Abstract
Formation of the mature HIV-1 reverse transcriptase (RT) p66/p51 heterodimer requires subunit-specific processing of the p66/p66′ homodimer precursor. Since the ribonuclease H (RH) domain contains an occult cleavage site located near its center, cleavage must occur either prior to folding or subsequent to unfolding. Recent NMR studies have identified a slow, subunit-specific RH domain unfolding process proposed to result from a residue tug-of-war between the polymerase and RH domains on the functionally inactive, p66′ subunit. Here, we describe a structural comparison of the isolated RH domain with a domain swapped RH dimer that reveals several intrinsically destabilizing characteristics of the isolated domain that facilitate excursions of Tyr427 from its binding pocket and separation of helices B and D. These studies provide independent support for the subunit-selective RH domain unfolding pathway in which instability of the Tyr427 binding pocket facilitates its release followed by domain transfer, acting as a trigger for further RH domain destabilization and subsequent unfolding. As further support for this pathway, NMR studies demonstrate that addition of an RH active site-directed isoquinolone ligand retards the subunit-selective RH′ domain unfolding behavior of the p66/p66′ homodimer. This study demonstrates the feasibility of directly targeting RT maturation with therapeutics.
INTRODUCTION
The infectivity of the human immunodeficiency virus is dependent on a reverse transcriptase that converts viral RNA into dsDNA (1,2). Current drug therapies target the mature p66/p51 RT heterodimer (1,3–6). However, in the virion RT undergoes a complex conformational maturation process that provides potentially useful points of chemotherapeutic intervention. The limited information available for this transformation has led to a proliferation of proposed pathways for formation of the p66/p51 RT heterodimer (7–20). Recent NMR studies have provided detailed information for the p51 and p66 monomer structures, and clarified some of the conformational transitions that convert p66 into the p66/p66′ homodimer precursor (21–23). The initially formed p66/p66′ homodimer exists as a structural heterodimer, in which the active polymerase and RNase H (RH) domains are on the p66 subunit, while the p66′ subunit contains a polymerase′ domain in an inactive fold as well as a second RH′ domain that is tethered to the polymerase′ by residues unraveled from its C-terminal αM′ helix. Dimerization sets up a competitive tug-of-war between the polymerase′ and RH′ domains for common residues located at the boundary. Importantly, the cleavage site targeted by HIV protease is located near the center of the RH domain, rendering it inaccessible to proteolytic cleavage in both the isolated RH domain and the p66 monomer. It thus has been postulated that destabilization of the p66′ RH′ domain resulting from the N-terminal residue transfer to helix αM′ in the p66′ polymerase′ domain above is sufficient to result in subunit-specific RH′ domain unfolding, so that the cleavage site becomes exposed (21). Nevertheless, the nature of this unfolding process remains undetermined, and other studies have contested the possibility that the domain is able to unfold sufficiently to expose the cleavage site (8).
It previously has been observed that bacterial expression of the isolated RT RH domain often results in both monomer and dimer species, and it was proposed that the RH dimer exists as a domain swapped structure (24). Domain swapped dimers are believed to arise as a result of the capture and stabilization of a partially unfolded protein conformation by formation of a set of homologous stabilizing interactions with a second molecule (25–27). Domain swapping generally occurs at a hinge region around which proteins tend to locally unfold prior to more complete unfolding (25). Since the monomer-dimer interconversion requires a transition through an unfolded or partially unfolded state, characterization of the domain swapped RH domain can provide fundamental insight related to the proposed unfolding of the RH′ domain, and hence the role of RH′ unfolding in the RT maturation process. Consequently we have investigated this structure in detail and, as outlined below, have identified intrinsic structural characteristics of the RH domain that facilitate both unfolding and domain swapping. Further, these results reveal interesting variations in the Tyr427 binding pocket that directly implicates Tyr427 as a key trigger of the unfolding process.
Building upon this further support for an RT maturation pathway involving subunit-selective RH domain unfolding, we also demonstrate that a tight-binding RH inhibitor can significantly retard unfolding of the supernumerary RH′ domain and hence interfere with the RT maturation pathway. This demonstration represents a new approach for the development of RT-directed therapy.
MATERIALS AND METHODS
Expression and purification of labeled and unlabeled RH constructs used in these studies
The expression and purification of RH and its mutants were identical to that described in the previous study (28). The single mutations were introduced using the QuickChange XL site directed mutagenesis kit (Agilent). The constructs used in these studies included RHmnel studied previously (28,29), corresponding to the HXB2 p66 residues 427–560 with an N-terminal MNEL leader sequence originally described by Becerra et al. (30). N-terminal truncated constructs included RHΔNT, corresponding to p66 residues 429–560 with an N-terminal Leu429Met mutation, an extended linker construct, RHΔNT-EL, in which an additional Pro-Asp-Gln sequence was introduced into RHΔNT immediately following Gln512, and RHΔNT(E514L) in which RHΔNT contains an E514L substitution in the αB-αD linker. Monomer and dimer fractions of the RH domain were separated on a Superdex 200 10/300 column or, for greater resolution, a HiLoad 26/60 chromatography column, as indicated. Samples were run at 4°C with run times of ∼1 h for the smaller column and ∼8 h for the larger column.
NMR Methods
For the kinetic studies, the lyophilized RNase H samples were dissolved in 25 mM Tris-HCl-d11 in D2O, pD 7.5, 100 mM KCl, 0.02% NaN3. Sample concentrations were 100 μM unless otherwise indicated. The amide H/D exchange studies utilized samples that were initially lyophilized and then dissolved in 25 mM Tris-HCl-d11 in D2O, pD 7.5, 100 mM KCl, 0.02% NaN3, and the protein concentrations were 200 μM. As indicated, some studies also included 1.5 mM of the RH inhibitor 2-hydroxyisoquinoline-1,3(2H,4H)-dione and 5 mM MgCl2. The 1H-15N HSQC experiments were collected during successive 40 min periods at 25°C. Synthesis of hydroxyisoquinolone (HIQ) is described by Billamboz et al. (31) and was obtained as a custom synthesis product from MRIGlobal (Kansas City, MO, USA).
All NMR spectra were acquired on Agilent INOVA 600 and 800 MHz NMR spectrometers equipped with 5 mm 1H[13C,15N] triple-resonance cryogenically cooled probes. The 1H-13C HMQC spectra were obtained at the temperatures indicated using Agilent's Biopack gChmqc experiment (Agilent, Santa Clara, CA) with 1024 X 96 complex points and sweep widths of 14 and 10 ppm in the1H and 13C dimensions (center 10 ppm), using a relaxation delay of 1 s. The 1H-15N HSQC RNase H spectra were acquired at 25 °C using Agilent's BioPack gNfhsqc experiment with 1024 X 128 complex points and sweep widths of 14 and 29.6 ppm in the 1H and 15N dimensions (center ∼ 120 ppm), using a relaxation delay of 1 s. All NMR data were processed by NMRpipe (32) and analyzed with NMRViewJ (33).
Backbone assignments of the domain swapped RH homodimer were performed on triply labeled U-[2H,13C,15N]RHmnel. Resonances were assigned using standard protocols and have been deposited in the BMRB under ID: 26718. Previous monomer assignments are available under codes 5347 and 5931 (29). See also Zheng et al. (28) for corrected assignments of Ile521 and Ile556.
Kinetic studies
Domain swap kinetics were determined from exponential fits of the normalized resonance intensities in the 1H-13C HMQC spectra starting from the initially purified dimer fractions of Ile-labeled RH domain and several variants using the relations:
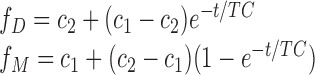 |
(1) |
so that the fractional probabilities vary from c1 to c2. In a few cases the analysis was limited to the resolved dimer resonance due to overlap. Samples were in 25 mM Tris HCl-d11, 100 mM KCl, pD 7.5 and run at 25°C and results are summarized in Table 1. Time constants were also estimated for RHmnel using time lapse NMR spectroscopy. The RHΔNT(E514L) mutant studied is apparently somewhat less stable than RHΔNT, and undergoes very slow precipitation over the course of the studies. Since the analysis of the fractional monomer and dimer intensities is normalized to the sum, this precipitation does not influence the analysis.
Table 1. RHΔNT Dimer decay time constants.
| Residue | Time constanta (Min) | Dimer fraction @ equilibrium |
|---|---|---|
| 482 | 25.2 | - |
| 505 | 24.5 | 0.13 |
| 506 | 24.5 | 0.12 |
| 521 | 20.0 | 0.20 |
| 522 | 26.1 | 0.16 |
| Mean | 24.1 ± 1.1 | 0.15 ± 0.02 |
| RHΔNT (PDQins) | undetectable | |
| RHΔNT(E514L) | 0.50 |
aDecay time constants were determined at 25°C and mean values include a standard error calculation.
Time dependent HMQC studies of p66 homodimer conformational evolution utilized [13CH3-Ile,U-2H]p66 and were performed at 35°C as described previously (23), except that the samples also contained 2 mM HIQ and 6 mM MgCl2.The results are thus directly comparable with those shown in Figure 7 of Zheng et al. (23).
