Abstract
Telomere integrity is essential to maintain genome stability, and telomeric dysfunctions are associated with cancer and aging pathologies. In human, the shelterin complex binds TTAGGG DNA repeats and provides capping to chromosome ends. Within shelterin, RAP1 is recruited through its interaction with TRF2, and TRF2 is required for telomere protection through a network of nucleic acid and protein interactions. RAP1 is one of the most conserved shelterin proteins although one unresolved question is how its interaction may influence TRF2 properties and regulate its capacity to bind multiple proteins. Through a combination of biochemical, biophysical and structural approaches, we unveiled a unique mode of assembly between RAP1 and TRF2. The complete interaction scheme between the full-length proteins involves a complex biphasic interaction of RAP1 that directly affects the binding properties of the assembly. These results reveal how a non-DNA binding protein can influence the properties of a DNA-binding partner by mutual conformational adjustments.
INTRODUCTION
Telomeres are specific nucleoprotein complexes that protect the chromosomes termini against degradation, checkpoint activation and illicit repair. Shelterin is a six-protein complex that constitutes the core of the telomere structure by binding double-stranded DNA through TRF1 and TRF2 (Telomeric Repeat-binding Factors 1 and 2) and single-stranded DNA through POT1. TIN2 bridges TRF1 and TRF2, whereas TPP1 links double-stranded DNA and single-stranded DNA through its interaction with POT1 and TIN2, and RAP1 tightly interacts with TRF2 (1,2). Shelterin functions depend on the recruitment of many associated factors (2,3), and on the formation of specific assemblies with DNA (4). Systematic study of the telomeric proteins interaction network reveals more than 1000 protein–protein interactions and telomere-associated proteins (5). The individual implication of these proteins in shelterin functions has been extensively studied, but the functional synergies upon complex formation remain poorly understood. Telomere repeats induce subnuclear compartments significantly enriched in shelterin proteins. The local high concentration of shelterin proteins implies that the description of high affinity interactions is not sufficient to properly understand the regulatory processes involved in telomere maintenance. It is known that weak-affinity (KD > 10−4 M) and transient interactions are equally important than high-affinity interactions (KD < 10−6 M) in the regulation of many cellular pathways (6). However, our understanding of these regulatory processes is limited by our ability to study short-lived interactions. Weak and transient interactions are particularly difficult to study in multifunctional proteins, which interact with numerous partners often through a common or similar interface.
Among shelterin proteins, TRF2 is a multifunctional protein that prevents non-homologous end joining (NHEJ) and ATM(ataxia telangiectasia mutated)-dependent checkpoint as well as DNA replication (2,3,7–9). Moreover, TRF2 binds several extra-telomeric sites where it regulates gene expression (10,11). TRF2 is composed of a TRFH domain involved in homodimer assembly, in recruitment of several proteins (12–18), in DNA condensation (19, Benarroch-Popivker et al., in press) and in ATM inhibition signaling (20), and a C-terminal Myb/telobox domain that specifically binds double-stranded DNA (21,22). The basic N-terminal region of TRF2 hosts binding sites for several telomere-interacting molecules including the telomeric RNA TERRA, provides a specific ability to bind Holliday junctions (HJ) and to protect them from resolvase cleavage, and helps TRF2 to condense DNA and chromatin (19,23–27). The long linker between the TRFH and the Myb/telobox domain contains the binding sites for the two shelterin proteins RAP1 (Repressor Activator Protein 1) and TIN2 (28,29), and prevents 53BP1 recruitment to telomeres (20). TRF2/RAP1 complex is involved in NHEJ protection in an in vitro assay (30). Previous studies have reported that RAP1 deletion does not seem to affect cell viability or telomere protection in vivo in a wild-type background (31,32). However, the use of a separation-of-function mutant of TRF2 revealed that RAP1 is involved in a backup mechanism that prevents NHEJ when DNA condensation-mediated telomere protection is impaired (Benarroch-Popivker et al., in press). Moreover, ChIP-seq analyses have shown that RAP1 can be found at chromatin sites in the absence of TRF2 (33,34). Cytoplasmic localization of RAP1 has also been reported, as well as its involvement in NF-κB signalling (35,36). A recent study shows that RAP1 can affect TRF2 interaction properties toward duplex DNA (37). In TeloPIN database (5), RAP1 and TRF2 interact respectively with 131 and 247 partners, among which 32 are common partners between RAP1, TRF2 and other shelterin proteins and 12 are common partners exclusively between RAP1 and TRF2. Therefore, whether or not RAP1/TRF2 interaction may affect/reinforce their respective properties and how DNA/TRF2 complex recruits its numerous protein partners compared to RAP1/TRF2/DNA complex remain to be fully characterized.
In order to address this central question, we initiated a comprehensive study of the parameters that describe the assembly of RAP1 and TRF2. We analysed the RAP1/TRF2complex both in terms of structure and binding properties. We observed that RAP1 affects TRF2 behavior in vitro toward particular DNA structures. Through a combination of SAXS, ITC, crystallography and biochemical approaches, we were able to delineate the complete interaction scheme between the full-length proteins. Finally, we confirmed that a construct of RAP1 deleted for its C-terminal high-affinity binding site is sufficient to modulate TRF2 properties toward Holliday junction and observed that the binding properties of TRF2/DNA or of RAP1/TRF2/DNA toward proteins from HeLa cells nuclear extracts are different.
MATERIALS AND METHODS
Protein constructs and peptides used in this study
All protein constructs used in this study are listed in Supplementary Table S1. All peptides are also listed in Supplementary Data.
HJ protection
A total of 5 nM of telomeric HJ (tHJ) labelled on one strand was incubated with different concentrations of proteins in 10 μl of binding buffer containing 10 mM MgAc2 and digested for 60 min using 2 nM of T7 Endonuclease I, MBP-CceI and MBP-RusA or 50 nM of GEN11–527. Reactions were performed at 4°C for Endo I or 37°C for MBP-CceI, MBP-RusA and GEN11–527. The reaction was stopped by addition of 6 μg of proteinase K followed by 20 min incubation at 4°C or 15 min at 37°C. Formamide was added to a final concentration of 60% (v/v) and the samples loaded on a 10% denaturing acrylamide (19:1, bis:mono) 1× TBE gel. After migration in 1× TBE, gels were analysed using a Phosphorimager TyphoonFLA9000 (GE Healthcare) and the Image Quant software (GE Healthcare).
EMSA
Proteins were incubated 15 min a 4°C with 5 nM DNA in 10 μl of 20 mM Tris-Acetate pH8, 0.1 mg ml−1 BSA, 1 mM DTT, 50 mM KAc, 30 mM NaCl, 5% (v/v) glycerol (binding buffer). Gel electrophoresis was performed as published (38) and gels were analysed as above.
AFM
A total of 50 nM of TRF2 dimer and either 100 nM of full length Rap1 or Rap1-RCT were pre-incubated at 0°C for 15 min in the binding buffer (150 mM KCl, 5 mM HEPES pH 7.4). DNA (final concentration 10 nM) was then incubated with a 1:1 molar ratio of TRF2 with the preincubated proteins for 20 min at 25°C in the same binding buffer. Samples were crosslinked with glutaraldehyde (0.1% final) for 30 min on ice and applied on freshly cleaved mica surfaces treated with 10 mM MgCl2. After 2 min, mica was washed with 1 ml of deionized water and dried with a gentle flux of nitrogen. Imaging was performed on a Nanoscope IIIa equipped with E-scanner (Digital Instruments Inc, Santa Barbara, CA, USA), in air under Tapping Mode using silicon tips. Images were recorded at 1.5–2.0 Hz over scan areas 1 μm wide (512 × 512 pixels). Raw scanning force microscopy images were flattened using the manufacturer's software and converted into TIF files. Contour lengths (CLs) were measured by the read-through length method using SigmaScan Pro software (SPSS Inc, Chicago, IL, USA). Volumes calculated as half-ellipsoids as published (38). Between 150 and 250 objects were scored for each condition. The 2D probability density map is a smoothed distribution that represents the convolution of the experimental distribution with a s1 × s2 gaussian pulse. To construct the 2D map, each protein/DNA complex with a raw DNA length Lraw and a complex volume Volcomplex contributes as gaussian distribution of respectively s1 and s2 width in Lraw and Volcomplex directions. Each pixel of the 2D map is the sum of the various gaussian contributions from the experimental data points. The resulting 2D map represents a 100 by 100 pixel image of the probability to find a protein/DNA complex with given raw DNA length and complex volume, the intensity of each pixel is given by the associated probability density colour scale.
Protein expression and purification
Protocols are detailed in supplementary Data.
SAXS
SAXS experiments were performed at SWING beamline, synchrotron SOLEIL. TRF2, RAP1 and RAP1-RCT were set in the buffer containing 20 mM Tris pH = 7.5, 150 mM NaCl, 5 mM 2-mercaptoethanol and 0.2 mM PMSF. TRF2 was concentrated to 215 μM (monomers), RAP1 to 326.7 μM, RAP1-RCT to 1015.7μM, TRF2 and RAP1 were mixed to 120 and 144μM respectively, and TRF2 and RAP1-RCT were mixed to 171.5 and 205.8 μM respectively, both corresponding to a RAP1/TRF2 molar ratio of 1.2. All data were collected at SWING beamline at SOLEIL synchrotron using either the sample changer (RAP1 full-length), or using the online HPLC system (39), equipped with a BIO-SEC3 column (Agilent) for all other proteins or complexes. After data collection, all leftovers from these preparations were analysed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) to ensure no degradation occurred during the experiments. The detailed analysis of the data is described in supplementary Data.
