Abstract
A number of studies have demonstrated that various components of the ATRX/DAXX/Histone H3.3 complex are important for heterochromatin silencing at multiple genomic regions. We provide an overview of the individual components (ATRX, DAXX and/or H3.3) tested in each study and propose a model where the ATRX/DAXX chaperone complex deposits H3.3 to maintain the H3K9me3 modification at heterochromatin throughout the genome.
INTRODUCTION
The DNA in eukaryotic cells is wrapped around histones which can be post-translationally modified to regulate transcription. Large genomes, such as the mouse and human genomes, are replete with repetitive sequences including tandem repeats at centromeres and telomeres, and interspersed repeats (e.g. LINEs, SINEs, Alu repeats and ERVs). In order to prevent aberrant transcription, these genomic repeats are packaged into constitutive heterochromatin, a condensed chromatin structure which restricts accessibility to transcriptional machinery. Aberrant transcription of these repeats can compromise genome integrity and maintaining the heterochromatic architecture at these sites is critical to genome stability (1). The passage of polymerases during DNA replication or transcription poses a threat to chromatin and the timely restoration of nucleosomes is important for maintaining epigenetic memory (2).
The replication of DNA during S-phase is accompanied by the deposition of newly synthesised histones as chromatin is rapidly reassembled behind the replication fork (3). The canonical histones, H3.1/H3.2, are specifically expressed during this period and incorporated into chromatin in a replication-coupled manner. In contrast, the histone variant H3.3 is expressed throughout the cell cycle and deposition of this variant is replication-independent (4–6). In addition to the distinct expression patterns, H3.3 differs from the canonical H3.1/H3.2 counterparts at five amino acid residues. These substitutions in H3.3 mediate interactions with chaperone complexes which are unique to H3.3 (7,8) and facilitate the replication-independent deposition of this variant (4). Two major H3.3-specific chaperones have been identified; the HIRA complex which deposits H3.3 at genic regions (5,6) and the ATRX/DAXX complex which is responsible for H3.3 deposition at repetitive regions of the genome (9–11).
The association of H3.3 with heterochromatin was first reported at the telomeres of mouse ES cells (12). This was closely followed by the identification of a chaperone complex comprised of ATRX and DAXX (9,10). ATRX is a chromatin remodeller which is thought to localise to telomeres via interactions with G-quadruplex secondary structures (13,14) while DAXX is an H3.3-specific chaperone (8). Both components of the ATRX/DAXX complex were found to be essential for the deposition of H3.3 at telomeres (9,10) and pericentric heterochromatin (PCH; 11) though the functional significance of this pathway was unclear. More recent studies have now demonstrated that ATRX/DAXX mediated deposition of H3.3 is not limited to telomeric and pericentric repeats but occurs at heterochromatin distributed throughout the genome of mouse ES cells (15–18), including endogenous retroviral repeats (ERVs; 16–18), imprinted differentially methylated regions (DMRs) and selected intragenic methylated CpG islands (CGIs; 15). ATRX/DAXX/H3.3 enrichment at these sites coincides with the H3K9me3 heterochromatin modification and disruption of ATRX, DAXX or H3.3 led to a loss of H3K9me3 at these regions (15,16,18–20). This review will consolidate the major findings of these studies and outline the evidence to support a model where ATRX/DAXX deposits H3.3 to maintain H3K9me3 heterochromatin in the genome.
