Abstract
MicroRNAs play a fundamental role in retinal development and function. To characterise the miRNome of the human retina, we carried out deep sequencing analysis on sixteen individuals. We established the catalogue of retina-expressed miRNAs, determined their relative abundance and found that a small number of miRNAs accounts for almost 90% of the retina miRNome. We discovered more than 3000 miRNA variants (isomiRs), encompassing a wide range of sequence variations, which include seed modifications that are predicted to have an impact on miRNA action. We demonstrated that a seed-modifying isomiR of the retina-enriched miR-124-3p was endowed with different targeting properties with respect to the corresponding canonical form. Moreover, we identified 51 putative novel, retina-specific miRNAs and experimentally validated the expression for nine of them. Finally, a parallel analysis of the human Retinal Pigment Epithelium (RPE)/choroid, two tissues that are known to be crucial for retina homeostasis, yielded notably distinct miRNA enrichment patterns compared to the retina. The generated data are accessible through an ad hoc database. This study is the first to reveal the complexity of the human retina miRNome at nucleotide resolution and constitutes a unique resource to assess the contribution of miRNAs to the pathophysiology of the human retina.
INTRODUCTION
MicroRNAs (miRNAs) are a class of short non-coding RNAs that act as post-transcriptional regulators of gene expression by inducing transcript degradation or translational inhibition of their target mRNAs (1). They control specific pathways by targeting networks of functionally correlated genes and represent key mediators of fundamental biological processes in both physiological and pathological conditions (1). Defects in miRNA function have profound effects on development (2–4). In humans, deregulation of miRNA expression, caused by mutations in either the miRNA itself or its target genes, has been correlated with a number of pathological conditions such as diabetes, neurodegenerative diseases, heart failure and genetic disorders (5,6). Growing evidence supports a key role of miRNAs in human cancers (7). MiRNAs are also emerging as disease biomarkers and possible therapeutic targets in human disorders (8).
The retina is a light-sensitive layer of the eye that converts light to neural signals and represents the tissue target of a considerable number of human inherited diseases (RetNet database, http://www.sph.uth.tmc.edu/Retnet/home.htm). Several lines of evidence support the importance of miRNAs in normal retinal development and function (9). Global perturbation of miRNA function in the eye of conditional Dicer mouse mutants impairs normal development of the retina, lens, cornea and optic chiasm (10–13). MiRNA contribution to retina function and homeostasis is equally crucial in post-developmental stages. Specific disruption of miRNA processing in cone photoreceptors (PRs) leads to dramatic functional impairments (14). Also the targeted ablation of individual retina-enriched miRNAs has profound effects on vertebrate eye development and function (15–20). Finally, we recently reported the first evidence of the pathogenic role of a miRNA in a retinal disease in humans (21).
To date, reports on the analysis of the miRNA transcriptome (miRNome) in the human retina are not available. Information on miRNA expression in the human retina is primarily inferred from the murine counterpart (22–25). The human and the murine retina have significant structural and functional differences (particularly concerning the number and organization of cones) hence, it is not surprising that there are many examples of identical gene lesions that induce different phenotypes in patients and mouse models (26). The above differences are expected to reflect also in the expression profiles of miRNAs. Therefore, a thorough characterization of the human retina miRNome is an essential step towards a deeper understanding of the physiopathology of this tissue.
In this work, we investigated the complexity of the human retina miRNome. We exploited the recent advances in sequencing technologies and analysis for the implementation of a hypothesis-free, unbiased study of unprecedented resolution and sensitivity. To achieve this goal, we analysed the expression levels of miRNAs and their variants in the neural retina of sixteen healthy individuals by small RNA-seq. This allowed us to obtain a highly reliable assessment of the miRNome composition of this tissue. Moreover, we identified fifty-one novel putative miRNAs and validated the expression of nine of them using an independent approach. In addition, we studied the miRNome of the RPE/choroid, because these tissues have a crucial role in the functional interplay that leads to retinal disease. The data generated in this work are publicly available through a user–friendly, web-accessible database (http://miretina.tigem.it). To date, this is the first systematic analysis of miRNA expression and variability in the human retina.