Crystallization and structure determination
Crystals of TRFH in complex with peptide RAP1-YLP 89ENRERLELEAYRLGPASA106 were obtained by hanging drop vapour diffusion at room temperature, with a concentration of TRFH of 34 mg/ml and a molar ratio of TRFH/Rap1-YLP of 1:3.5 in a buffer containing 12.5 mM Tris pH 8.0, 100mM NaCl and DTT 2.5 mM. The most suitable crystals grew in 16% PEGmme 550, 50 mM Hepes pH 8.0, 5 mM MgCl2, 5 mM DTT. Native diffraction data were collected on PX1 beamline at SOLEIL synchrotron, and reduced with XDS (40). The structure was solved by molecular replacement with PHASER (41), using one TRFH monomer from 3BUA PDB entry: as model probe (15), and refined with BUSTER5 (42).
Protein footprinting and Mass spectrometry analysis
The setup of protein footprinting protocols and mass spectrometry analysis are detailed in Supplementary Data.
ITC
Molecular interactions (see Supplementary Table S2) were investigated by Isothermal Titration Calorimetry using a VP-ITC and an iTC200 instruments (GE Healthcare). The solutions were degassed prior to use, at the same temperature as those used in the experiments. Integration of the peaks corresponding to each injection, correction for the baseline, and the fit were done using Origin-based software provided by the manufacturer and enabled to calculate the equilibrium dissociation constants of the interactions presented in this study. The experimental conditions of all ITC experiments are indicated in Supplementary Table S2. Blank experiments (heats of dilution from titrants into buffer) were also performed.
SEC-MALS
TRF2 protein at a concentration of 100 μM (in a buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM 2-mercaptoethanol) was applied to Bio SEC-3 column (Agilent) on a HPLC Shimadzu system coupled to MALS/QELS/UV/RI (Wyatt Technology). Data were analysed using ASTRA software after calibration with a bovine serum albumin standard.
Cell culture
HeLa cells were kindly provided by Dr Carl Mann (I2BC), and grown in 15-cm dishes in DMEM medium supplemented with 10% v/v fetal bovine serum and a mix of penicillin and streptomycin to prevent bacterial contamination. Cells were amplified to 30 15-cm dishes at 80–90% confluence, and finally harvested by scraping in cold PBS just prior to the preparation of nuclear extracts.
Preparation of nuclear extracts
Nuclear extracts were prepared following the protocol described in (43), with minor modifications detailed in Supplementary Data.
Streptavidin-DNA pull down experiments
The following oligonucleotides were annealed to make a biotinylated duplex DNA molecule displaying a TRF2 binding site (two tandem TAGGG motifs): biotin-5′-TACTAGGGTTAGGGTTAGCA-3′ and complementary 5′-ATGCTAACCCTAACCCTAGT-3′ yielding a single base 5′-overhang at each end. A total of 9 nmoles of biotinylated DNA were loaded at 0.1 ml/min on a HiTrap Streptavidin HP 1 ml column (GE Healthcare) first equilibrated in binding buffer (Tris–HCl 20 mM pH 7.5; NaCl 150 mM; 2-mercaptoethanol 5 mM; EDTA 0.2 mM; PMSF 0.1 mM). A peak in A260 indicated that the column was loaded with a saturating amount of biotinylated DNA. The column was then loaded at 0.2 ml/min with either TRF2, preformed TRF2/RAP1 complex, preformed TRF2/RAP1-ARAGA complex, or nothing (control experiment) and washed with binding buffer until A280 returned to a flat baseline. In each case, non-zero A280 of the flow through indicated that the column was loaded with a saturating amount of protein. A total of 490 μg of nuclear extracts were loaded at 0.2 ml/min on the column. When A280 returned to a flat baseline, bound proteins were eluted at 0.5 mL/min with elution buffer (same as binding buffer except NaCl 1 M). Between each experiment, the column was washed with elution buffer until A280 returned to a flat baseline, and equilibrated again in binding buffer. Eluted samples were analysed by SDS-PAGE.
Model building and conservation analysis
The BRCT domain of RAP1 was modelled using MODELLER software (44) and the structure of the BRCT domain of Rap1 from S. cerevisiae (45, PDB entry 2L42). The resulting model and Myb domain of RAP1 (46) were then manually positioned along the TRFH domain of TRF2 with Pymol (47, http://www.pymol.org) based on the TRFH/RAP1-peptide crystal structure and the protein footprinting results. Conservation analysis was performed with CONSURF software (48) using 14 sequences for both TRF2 and RAP1 from the same species of vertebrates (See sequence codes in Supplementary Data).
RESULTS
RAP1 affects TRF2 binding properties toward DNA
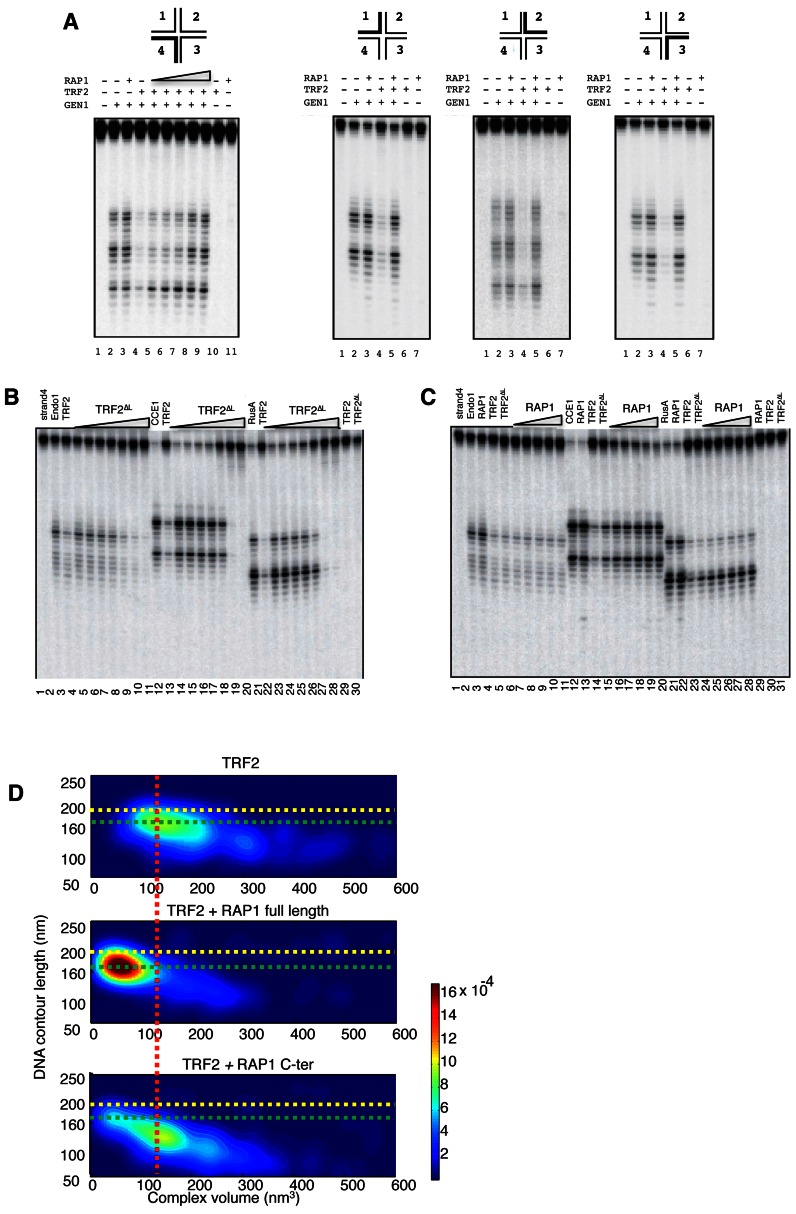
As RAP1 is a key partner of TRF2 probably implicated in TRF2 multi-functionality, we first measured its effect on TRF2 ability to interact with various DNA structures. In addition to linear double stranded TTAGGG (T2AG3) DNA, TRF2 recognizes branched DNA structures like HJ, the four-stranded DNA intermediates of homologous recombination (24,25). Interestingly and in contrast with what was previously observed (49), ITC (Isothermal Titration Calorimetry) and EMSA (Electrophoretic Mobility Shift Assay) experiments show that RAP1 does not bind linear double stranded TTAGGG (T2AG3) DNA, and does not prevent TRF2/DNA or TRF2/HJ binding (Supplementary Figure S1). Among different possibilities, one difference with the RAP1 construct used by Griffith et al. (49) is the removal of the positively charged His-tag in our conditions, which presence could lead to unspecific interaction with the negatively charged DNA. Binding of TRF2 to HJ has been shown to protect this structure against the cleavage of various archetypical resolvases: human GEN1, T7 Endonuclease I, yeast mitochondrial CCEI and the Escherichia coli RusA enzyme. Interestingly, we observe that RAP1 inhibits TRF2 protection against these enzymes in a concentration dependent manner (Figure 1A, Supplementary Figure S2). This is not due to a removal of TRF2 from the HJ (Supplementary Figure S1F) and we do not observe a direct effect of RAP1 on the resolvases (Figure 1A, Supplementary Figures S2 and S3), nor an increase in cleavage when we used the protection-deficient mutant of TRF2, TRF2ΔB, deleted for the N-terminus B-domain (Supplementary Figure S3) (25). These results suggest that this ‘recovery’ of resolvase activity is not caused by a RAP1-dependent increase in the enzyme activity but rather by a decrease in TRF2-dependent protection. This effect of RAP1 was not expected since TRF2 HJ protection originates from the binding of the N-terminal basic domain of TRF2 on the centre of the HJ (25), and the known RAP1 Binding Motif (TRF2-RBM) is located at residue 275–316 (28). Intriguingly, when we use a TRF2 mutant lacking the whole linker (or hinge) domain and thus the TRF2-RBM (TRF2ΔL, Figure 1B), we still observe this RAP1 effect of ‘anti-protection’ for CCE1 and RusA, although higher concentration of RAP1 would be needed (Figure 1C). Therefore, we conclude that, in our conditions, RAP1 does not directly interact with DNA as linear double stranded form, does not interact with DNA as HJ and does not modify the capacity of TRF2 to interact with linear or branched DNA. But interestingly, the presence of RAP1 prevents TRF2 protection of HJ independently of the presence of TRF2-RBM.