The ATRX/DAXX complex localises to heterochromatin
The interaction between the ATRX/DAXX complex and histone H3.3 was first described at telomeres and PCH (9–11). These highly repetitive regions of the genome are archetypal constitutive heterochromatin, and well documented as being enriched for H3K9me3, H4K20me3 and DNA methylation. More recent ChIP-seq studies have demonstrated that ATRX binding sites across the genome are generally associated with heterochromatic modifications (H3K9me3, H4K20me3, DNA methylation; 15). Further supporting the idea that ATRX localises to heterochromatin, a number of studies detected ATRX (15–18) and DAXX (16,18) enrichment at ERV repeats, particularly at IAP repeats (Intracisternal A particle; 16,18). ERVs are transposable elements which are known to be modified as silent heterochromatin, as aberrant expression results in genome instability (1). Consistent with this, ATRX/DAXX binding at ERVs coincided with enrichment of H3K9me3 and the co-repressor complex KAP1 (aka Trim28) and SETDB1 (aka ESET; 16,17), a lysine methyltransferase which catalyses H3K9me3. It is likely that the heterochromatin modifications at these regions direct ATRX binding as the ADD domain of ATRX has been demonstrated to specifically recognise H3K9me3 (21–23). This interaction has been confirmed at the cellular level where ATRX localisation to PCH was demonstrated to be dependent on H3K9me3 catalysed by SUV39H (23). Furthermore the monoallelic localisation of ATRX to the methylated, heterochromatic allele of imprinted DMRs (15) demonstrates that chromatin modifications play an important role in directing ATRX localisation.
ATRX/DAXX is required for H3.3 deposition at heterochromatin
The ATRX/DAXX complex is known to be important for deposition of H3.3 at telomeres and PCH. ChIP-seq of endogenous H3.3 in WT, ATRX KO (15,16) and DAXX KO (16) cells demonstrated that this was also true for the methylated allele of imprinted genes (15) as well as certain tandem repeats and intragenic CGIs (15). H3.3 was found to be generally enriched at ATRX binding sites in WT cells and this enrichment was lost in ATRX KO (15,16) and DAXX KO cells (16). H3.3 was also found to be enriched at IAP/ERV retrotransposons (15–17) although there is some disagreement about the requirement of ATRX/DAXX for H3.3 deposition at these sites. Two studies reported that ATRX/DAXX KO results in loss of H3.3 at IAP/ERVs (15,16) while one study reported increased H3.3 at IAP/ERVs in ATRX KO cells (17). The primary difference between these studies is that ChIP was performed either against endogenous H3.3 (15,16) or an YFP-tagged H3.3 (9).
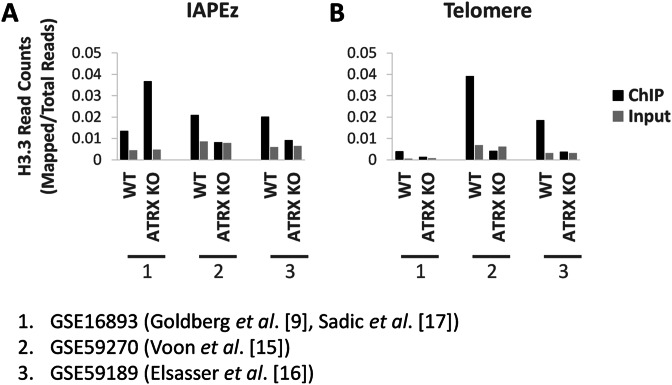
We have eliminated bioinformatics analysis as a possible variable by using a single method to re-align all three data sets. Similar to Sadic et al. (17), when we mapped the YFP-tagged H3.3 data set (9) to IAPEz repeats (Figure 1), we detected a gain of YFP-H3.3 at this ERV in ATRX KO cells. However, the two H3.3 data sets generated using an antibody against endogenous H3.3 (15,16) did not recapitulate this result and H3.3 enrichment at IAPEz ERVs was consistently lost in ATRX KO cells (Figure 1). H3.3 ChIP-seq reads from all three data sets were also mapped to the telomeres as a control. ATRX KO led to decreased H3.3 incorporation at telomeres in all three data sets although telomere enrichment was clearer for endogenous H3.3, compared to the YFP-tagged H3.3, in normal cells (Figure 1). Several lines of evidence appear to support ChIP data sets of endogenous H3.3. The relative enrichment profile of H3.3 at ERVs compared to telomeres in the endogenous H3.3 ChIP data sets are in agreement with immunofluorescence analyses (10,12). In addition, upregulated IAP expression was detected in both ATRX KO (17) and H3.3 KO cells (16; see below), supporting a role for ATRX/DAXX-dependent deposition of H3.3 at ERVs, in agreement with the endogenous H3.3 ChIP-seq data.
Figure 1.