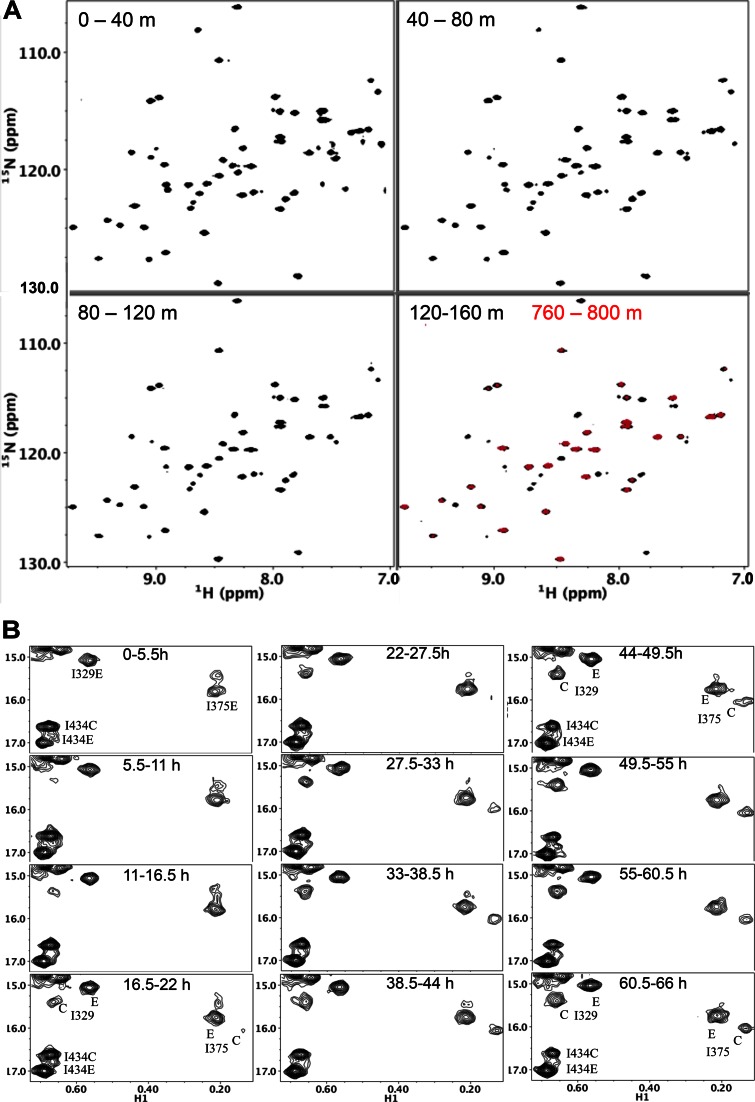
Figure 7.
Effect of an RH active site-directed ligand on the H/D exchange kinetics of RHΔNT and on the maturation behavior of the p66 homodimer. (A) 1H-15N HSQC spectra of RHΔNT in the presence of 5 mM MgCl2 and 1.5 mM HIQ after an H2O/D2O buffer change, obtained during the time periods indicated. The red spectrum in the lower right hand panel was obtained over the 760–800 min accumulation period after changing to the D2O buffer. Other conditions as in Figure 5B. (B) Time dependent changes of connection′ (Ile329 and Ile375) and RH′ domain (Ile434) Ile resonances in the [13CH3-Ile,U-2H]p66/[13CH3-Ile,U-2H]p66′ homodimer in the presence of a Mg-HIQ RH-directed ligand. The ‘E’ and ‘C’ resonance labels indicate assignments to the extended and compact polymerase conformations that are present in the p66 and p66′ subunits, respectively. Conditions favoring dimerization were introduced at t = 0, and the samples also contained 6 mM MgCl2, 1 mM HIQ in 25 mM Tris HCl-d11, pD 7.5, 100 mM KCl and 0.02% NaN3. Each spectrum corresponds to a sequential 5.5 h accumulation period. Samples were run on an Agilent 800 MHz NMR spectrometer at 35°C, under conditions identical to those reported previously without the addition of Mg-HIQ (23).
X-ray crystallography
Crystals of the RHmnel construct of the RNase H domain of RT were grown using sitting drop vapor diffusion. Drops consisted of 0.125 μl protein solution (16 mg/ml in water) with 0.375 μl of the reservoir solution which contained 100 mM ammonium acetate, 100 mM bis-Tris pH 5.5 and 17% PEG 10K. For data collection, the reservoir solution was exchanged with 100 mM ammonium acetate, 100 mM bis-Tris pH 5.5 and 30% PEG 10K and the crystals were allowed to equilibrate overnight. Crystals were harvested and flash frozen in liquid nitrogen. A low resolution data set was collected on an in-house rotating anode source at 2.45 Å and processed using HKL2000 (34). Molecular replacement was carried out using the coordinates, pdb ID code: 3K2P of the RNase H domain, with the program Phaser (35) and partially refined within PHENIX (36). This model was then used for molecular replacement against a higher resolution data set (2.05 Å) collected on the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. The higher resolution data were processed in HKL2000 and refined using PHENIX. The X-ray parameters are summarized in Table 2, and the structure has been deposited in the protein data bank under ID: 5DZM.
Table 2. Crystallographic data statistics.
| Crystallographic data statistics | ||
|---|---|---|
| Source | Rotating anode | (SER-CAT) 22-ID |
| unit cell | a = b = 56.21 Å, c = 132.61 Å; | a = b = 56.14 Å, c = 133.75 Å; |
| α = β = 90°, γ = 120° | α = β = 90°, γ = 120° | |
| Space Group | P32 | P32 |
| Resolution (Å) | 50.0–2.45 | 50–2.05 |
| # of observations | 66 064 | 155 656 |
| unique reflections | 17 190 | 29 211 |
| Rsym(%)(last shell)a | 6.1 (51.0) | 4.3 (38.0) |
| I/σI (last shell) | 15.7 (2.7) | 10.8 (2.6) |
| Mosaicity range | 1.26–1.31 | 0.22–0.43 |
| completeness(%) (last shell) | 99.9 (100.0) | 98.3 (79.6) |
| Refinement statistics | ||
| Rcryst(%)b | 17.8 | |
| Rfree(%)c | 22.5 | |
| # of waters | 227 | |
| Overall Mean B (Å) | ||
| Chains A, B, C, D | 27.2, 30.5, 27.6, 30.92 | |
| Solvent | 30.25 | |
| r.m.s. deviation from ideal values | ||
| bond length (Å) | 0.004 | |
| bond angle (°) | 0.83 | |
| dihedral angle (°) | 14.12 | |
| Ramachandran Statisticsd | ||
| residues in: | ||
| favored (98%) regions (%) | 98.7 | |
| allowed (>99.8%) regions (%) | 1.3 | |
a Rsym = ∑ (| Ii - < I>|) / ∑(Ii) where Ii is the intensity of the ith observation and <I> is the mean intensity of the reflection.
b Rcryst = ∑∥ Fo| - | Fc ∥ / ∑| Fo| calculated from working data set.
c Rfree was calculated from 5% of data randomly chosen not to be included in refinement.
d Ramachandran results were determined by MolProbity.
RESULTS
Stability of the RH monomer and homodimer
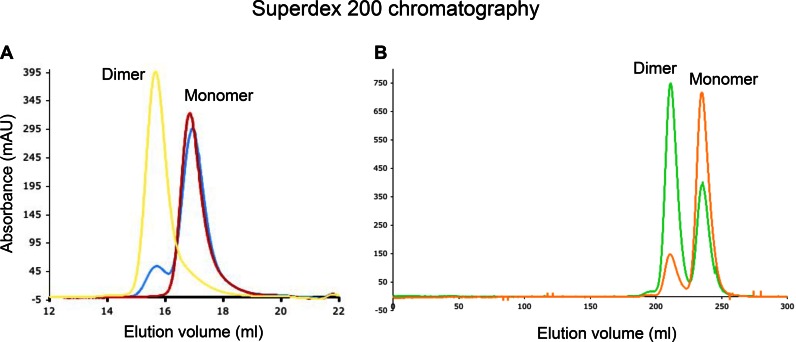
Purification of bacterially overexpressed RT RH domain with a size exclusion column yielded two major fractions for which the retention volumes were consistent with monomeric and dimeric forms. Since the RH domain lacks cysteine residues, the dimeric form is not dependent on formation of disulfide bonds, but reflects an alternate stabilization mechanism. An RNase H domain monomer construct, RHmnel, corresponding to p66 residues 427–560 and containing an MNEL leader sequence, was expressed and incubated overnight at either 25°C or 37°C. No significant equilibration to the dimer was observed at 25°C, while some dimer was present after the 37°C incubation (Figure 1A). NMR kinetic studies starting with the purified, Ile-labeled RHmnel dimer indicated a time dependent monomer formation at 37°C with a mean time constant of 6.3 days (Supplementary Figure S1).