Figure 1.
RAP1 affects TRF2 properties towards linear and branched DNA. (A) A total of 5 nM of telomeric Holliday Junction (tHJ) labelled on strand indicated at the top was incubated with 200 nM of TRF2 with and without increasing amount of RAP1 prior to cleavage with GEN1[1–527]. tHJ is a mobile Holliday junction that contains 4 (TTAGGG) repeats (one in each arm of the junction). The thick line of each representation corresponds to the labelled arms. Concentrations of RAP1 used were 100, 200, 400, 1000, 1500 nM (experiment on the left) or at 1500 nM for the other three experiments. (B) 5 nM of tHJ labelled on strand 4 was incubated with increasing amounts of TRF2ΔL prior to cleavage with either Endonuclease I, CCE1 or RusA. Concentrations of TRF2ΔL used were 10, 20, 50, 100, 200, 500, 1000 nM. In lanes 3, 12 and 21 the junction is incubated with 200 nM of TRF2 prior to cleavage. In lanes 29 and 30 the junction is incubated with 1000 nM of TRF2 and TRF2ΔL. (C) 5 nM of tHJ labelled on strand 4 was incubated with 200 nM of TRF2ΔL with and without increasing amount of RAP1 prior to cleavage with either Endonuclease I, CCE1 or RusA. Concentrations of RAP1 used were 100, 200, 400, 1000, 1500 nM. In lanes 3, 12 and 21 the junction is incubated with 1500 nM of RAP1 prior to cleavage. In lanes 4, 13 and 22 the junction is incubated with 200 nM of TRF2 prior to cleavage. In lanes 29, 30 and 31 the junction is incubated with 1500 nM of RAP1, 200 nM of TRF2 and 200 nM of TRF2ΔL. (D) Two-dimensional probability maps calculated from AFM experiments showing the binding of TRF2 (10 nM of dimers) on a 650 bp telomeric DNA (10 nM) either alone (top panel) or in the presence of 40 nM (monomer) of either full-length RAP1 (middle panel) or RAP1-RCT (bottom panel). Yellow dotted lines show the contour length of unbound DNA. Green and red dotted lines show the contour length and volume of the main TRF2–DNA complex respectively. Colour scale of the probability on the right of the maps.
Table 1.
Table 1. X-ray diffraction data collection and refinement statistics.
| Wavelength (Å) | 0.98 |
| Space-group | P21 |
| Diffraction limits (last shell) | 2.44 Å (2.63 Å–2.44 Å) |
| Unit cells (axbxcxαxβxγ) | 53.8 × 104.8 × 85.3 |
| 90 × 94.4 × 90 | |
| Rmerge | 0.068 (0.664) |
| Number of unique reflections | 33 979 (5718) |
| I/σ | 1429 (2.26) |
| Completeness | 0.965 (0.794) |
| Wilson B-factor (Å2) | 51.6 |
| Molecular Replacement (LLG) | 1391 |
| Refinement | |
| Resolution | 2.44 Å |
| Rwork | 0.193 |
| Rfree | 0.237 |
| Number of non-solvent atoms | 6938 |
| Number of water molecules | 94 |
| Figure of merit | 0.7813 |
| RMSD bond lengths | 0.004 |
| RMSD bond angles | 0.976 |
| RMSD chirality | 15.25 |
| PDB entry | 4RQI |
LLG: log of likelihood gain.
In addition to its ability to bind particular DNA structures, TRF2 has been shown to condense linear DNA by promoting positive supercoils in the bound DNA (19,38, Benarroch-Popivker et al., in press). As previously described (19), we drew 2D-probability maps giving the probability of any TRF2/DNA complex to exhibit a given volume in nm3 for a given contour length in nm as measured by Atomic Force Microscopy (AFM). As previously observed (19), TRF2 shortens DNA and drives the formation of complexes that become more and more condensed when the ratio of TRF2 versus DNA increases (Figure 1D, top). In the presence of a small excess of full-length RAP1, DNA is still condensed in the complex compared to unbound DNA, but the highly condensed complexes largely disappear (Figure 1D, middle). Interestingly, the use of the known RAP1 C-terminal TRF2 high-affinity binding domain (RAP1-RCT) instead of full-length RAP1 leads to an increased condensation of DNA (Figure 1D, bottom). Therefore, RAP1 does not prevent TRF2 to condense linear DNA, but alters the level of condensation by blocking high degrees of oligomerization in the TRF2/DNA complexes. But interestingly, RAP1-RCT domain, which tightly binds TRF2, is not sufficient to block this oligomerization.
Altogether, these data show that RAP1 does not prevent TRF2 interaction with linear or branched DNA, but it clearly alters some of the properties of TRF2 toward DNA, in particular by relieving the protective action of TRF2 on HJ cleavage by resolvase enzymes. Strikingly, this effect of RAP1 is maintained despite the deletion of the high affinity RAP1-RCT binding-site of TRF2. In the same way, the isolated RAP1-RCT domain, which contains the high affinity TRF2 binding-site, is not sufficient to prevent TRF2 to form highly condensed complexes with linear DNA. Therefore, these properties of RAP1 necessarily involve additional regions than the described TRF2 high affinity binding area.
Interaction of RAP1 with TRF2 is associated to conformational adjustment
The effect of RAP1 on TRF2 properties toward DNA raised questions on the structure of the TRF2/RAP1 complex. In particular, we wanted to investigate whether the modified behaviour of TRF2 in the presence of RAP1 resulted from a conformational adjustment.
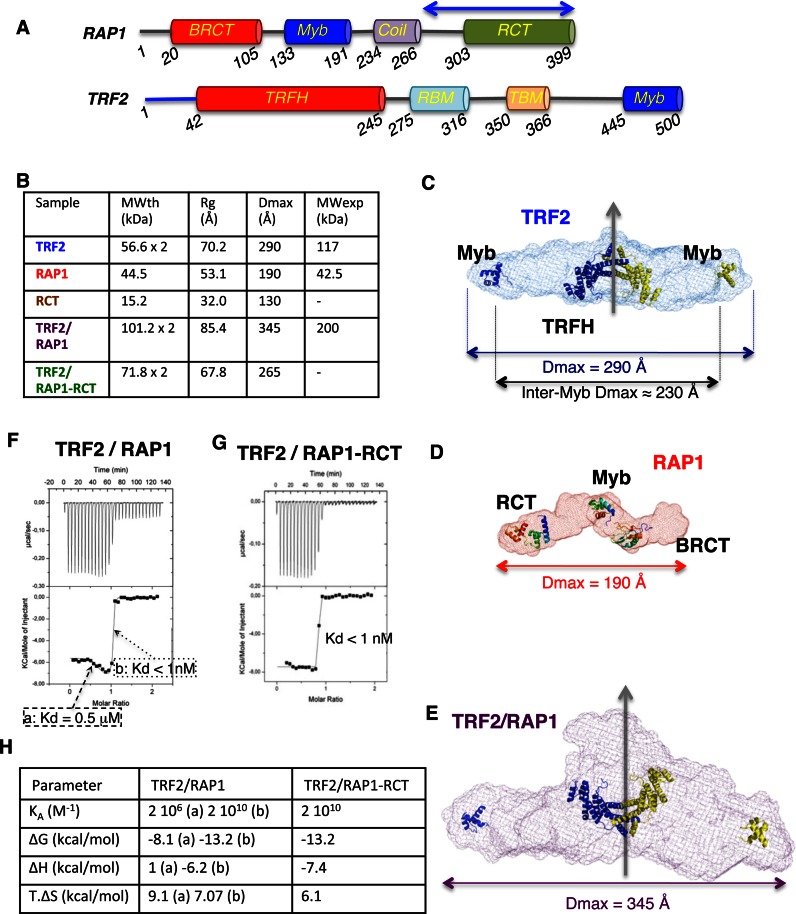
TRF2 and RAP1 are each made of globular domains linked by long unstructured regions, including residues 1–42 and 245–445 in the case of TRF2 that correspond to almost 50% of the sequence, and residues 1–20, 245–275 and 316–445 in the case of RAP1 that correspond to 45% of the sequence. This suggests a particularly high flexibility of the individual molecules as well as of the RAP1/TRF2 complex (Figure 2A), which is not compatible with X-ray crystallography or electron microscopy approaches. Moreover, the size of the complex (around 200 kDa) impedes a straightforward nuclear magnetic resonance study. On the opposite, Small Angle X-ray Scattering (SAXS) analysis is particularly appropriate to gain information about the structuration and the average conformation of flexible proteins and their assembly in solution (50). For each of the isolated full length TRF2, full length RAP1 and RAP1/TRF2 complex, RAP1-RCT construct containing residues 270–399 (blue arrow on Figure 2A), and RAP1-RCT/TRF2 complex, and after careful control of the high purity level (>95%) and structural integrity of our tag-free samples (Supplementary Figure S4), we collected SAXS curves on SWING beamline at SOLEIL synchrotron, using the online HPLC to avoid any sample aggregation (39). The experimental molecular weight derived from the SAXS curves revealed that TRF2 is a dimer, that RAP1 is a partially unstructured monomer and that RAP1/TRF2 contains one TRF2 dimer and two RAP1 monomers (Figure 2B). The p(R) functions and normalized Kratky plots (51) show that proteins alone or in complex behave as elongated molecules and adopt either partially unfolded (in the case of RAP1) or pearl-necklace-like conformation (Supplementary Figure S5). The high flexibility was not compatible with calculation of reliable 3D models and we chose to calculate ab initio envelopes defined as chain-like ensembles of dummy residues (see Supplementary Data). TRF2 dimer is highly extended with a high degree of flexibility and a distance between the Myb DNA-binding domains of each monomer that can reach 230 Å (Figure 2C). TRF2, TRF2/RAP1 and TRF2/RAP1-RCT complexes all display a pseudo two-fold symmetry although no symmetry constraint was used during calculation process (Figure 2E and Supplementary Figure S5G). In the case of the short construct RAP1-RCT, the complex formed with TRF2 does not involve particular conformational adjustment (Supplementary Figure S6A). On the contrary, the superimposition of TRF2 and RAP1/TRF2 envelopes shows a spreading for the complex compared to TRF2 alone, in agreement with the increased value of Dmax between the two p(R) functions (Supplementary Figure S6B). In addition, the shape of RAP1/TRF2 envelope is not compatible with a complete superimposition with TRF2 envelope and two RAP1 envelopes (Supplementary Figure S6C and D). Therefore, the formation of TRF2/RAP1 complex involves a major conformational adjustment of both proteins.
