari W., et al. Essential role of eIF5-mimic protein in animal development is linked to control of ATF4 expression. Nucleic Acids Res. 2014;42:10321–10330. doi: 10.1093/nar/gku670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bavli-Kertselli I., Melamed D., Bar-Ziv L., Volf H., Arava Y. Overexpression of eukaryotic initiation factor 5 rescues the translational defect of tpk1w in a manner that necessitates a novel phosphorylation site. FEBS J. 2015;282:504–520. doi: 10.1111/febs.13158. [DOI] [PubMed] [Google Scholar]
- 60.Boex-Fontvieille E., Daventure M., Jossier M., Zivy M., Hodges M., Tcherkez G. Photosynthetic control of Arabidopsis leaf cytoplasmic translation initiation by protein phosphorylation. PLoS One. 2013;8:e70692. doi: 10.1371/journal.pone.0070692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dennis M.D., Browning K.S. Differential phosphorylation of plant translation initiation factors by Arabidopsis thaliana CK2 holoenzymes. J. Biol. Chem. 2009;284:20602–20614. doi: 10.1074/jbc.M109.006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Homma M.K., Homma Y. Regulatory role of CK2 during the progression of cell cycle. Mol. Cell. Biochem. 2005;274:47–52. doi: 10.1007/s11010-005-3111-3. [DOI] [PubMed] [Google Scholar]
- 63.Majumdar R., Bandyopadhyay A., Deng H., Maitra U. Phosphorylation of mammalian translation initiation factor 5 (eIF5) in vitro and in vivo. Nucleic Acids Res. 2002;30:1154–1162. doi: 10.1093/nar/30.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maiti T., Bandyopadhyay A., Maitra U. Casein kinase II phosphorylates translation initiation factor 5 (eIF5) in Saccharomyces cerevisiae. Yeast. 2003;20:97–108. doi: 10.1002/yea.937. [DOI] [PubMed] [Google Scholar]
- 65.Homma M.K., Wada I., Suzuki T., Yamaki J., Krebs E.G., Homma Y. CK2 phosphorylation of eukaryotic translation initiation factor 5 potentiates cell cycle progression. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15688–15693. doi: 10.1073/pnas.0506791102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi S.K., Lee J.H., Zoll W.L., Merrick W.C., Dever T.E. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- 67.Hiraishi H., Shin B.S., Udagawa T., Nemoto N., Chowdhury W., Graham J., Cox C., Reid M., Brown S.J., Asano K. Interaction between 25S rRNA A loop and eukaryotic translation initiation factor 5B promotes subunit joining and ensures stringent AUG selection. Mol. Cell. Biol. 2013;33:3540–3548. doi: 10.1128/MCB.00771-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin-Marcos P., Hinnebusch A.G., Tamame M. Ribosomal protein L33 is required for ribosome biogenesis, subunit joining, and repression of GCN4 translation. Mol. Cell. Biol. 2007;27:5968–5985. doi: 10.1128/MCB.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shin B.S., Maag D., Roll-Mecak A., Arefin M.S., Burley S.K., Lorsch J.R., Dever T.E. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell. 2002;111:1015–1025. doi: 10.1016/s0092-8674(02)01171-6. [DOI] [PubMed] [Google Scholar]
- 70.Terenin I.M., Dmitriev S.E., Andreev D.E., Shatsky I.N. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 71.Lee S., Truesdell S.S., Bukhari S.I., Lee J.H., LeTonqueze O., Vasudevan S. Upregulation of eIF5B controls cell-cycle arrest and specific developmental stages. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E4315–E4322. doi: 10.1073/pnas.1320477111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang F., Saini A.K., Shin B.S., Nanda J., Hinnebusch A.G. Conformational changes in the P site and mRNA entry channel evoked by AUG recognition in yeast translation preinitiation complexes. Nucleic Acids Res. 2015;43:2293–2312. doi: 10.1093/nar/gkv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheung Y.N., Maag D., Mitchell S.F., Fekete C.A., Algire M.A., Takacs J.E., Shirokikh N., Pestova T., Lorsch J.R., Hinnebusch A.G. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alone P.V., Cao C., Dever T.E. Translation initiation factor 2gamma mutant alters start codon selection independent of Met-tRNA binding. Mol. Cell. Biol. 2008;28:6877–6888. doi: 10.1128/MCB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.