MATERIALS AND METHODS
Human retina sample collection
In collaboration with the Eye Bank of Venice, we collected retina samples from sixteen different donors (for a description of donors see Supplementary Table S1) in compliance with the tenets of the Declaration of Helsinki and after obtaining the informed consent from the donor's next of kin. The donors were not reported to have pre-existing retinal pathologies. The average post-mortem time of the samples was 22 h 40 min (ranging from 6 to 26 h). Bulbs deriving from multi-organ donors were excluded from the analysis. We used our established protocol for the dissection of the retina from the human eye bulbs (27). Briefly, an incision was performed on the sclera and the scleral tissue was cut and removed leaving the rest of the eye bulb intact. The RPE/choroid was then removed as an intact sheet, revealing the neural retina. The latter was gently pulled away from the underlying vitreous body that remained intact. The dissected retinal tissue was visually inspected to exclude any cross contamination with the pigmented RPE/choroid and was immediately submerged in RNA Stabilisation Reagent (RNA later; Qiagen).
RNA extraction
Total RNA was extracted from the 16 human retina and 2 RPE/choroid samples using the miRNeasy Kit (Qiagen), following the manufacturer′s instructions. RNA was quantified using a NanoDrop ND-8000 spectrophotometer (NanoDrop Technologies) and the integrity was evaluated using an RNA 6000 Nano chip on a Bioanalyzer (Agilent Technologies). The RNA of all 16 samples was highly intact with an average RNA Integrity Number (RIN) of 8.9 ± 0.6. The RIN value of the two RPE/choroid samples was 7.9 and 7.1. Total RNA from human liver and muscle used for quantitative reverse transcription polymerase chain reaction (PCR) experiments (see below) was isolated as above, whereas total RNA from human blood was extracted using TRIzol (Invitrogen) according to the manufacturer's protocol. Total human brain RNA was obtained from a commercial source (FirstChoice, Ambion).
Library preparation and deep sequencing
For small RNA-seq analysis, small RNA libraries were constructed using a TruSeq small RNA sample preparation kit (Illumina), following the manufacturer′s protocol. We generated small RNA-seq libraries using 1 μg of total RNA as input from each sample. Using multiplexing, we combined up to 12 samples into a single lane in order to obtain sufficient coverage and ran two technical replicates for each library. The sequencing was carried out in collaboration with the Next Generation Sequencing (NGS) Facility at the Telethon Institute of Genetics and Medicine (TIGEM). Cluster generation was performed on Flow Cell v3 (TruSeq SR Cluster Kit v3; Illumina) using cBOT. Sequencing was performed on the HiSeq1000 platform. Each library was loaded at a concentration of 8–10 pM, which we had previously established as optimal.
Computational analysis of deep sequencing data
To determine the miRNA expression levels across samples, the raw data were analysed using the pipeline already established at the Bioinformatics and Statistics Core Facility at TIGEM. Briefly, the reads were trimmed to remove adapter sequences and low quality ends and the resulting sequences shorter than 16 nucleotides (nt) were discarded. Reads mapping to contaminating sequences (e.g. ribosomal RNA, phIX control) were filtered-out. Filtered reads were aligned both to the human genome (hg19) and to human mature and precursor (hairpin) miRNAs (miRBase v.20) (28), using the CASAVA software (Illumina). Briefly, the reads were aligned and grouped on the sequence of the mature miRNAs allowing up to two mismatches within the exact length of the reference mature sequence (i.e. excluding trimming or extension variants). The number of reads mapping on each mature miRNA was counted and then normalized using either the Trimmed Mean of M-values (TMM) method (29) or reads per million (CPM). The ranked lists were obtained using the Borda rule.
isomiR identification
To obtain a survey on miRNA sequence variability in the samples, trimmed reads were processed with a pipeline containing the software SeqBuster-mirAligner (30). SeqBuster identifies different types of miRNA variability that include 5′- and 3′-end modifications, 3′ addition variants and nucleotide substitution variants. The mapping step was performed using as reference the miRBase (v.20) entries. Default parameters were applied (i.e. one mismatch, 3 nt for the 3′ or 5′ trimming variants, 3 nt for the 3′-end addition variants).
miRNA target prediction
Prediction of miRNA targets was performed using microT (http://diana.cslab.ece.ntua.gr/) (31) and TargetScan custom (http://www.targetscan.org/) (32) software. The full sequence of the miRNA and its isomiR was used as query in microT, while in TargetScan predictions were performed using their respective seed sequences as query.