Raw read files of YFP-H3.3 ChIP (9), endogenous H3.3 ChIP (15,16) and matched input sequencing from WT and ATRX KO cells were downloaded from GEO (35,36) and mapped to repeats with Repeat Enrichment Estimator (37). Samples were normalised for total read counts by dividing mapped reads against total mappable reads. Results show normalised reads counts of H3.3 ChIP and matched input samples for each data set at (A) IAPEz repeats and (B) telomere repeats. YFP-H3.3 was increased at IAPEz repeats in ATRX KO relative to WT cells. Endogenous H3.3 was decreased at IAPEz repeats in ATRX KO relative to WT cells. All three data sets showed decreased H3.3/YFP-H3.3 at telomeres in ATRX KO relative to WT cells.
It is possible that the discrepancy between the endogenous H3.3 and YFP-H3.3 has arisen due to the introduction of the YFP tag. For example, H3.3 turnover has been demonstrated to be important for regulation (24) and the YFP tag may interfere with this process. However, it is clear that further experiments are required to reconcile the discrepancies between these data sets. Future analysis using a different epitope tag, a different antibody or H3.3 K9 mutant cells would be useful for clarifying the differences between these studies. Nonetheless, when taken together, these results suggest that ATRX/DAXX localises to selected heterochromatic regions in the genome and deposits H3.3. This pathway has now been extended from telomeres and PCH to also include methylated imprinted DMRs, ERVs/IAPs and selected short tandem repeats.
ATRX/DAXX mediated deposition of H3.3 is required for maintaining H3K9me3
In addition to the loss of H3.3, disruption of the ATRX/DAXX complex in mouse ES cells also led to the loss of H3K9me3 at some genomic regions. ATRX KO led to a reduction in H3K9me3 at methylated imprinted DMRs, intragenic CGIs (15) and IAP repeats (16) as detected by ChIP-qPCR and ChIP-seq. Similarly, a reduction in H3K9me3 was also detected at IAPs (16) and telomeres (18) in DAXX KO cells. In addition, direct knockout of H3.3 also led to decreased H3K9me3 at IAPs (16) and telomeres (19,20). ChIP-reChIP assays of H3.3 and H3K9me3 on a genome-wide basis (16) and specifically at telomeres (19) demonstrated that H3.3 at these heterochromatic sites were directly modified with K9me3 (H3.3 K9me3). Overall, these data support a model where ATRX/DAXX mediates deposition of H3.3 which can be modified with K9me3 to facilitate the maintenance of heterochromatin.
SUV39H and SETDB1 catalyse K9me3 on H3.3
The H3K9me3 modification is catalysed by SUV39H1/2 at telomeres and PCH (25,26) while SETDB1 acts in a complex with KAP1 to catalyse H3K9me3 at ERVs (27,28) and methylated imprinted DMRs (28,29). The KAP1/SETDB1 complex was found to co-localise with ATRX at ERVs (17) and generally with ATRX/DAXX/H3.3/H3K9me3 across the genome (16). Consistent with previous studies, depletion of SETDB1 led to reduced H3K9me3 at IAPs/ERVs (16,17) and also at telomeres (19), albeit to a lesser degree. A direct interaction between DAXX and KAP1 has been identified by co-immunoprecipitation (16) and suggests that DAXX contributes to KAP1 binding at some genomic sites. Although KRAB-ZNFs, such as ZFP809 at ERVs (30) and ZFP57 at imprinted genes (29,31), are known to be the main determinant for KAP1 localisation, it is possible that DAXX may help to further stabilise these interactions. Overall, these studies suggest that the KAP1/SETDB1 complex is able to catalyse H3.3K9me3 at these sites. In addition, DAXX (18) and H3.3 (19) were also found to interact with SUV39H and depletion of SUV39H led to a loss in H3.3 K9me3 at telomeres (19). Combined, these results demonstrate that both SETDB1 and SUV39H are able to catalyse the trimethylation of lysine 9 on H3.3.