Figure 2.
Formation of TRF2 complex with RAP1 is associated to conformational adjustment and biphasic signal of interaction. (A) Domains organization of RAP1 and TRF2. The blue arrow shows the limit of RAP1-RCT construct. (B) Table of theoretical molecular weight (MWth), giration radius (Rg), maximal distances (Dmax) and experimental molecular weight (MWexp) from Guinier calculation performed with each corresponding SAXS curve. (C–E) Ab initio averaged envelopes of TRF2 (C), RAP1 (D) and TRF2/RAP1 (E) with superimposed globular domains as references. (F and G) ITC characterization of the interaction between TRF2 and RAP1 (F) or RAP1-RCT (G). (H) thermodynamic parameter associated to the ITC experiment performed for TRF2/RAP1 (F), with parameters associated to the first (a) and second (b) transition and for TRF2/RAP1-RCT.
In order to further characterize and quantify TRF2/RAP1 mode of assembly, we measured the interaction between full-length RAP1 and TRF2 by ITC. We chose this method because it is the only one able to provide affinity constant and thermodynamic parameters of the interaction between unlabelled molecules in solution. As compared to previous studies, we used high protein concentration, and low temperature, which favour low affinity interaction (37, Supplementary Figure S7A). In these particular conditions, we were able to observe a reproducible biphasic isotherm. The non-linear least squares fit of the data to a two-independent sites binding model reveals a dissociation constant of less than 1 nM for the high affinity signal, and a dissociation constant of 0.5 μM for the lower affinity signal (Figure 2F). The high affinity signal observed in the biphasic curve of the full-length proteins is equivalent to the one measured between RAP1-RCT and TRF2 (Figure 2G). The thermodynamic parameters demonstrate that the high-affinity transition corresponds to the known interaction between RAP1-RCT and TRF2-RBM (Figure 2H, 28). The presence of a second signal also suggests that the interaction between RAP1 and TRF2 full length involves additional regions from both partners than the previously described RAP1-RCT and TRF2-RBM domains (28).
Complete interaction of RAP1 with TRF2 extends from BRCT to C-terminal domain
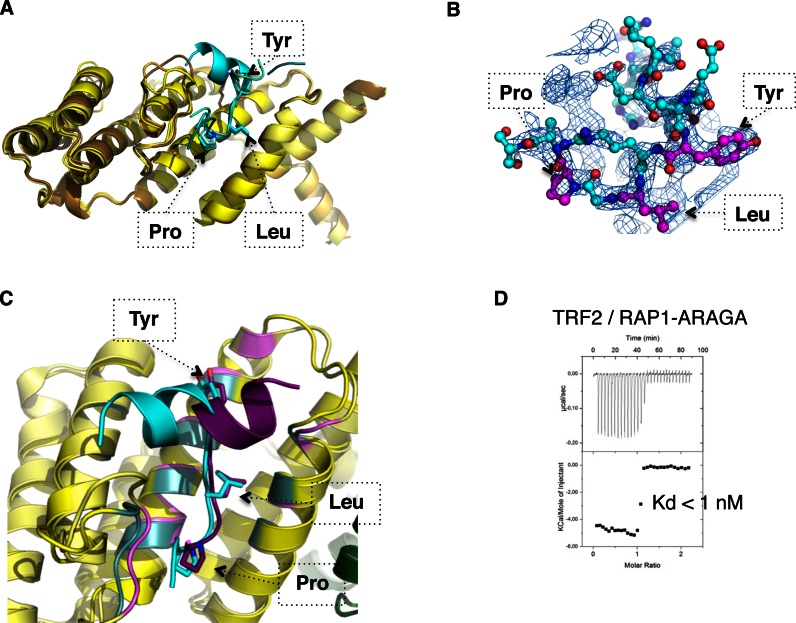
Previous structural studies have shown that TRF2 dimerization domain (TRFH) specifically interacts with proteins containing the consensus sequence YxLxP or YRL (15,17). This motif belongs to numerous TRF2 partners among which the protein Apollo. Indeed a peptide containing the consensus sequence, Apollo-YLP (494ATEFRGLALKYLLTPVNF511), interacts with the TRFH domain with an affinity of 120 nM (15), or 59 nM in our hands (Supplementary Figure S7B). This motif is present in RAP1 sequence at position 99YRLGP103, located at the C-terminus of the BRCT domain (Figure 2A). The corresponding peptide 89ENRERLELEAYRLGPASA106 (RAP1-YLP) interacts with TRFH with a dissociation constant of 13.0 μM (Supplementary Figure S7C). In order to delineate the structural determinants of this interaction we solved the crystal structure of the RAP1-YLP/TRFH complex at 2.4Å resolution (Figure 3A and B). The superimposition of our structure with this of TRFH in complex with the Apollo-YLP peptide (PDB entry: 3BUA, 15) shows that the structures are similar, although the N-terminal part of Apollo-YLP and RAP1-YLP peptides are oriented in opposite directions (Figure 3C). In the case of the Apollo peptide, substitution of Tyr, Leu or Pro into Ala affects the interaction with the TRFH domain, although the effect was not equivalent for each position (15). We mutated the region 99YRLGP103 into 99ARAGA103 in RAP1 full length and, after checking the structural integrity of this RAP1-ARAGA mutant by circular dichroism (Supplementary Figure S4C–E), we measured its interaction with TRF2. The high affinity ITC transition is maintained with the same dissociation constant than for wild-type RAP1, but the low affinity signal becomes not significant (Figure 3D), as in the case of RAP1-RCT (Figure 2G). Altogether, our data indicate that the sequence YRLGP99–103 from RAP1 interacts with the TRFH domain, is essential for the secondary interaction to occur with TRF2, and is necessary for a complete interaction of RAP1 with TRF2, although the residual secondary signal suggests that additional regions may participate.
Figure 3.
Complete interaction of RAP1 with TRF2 involves YXLXP motif and TRF2 dimerization domain. (A) Superimposition of the four TRFH monomers observed in the crystal asymmetric unit (from pale to dark yellow) and of the four corresponding RAP1-YLP peptides (from pale to dark cyan). (B) 2Fo-Fc electron density map at 1sigma level (blue mesh) around one RAP1 peptide represented in ball and stick cyan, with motif Tyr-Leu-Pro in magenta. (C) Superimposition of TRFH (yellow) in complex with Apollo-YLP (dark purple) or RAP1-YLP peptide (cyan). The region of TRFH involved in the interaction with Apollo-YLP is shown in violet, and with RAP1-YLP in light cyan. The rms deviation calculated on C-alpha of each of the four TRFH monomer/RAP1-YLP and of TRFH monomer/Apollo-YLP is between 0.54 and 0.88 Å. (D) ITC characterization of the interaction between TRF2 with RAP1-ARAGA.