Luciferase reporter assay
The firefly luciferase (pFL) vector containing the 3′ UTR of the human CDH11 gene (amplified using the primers CDH11_UTR_F_SpeI: 5′- ggactagtGAACTGTGTCTGGCGTTCTC-3′ and CDH11_UTR_R_SpeI: 5′-ggactagtAGATTGCTTCTTATATTGAAGC-3′; pFL-CDH11-wt-3'UTR) was used in luciferase assays. A vector containing the 3′ UTR of the human CDH11 gene (pFL-CDH11-mut-3'UTR) with a modified seed-binding sequence, was generated using the following primer sequences: CDH11_F_MUT (5′-GTAATTTCTAAGGCCACTCTGGACACTCTATATG-3′) and CDH11_R_MUT (5′-CCAGAGTGGCCTTAGAAATTACAGCTCAGAACTG-3′). A Renilla luciferase vector (pRL; Promega) was used to account for the transfection efficiency. Cells were transfected sequentially with the vectors and 50 nM miRIDIAN miRNA Mimics (Dharmacon), using respectively PolyFect (Qiagen) and INTERFERin (Polyplus), according to the manufacturer's recommendations. The sequence of the custom mimic for the isomiR-124-3p was: 5′-UUAAGGCACGCGGUGAAUGCC-3′. Renilla and firefly luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega), according to the manufacturer's protocol. Each assay was performed in triplicate, and the results are means of at least three independent assays. Error bars represent the standard error of the mean. The Student's t-test was used to compare the data between the selected pairs.
Identification of novel miRNAs
We used the miRDeep2 prediction algorithm (33) as a suite of different modules. First, to process the reads and map them to the reference genome, we used the mapper module (mapper.pl) with the following parameters: -e/c -p hg19 -h -j -k TGGAATTCTCGGGTGCCAAGG -l 18 -m -s/t. We then ran miRDeep2 at default parameters. We analysed each sample individually and retained only the predicted precursors that were assigned a miRDeep score equal or greater to 1. As a final step, we used an in-house pipeline to filter the miRDeep2 output and to assemble the results in a presence/absence matrix with rows containing genomic coordinates and columns containing sample identifiers.
Quantitative reverse transcription PCR
Quantitative Reverse Transcription (qRT-) PCR analysis of the novel predicted mature miRNAs was performed using the miRCURY LNA microRNA PCR System (Exiqon) and custom-designed assays starting with 20 ng of total RNA. Real-time PCR amplification on the cDNA template was performed with miRNA-specific, LNA optimized primer sets. The ubiquitous miR-26a was used as a reference endogenous control for normalization in the context of each tissue. All reactions were performed in triplicate. The quantification of relative expression was performed as previously described (15).
RNA in situ hybridization (ISH)
Custom miRCURY LNA Detection probes with double digoxigenin (DIG)-labelling at the 5′ and 3′-end were synthesized by Exiqon. Their sequence was /5DigN/AGACAGCAGCCACTAGGCACATA/3Dig_N (for nrm7, overlapping with an intron of WASF3) and /5DigN/AGGCTGTTTAAACACATATGCG/3Dig_N (for nrm29, overlapping with an intron of OTX2). Their calculated RNA Tm was 83°C and 82°C, respectively. RNA ISH experiments on human adult eye sections using LNA-modified probes were performed using the protocol recommended by Exiqon with minor modifications. To this purpose, an eye bulb of a cornea donor was obtained through collaboration with the Eye Bank of Venice (Sample 51 in Supplementary Table S1). Following excision of the cornea, the eye bulb was fixed with 4% paraformaldehyde (PFA) in PBS for 48 h, cryoprotected by treatment with 30% sucrose in phosphate buffered saline (PBS) and embedded in OCT. Twenty-micrometer cryosections were collected on slides (Superfrost Plus; Fisher Scientific, Pittsburgh, PA) and postfixed with 4% PFA in PBS for 15 min. Sections were treated with 5 μg/ml proteinase K for 15 min at room temperature and the reaction was stopped with washes in 2 mg/ml glycine. ISH of 40nM of probe was performed at 52°C for 2 h using the miRCURY LNA microRNA ISH Optimisation kit (FFPE). A double-DIG-labelled LNA scrambled probe was hybridized in parallel at the same concentration and optimal hybridization temperature, as negative control. After blocking, sections were incubated with alkaline phosphatase (AP)-labelled anti-digoxigenin antibody (anti-DIG; 1:1000; Roche) and exposed to the substrate for AP, nitroblue tetrazolium and 5-bromo-4-chloro-3-indoyl phosphate (NBT-BCIP; Roche) for 2 h at 30°C. Slides were dehydrated and mounted (Eukitt Mounting Medium; EMS, Fort Washington, PA).