Disruption of ATRX/DAXX/H3.3K9me3 leads to aberrant transcription
Previous studies have shown that ATRX KO leads to increased TERRA transcription at telomeres (9) and this was verified by siRNA depletion of ATRX and DAXX (18). Knockout of histone H3.3 similarly led to increased TERRA transcription (19) confirming that the ATRX/DAXX deposition of H3.3 is important for suppressing telomere transcription. This has now been extended to other genomic loci; ATRX KO cells failed to silence a mini-IAP transgene (17) and resulted in aberrant allelic expression of imprinted genes (15) while H3.3 KO led to increased ERV transcription in mouse ES cells (16). It should be noted that multiple mechanisms may mediate transcriptional silencing at these genomic regions and the ATRX/DAXX/H3.3 complex is one of many heterochromatin factors which maintain silencing. The degree of transcriptional deregulation varies depending on which heterochromatin pathway is disrupted, however, aberrant transcription is a reliable indicator of alterations in the underlying chromatin structure. The importance of ATRX/H3.3 in heterochromatin maintenance is further demonstrated by the decrease in other silent chromatin modifications including H4K20me3 at telomeres (19) and DNA methylation at DMRs of imprinted genes (15). Taken together, these studies demonstrate that ATRX/DAXX deposition of H3.3 K9me3 is important for maintaining silent heterochromatin at a number of genomic sites.
CONCLUSIONS
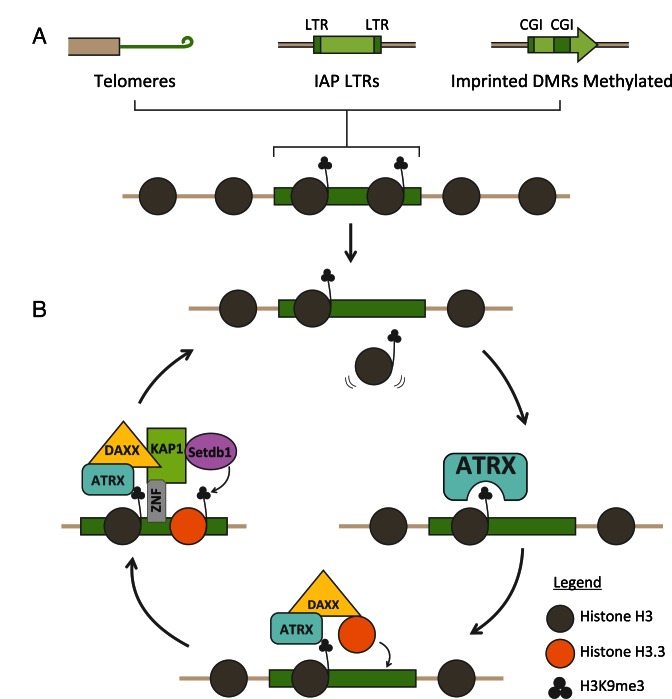
A substantial proportion of mammalian genomes is comprised of repetitive DNA which includes telomeres, centromeres and ERVs. Aberrant transcription of these repeats is deleterious to the organism and maintaining heterochromatic silencing at these repeats is critical for protecting genomic integrity. Chromatin modifications, such as DNA methylation, H3K9me3 and H4K20me3, are known to be important for maintaining silencing. In addition, a recent set of studies have now also identified the histone variant H3.3 as an important component of heterochromatin in mouse ES cells. The complex which deposits H3.3 at heterochromatin is comprised of DAXX, an H3.3 specific chaperone, and a chromatin remodeller, ATRX. This chaperone complex both targets H3.3 to specific heterochromatic sites and interacts with histone modifiers to maintain the K9me3 heterochromatin mark. The ADD domain of ATRX preferentially interacts with H3K9me3 which likely drives the localisation of ATRX/DAXX/H3.3 to heterochromatin modified regions including telomeres, ERVs and the methylated imprinted DMRs. DAXX is then able to interact with either the KAP1/SETDB1 co-repressor complex or SUV39H, both of which catalyse H3K9me3. ATRX/DAXX is therefore able to target heterochromatin for deposition of H3.3 and recruit methyltransferases to mediate direct modification of K9me3 on H3.3. This represents a self-reinforcing pathway which is able to protect heterochromatin integrity throughout the cell cycle (Figure 2).
Figure 2.