Figure 1.
Stability of RH monomer and dimer. (A) Incubation of purified RHmnel monomer for a period of ∼20 h at 25°C yielded only the monomer (red curve); incubation of purified RHmnel monomer at 37°C over a similar period yielded a monomer – dimer mixture (blue curve). The position of the purified dimer corresponds to the yellow curve. Fractions were separated on a Superdex 200 10/300 column. (B) Purified samples of RHΔNT monomer (gold curve) or dimer (green) rapidly reloaded on the Superdex 200 column HiLoad 26/60, eluted as monomer-dimer mixtures. Chromatogrpahic separations were run at 4°C using 50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA as the running buffer.
We previously introduced RHΔNT as a model for the destabilized RH domain that is formed as a result of the transfer of N-terminal residues to the polymerase domain (21). In this construct, residues Tyr427 and Gln428 have been deleted and a Leu429Met mutation becomes the N-terminus. Expression of the RHΔNT construct also yielded monomer and dimer species, however, the monomer-dimer interconversion rate is much greater. As shown in Figure 1B, reloading either the monomer (orange curve) or the dimer (green curve) on a Superdex 200 HiLoad column yielded a monomer-dimer mixture after chromatographic separation. The greater stability of the RHmnel construct made it suitable for crystallization.
Crystallographic characterization
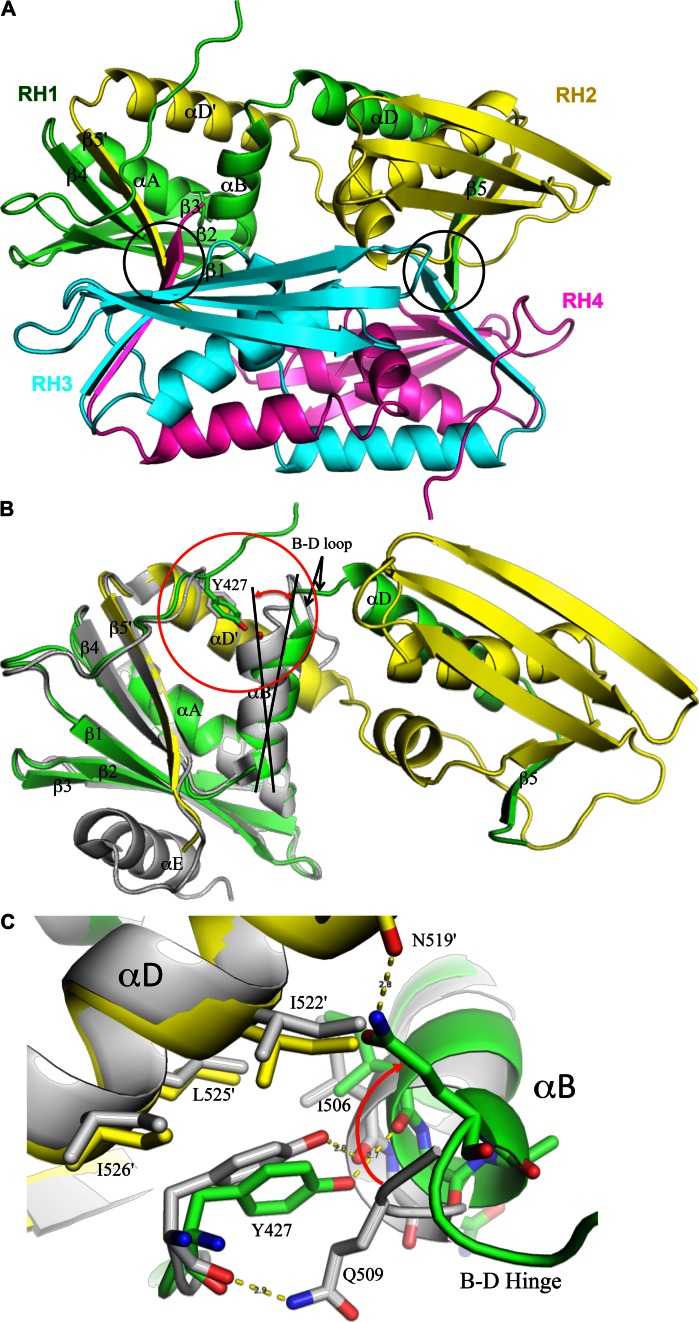
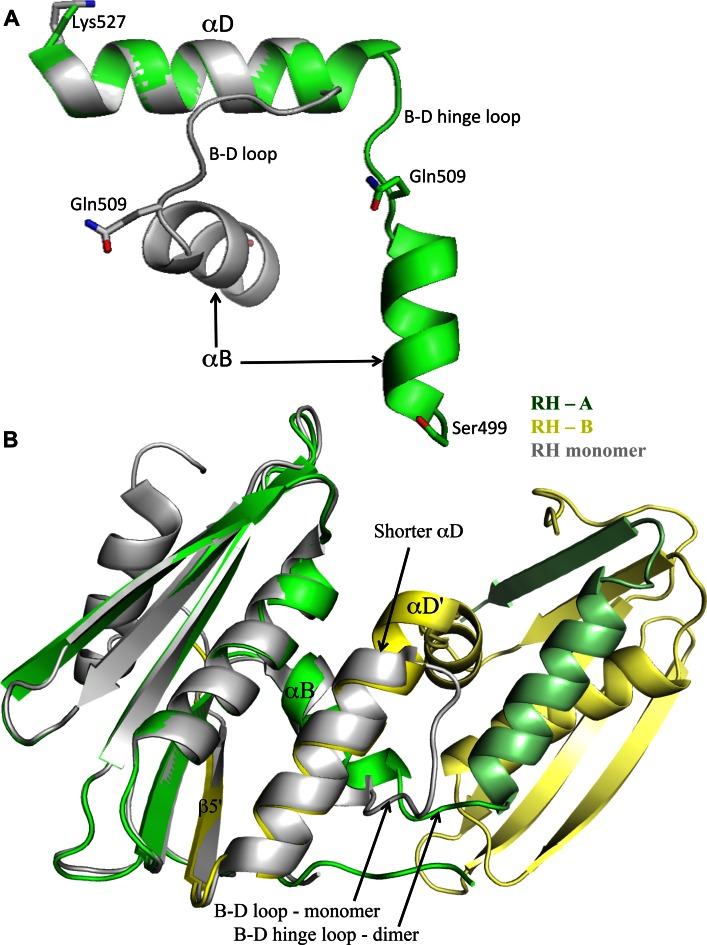
The purified RHmnel dimer was crystallized as described in Methods, and found to correspond to a domain swapped homodimer. The unit cell contains a pair of domain-swapped homodimers for a total of four molecules in the asymmetric unit, with the two dimers in a perpendicular orientation relative to the long axis of the dimer (Figure 2A). As illustrated in Figure 2B, the fold of each peptide follows the same path as the monomer from the N-terminus through helix B, after which the B-D loop acts as a hinge, connecting to helix αD and continuing through β5 in the second molecule of the dimer. The C-terminal helix αE was not observed in these structures, consistent with frequent observations that the αE helix is dynamic in solution (28,37–40). Comparison with the monomer structure indicates that there would be some steric conflict between residues at the N-terminus of helix E and the N-terminal segment arising from a molecule in the second dimer. Overall, the interactions of αD and β5 with the second molecule of the dimer mirror the corresponding interactions in the monomer. Combining elements β1-β2-β3-αA-β4-αB from one monomer with αD and β5 from its domain swapped partner forms a globular structure that overlays well with the isolated RH monomer (pdb: 3K2P, (41) (Figure 2B).
Figure 2.
Crystal structure of domain swapped Rnase H (RH). (A) The unit cell contains two pairs of domain swapped dimers: RH1-RH2 and RH3-RH4, in an approximately perpendicular orientation (color coding: green+yellow; cyan+magenta). Each globular structure contains elements β1-β2-β3-αA-β4-αB derived from a single peptide chain, and αD and β5 derived from its domain swapped partner. The circled regions correspond to the β-sheet formed from Trp535 residues that link the two domain swapped dimers. (B) Expanded ribbon representation of a domain swapped dimer (green, yellow) overlaid with an RH domain monomer structure (gray, pdb: 3K2P, 41). The β1-β2-β3-αA-β4 from molecule RH1 (green) and the αD-β5 structural elements from molecule RH2 (yellow) agree closely with the monomer, however, αB exhibits a relative change in orientation. (C) Expanded view of the monomer-dimer structural variation in the Tyr427 binding pocket. The Tyr427 sidechain remains H-bonded to Ile506 in both structures, however in RH2, αB is tilted even further so that this H-bond interaction is not present.
The two domain swapped dimers in the asymmetric unit interact via an anti-parallel β-sheet formed from the backbones of pairs of Trp535 residues on molecules RH1-RH3 and RH2-RH4. The additional stabilization of the tetramer by a number of hydrophobic and H-bond interactions suggests that this species might also be present in solution, however based on the chromatography data, it would correspond to a very small fraction of the total RH molecules at the concentration ranges studied.