RAP1 stretches along TRF2 dimerization domain
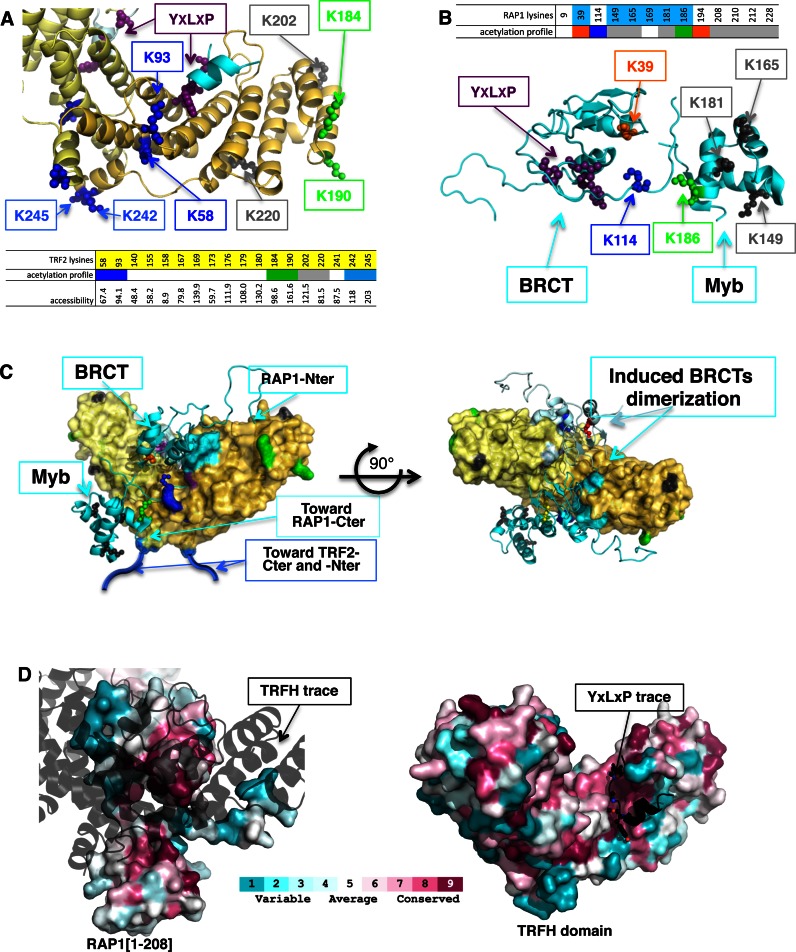
In order to probe the protein surfaces involved in TRF2/RAP1 assembly, we performed protein footprinting using in vitro lysine acetylation by N-hydroxysulfosuccinimide acetate. This compound can acetylate lysines that are solvent-accessible but neither those buried in the structure nor those protected by interactions with another molecule. Through analysis of acetylation profiles using mass spectrometry, it is thus possible to determine which lysines are protected/buried in a protein. TRF2 monomer and RAP1 contain respectively 44 and 25 lysines among which we observed acetylation for 24 lysines from TRF2 and 14 from RAP1 (Supplementary Figures S8 and 9). We compared the lysine acetylation profiles of TRF2, RAP1 and RAP1-ARAGA alone, from these of TRF2/RAP1 and TRF2/RAP1-ARAGA complexes. The comparison of lysine acetylation profiles between protein alone or in complex highlights various lysine populations that are: (i) acetylated in all conditions, indicating constantly accessible regions that are therefore not involved in the interactions; (ii) acetylated in the proteins alone and not in the complex with RAP1 wild type or with RAP1-ARAGA, indicating regions that participate in the interaction; or (iii) acetylated in the proteins alone and in the complex with RAP1-ARAGA but not with wild-type RAP1, showing that the mutation ARAGA affects the interaction in this region; (iv) not acetylated in the proteins alone and acetylated in either both complexes or in TRF2/RAP1 complex only, pointing regions that become accessible upon complex formation, probably due to an opening of the corresponding region (Supplementary Data, Supplementary Figures S8 and 9). Taken together, the modifications in acetylation profiles allowed us to map the region of TRFH domain and RAP1 N-terminal region involved or not in TRF2/RAP1 complex and affected or not by the ARAGA mutation (Figure 4A and B, and Supplementary Figure S10).
Figure 4.
Complete interaction of RAP1 with TRF2 involves large area from RAP1 N-terminal moiety and TRF2 dimerization domain. (A and B) Acetylation profile of lysine residues in TRF2, RAP1 or RAP1-ARAGA alone, compared to TRF2/RAP1 or TRF2/RAP1-ARAGA. Lysine residues are represented in small sphere and coloured differently if they are protected (blue, or light blue if the precise acetylation position was not determined), more accessible (red), or not affected (black) in both TRF2/RAP1 and TRF2/RAP1-ARAGA complex, or protected (green) or more accessible (orange) only in TRF2/RAP1 complex. Residues Y-L-P are shown as purple spheres. The inserted tables show the lysine number in the sequence, highlighted in blue in RAP1 globular domain, in yellow in TRF2 globular domain. The acetylation profile follows the same colour code as in the structure representation. (A) TRFH/RAP1-YLP peptide crystal structure; (B) Juxtaposition of RAP1[1–121] 3D model calculated with Modeller (41), and RAP1-Myb nuclear magnetic resonance structure (PDB entry: 1FEX). (C) Positioning of RAP1 BRCT and Myb domains on TRFH surface based on combination of the crystal structure and footprinting experiments, showing on the right side the induced dimerization of BRCT domain (colour code identical than in Figure 3). (D) Surface representation of RAP1[1–208] coloured upon the conservation score of CONSURF software (48) with the trace of the TRFH domain in black shadow (left); Surface representation of TRFH domain of TRF2 coloured upon the conservation score of CONSURF software (48) with the RAP1-YLP peptide in black (right). All structure representations were drawn with software Pymol (47).
Altogether, our results not only confirm the interaction of the N-terminal moiety of RAP1 with TRFH, but also enable us to draw an interaction path along TRFH and RAP1-Nter, and to propose an interaction scheme (Figure 4C, and Supplementary video). When the TRFH dimer is oriented with the inner side on top, RAP1 interaction path goes through the area facing us of the right monomer, the symmetrical surface still being accessible for the binding of a second RAP1 molecule without any steric hindrance and ends at both the N- and C-terminal ends at the bottom of the TRFH domain (Figure 4C, left). Interestingly, the location of the YxLxP peptide at one extremity of RAP1 BRCT domain implies that RAP1/TRF2 assembly induces a dimerization of the BRCT domain (Figure 4C, right). Therefore, RAP1 complete interaction with TRF2 anchors both at the C-terminal domain and at the YxLxP sequence from the BRCT domain, and involves large additional regions from both proteins. Conservation analysis of TRF2 and RAP1 among vertebrates shows that the region of the TRFH domain from TRF2 that binds the motif YxLxP is highly conserved in agreement with its known crucial implication in TRF2 function (Figure 4D, right). In the case of RAP1, the YRL motif (17) is 100% conserved among vertebrates, and the surface from the N-terminal moiety involved in the interaction with TRFH domain from TRF2 is highly conserved, in agreement with an important function among vertebrates (Figure 4D, left).
Comparison of RAP1 and RAP1-ARAGA effect on TRF2 protein binding properties
The partial overlap between RAP1-YLP and Apollo-YLP binding sites on the TRFH domain raises the question of a possible competition between both proteins for TRF2 binding. The strong difference in affinities for TRFH domain exhibited by RAP1-YLP and Apollo-YLP (13 μM versus 59 nM respectively) suggests that Apollo-YLP should be able to interact with TRF2 in the presence of RAP1. On the other hand, the high-affinity site provided by the RAP1-RCT/ TRF2-RBM anchor (1 nM) brings the two molecules together thus increasing local concentration of the RAP-YLP. To choose between these two hypotheses, we measured both interactions of the Apollo-YLP peptide on a pre-formed TRF2–RAP1 complex and of RAP1 on a pre-formed TRF2/Apollo-YLP peptide complex (Supplementary Figure S7C–E). In the first case, we observed a binding of the Apollo-YLP peptide with an affinity of 47 nM, similar to this of 46 nM obtained in the absence of RAP1. In the case of pre-formed TRF2/Apollo-YLP complex, we still observed a biphasic isotherm, although the low affinity signal was too low to be quantified. This suggests that the formation of RAP1/TRF2 complex does not affect TRF2 binding properties toward Apollo protein, in agreement with the in vivo described capacity of Apollo to interact with TRF2 in the presence of RAP1 (13).