RESULTS
Sample collection and overview of the small RNA-seq data
We employed a Next Generation Sequencing (NGS)-based approach to determine the miRNome of the human retina. To this end, we collected human retina samples from the eye bulbs of sixteen cornea donors, in collaboration with the Eye Bank of Venice (27). Donors’ main descriptors are detailed in Supplementary Table S1. To minimise the effects of long post-mortem intervals (i.e. from death to the storage of the dissected tissue in a RNA stabilising solution) on RNA integrity, we analysed samples in which the total post-mortem time did not exceed 26 h. Moreover, the absence of pigmentation, ascertained by visual inspection, was used as an indication of the lack of potential Retinal Pigment Epithelium (RPE) crossover contamination. An average of 28.5 million single-end reads were obtained per sample after trimming, ranging from 21 to 40 million. Supplementary Table S2 provides the read-mapping report of the sequencing output per sample.
The filtered reads were aligned in parallel both to known human miRNAs (miRBase v.20) (28) and genome (hg19 release). On average, 82% of the total reads from the retina samples mapped to mature miRNAs or to known miRNA precursors, while 4–10% of reads mapped to genomic locations that did not overlap with any known mature or precursor sequence (Supplementary Table S2). We found a prevalence of reads mapping to miRNAs deriving from the 5p arm of the precursor hairpins (Supplementary Table S2). This prevalence may mainly reflect the predominant presence of 5p-derived miRNAs at the top ranking positions in the human retina (see below). Following pre-processing, filtering and quality control, the resulting reads were used for subsequent expression ranking, isomiR analysis and novel miRNA discovery.
Global analysis of known miRNA expression in the human retina
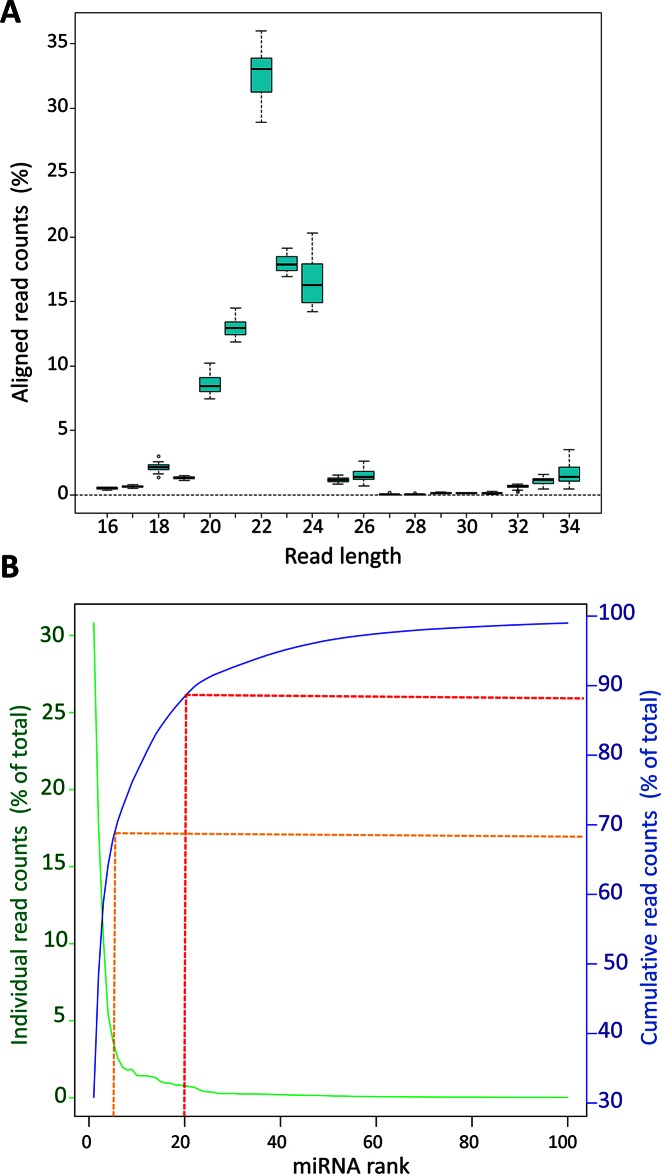