Although the structure of the RH1-RH2 and RH3-RH4 domain swapped pairs is identical, small conformational differences were observed between the two RH molecules of each domain swapped pair (Supplementary Figure S2). This appears to result primarily from differences in the lengths and conformations of the B-D linkers, so that a slightly more extended linker in RH2 results in a displacement of C-terminal αD-β5 relative to the N-terminal β1-β2-β3-αA-β4-αB structural elements. The main variations in backbone dihedral angles occur near Pro510. This in turn produces a relative displacement of the two pseudo-monomer structures that form the dimer. This variability may result from constraints imposed by formation of the tetramer interface, or perhaps by other lattice contacts. In contrast, solution NMR studies of the Ile-labeled RH dimer did not reveal evidence of doubled resonances, as was also the case in the 1H-15N HSQC NMR studies reported by Christen et al. (24). Hence, it appears that either the structural differences between the two molecules of the homodimer are selected by lattice contacts, or the interconversion barriers between the two conformations are insufficient to result in slow exchange, which would be required for the observation of multiple resonances.
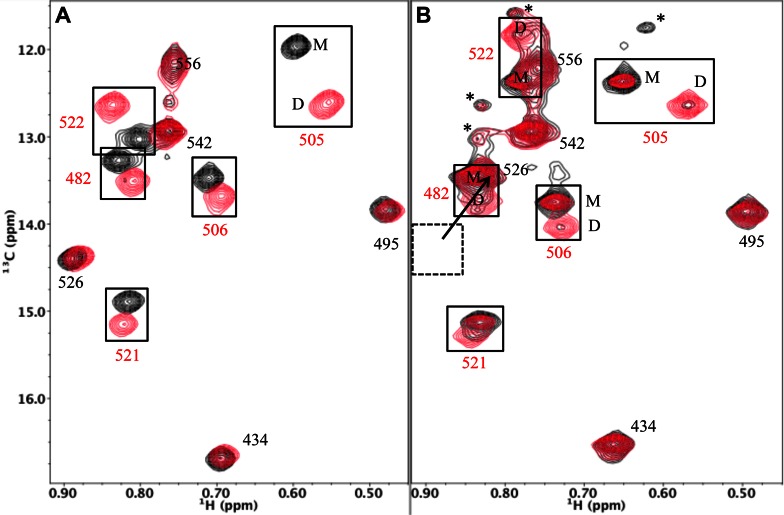
In contrast with the lack of resonance variations between the two molecules of the domain swapped dimer, there are significant differences between the resonances of the dimer and the RH monomer. Christen et al. (24) reported that 24 amide resonances showed significant monomer/dimer shift changes, and we observed that 5 of the 10 Ile δ-methyl resonances exhibit significant monomer-dimer shift variations (Figure 3). The perturbed residues are located on helix A (Ile482), helix B (Ile505, Ile506) and helix D (Ile522, Ile526). In both studies, the most significant shift variations correspond to residues in the helix B-helix D region of the protein.
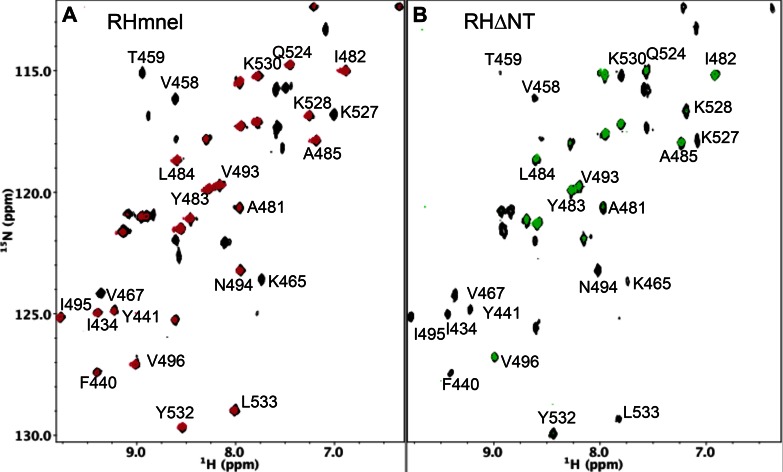
Figure 3.
1H-13C HMQC spectra of [13CH3-Ile]RH constructs. (A) 1H-13C HMQC spectra of [13CH3-Ile]RHmnel monomer (black) overlaid with 1H-13C HMQC spectra of [13CH3-Ile]RHmnel dimer (red). (B) 1H-13C HMQC spectra of [13CH3-Ile]RHΔNT monomer (black) overlaid with 1H-13C HMQC spectra of [13CH3-Ile]RHΔNT dimer (red). As a consequence of faster monomer-dimer interconversion rate of RHΔNT, purified monomer or dimer samples show some contamination with the dimer and monomer resonances, respectively. As a result of the absence of Tyr427, the Ile526 and Ile522 resonances are both shifted to lower δ13C and δ1H values, so that the Ile526 resonances overlap the Ile482 monomer resonance, and the Ile556 resonances partially overlap the Ile522 monomer peak. The spectra of [13CH3-Ile]RHΔNT contain several additional small resonances indicated with an asterisk that have not been assigned. Spectra were obtained at 25°C and samples were in Tris-HCl-d11 in D2O (pD 7.5), 100 mM KCl.
Monomer/dimer structural variations of helix B-helix D
A structural comparison of the domain swapped dimer with the monomer reveals several interesting variations, particularly involving helices B and D, that appear related to molecular stability and to the monomer-dimer interconversion process. As illustrated in Figure 2C, Tyr427 sits in a hydrophobic pocket bounded by αB on one side and αD on the other and thus contributes significantly to the interaction between the two helices. The interface with αD involves primarily hydrophobic contacts with Leu525 and other hydrophobic residues. In the monomer structure (pdb: 3K2P), interactions with αB include a hydrophobic interaction with the Gln509 sidechain involving the opposite face of the phenol ring, an H-bond of the Tyr427 carbonyl oxygen with the Gln509 amide sidechain, and an H-bond of the tyrosyl hydroxyl with the Ile506 carbonyl oxygen.
As indicated in Figure 2B, C, the orientation of αB differs in the two structures, exhibiting a relative tilt of ∼12°. An even greater orientational variation is present in the second (RH2) molecule of the dimer, which is probably the dominant species in solution (Supplementary Figure S2). In the domain swapped structure, this altered orientation of αB alters the Tyr427 binding pocket. The Tyr427 sidechain is pulled away from αD, and Gln509 no longer interacts with Tyr427 (Figure 2C). The H-bond between Tyr427 and Ile506 is maintained in one molecule of the dimer, but is weak or absent in the second dimer molecule. Since formation of the domain swapped structure involves separation of helices B and D, weakening of the Tyr427-αD interaction is predicted to facilitate this conversion. This prediction is supported by the kinetic measurements summarized below, indicating that the monomer-dimer interconversion is greatly accelerated in the RHΔNT construct that lacks both Tyr427 and Gln428.
A second significant monomer/dimer structural difference involves a transfer of residues from the B-D loop to extend the length of helix αD. In the RH monomer, seven loop residues, Gln509-Ser515, link helices B and D. In the domain swapped structure, the B-D loops become hinges and, although the hinge length differs slightly between the two molecules that constitute the domain swapped dimer, both hinge loops are considerably shorter than the monomer loop. The B-D hinge in RH1 includes only two residues, 510–511 and in RH2 includes residues 508–511. In both molecules of the dimer, αD is extended by a complete helical turn, and αB is also extended in RH1. The extension of αD in the dimer is illustrated by the monomer-dimer helix D overlay shown in Figure 4A, and reduction in the B-D linker length in the dimer is illustrated in Figure 4B. This comparison suggests that the longer B-D loop and helix unwinding present in the monomer is required to relieve conformational strain that is largely alleviated in the dimer. Similar relationships have been observed in other domain swapped proteins (27,42–46).
Figure 4.
Shortening of the B-D hinge loop. (A) Ribbon representations of the αB-αD regions of one molecule of the dimer (green) and the RH monomer (gray) in which the αD helices have been aligned. As is apparent from the figure, several residues that are part of the B-D loop in the monomer are incorporated into an extended αD in the dimer. The reduction in length of the dimer B-D linker is also apparent in Figure 2B. (B) Ribbon diagram overlay of the RH monomer (gray, pdb: 3K2P) with the domain swapped dimer illustrating the change in length of αD and the B-D linker.