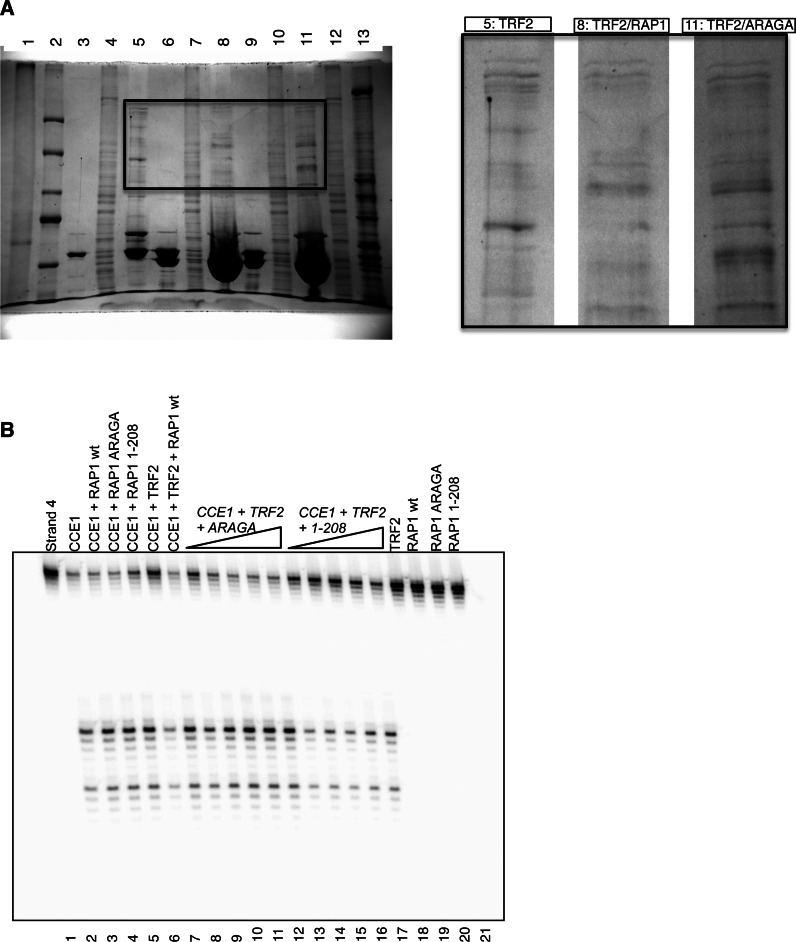
In order to further verify how RAP1–YLP interaction may affect the binding properties of TRF2, we designed a streptavidin-DNA pull down experiment, using a biotinylated (T2AG3)2 oligo-nucleotide immobilized on a streptavidin column. After saturation of the immobilized oligonucleotide with TRF2, TRF2/RAP1 or TRF2/RAP1-ARAGA, we injected HeLa cells nuclear extract, washed, eluted bound proteins by increasing ionic strength and compared both chromatography elution and SDS-PAGE profiles. We first observe that the chromatography elution profiles are different in the case of TRF2, TRF2/RAP1 and TRF2/RAP1-ARAGA (Supplementary Figure S12). We performed our comparison on the region of the gel corresponding to molecular weight higher than this of TRF2 or RAP1 in order to avoid contamination with degradation products of TRF2, RAP1 or RAP1-ARAGA. We first observed that a large number of proteins are retained by TRF2, TRF2/RAP1 or TRF2/RAP1-ARAGA (lanes 5, 8 and 11 respectively in Figure 5A). Our comparison focuses on the presence or absence of specific bands, and not of their intensity, which would not be significant enough. We observed that the presence of RAP1 or RAP1-ARAGA is associated with supplementary bands compared to TRF2 alone, which agrees with TeloPIN. As RAP1 does not directly interact with DNA, we didn't compare the binding properties of RAP1 to these of RAP1/TRF2. On the contrary, comparison of RAP1/TRF2 and RAP1-ARAGA/TRF2 provides pieces of information on the effect of RAP1–YLP interaction with TRF2. Interestingly, we observed that the SDS-PAGE patterns are different between RAP1/TRF2 and RAP1-ARAGA/TRF2 (lanes 8 and 11 in Figure 5B). Therefore, the binding properties of TRF2/RAP1 or TRF2/RAP1-ARAGA, loaded on (T2AG3)2 oligonucleotide, are different. We conclude that the mutation of the YxLxP motif associated to the complete interaction of RAP1 with TRF2 dimerization domain directly affects the binding properties of the assembly formed with TRF2.
Figure 5.
Interaction of RAP1 with TRF2 is associated to different binding properties. (A) Left panel, SDS-PAGE analysis of a typical streptavidin-DNA pulldown experiment (Coomassie blue stained). Lanes contain: (1) input nuclear extract, (2) molecular weight marker (Weights from top to bottom: 250, 150, 100, 80, 60 kDa); (3) input purified TRF2, (4) nuclear extract flow through, (5) elution of TRF2-bound proteins; (6) input purified TRF2/RAP1 complex, (7) nuclear extract flow through, (8) elution of TRF2/RAP1-bound proteins; (9) input purified TRF2/RAP1-ARAGA complex, (10) nuclear extract flow through, (11) elution of TRF2/RAP1-ARAGA-bound proteins; (12) nuclear extract flow through in absence of telomeric proteins, (13) elution of DNA-bound proteins (control). Right panel, enlarged view of lanes 5, 8 and 11 of the SDS-PAGE. (B) 5 nM of tHJ labelled on strand 4 was incubated with 200 nM of TRF2 with and without increasing amount of RAP1-ARAGA or RAP1[1–208] prior to cleavage with CCE1. Concentrations of RAP1-ARAGA used were 200, 400, 1000, 1500, 3000 nM (lanes 8–12 and 3000 nM in lanes 4 and 20. Concentrations of RAP1[1–208] used were 100, 200, 400, 1000, 1500 nM (lanes 13–17), and 1500 nM in lanes 5 and 21. Concentration of RAP1-wt in lanes 3, 7 and 19 is 1500 nM. In lanes 3, 12 and 21 the junction is incubated with 1500 nM of RAP1 prior to cleavage.
Effect of RAP1[1–208] on TRF2 HJ protection
In the initial steps of this study, we observed that RAP1 affect the ability of TRF2 to protect HJ from resolvase activity even after deletion of RAP1 binding motif of TRF2 (Figure 1A–C). Acetylation profile shows that both RAP1-wt and RAP1-ARAGA protects from acetylation lysines located near the N-terminal region of TRF2: Lys58, Lys93, Lys242 and Lys245. On the contrary, RAP1-wt protects Lys184 and Lys190 but RAP1-ARAGA does not (Figure 4A and C, left). It has been previously observed that the HJ protection involves the N-terminal region of TRF2 (25), located close to the region protected by both RAP1-wt and RAP1-ARAGA, but on the opposite side from the region that binds RAP1 YxLxP motif or from the region protected by RAP1-wt only. Therefore, we expect that a construct of RAP1 lacking the high-affinity C-terminal TRF2 binding site should have similar effect on HJ protection by TRF2 than this observed with RAP1 on TRF2 ΔL, and that RAP1 or RAP1-ARAGA should affect in the same way HJ protection by TRF2.
We tested the effect of RAP1-ARAGA and of RAP1[1–208] on HJ protection by TRF2. We observed that RAP1-ARAGA affects TRF2 HJ protection in a similar manner than RAP1-wt. The effect of RAP1[1–208] on TRF2 HJ protection is similar to this observed with RAP1 full length on TRF2ΔL mutant, a higher quantity of RAP1[1–208] is necessary (Figure 5A) in the same way than a higher quantity of RAP1 was necessary with TRF2ΔL (Figure 1A,B and C). Therefore, the interaction of RAP1[1–208] with a region of TRF2 located at the bottom of the TRFH domain near the N-terminal region directly affects the property of TRF2 to protect HJ from resolvase. We conclude that this particular region is involved in HJ protection by TRF2 (Figure 4C).
DISCUSSION
Our comprehensive study of RAP1, TRF2 and the effect of RAP1/TRF2 complex formation on their conformation and interaction properties reveal new elements that strongly deepen our knowledge of these two proteins and their inter-relation. First of all, it is essential to remind how experimental conditions and sample preparation may affect the results. Although analyses of crystal structures show little effect of the presence of His-tag (52), it has been shown that His-tag can affect protein function and interaction properties (53,54). We therefore chose to exclusively work with tag-free constructs of our proteins. We first observed that RAP1 affects both the ability of TRF2 to form condensed multimeric complexes with DNA and to protect Holliday junction from resolvases (Figure 1), that the formation of RAP1/TRF2 assembly is more complex than the known RAP1-RCT/TRF2-RBM interaction (15), and that the assembled molecules form a distinct entity. The combination of SAXS and ITC analysis confirms that the formation of RAP1/TRF2 complex involves a profound conformational adjustment (SAXS), associated with a two-transition mode of interaction (ITC) (Figure 2). Moreover, the isolated RAP1-RCT domain is not sufficient to induce a conformational adjustment, or a secondary ITC signal (Figure 2, Supplementary Figure S5G). A YXLXP motif (or YRL motif), specific for the interaction with the TRF2 dimerization domain, TRFH (15,17), is located at the C-terminus of the BRCT domain of RAP1 at position 99–103. The crystal structure of TRFH in complex with a peptide of RAP1 containing the YXLXP motif show that the molecular determinants of the interaction involve the YXLXP motif and is similar to that previously described between TRFH and Apollo-YLP peptide (Figure 3A–C, 15). Moreover, the similar ITC interaction profiles between RAP1-ARAGA and TRF2 (Figure 3C) and between RAP1 and the pre-formed TRF2/Apollo-YLP complex (Supplementary Figure S6F) show that the Apollo-YLP peptide compete with RAP1 for binding on TRF2 TRFH domain. This competition is centred on the YXLXP site, although it is not documented whether additional regions of full length Apollo might interact with full length TRF2. The residual secondary signal in both cases also suggests that additional regions of RAP1 are involved in the secondary binding site (Figure 3C and Supplementary Figure S6F). Protein footprinting experiments allowed us to complete the RAP1/TRF2 interaction scheme and to model the assembly formed by RAP1 N-terminus region, comprising residues 1–211, and TRFH. In this model, the N-terminus moiety of RAP1 follows one face of the TRFH domain, interacts through YXLXP sequence in the inner part of the TRFH, and ends at the external side of the domain (Figure 4C, left). Looking from the top of the TRFH domain, we observe that this interaction of the N-terminal region of RAP1 induces an unexpected dimerization of the BRCT domain (Figure 4C, right). The high conservation among vertebrates of the known YXLXP binding site of TRF2, of the YRLXP motif of RAP1 and of the surface of RAP1 involved in the secondary interaction also reveals that this secondary interaction of RAP1 and the capacity of TRF2 to interact with YRLXP motif containing proteins are of equal importance.