The predominant read length after trimming was 22 nt. On average, ≈80% of the reads ranged between 21 and 24 nt (Figure 1A), indicating that the most abundant class of small RNAs detected were miRNAs. Reads were then mapped to the 2555 unique mature human miRNA sequences annotated in the miRBase (v.20). We identified a total of 480 miRNAs with a minimum expression level of 9 reads across all 16 retina samples considered (Supplementary Table S3). Principal component analysis of miRNA expression profiles did not reveal any evident clustering of the donors related to the factors considered (age, gender, cause of death and post-mortem time of the donors) (Supplementary Figure S1). We also looked at the variability of expression of individual miRNAs across all 16 samples by calculating the coefficient of variation (Supplementary Table S3). The variability was low across samples with an expected higher trend for miRNAs expressed at lower levels (Supplementary Figure S2).
Figure 1.
Length distribution analysis of aligned reads from retina samples and distribution of reads across the 100 top-ranked miRNAs. (A) Boxplot summarising the distribution of the total number of aligned reads from the human retina samples (represented as % of total reads) across various lengths. (B) Abundance of reads for each individual miRNA (green line) and cumulative contribution of the reads (shown as % of total read count; blue line) across the 100 top-ranked miRNAs. Red and orange dotted lines show the cumulative contribution of the top 20 and top 5 miRNAs, respectively.
We ranked the expressed miRNAs based on their abundance (Table 1 and Supplementary Table S3). The twenty top-ranked miRNAs accounted for almost 90% of the total reads (Figure 1B; dotted red line) and the top five for 70% (Figure 1B; dotted orange line). Interestingly, the sum of reads mapped on the three members of the miRNA cluster miR-183/96/182 represented, on average, almost half (44%) of the total reads of each sample, consistent with their important role in photoreceptor function (14,34).
Table 1. The 40 most highly expressed miRNAs in the human retina.