Effect of N-terminal deletion on monomer-dimer interconversion kinetics
We previously introduced the RHΔNT construct as a model for the destabilized RH domain intermediate that results after the transfer of the N-terminal Tyr427-Gln428 residues to the polymerase domain (21). The structural comparison presented above indicates that this construct would be expected to exhibit an enhanced monomer-dimer interconversion rate, since one of the important contributors to αB-αD stability is eliminated. The 1H-13C HMQC spectrum of [13CH3-Ile]RHΔNT exhibits large perturbations of the Ile522 and Ile526 resonances corresponding to residues located near the Tyr427 binding pocket, as well as smaller but significant perturbations for several other Ile resonances, consistent with a more subtle, global structural perturbation. As a result of the faster monomer-dimer interconversion rate, even the initial spectrum obtained for the purified dimer sample exhibited significant monomer resonances (Figure 3B). The transfer of resonance intensity from dimer to monomer peaks for several Ile residues was determined starting from an initially purified RHΔNT dimer sample as described in Methods. The time constants and equilibrium dimer fractions obtained are summarized in Table 1 (See also Supplementary Figure S3). Time-dependent decay of the RHΔNT dimer was determined from intensity measurements of the Ile482, Ile505, Ile506, Ile521 and Ile522 resonances at 25°C, giving a mean time constant of 24.1 ± 2.0 min (Table 1). In contrast, equilibration of the RHmnel construct is a much slower process, exhibiting a mean decay time of 152 h determined at higher temperature (37°C) (Supplementary Figure S1) and indeed the crystallization results shown above are based on the isolation and crystallization of the long-lived RHmnel dimer. Since monomer-dimer inerconversion requires a substantial degree of unfolding, this result provides strong evidence that loss of only a few RH domain N-terminal residues can substantially increase the RH unfolding rate.
Effect of N-terminal deletion on H/D exchange
As shown previously (21), the 1H-15N HSQC spectrum of RHΔNT indicates that this construct retains the same general fold as the wt domain, and the Phe440 and Tyr441 amide resonances are still readily observed in this construct. In order to further evaluate the dynamic perturbation of RHΔNT, we performed H/D exchange studies as described in Methods. Figure 5 compares the 1H-15N HSQC spectra of RHmnel (panel A) and RHΔNT (panel B) obtained during the intervals indicated. During the first 40 min accumulation period, ∼57 out of 128 amide resonances remain in RHmnel, while ∼47 out of 128 resonances are observed in RHΔNT, although many of the peaks in the truncated construct are substantially weaker (Supplementary Figure S4). Amide resonances for cleavage site residues Phe440 and Tyr441 are readily observed in both spectra. These residues remain in a well-protected environment in RHmnel, and give strong signals after 6 h in the D2O buffer (Figure 5A). In sharp contrast, the Phe440 and Tyr441 amide resonances in the RHΔNT construct are no longer visible even over the second 40 min accumulation period. The amide groups on strand β5, residues 530–536, also show considerably less protection in the RHΔNT construct. The other protected amides are mostly within the helices, and in a few cases connect β1, β2, β3 and β4. These comparisons provide a graphic illustration of the significant dynamic instability and resulting solvent accessibility introduced by the loss of the N-terminal Tyr427 and Gln428 residues. The effect on the Phe440 and Tyr441 resonances indicates that this instability extends to the central β-sheet structure.
Figure 5.
H/D exchange behavior of RH and RHΔNT. (A) 1H-15N HSQC spectrum of RHmnel obtained during the first 40-min period after replacement of the H2O buffer with D2O buffer (black spectrum), overlaid with a spectrum obtained during the 40-min period starting at 6.6 h after buffer replacement (red spectrum). (B) 1H-15N HSQC spectra of RHΔNT obtained during the first 40-min accumulation period after buffer exchange (black spectrum) and the second 40-min period (green spectrum). The amides protons of the cleavage site residues F440 and Y441 in RHmnel are readily observed after 6.6 h, while in RHΔNT they have been fully exchanged. Exchange studies were performed at 25 °C in 25 mM Tris-HCl-d11 in D2O (pD 7.5), 100 mM KCl.
Tests of the linker length hypothesis
As outlined above, structural comparisons suggest that a B-D linker that is too short to support a stable αB-αD connection contributes to destabilization of the RH monomer and is a driving force for the unfolding required to form the domain swapped dimer. Similar relationships between linker length and dimer/monomer ratios have been observed in other studies of domain swapped proteins (25,27,42,44). In order to further evaluate this relationship for the RH domain, a construct containing an extended linker RH (RHΔNT-EL) was constructed by inserting an additional Pro-Asp-Gln sequence after Gln512, effectively allowing the repeated PDQ sequence to be in two places at once. The additional residues were inserted into the linker of the RHΔNT construct for this study because it rapidly equilibrates, facilitating determination of the equilibrium dimer/monomer ratio. No Ile resonances attributable to the dimer form of this extended linker construct were observed in the 1H-13C HMQC spectra of [13CH3-Ile]RHΔNT-EL, despite using a relatively high concentration of 450 μM and allowing 24 h for monomer/dimer equilibration at 37°C (Supplementary Figure S5A). This compares with a mean dimer fraction of 15% for RHΔNT based on five Ile resonance intensity ratios (Table 1). This result thus supports the hypothesis that the RH domain is destabilized by a B-D linker that is too short to support a more stable monomer fold.
As a second test of this hypothesis, we introduced an E514L mutation predicted to favor incorporation of linker residue 514 into helix αD, and thus to increase the relative stability of the dimer. To the extent that the mutated E514L residue becomes incorporated into an extended αD helix, the mutated residue comes in contact with Ile505 so that significant shift perturbations are predicted, and these were observed for both the monomer and dimer resonances in the spectra of RHΔNT(E514L) (Supplementary Figure S5B). Importantly, the dimer/monomer equilibrium ratio is shifted to ∼1.0, representing an increase from the value of ∼0.18 observed for RHΔNT. Thus, both mutational strategies altered the dimer/monomer ratio consistent with the predicted effects of the B-D linker.
The monomer-dimer transition
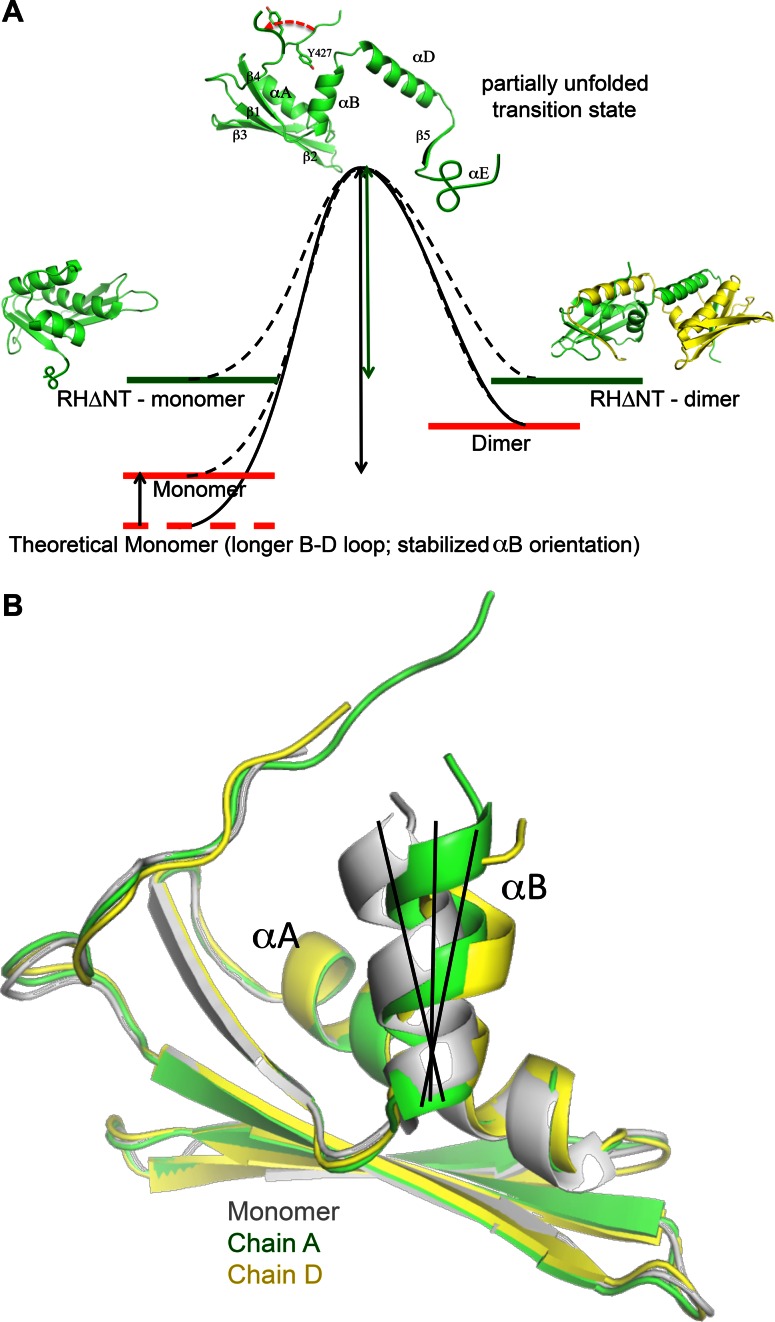
The studies summarized above lead to a qualitative monomer-dimer transition state energy diagram shown in Figure 6A. The transition state is proposed to involve separation of αB and αD as is observed in the domain swapped structure. Separation of the two helices also requires additional disorder since the interactions of β5, and αE with other structural elements will also be disrupted. Comparison of the RH monomer and dimer structures indicates that the monomer is characterized by several intrinsic destabilizing features. (i) The B-D linker creates strain in the monomer structure that is relaxed in the domain swapped dimer. The predicted relationship between the length of the B-D linker and the dimer/monomer ratio is supported by the mutation studies summarized in the previous section. (ii) Weak orientational constraints on αB destabilize the Tyr427 binding pocket. This helix lies on the surface of the domain and lacks sidechain H-bond interactions that can constrain the helix orientation more effectively than less specific hydrophobic interactions. This orientational instability is most clearly illustrated by an overlay of ribbon diagrams corresponding to residues 430–495 of the monomer and each of the two molecules (RH1 and RH2) of the dimer (Figure 6B). One consequence of this variability is that the Ile506-Tyr427 H-bond is maintained in only one of the two molecules of the dimer. The low orientational stability of αB significantly compromises the Tyr427 binding pocket, which includes many fewer interactions than are present in the monomer, and the tyrosyl sidechain is pulled away from stabilizing hydrophobic contacts with αD residues (Figure 2C). Thus, we conclude that the RH domain is intrinsically less stable than a theoretical domain containing a longer B-D loop and a more constrained αB helix. Nevertheless, it is important to emphasize that the destabilizing effects noted above are limited, since if the RH domain were too unstable it could have a tendency to unfold in the p66 monomer, which could lead to more extensive proteolytic processing prior to dimer formation, and hence to formation of low activity p51/p51′ homodimers.