Model for ATRX/DAXX/H3.3 in maintaining heterochromatin. (A) Heterochromatic regions such as telomeres, IAP LTRs and methylated imprinted DMRs are distributed throughout the genome and enriched for H3K9me3. (B) ATRX recognises H3K9me3 and acts with DAXX to deposit H3.3 to replace histones which are lost. DAXX interacts with KAP1 and SETDB1 to catalyse K9me3 on newly deposited H3.3. ATRX/DAXX/H3.3 are able to act continuously through the cell cycle to ensure constant maintenance of H3K9me3 heterochromatin.
These studies have identified the ATRX/DAXX/H3.3 complex as an important contributor to heterochromatin silencing in mouse ES cells however, a number of outstanding questions remain. Perhaps the most puzzling of these is why H3.3 is required at all. One possible clue could lie in the replication-independent expression and incorporation of the H3.3 variant. This would suggest that ATRX/DAXX/H3.3 are particularly important outside of S-phase when the canonical histones are unavailable. Future analyses with highly quantitative time-lapse imaging experiments would provide useful information of the dynamics of H3.3, including turnover and deposition of newly synthesised H3.3, and recruitment of other chromatin repressors at these sites. Furthermore, while it is evident that depletion of ATRX/DAXX/H3.3 leads to loss of heterochromatin, the overall phenotypic consequences remain unclear. This is particularly pertinent to ALT cancers where ATRX mutations have been strongly linked to increased recombination at telomeres (32,33). Several studies have demonstrated that disruption of heterochromatin facilitates aberrant telomere recombination (34). Detailed analyses of how heterochromatin structure, including H4K20me3 and DNA methylation, is disrupted in ATRX/DAXX/H3.3 deficient cells would provide insights into how this pathway might promote genome instability in cancer.
Acknowledgments
The authors wish to thank Joey Man for drawing Figure 2.
FUNDING
Australia Research Council (ARC) Future Fellowship Award from the ARC, Australia [to L.H.W.]. Funding for open access charge: Australia Research Council FT140100128.
Conflict of interest statement. None declared.
REFERENCES
- 1.Leung D.C., Lorincz M.C. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem. Sci. 2012;37:127–133. doi: 10.1016/j.tibs.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Jasencakova Z., Scharf A.N., Ask K., Corpet A., Imhof A., Almouzni G., Groth A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Groth A., Rocha W., Verreault A., Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad K., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 5.Ray-Gallet D., Quivy J.P., Scamps C., Martini E.M., Lipinski M., Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 6.Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 7.Daniel Ricketts M., Frederick B., Hoff H., Tang Y., Schultz D.C., Singh Rai T., Grazia Vizioli M., Adams P.D., Marmorstein R. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 2015;6:7711. doi: 10.1038/ncomms8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsasser S.J., Huang H., Lewis P.W., Chin J.W., Allis C.D., Patel D.J. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–565. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldberg A.D., Banaszynski L.A., Noh K.M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X., et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong L.H., McGhie J.D., Sim M., Anderson M.A., Ahn S., Hannan R.D., George A.J., Morgan K.A., Mann J.R., Choo K.H. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drane P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong L.H., Ren H., Williams E., McGhie J., Ahn S., Sim M., Tam A., Earle E., Anderson M.A., Mann J., et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Law M.J., Lower K.M., Voon H.P., Hughes J.R., Garrick D., Viprakasit V., Mitson M., De Gobbi M., Marra M., Morris A., et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Levy M.A., Kernohan K.D., Jiang Y., Berube N.G. ATRX promotes gene expression by facilitating transcriptional elongation through guanine-rich coding regions. Hum. Mol. Genet. 2015;24:1824–1835. doi: 10.1093/hmg/ddu596. [DOI] [PubMed] [Google Scholar]