| Rank | Exactly matching the annotated sequence | Including isomiRs | ||||
|---|---|---|---|---|---|---|
| miRNA Name | Mean Expression1 | miRNA Name | Mean Expression1 | Rank variation2 | Variation in reads (%)3 | |
| 1 | hsa-miR-182-5p | 304344 | hsa-miR-182-5p | 310414 | - | +2 |
| 2 | hsa-miR-183-5p | 131095 | hsa-miR-183-5p | 177857 | - | +36 |
| 3 | hsa-miR-26a-5p | 108111 | hsa-miR-181a-5p | 104145 | +1 | +11 |
| 4 | hsa-miR-181a-5p | 94166 | hsa-miR-26a-5p | 54470 | -1 | -50 |
| 5 | hsa-miR-204-5p | 64600 | hsa-miR-204-5p | 37292 | - | -42 |
| 6 | hsa-miR-22-3p | 39597 | hsa-miR-30a-5p | 26149 | New entry | +1617 |
| 7 | hsa-let-7a-5p | 22123 | hsa-miR-22-3p | 19998 | -1 | -49 |
| 8 | hsa-miR-191-5p | 21971 | hsa-miR-30d-5p | 18513 | +24 | +619 |
| 9 | hsa-miR-124-3p | 16883 | hsa-miR-124-3p | 18105 | +1 | +7 |
| 10 | hsa-miR-9-5p | 17074 | hsa-let-7a-5p | 14516 | -3 | -34 |
| 11 | hsa-miR-127-3p | 12907 | hsa-miR-9-5p | 14355 | -1 | -16 |
| 12 | hsa-miR-192-5p | 12726 | hsa-miR-143-3p | 14389 | +12 | +284 |
| 13 | hsa-let-7f-5p | 11418 | hsa-miR-27b-3p | 13769 | +1 | +45 |
| 14 | hsa-miR-27b-3p | 9497 | hsa-miR-191-5p | 13035 | -6 | -41 |
| 15 | hsa-miR-96-5p | 7380 | hsa-miR-127-3p | 10488 | -4 | -19 |
| 16 | hsa-miR-26b-5p | 6399 | hsa-miR-101-3p | 9660 | New entry | +721 |
| 17 | hsa-miR-30b-5p | 5196 | hsa-miR-192-5p | 9464 | -5 | -26 |
| 18 | hsa-miR-92a-3p | 5251 | hsa-miR-125a-5p | 8437 | +11 | +197 |
| 19 | hsa-miR-99b-5p | 5556 | hsa-let-7f-5p | 8237 | -6 | -28 |
| 20 | hsa-miR-125b-5p | 4571 | hsa-miR-96-5p | 7776 | -5 | +5 |
| 21 | hsa-miR-151a-5p | 4536 | hsa-miR-99b-5p | 7344 | -2 | +32 |
| 22 | hsa-miR-211-5p | 4452 | hsa-miR-92a-3p | 6798 | -4 | +29 |
| 23 | hsa-miR-126-5p | 3792 | hsa-miR-181b-5p | 5317 | +5 | +73 |
| 24 | hsa-miR-143-3p | 3748 | hsa-miR-30e-5p | 4137 | New entry | +1918 |
| 25 | hsa-miR-16-5p | 3562 | hsa-miR-125b-5p | 3830 | -5 | -16 |
| 26 | hsa-let-7g-5p | 3317 | hsa-miR-100-5p | 3550 | +8 | +16 |
| 27 | hsa-miR-148a-3p | 3066 | hsa-miR-30b-5p | 2792 | -10 | -46 |
| 28 | hsa-miR-181b-5p | 3078 | hsa-miR-26b-5p | 2714 | -12 | -58 |
| 29 | hsa-miR-125a-5p | 2837 | hsa-miR-181c-5p | 2770 | +3 | +2 |
| 30 | hsa-miR-92b-3p | 3029 | hsa-miR-151a-5p | 2777 | -9 | +4 |
| 31 | hsa-miR-181a-2-3p | 2664 | hsa-miR-151a-3p | 2528 | +7 | -7 |
| 32 | hsa-miR-181c-5p | 2707 | hsa-miR-148a-3p | 2522 | -5 | -18 |
| 33 | hsa-miR-30d-5p | 2574 | hsa-miR-16-5p | 2444 | -8 | -31 |
| 34 | hsa-miR-100-5p | 2285 | hsa-miR-92b-3p | 2556 | -4 | -16 |
| 35 | hsa-let-7c-5p | 2125 | hsa-miR-211-5p | 2421 | -13 | +23 |
| 36 | hsa-miR-103a-3p | 1968 | hsa-let-7g-5p | 2356 | -10 | -29 |
| 37 | hsa-miR-29b-3p | 1833 | hsa-miR-486-5p | 2316 | New entry | +44 |
| 38 | hsa-miR-151a-3p | 1732 | hsa-miR-29a-3p | 2193 | New entry | +86 |
| 39 | hsa-miR-186-5p | 1750 | hsa-miR-126-5p | 2075 | -16 | -45 |
| 40 | hsa-miR-21-5p | 1729 | hsa-miR-30c-5p | 2005 | New entry | +93 |
1TMM normalization.
2Variation in the ranking of each miRNA following isomiR inclusion.
3Variation in the mean expression values (shown as % of reads).