The similar ITC interaction profiles between TRF2 and Apollo-YLP and between the preformed TRF2/RAP1 complex and Apollo-YLP also suggests that RAP1 interaction may probably not hinder Apollo interaction (Supplementary Figure S7C and D). Similar analysis could apply to SLX4, which contains a TRFH binding motif (1014RGLEVHRLAPW1028), is characterized by a dissociation constant of 750 nM with TRF2, and adopts the same conformation than Apollo-YLP peptide in complex with TRFH (PDB entry: 4M7C, 16). In TRF1 dimerization domain, the region equivalent to this of TRF2 specifically interacts with the TIN2 protein. A TIN2 peptide248THPEPLAGRHFNLAPLGR265, specific for TRF1 dimerization domain, interacts in vitro with TRF2 TRFH with a dissociation constant of 6 μM (15), in the same order of magnitude than this of RAP1-YLP peptide on TRF2 (13 μM). The variable affinity measured for the YXLXP-containing peptides suggests that RAP1 secondary interaction might (i) impede inappropriate binding on the TRFH of TRF2 in case of unspecific binding to TRF2 TRFH of FxLxP containing proteins like TIN2 which are specific for TRF1 TRFH; (ii) vary depending on the chemical environment. In this way, our streptavidin-DNA pull down reveals that, when bound to a (T2AG3)2 oligonucleotide, RAP1/TRF2 or RAP1-ARAGA/TRF2 do not retain the same proteins from a HeLa nuclear extract. Therefore, the capacity of RAP1 to form or not a complete interaction with TRF2 through the YXLXP motif is associated with different binding properties (Figure 5A).
In addition to this possible regulatory function, we have seen that RAP1 does not modify TRF2 property to condense DNA (Figure 1), but it does affect the oligomerization level that we observed in AFM experiments. This must be a consequence of the complete RAP1 interaction since RAP1-RCT alone is not able to do so. The mechanism behind could involve steric hindrance or hiding of an oligomerization-prone region of TRF2, located at or close to the TRFH domain. At last, RAP1 affects the basic domain-dependent ability of TRF2 to protect HJ against resolvases. Although the precise mechanism remains to be determined, our data show that the interaction of RAP1 with the region of TRF2 located close to the N-terminus of the TRFH domain is part of the mechanism behind the alteration in the protection of HJ against resolvases (Figure 5B).
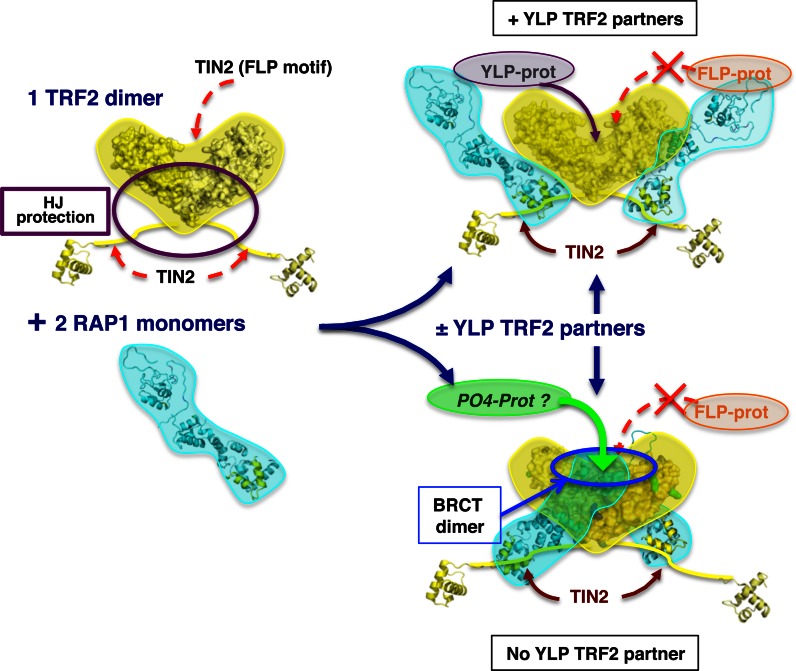
Based on our results, we propose an overall scheme of the formation of the RAP1/TRF2 complex and the possible effect on their binding properties (Figure 6). RAP1 alone is highly flexible and extended, according to the Dmax and normalized Kratky plot for the protein in solution. In the absence of RAP1, TRF2 interacts with multiple partners, with no site discrimination in the case of TIN2 (Figure 6, left). RAP1 conformation is clearly stabilized upon RAP1/TRF2 complex assembly, as shown by the normalized Kratky plots (Supplementary Figure S5E). The complex could adopt alternative open or closed conformation depending on the presence or absence of TRFH high-affinity TBM-containing protein partners (such as Apollo or SLX4) that compete with RAP1 N-terminal region, or depending on the chemical environment that may favour or not secondary interaction (Figure 6, right). RAP1/TRF2 closed conformation inhibits additional illicit interaction with TRFH (such as the one with TIN2), induces RAP1 BRCT domain dimerization (Figure 4C), and forms a new entity that may interact with other specific telomeric or non-telomeric partners, whereas RAP1/TRF2 open conformation has different binding properties (Figure 6). Of note, TRF2 alone, RAP1 alone and closed RAP1/TRF2 models are in agreement with the combination of our results. The open form model of RAP1/TRF2 complex is proposed to account for the results of ITC and streptavidin-DNA pull down experiments performed with RAP1-ARAGA as compared to RAP1.
Figure 6.
Model of binding properties of TRF2/RAP1 assembly. Proposed model of the assembly of TRF2/RAP1 and the effect of its malleability on its binding properties. The presence of RAP1 (cyan) prevents the binding of non-specific proteins like FLP containing proteins (orange) in the TBM area of TRF2 (yellow), but does not prevent the binding of high-affinity YLP containing proteins (purple). In the absence of YLP containing proteins, RAP1 complete interaction induces the formation of BRCT dimer (blue circle) that could recruit new phosphorylated partners (green).
Our comprehensive study shows that RAP1 affects TRF2 conformation and function and that TRF2 also affects RAP1 conformation. The induced dimerization of RAP1 BRCT domain upon full interaction with TRF2 is of particular interest (Figure 4C, right). The lysine acetylation profile of RAP1-ARAGA in the footprinting experiments shows that mutation of the YxLxP motif affects RAP1 interaction with TRFH, and particularly the induced dimerization of the BRCT domain. In addition, the streptavidin-DNA pull down shows that the capacity of RAP1 to form a complete interaction with TRF2 or not is associated with different binding properties. This provides both a clue about the functional role of RAP1 BRCT domain, and for the first time a possible explanation for the high conservation of this domain among Rap1 proteins from yeast to human. Indeed, BRCT domains are found in a wide array of prokaryotic and eukaryotic proteins. In human, at least 23 genes coding for proteins with BRCT domains have been listed, most of them implicated in DNA damage response. The induced dimerization mimics a BRCT tandem, known for its propensity to interact with phosphorylated proteins (55). The induced dimerization of RAP1 BRCT upon interaction with TRF2 suggests that RAP1 functions could be quite different whether RAP1 co-localizes with TRF2 or not. A recent study using a separation-of-function mutant of TRF2 has highlighted that RAP1 is involved in TRF2-mediated anti-NHEJ function as a complementary pathway (Benarroch-Popivker et al., in press). The precise role of the BRCT domain in this function of RAP1 remains to be addressed.
Altogether, our data indicate that formation of the RAP1/TRF2 complex needs the presence of several domains of the proteins. Importantly, we show that the RAP1/TRF2 complex forms a structural entity with different competences than individual RAP1 and TRF2. We have shown that the available accessible surface of TRF2 or RAP1/TRF2 determines binding preferences for other partners of TRF2. Determining which proteins are more specific for TRF2 or for RAP1/TRF2 should provide a key to the regulation of TRF2 and RAP1 functions. In a general manner, one should consider RAP1, TRF2 and RAP1/TRF2 as three distinct, although related, entities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors wish to thank Stephen CY Ip, Stephen C West, Anne-Cécile Déclais and David MJ Lilley for kindly providing resolving enzymes or plasmids to produce them. We thank Dr Carl Mann for his help with cell culture and for the HeLa cells he provided, Magali Aumont-Nicaise for her support in the use of ITC instruments at IBBMC supported by the French Infrastructure for Integrated Structural Biology (FRISBI), Valérie Campanacci for her help in SEC-MALS experiments and Centrino Moscalenko for her help in drawing the AFM 2D maps. Initial discussions with Delphine Benarroch-Popivker and Bianca Sclavi were very useful for the design of the protein footprinting experiments. We are very grateful to PX1, PX2 and SWING beamlines local contacts from synchrotron SOLEIL for their help during data acquisition.
Author contributions: G.G., S.M., S.P., Y-VLB, R.T., M-.J.G-.P., E.G. and M-.H.L.D. conceived and designed the experiments. G.G., S.M., S.P., R.B., Y-.V.L.B., C.T.L., W.M., P.R., B.G., R.T., M-.J.G-.P. and M-.H.L.D. performed the experiments. G.G., S.M., S.P., R.B., Y-.V.L.B., C.T.L., W.M., P.R., B.G., R.T., M-.J.G-.P., E.G. and M-.H.L.D. analyzed the data. G.G., S.M., S.P., M-.J.G-.P., E.G. and M-.H.L.D. wrote the manuscript.
Footnotes
Present address:
Rémi Buisson, Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston, MA 02129, USA.
Yann-Vaï Le Bihan, Division of Cancer Therapeutics, The Institute of Cancer Research, London, UK.