Figure 6.
Monomer-dimer transition state diagram. (A) The monomer-dimer interconversion is suggested to proceed via a transition state that loosely resembles either molecule of the dimer. As outlined in the text, the monomer stability is proposed to be slightly compromised by intrinsic structural features, particularly the lack of constraints on αB orientation and the truncation of αD. Loss of several N-terminal residues resulting from the inter-domain tug-of-war further destabilizes the monomer. (B) Overlaid ribbon diagrams for residues 427–506 (i.e. showing β1- β2- β3-αA-β4-αB) in the monomer (gray) and each molecule of the domain swapped dimer (green, yellow). The molecules were aligned from residues 427–495 to illustrate the variable positioning of helix αB.
The absence of Tyr427 in the RHΔNT model construct clearly removes an important mediator of the αB-αD interaction, and is expected to facilitate separation of the B and D helices as is observed in the domain swapped dimer. The destabilizing effect of this truncation is apparent from the reduction in ΔG by 2.0 ± 0.4 kcal/mol determined from urea denaturation studies (21), the faster dimer decay rate of the domain swapped dimer (Table 1, Supplementary Figure S3), and the faster H/D exchange observed for this construct (Figure 5). In the isolated domain, the structure spends most of its time in either of the alternately folded forms, however in the p66/p66′ homodimer, the unfolded residues will be subject to many additional interactions so that refolding becomes much less probable.
Targeting RT maturation via the supernumerary RH domain
As outlined above and in previous studies (21,23), the mature RT p66/p51 heterodimer is formed from a p66/p66′ homodimer that contains two folded RH domains. Proteolysis of the supernumerary RH′ domain on the p66′ subunit is likely completed within the virion, so that the invading virus can release an active reverse transcription complex into the host cell (47). Based on the studies presented above and previously, RH′ domain proteolysis is a fairly slow process that is rate limited by RH′ unfolding and exposure of its buried proteolytic site. Stabilizing the supernumerary RH′ domain of the p66′ subunit thus represents one approach to inhibiting formation of the mature heterodimer. Several groups have described the development of active site-targeted RH inhibitors designed to interfere with the ribonuclease H activity in the p66 subunit (41,48,49). In principle, these inhibitors also would be expected to interfere with RH′ unfolding and hence to retard or block RT maturation.
We previously demonstrated that the active site-directed RH inhibitor 2-hydroxyisoquinoline-1,3(2H,4H)-dione (HIQ) (31) stabilizes RH helix E (28). The effect of Mg-HIQ on the H/D exchange behavior of the RHΔNT construct is shown in Figure 7A. Comparison of the initial 1H-15N HSQC spectra with those shown in Figure 5 indicates a significant increase in the number of observed amide resonances, consistent with stabilization of helix E as well as the entire active site region of the domain. More importantly HIQ dramatically reduces the rates of H/D exchange for most of the amide peaks. Thus, more amide resonances of the RHΔNT-Mg-HIQ complex remain after 800 min than are observed in the 1H-15N HSQC spectrum of RHΔNT in the absence of HIQ obtained over the second 40 min accumulation period (Figure 5B).
The effect of Mg-HIQ on maturation of the p66/p66′ homodimer was investigated using isoleucine labeled [13CH3-Ile,2H]p66, as described previously (23). A region of the 1H-13C HMQC spectra showing the Ile434, Ile329 and Ile375 δ-methyl resonances as a function of time after the initiation of dimerization conditions is shown in Figure 7B. As in our previous studies, resonances arising from the p66 subunit are labeled ‘E’ corresponding to the extended conformation, and resonances arising from the p66′ subunit that will develop into the p51 subunit are labeled ‘C’ corresponding to the compact conformation. These spectra were obtained under conditions similar to those recently used in a similar dimerization study (23), but with the addition of 6 mM MgCl2 and 1 mM HIQ. A comparison of the spectra shown in Figure 7B with those shown in Figure 7A of Zheng et al. (23), shows a significant lengthening of the time constants characterizing the disappearance of the RH′ Ile resonance (Ile434C) and the appearance of connection′ domain Ile resonances (Ile329C and Ile375C). In the previous study, the Ile434C resonance has largely disappeared and the Ile329C and Ile375C resonances are near their equilibrium intensities by 22 h. In the presence of Mg-HIQ, there is still significant intensity of the Ile434C resonance at 66 h and the Ile329 and Ile375 resonances have not yet attained their equilibrium intensity. Analysis of the time-dependent intensities of the Ile434C, Ile495C and Ile521C resonances arising from the RH′ domain gives a mean time constant of 81.2 ± 7.1 h, compared with a mean decay time constant of 6.5 h obtained previously (21). Thus, the Mg-HIQ significantly interferes with RH′ unfolding and with maturation of the connection′ domain in the p66′ subunit. We further note that the corresponding Ile434E, Ile329E and Ile375E resonances in the p66 subunit are near their equilibrium values by the second accumulation period (Figure 7B), as observed in the previous study. These results demonstrate that it is possible to target RT maturation with agents that stabilize the supernumerary RH′ domain.
DISCUSSION
Formation of the RT heterodimer from its p66 precursor protein has been one of the most poorly understood aspects of viral maturation (7–19). However, recent studies have provided structural information for the p66 monomer as well as time-dependent spectra for the p66/p66′ homodimer after initiation of conditions favoring dimerization that provide substantial insight into the maturation process (21–23). These studies demonstrate that the initially formed p66/p66′ homodimer is a structural heterodimer that contains two folded RH domains: a functional RH domain on the p66 subunit and a supernumerary RH′ domain on p66′. Labeled Ile resonances arising from RH′ gradually decay, while those arising from the p66 RH domain are approximately constant, consistent with a subunit-selective unfolding process that exposes the proteolytically labile site initially buried within the molecule. The unfolded RH′ domain will then be subject to proteolytic attack. Nevertheless, that pathway remains controversial and other recent NMR studies concluded that maturation models that involve a completely or predominantly unfolded RH domain are unlikely (8).
Domain swapped dimers provide a snapshot of a partially unfolded monomer which has become stabilized by formation of a dimeric structure with a second, similarly unfolded molecule. Such structures are generally considered to provide useful insights into protein folding and unfolding pathways (25–27,42–46), and thus can provide insight into the RT maturation process. The studies reported above indicate that the monomer and dimer forms of the isolated RH domain interconvert on a time scale of days at 37°C, but removal of the Tyr427-Gln428 residues accelerates the dimer decay rate to under 0.5 h. This acceleration is accompanied by large increases in the H/D exchange rates for many of the amide resonances.
Structural comparison with the RH monomer allows identification of features that facilitate RH domain unfolding, illuminating the basis for intermittent release of the N-terminal Tyr427 residue, so that it becomes available for incorporation into the contiguous polymerase domain as part of a molecular tug-of-war. Based on the results presented above, strain in the B-D linking segment that connects helices B and D, as well as orientational instability of helix B, destabilize the Tyr427 binding pocket, enhancing the frequency of breathing motions in which Tyr427 makes excursions out of this pocket. Conversely, loss of Tyr427 eliminates an important mediator of the αB-αD interaction that further destabilizes the RH monomer structure. Interestingly, Tyr427 also becomes displaced from its binding pocket in the reported structures of the HIV-1 – Escherichia coli RH chimera (3HYF, (49); 3QIO, (50)) due to conflicts with Ile and Trp sidechains derived from the grafted E. coli segment (Supplementary Figure S6).