Finally, we compared the list of miRNAs expressed in the human retina with the corresponding one previously reported by Soundara Pandi et al. for the murine counterpart (25). We found that one third of the miRNAs expressed in the human retina were also detected in mouse (Supplementary Table S4). Interestingly, the miRNAs detected in the retina of both species accounted for more than 95% of the total reads (96.6% and 97.9% in the human and mouse retina, respectively) (Supplementary Table S4).
IsomiR identification and expression analysis in the human retina
To characterise the variability of miRNA biogenesis in the human neural retina we investigated isomiR diversity. The majority of miRNAs comprise multiple sequence isoforms (termed isomiRs) (35–37). These sequence variants differ from the canonical mature miRNA sequence deposited in miRBase for the addition or trimming of nucleotides at either end and may also bear nucleotide substitutions.
We used the mirAligner algorithm to determine the extent of isomiR representation in the human retina for all miRNAs passing the threshold of 9 reads previously applied. To increase reliability, we selected only the isomiRs that were represented with a frequency of at least 1% with respect to the abundance of the reference miRNA. By applying these criteria, we identified 3350 ± 474 (average ± SD) different isomiRs in each human retina sample. Supplementary Table S5 reports the complete list of isomiRs identified in this study. However, for all subsequent analyses, we focused our attention only on isomiRs that were detected in all sixteen retinal samples (n = 2026) (Supplementary Table S5) to exclude variations due to DNA polymorphisms among donors.
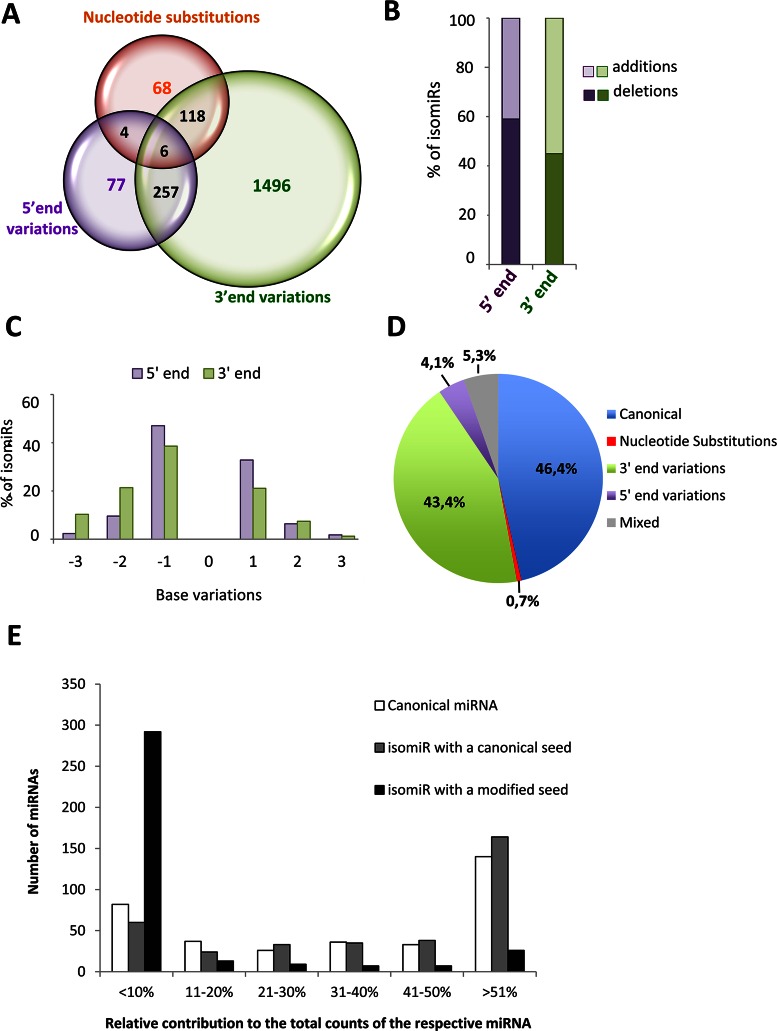
In our data set, 92% of the retina-expressed miRNAs showed sequence variants. The contribution of isomiRs to the expression levels of the different miRNAs was extremely variable, ranging from 0% to 100% of the total reads (Supplementary Table S5). By comparison to the canonical sequences deposited in the miRBase, we found all reported sources of sequence variation, i.e. trimming and additions at either the 5′- or 3′-end, and nucleotide substitutions (Supplementary Table S5). Out of the 2026 isomiRs detected in common across all retina samples, 3′-end variations were the most common modifications found (92.6%, 1877/2026) (Figure 2A). Variations at the 5′-end were less frequent (16.9%, 344/2026) and substitutions within the mature miRNA sequence were the least common variants observed (9.4%, 196/2026) (Figure 2A). Almost one fifth of the isomiRs displayed more than one modification, the most frequent being the presence of both 5′-end and 3′-end variations (Figure 2A and Supplementary Table S5). A very limited number of isomiRs had mixed variations of the three above-mentioned types (i.e. combinations of nucleotide substitutions, 5′-end variations and 3′-end variations) (Figure 2A and Supplementary Table S5). Nucleotide deletions (trimming) were the most frequent modification events at the 5′-end (59%; Figure 2B). Instead, additions were the most common modifications at the 3′-end (55%; Figure 2B), due to the fact that besides templated changes, there was a large number of untemplated additions, believed to occur post-transcriptionally, such as the addition of an adenine (38). The majority of either the 3′ or 5′-end trimming and templated additions involved a single base change (Figure 2C). In terms of expression levels (i.e. contribution to total read counts), canonical miRNA sequences represented the most abundant category, closely followed by isomiRs with variations at the 3′-end (Figure 2D).