FUNDING
Agence Nationale de la Recherche (TELOLOOP grant) [ANR-1582-30020690]; Ligue contre cancer (EG équipe labellisée); INCa (TELOCHROM grant); Essonne committee of the Ligue contre le cancer (grant M18756 and M22897); Foundation ARC pour la Recherche sur le Cancer (grant SFI20121205503); Commissariat à l'Energie Atomique et aux Energies Alternatives (CEA); French Infrastructure for Integrated Structural Biology (FRISBI); Centre National de la Recherche Scientifique (CNRS); Institut National de la Santé et de la Recherche Médicale (INSERM). Funding for open access charge: CEA; ANR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Liu D., O'Connor M.S., Qin J., Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 3.Songyang Z., Liu D. Inside the mammalian telomere interactome: regulation and regulatory activities of telomeres. Crit. Rev. Eukaryot. Gene Expr. 2006;16:103–118. doi: 10.1615/critreveukargeneexpr.v16.i2.10. [DOI] [PubMed] [Google Scholar]
- 4.Giraud-Panis M.-J., Pisano S., Benarroch-Popivker D., Pei B., Le Du M.-H., Gilson E. One identity or more for telomeres. Front. Oncol. 2013;3:48. doi: 10.3389/fonc.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo Z., Dai Z., Xie X., Feng X., Liu D., Songyang Z., Xiong Y. TeloPIN: a database of telomeric proteins interaction network in mammalian cells. Database (Oxford) 2015:bav018. doi: 10.1093/database/bav018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J., Gronenborn A.M. Weak protein complexes: challenging to study but essential for life. FEBS J. 2014;281:1948–1949. doi: 10.1111/febs.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilson E., Géli V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 8.Verdun R.E., Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 9.Ye J., Lenain C., Bauwens S., Rizzo A., Saint-Léger A., Poulet A., Benarroch D., Magdinier F., Morere J., Amiard S., et al. TRF2 and Apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell. 2010;142:230–242. doi: 10.1016/j.cell.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 10.Biroccio A., Cherfils-Vicini J., Augereau A., Pinte S., Bauwens S., Ye J., Simonet T., Horard B., Jamet K., Cervera L., et al. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat. Cell Biol. 2013;15:818–828. doi: 10.1038/ncb2774. [DOI] [PubMed] [Google Scholar]
- 11.Ye J., Renault V.M., Jamet K., Gilson E. Transcriptional outcome of telomere signalling. Nat. Rev. Genet. 2014;15:491–503. doi: 10.1038/nrg3743. [DOI] [PubMed] [Google Scholar]
- 12.Lenain C., Bauwens S., Amiard S., Brunori M., Giraud-Panis M.-J., Gilson E. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr. Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 13.van Overbeek M., de Lange T. Apollo, an artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr. Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Fairall L., Chapman L., Moss H., de Lange T., Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell. 2001;8:351–361. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y., Yang Y., van Overbeek M., Donigian J.R., Baciu P., de Lange T., Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 16.Wan B., Yin J., Horvath K., Sarkar J., Chen Y., Wu J., Wan K., Lu J., Gu P., Yu E.Y., et al. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Rep. 2013;4:861–869. doi: 10.1016/j.celrep.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H., Lee O.-H., Xin H., Chen L.-Y., Qin J., Chae H.K., Lin S.-Y., Safari A., Liu D., Songyang Z. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat. Struct. Mol. Biol. 2009;16:372–379. doi: 10.1038/nsmb.1575. [DOI] [PubMed] [Google Scholar]
- 18.Sarek G., Vannier J.B., Panier S., Petrini J.H., Boulton S.J. TRF2 recruits RTEL1 to telomeres in S phase to promote T-loop unwinding. Mol. Cell. 2015;57:622–635. doi: 10.1016/j.molcel.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulet A., Pisano S., Faivre-Moskalenko C., Pei B., Tauran Y., Haftek-Terreau Z., Brunet F., Le Bihan Y.-V., Ledu M.-H., Montel F., et al. The N-terminal domains of TRF1 and TRF2 regulate their ability to condense telomeric DNA. Nucleic Acids Res. 2012;40:2566–2576. doi: 10.1093/nar/gkr1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto K., Bartocci C., Ouzounov I., Diedrich J.K., Yates J.R. 3rd, Denchi E.L. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bilaud T., Brun C., Ancelin K., Koering C.E., Laroche T., Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 22.Court R., Chapman L., Fairall L., Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palm W., de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 24.Fouche N., Cesare A.J., Willcox S., Ozgur S., Compton S.A., Griffith J.D. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J. Biol. Chem. 2006;281:37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- 25.Poulet A., Buisson R., Faivre-Moskalenko C., Koelblen M., Amiard S., Montel F., Cuesta-Lopez S., Bornet O., Guerlesquin F., Godet T., et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker A.M., Fu Q., Hayward W., Victoria S., Pedroso I.M., Lindsay S.M., Fletcher T.M. The telomere binding protein TRF2 induces chromatin compaction. PLoS One. 2011;6:e19124. doi: 10.1371/journal.pone.0019124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y., Rai R., Zhou Z.-R., Kanoh J., Ribeyre C., Yang Y., Zheng H., Damay P., Wang F., Tsujii H., et al. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat. Struct. Mol. Biol. 2011;18:213–221. doi: 10.1038/nsmb.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B., Oestreich S., de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 30.Bae N.S., Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 31.Kabir S., Sfeir A., de Lange T. Taking apart Rap1: an adaptor protein with telomeric and non-telomeric functions. Cell Cycle Georget. Tex. 2010;9:4061–4067. doi: 10.4161/cc.9.20.13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabir S., Hockemeyer D., de Lange T. TALEN gene knockouts reveal no requirement for the conserved human shelterin protein Rap1 in telomere protection and length regulation. Cell Rep. 2014;9:1273–1280. doi: 10.1016/j.celrep.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez P., Thanasoula M., Carlos A.R., Gómez-López G., Tejera A.M., Schoeftner S., Dominguez O., Pisano D.G., Tarsounas M., Blasco M.A. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat. Cell Biol. 2010;12:768–780. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang D., Xiong Y., Kim H., He Q., Li Y., Chen R., Songyang Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011;21:1013–1027. doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teo H., Ghosh S., Luesch H., Ghosh A., Wong E.T., Malik N., Orth A., de Jesus P., Perry A.S., Oliver J.D., et al. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat. Cell Biol. 2010;12:758–767. doi: 10.1038/ncb2080. [DOI] [PubMed] [Google Scholar]
- 36.Takai K.K., Hooper S., Blackwood S., Gandhi R., de Lange T. In vivo stoichiometry of shelterin components. J. Biol. Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janoušková E., Nečasová I., Pavloušková J., Zimmermann M., Hluchý M., Marini V., Nováková M., Hofr C. Human Rap1 modulates TRF2 attraction to telomeric DNA. Nucleic Acids Res. 2015;43:2691–2700. doi: 10.1093/nar/gkv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amiard S., Doudeau M., Pinte S., Poulet A., Lenain C., Faivre-Moskalenko C., Angelov D., Hug N., Vindigni A., Bouvet P., et al. A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 39.David G., Pérez J. Combined sampler robot and high-performance liquid chromatography: a fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. J. Appl. Crystallogr. 2009;42:892–900. [Google Scholar]
- 40.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Smart O.S., Vonrhein C., Womack T.O. BUSTER, version 2.8. 0. Camb. U. K. Glob. Phasing Ltd; 2011. [Google Scholar]
- 43.Abmayr S.M., Yao T., Parmely T., Workman J.L. Current Protocols in Molecular Biology. Vol. 75. John Wiley & Sons, Inc; 2006. Preparation of nuclear and cytoplasmic extracts from mammalian cells. 12.1, 12.1.1–12.1.10. [DOI] [PubMed] [Google Scholar]
- 44.Eswar N., Eramian D., Webb B., Shen M.-Y., Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. Clifton N.J. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W., Zhang J., Zhang X., Xu C., Tu X. Solution structure of Rap1 BRCT domain from Saccharomyces cerevisiae reveals a novel fold. Biochem. Biophys. Res. Commun. 2011;404:1055–1059. doi: 10.1016/j.bbrc.2010.12.109. [DOI] [PubMed] [Google Scholar]
- 46.Hanaoka S., Nagadoi A., Yoshimura S., Aimoto S., Li B., de Lange T., Nishimura Y. NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J. Mol. Biol. 2001;312:167–175. doi: 10.1006/jmbi.2001.4924. [DOI] [PubMed] [Google Scholar]
- 47.Schrödinger LLC. The PyMOL Molecular Graphics System. Version 1.7.4. 2010 [Google Scholar]
- 48.Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arat Ö.N., Griffith J.D. Human Rap1 interacts directly with telomeric DNA and regulates TRF2 localization at the telomere. J. Biol. Chem. 2012;287:41583–41594. doi: 10.1074/jbc.M112.415984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rambo R.P., Tainer J.A. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr. Opin. Struct. Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Durand D., Vivès C., Cannella D., Pérez J., Pebay-Peyroula E., Vachette P., Fieschi F. NADPH oxidase activator p67phox behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 2010;169:45–53. doi: 10.1016/j.jsb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 52.Carson M., Johnson D.H., McDonald H., Brouillette C., DeLucas L. His-tag impact on structure. Acta Cryst. 2007;D63:295–301. doi: 10.1107/S0907444906052024. [DOI] [PubMed] [Google Scholar]
- 53.Sabaty M., Grosse S., Adryanczyk G., Boiry S., Biaso F., Arnous P., Pignol D. Detrimental effect of the 6 His C-terminal tag on YedY enzymatic activity and influence of the TAT signal sequence on YedY synthesis. BMC Biochem. 2013;14:28. doi: 10.1186/1471-2091-14-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noirclerc-Savoye M., Flayhan A., Pereira C., Gallet B., Gans P., Ebel C., Breyton C. Tail proteins of phage T5: investigation of the effect of the His6-tag position, from expression to crystallisation. Protein Expr. Purif. 2015;109:70–78. doi: 10.1016/j.pep.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Gerloff D.L., Woods N.T., Farago A.A., Monteiro A.N.A. BRCT domains: a little more than kin, and less than kind. FEBS Lett. 2012;586:2711–2716. doi: 10.1016/j.febslet.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.