In the p66 monomer, the excursion of Tyr427 from its binding pocket is presumably transient, limiting the destabilization effect. However, in the p66/p66′ homodimer, the polymerase′ and RH′ domains located on the p66′ subunit compete for Tyr427′ and other nearby residues (21,23). Incorporation of these residues in the polymerase′ domain competes with the rapid re-incorporation of these residues into RH′, so that the destabilized RH′ domain persists for a much more extended period. In isolation, the RH domain apparently undergoes reversible unfolding and refolding, alternating between monomer and dimer. In p66/p66′, the unfolded RH′ domain is subject to many additional interactions, so that it presumably exists as an ensemble of unfolded or partially folded structures in which the Phe440-Tyr441 cleavage site is exposed and hence sensitive to proteolytic cleavage. Based on the disappearance of the RH′ resonances in the heterodimer, this longer lifetime of the destabilized RH′ is sufficient to result in unfolding a significant fraction of the time. Nevertheless, this is a slow process with a time constant of ∼6.5 h under the conditions of the previous study. Dynamic processes on this time scale are generally observable only using time lapse spectroscopy or its equivalent, and will not be detected using shorter time scale NMR methods. Peng and Li (51) have described a model system for such a folding tug-of-war, and concluded that the folding of a protein can generate enough mechanical strain to unravel a second protein. Structural data for the domain swapped dimer summarized above suggest a somewhat less active process in the p66/p66′ homodimer that is dependent on intrinsic instability of the binding pocket for the RH′ N-terminal Tyr427′ residue combined with the tendency of the polymerase′ domain to exploit the availability of this residue.
These studies lend further support to the conclusion that RH′ domain unfolding is the slowest, rate-limiting step in maturation of the RT heterodimer, and suggest that agents that stabilize the RH′ domain represent a potential strategy for inhibiting RT maturation. In the present study, we have specifically demonstrated that this rate-limiting step can be targeted using an RH domain active site-directed HIQ ligand. Addition of HIQ to the isolated, N-terminal truncated RH domain dramatically reduces H/D exchange rates, and addition of HIQ to an Ile-labeled p66 sample after initiation of conditions favoring dimerization greatly reduces the decay rates of the RH′ resonances. This study represents the first demonstration of the feasibility of targeting RT maturation rather than directly inhibiting RT heterodimer function. Inhibition of RT maturation can potentially interfere with RT function in several ways: (i) the polymerase activity of the p66/p66′ homodimer is lower than that of the RT heterodimer (52); (ii) the supernumerary RH′ domain can potentially interfere with interactions of RT with other cellular proteins; and (iii) failure of RH′ to unfold limits the irreversible proteolysis step, allowing dissociation back to the monomer form.
In summary, these studies provide independent evidence for the molecular tug-of-war that has been proposed to explain the subunit-selective unfolding of the RH′ domain (21), and for the central role of Tyr427 in determining the outcome. Further evaluations of the effectiveness and feasibility of targeting viral maturation will require infectivity studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH and National Institute of Environmental Health Sciences (NIEHS). The authors are grateful to Dr. Koteppa Pari, Dr. Thomas Transue and Dr. Juno Krahn for useful suggestions and input, and to Dr. Robin Stanley and Dr. Bret Freudenthal for critical reading of the manuscript.
FUNDING
Intramural Research Program of the NIH and National Institute of Environmental Health Sciences (NIEHS) [Research Project Number Z01-ES050147 to R.E.L.]; E.F.D. is supported by NIH and NIEHS under delivery order HHSN273200700046U; L.C.P. is supported by Research Project Number ZIA ES102645. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. Funding for open access charge: Intramural Research Program of the NIH and National Institute of Environmental Health Sciences (NIEHS) [Research Project Number Z01-ES050147 to R.E.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Sarafianos S.G., Marchand B., Das K., Himmel D.M., Parniak M.A., Hughes S.H., Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herschhorn A., Hizi A. Retroviral reverse transcriptases. Cell. Mol. Life. Sci. 2010;67:2717–2747. doi: 10.1007/s00018-010-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jochmans D. NovelHIV-1 reverse transcriptase inhibitors. Virus Res. 2008;134:171–185. doi: 10.1016/j.virusres.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Ibe S., Sugiura W. Clinical significance of HIV reverse-transcriptase inhibitor-resistance mutations. Future Microbiol. 2011;6:295–315. doi: 10.2217/fmb.11.7. [DOI] [PubMed] [Google Scholar]
- 5.Menendez-Arias L. Molecular basis of human immunodeficiency virus drug resistance: an update. Antiviral Res. 2010;85:210–231. doi: 10.1016/j.antiviral.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Ren J., Stammers D.K. Structural basis for drug resistance mechanisms for non-nucleoside inhibitors of HIV reverse transcriptase. Virus Res. 2008;134:157–170. doi: 10.1016/j.virusres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava S., Sluis-Cremer N., Tachedjian G. Dimerization of human immunodeficiency virus type 1 reverse transcriptase as an antiviral target. Curr. Pharm. Des. 2006;12:1879–1894. doi: 10.2174/138161206776873590. [DOI] [PubMed] [Google Scholar]
- 8.Sharaf N.G., Poliner E., Slack R.L., Christen M.T., Byeon I.J., Parniak M.A., Gronenborn A.M., Ishima R. The p66 immature precursor of HIV-1 reverse transcriptase. Proteins. 2014;82:2343–2352. doi: 10.1002/prot.24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma S.K., Fan N.S., Evans D.B. Human-Imunodeficiency-Virus Type-1 (Hiv-1) recombinant reverse-transcriptase - asymmetry in P66 subunits of the P66/P66 homodimer. FEBS Lett. 1994;343:125–130. doi: 10.1016/0014-5793(94)80303-x. [DOI] [PubMed] [Google Scholar]
- 10.Jacobo-Molina A., Arnold E. HIV reverse-transcriptase structure-function-relationships. Biochemistry. 1991;30:6351–6361. doi: 10.1021/bi00240a001. [DOI] [PubMed] [Google Scholar]
- 11.Sluis-Cremer N., Arion D., Abram M.E., Parniak M.A. Proteolytic processing of an HIV-1 pol polyprotein precursor: insights into the mechanism of reverse transcriptase p66/p51 heterodimer formation. Int. J. Biochem. Cell Biol. 2004;36:1836–1847. doi: 10.1016/j.biocel.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Tomasselli A.G., Sarcich J.L., Barrett L.J., Reardon I.M., Howe W.J., Evans D.B., Sharma S.K., Heinrikson R.L. Human-Immunodeficiency-Virus Type-1 reverse-transcriptase and ribonuclease-H as substrates of the viral protease. Protein Sci. 1993;2:2167–2176. doi: 10.1002/pro.5560021216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venezia C.F., Meany B.J., Braz V.A., Barkley M.D. Kinetics of association and dissociation of HIV-1 reverse transcriptase subunits. Biochemistry. 2009;48:9084–9093. doi: 10.1021/bi9010495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abram M.E., Parniak M.A. Virion instability of human immunodeficiency virus type 1 reverse transcriptase (RT) mutated in the protease cleavage site between RT p51 and the RT RNase H domain. J. Virol. 2005;79:11952–11961. doi: 10.1128/JVI.79.18.11952-11961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe D.M., Aitken A., Bradley C., Darby G.K., Larder B.A., Powell K.L., Purifoy D.J.M., Tisdale M., Stammers D.K. HIV-1 reverse-transcriptase - crystallization and analysis of domain-structure by limited proteolysis. Biochemistry. 1988;27:8884–8889. doi: 10.1021/bi00425a002. [DOI] [PubMed] [Google Scholar]
- 16.Braz V.A., Holladay L.A., Barkley M.D. Efavirenz binding to HIV-1 reverse transcriptase monomers and dimers. Biochemistry. 2010;49:601–610. doi: 10.1021/bi901579y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Restle T., Muller B., Goody R.S. Dimerization of Human-Immunodeficiency-Virus Type-1 reverse-transcriptase - a target for chemotherapeutic intervention. J. Biol. Chem. 1990;265:8986–8988. [PubMed] [Google Scholar]