- 15.Voon H.P., Hughes J.R., Rode C., De La Rosa-Velazquez I.A., Jenuwein T., Feil R., Higgs D.R., Gibbons R.J. ATRX Plays a Key Role in Maintaining Silencing at Interstitial Heterochromatic Loci and Imprinted Genes. Cell Rep. 2015;11:405–418. doi: 10.1016/j.celrep.2015.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elsasser S.J., Noh K.M., Diaz N., Allis C.D., Banaszynski L.A. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadic D., Schmidt K., Groh S., Kondofersky I., Ellwart J., Fuchs C., Theis F.J., Schotta G. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep. 2015;16:836–850. doi: 10.15252/embr.201439937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Q., Kim H., Huang R., Lu W., Tang M., Shi F., Yang D., Zhang X., Huang J., Liu D., et al. The Daxx/Atrx Complex Protects Tandem Repetitive Elements during DNA Hypomethylation by Promoting H3K9 Trimethylation. Cell Stem Cell. 2015;17:273–286. doi: 10.1016/j.stem.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Udugama M., Chang F.T., Chan F.L., Tang M.C., Pickett H.A., McGhie J.D., Mayne L., Collas P., Mann J.R., Wong L.H. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 2015;43:10227–10237. doi: 10.1093/nar/gkv847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang C.W., Shibata Y., Starmer J., Yee D., Magnuson T. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 2015;29:1377–1392. doi: 10.1101/gad.264150.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhayalan A., Tamas R., Bock I., Tattermusch A., Dimitrova E., Kudithipudi S., Ragozin S., Jeltsch A. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum. Mol. Genet. 2011;20:2195–2203. doi: 10.1093/hmg/ddr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eustermann S., Yang J.C., Law M.J., Amos R., Chapman L.M., Jelinska C., Garrick D., Clynes D., Gibbons R.J., Rhodes D., et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 2011;18:777–782. doi: 10.1038/nsmb.2070. [DOI] [PubMed] [Google Scholar]
- 23.Iwase S., Xiang B., Ghosh S., Ren T., Lewis P.W., Cochrane J.C., Allis C.D., Picketts D.J., Patel D.J., Li H., et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 2011;18:769–776. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maze I., Wenderski W., Noh K.M., Bagot R.C., Tzavaras N., Purushothaman I., Elsasser S.J., Guo Y., Ionete C., Hurd Y.L., et al. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron. 2015;87:77–94. doi: 10.1016/j.neuron.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benetti R., Gonzalo S., Jaco I., Schotta G., Klatt P., Jenuwein T., Blasco M.A. Suv4–20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters A.H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schofer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 27.Karimi M.M., Goyal P., Maksakova I.A., Bilenky M., Leung D., Tang J.X., Shinkai Y., Mager D.L., Jones S., Hirst M., et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung D., Du T., Wagner U., Xie W., Lee A.Y., Goyal P., Li Y., Szulwach K.E., Jin P., Lorincz M.C., et al. Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. Proc. Natl. Acad. Sci. U.S.A. 2014;111:6690–6695. doi: 10.1073/pnas.1322273111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quenneville S., Verde G., Corsinotti A., Kapopoulou A., Jakobsson J., Offner S., Baglivo I., Pedone P.V., Grimaldi G., Riccio A., et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf G., Yang P., Fuchtbauer A.C., Fuchtbauer E.M., Silva A.M., Park C., Wu W., Nielsen A.L., Pedersen F.S., Macfarlan T.S. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015;29:538–554. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuo X., Sheng J., Lau H.T., McDonald C.M., Andrade M., Cullen D.E., Bell F.T., Iacovino M., Kyba M., Xu G., et al. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J. Biol. Chem. 2012;287:2107–2118. doi: 10.1074/jbc.M111.322644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heaphy C.M., de Wilde R.F., Jiao Y., Klein A.P., Edil B.H., Shi C., Bettegowda C., Rodriguez F.J., Eberhart C.G., Hebbar S., et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovejoy C.A., Li W., Reisenweber S., Thongthip S., Bruno J., de Lange T., De S., Petrini J.H., Sung P.A., Jasin M., et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conomos D., Pickett H.A., Reddel R.R. Alternative lengthening of telomeres: remodeling the telomere architecture. Front. Oncol. 2013;3:27. doi: 10.3389/fonc.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Day D.S., Luquette L.J., Park P.J., Kharchenko P.V. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol. 2010;11:R69. doi: 10.1186/gb-2010-11-6-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]