- 18.Divita G., Rittinger K., Geourjon C., Deleage G., Goody R.S. Dimerization kinetics of HIV-1 and HIV-2 reverse-transcriptase - a 2-step process. J. Mol. Biol. 1995;245:508–521. doi: 10.1006/jmbi.1994.0042. [DOI] [PubMed] [Google Scholar]
- 19.Davies J.F., Hostomska Z., Hostomsky Z., Jordan S.R., Matthews D.A. Crystal-structure of the ribonuclease-H domain of HIV-1 reverse-transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 20.Hostomska Z., Matthews D.A., Davies J.F., Nodes B.R., Hostomsky Z. Proteolytic release and crystallization of the Rnase-H domain of Human-Immunodeficiency-Virus Type-1 reverse-transcriptase. J. Biol. Chem. 1991;266:14697–14702. [PubMed] [Google Scholar]
- 21.Zheng X., Pedersen L.C., Gabel S.A., Mueller G.A., Cuneo M.J., DeRose E.F., Krahn J.M., London R.E. Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation. Nucleic Acids Res. 2014;42:5361–5377. doi: 10.1093/nar/gku143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng X.H., Mueller G.A., Cuneo M.J., DeRose E.F., London R.E. Homodimerization of the p51 subunit of HIV-1 reverse transcriptase. Biochemistry. 2010;49:2821–2833. doi: 10.1021/bi902116z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng X., Perera L., Mueller G.A., DeRose E.F., London R.E. Asymmetric conformational maturation of HIV-1 reverse transcriptase. Elife. 2015;4:e06359. doi: 10.7554/eLife.06359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christen M.T., Menon L., Myshakina N.S., Ahn J., Parniak M.A., Ishima R. Structural basis of the allosteric inhibitor interaction on the HIV-1 reverse transcriptase RNase H domain. Chem. Biol. Drug Des. 2012;80:706–716. doi: 10.1111/cbdd.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding F., Prutzman K.C., Campbell S.L., Dokholyan N.V. Topological determinants of protein domain swapping. Structure. 2006;14:5–14. doi: 10.1016/j.str.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Byeon I.J., Louis J.M., Gronenborn A.M. A captured folding intermediate involved in dimerization and domain-swapping of GB1. J. Mol. Biol. 2004;340:615–625. doi: 10.1016/j.jmb.2004.04.069. [DOI] [PubMed] [Google Scholar]
- 27.Rousseau F., Schymkowitz J.W., Itzhaki L.S. The unfolding story of three-dimensional domain swapping. Structure. 2003;11:243–251. doi: 10.1016/s0969-2126(03)00029-7. [DOI] [PubMed] [Google Scholar]
- 28.Zheng X., Mueller G.A., Derose E.F., London R.E. Metal and ligand binding to the HIV-RNase H active site are remotely monitored by Ile556. Nucleic Acids Res. 2012;40:10543–10553. doi: 10.1093/nar/gks791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pari K., Mueller G.A., DeRose E.F., Kirby T.W., London R.E. Solution structure of the RNase H domain of the HIV-1 reverse transcriptase in the presence of magnesium. Biochemistry. 2003;42:639–650. doi: 10.1021/bi0204894. [DOI] [PubMed] [Google Scholar]
- 30.Becerra S.P., Clore G.M., Gronenborn A.M., Karlstrom A.R., Stahl S.J., Wilson S.H., Wingfield P.T. Purification and characterization of the Rnase H domain of HIV-1 reverse-transcriptase expressed in recombinant Escherichia Coli. FEBS Lett. 1990;270:76–80. doi: 10.1016/0014-5793(90)81238-j. [DOI] [PubMed] [Google Scholar]
- 31.Billamboz M., Bailly F., Barreca M.L., De Luca L., Mouscadet J.F., Calmels C., Andreola M.L., Witvrouw M., Christ F., Debyser Z., et al. Design, synthesis, and biological evaluation of a series of 2-hydroxyisoquinoline-1,3(2H,4H)-diones as dual inhibitors of human immunodeficiency virus type 1 integrase and the reverse transcriptase RNase H domain. J. Med. Chem. 2008;51:7717–7730. doi: 10.1021/jm8007085. [DOI] [PubMed] [Google Scholar]
- 32.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. Nmrpipe - a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Johnson B.A., Blevins R.A. Nmr View - a computer-program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers R., Clore G.M., Stahl S.J., Wingfield P.T., Gronenborn A. Analysis of the backbone dynamics of the ribonuclease-H domain of the Human-Immunodeficiency-Virus reverse-transcriptase using N-15-relaxation measurements. Biochemistry. 1992;31:9150–9157. doi: 10.1021/bi00153a006. [DOI] [PubMed] [Google Scholar]
- 38.Mueller G.A., Pari K., DeRose E.F., Kirby T.W., London R.E. Backbone dynamics of the RNase H domain of HIV-1 reverse transcriptase. Biochemistry. 2004;43:9332–9342. doi: 10.1021/bi049555n. [DOI] [PubMed] [Google Scholar]
- 39.Kern G., Handel T., Marqusee S. Characterization of a folding intermediate from HIV-1 ribonuclease H. Protein Sci. 1998;7:2164–2174. doi: 10.1002/pro.5560071014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powers R., Clore G.M., Bax A., Garrett D.S., Stahl S.J., Wingfield P.T., Gronenborn A.M. Secondary structure of the ribonuclease H domain of the human immunodeficiency virus reverse transcriptase in solution using three-dimensional double and triple resonance heteronuclear magnetic resonance spectroscopy. J. Mol. Biol. 1991;221:1081–1090. doi: 10.1016/0022-2836(91)90920-2. [DOI] [PubMed] [Google Scholar]
- 41.Himmel D.M., Maegley K.A., Pauly T.A., Bauman J.D., Das K., Dharia C., Clark A.D., Ryan K., Hickey M.J., Love R.A., et al. Structure of HIV-1 reverse transcriptase with the Inhibitor beta-Thujaplicinol bound at the RNase H active site. Structure. 2009;17:1625–1635. doi: 10.1016/j.str.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rousseau F., Schymkowitz J., Itzhaki L.S. Implications of 3D domain swapping for protein folding, misfolding and function. Adv. Exp. Med. Biol. 2012;747:137–152. doi: 10.1007/978-1-4614-3229-6_9. [DOI] [PubMed] [Google Scholar]
- 43.O'Neill J.W., Kim D.E., Johnsen K., Baker D., Zhang K.Y. Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus. Structure. 2001;9:1017–1027. doi: 10.1016/s0969-2126(01)00667-0. [DOI] [PubMed] [Google Scholar]
- 44.Schymkowitz J.W.H., Rousseau F., Wilkinson H.R., Friedler A., Itzhaki L.S. Observation of signal transduction in three-dimensional domain swapping. Nat. Struct. Biol. 2001;8:888–892. doi: 10.1038/nsb1001-888. [DOI] [PubMed] [Google Scholar]
- 45.Newcomer M.E. Protein folding and three-dimensional domain swapping: a strained relationship. Curr. Opin. Struct. Biol. 2002;12:48–53. doi: 10.1016/s0959-440x(02)00288-9. [DOI] [PubMed] [Google Scholar]
- 46.Dehouck Y., Biot C., Gilis D., Kwasigroch J.M., Rooman M. Sequence-structure signals of 3D domain swapping in proteins. J. Mol. Biol. 2003;330:1215–1225. doi: 10.1016/s0022-2836(03)00614-4. [DOI] [PubMed] [Google Scholar]
- 47.Warrilow D., Stenzel D., Harrich D. Isolated HIV-1 core is active for reverse transcription. Retrovirology. 2007;4:77. doi: 10.1186/1742-4690-4-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Billamboz M., Bailly F., Lion C., Touati N., Vezin H., Calmels C., Andreola M.L., Christ F., Debyser Z., Cotelle P. Magnesium Chelating 2-Hydroxyisoquinoline-1,3(2H,4H)-diones, as Inhibitors of HIV-1 Integrase and/or the HIV-1 Reverse Transcriptase Ribonuclease H Domain: Discovery of a Novel Selective Inhibitor of the Ribonuclease H Function. J. Med. Chem. 2011;54:1812–1824. doi: 10.1021/jm1014692. [DOI] [PubMed] [Google Scholar]
- 49.Kirschberg T.A., Balakrishnan M., Squires N.H., Barnes T., Brendza K.M., Chen X.W., Eisenberg E.J., Jin W.L., Kutty N., Leavitt S., et al. RNase H active site inhibitors of Human Immunodeficiency Virus Type 1 reverse transcriptase: design, biochemical activity, and structural information. J. Med. Chem. 2009;52:5781–5784. doi: 10.1021/jm900597q. [DOI] [PubMed] [Google Scholar]
- 50.Lansdon E.B., Liu Q., Leavitt S.A., Balakrishnan M., Perry J.K., Lancaster-Moyer C., Kutty N., Liu X.H., Squires N.H., Watkins W.J., et al. Structural and binding analysis of pyrimidinol carboxylic acid and N-Hydroxy quinazolinedione HIV-1 RNase H inhibitors. Antimicrob. Agents Chemother. 2011;55:2905–2915. doi: 10.1128/AAC.01594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng Q., Li H.B. Direct observation of tug-of-war during the folding of a mutually exclusive protein. J. Am. Chem. Soc. 2009;131:13347–13354. doi: 10.1021/ja903480j. [DOI] [PubMed] [Google Scholar]
- 52.Fletcher R.S., Holleschak G., Nagy E., Arion D., Borkow G., Gu Z., Wainberg M.A., Parniak M.A. Single-step purification of recombinant wild-type and mutant HIV-1 reverse transcriptase. Protein Expr. Purif. 1996;7:27–32. doi: 10.1006/prep.1996.0004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.