Figure 2.
Summary of isomiR heterogeneity and relative contribution of isomiRs with modified or canonical seed to miRNA expression in the human retina. (A) Venn diagram illustrating the number of retinal miRNA variants, classified in three types of sequence variation (5′-end variations, 3′-end variations, nucleotide substitutions). (B) Bar-graph indicating the percentage of 5′ or 3′ modifications that correspond to nucleotide additions (light colour) or trimming events (dark colour). (C) Percentage of individual isomiRs with templated additions or trimming of up to 3 nucleotides at the 5′ or 3′-end compared to the corresponding canonical sequences. (D) Pie-chart illustrating the average contribution of the different categories of isomiRs to miRNA expression in the human retina. (E) Contribution of canonical miRNAs (white bars) and of isomiRs with a canonical (grey bars) or modified seed (black bars) to overall miRNA expression in the human retina. The bars report the number of miRNAs for which the expression of each sequence class, relative to total counts, is within the five categories indicated on the x-axis. The data reported in this figure refer to the analysis of isomiRs detected in all retina samples (n = 16).
Considering that isomiRs can significantly contribute to miRNA representation, we repeated the ranking of the retinal miRNAs by adding the reads of all isomiRs to the reads of the corresponding canonical mature sequence (Table 1 and Supplementary Table S3). We identified a total of 356 miRNAs with a minimum expression level threshold of 9 reads, following count normalization (Supplementary Table S3). We noted that in some instances a given isomiR was by far more abundant compared to the corresponding canonical miRNA deposited in the miRBase (Supplementary Figure S3). For example, an isomiR of hsa-miR-30a-5p, bearing a two nucleotide trimming at the 3′-end compared to the reference sequence, was almost 33-fold more abundant than the canonical miRNA (Supplementary Table S5). Furthermore, by taking into consideration all sequence variants, the number of total reads mapping on miR-30a-5p was 16-fold higher with respect to the canonical one. Since inclusion of isomiRs changed significantly the relative abundance of individual miRNAs, we relied on the isomiRs-including ranking for subsequent analyses.
IsomiRs with a modified seed: characterization of an isomiR of miR-124-3p
Sequence variations at the 5′-end as well as nucleotide substitutions within the seed sequence (i.e. between nucleotides 2–8 of mature sequences) have been shown to have functional consequences as they may confer different targeting properties to a given miRNA (39). We therefore focused our analysis on changes between nucleotides 2–8 of mature miRNAs, because these are expected to give rise to a modified seed sequence. On average, across the 16 retina samples analysed, around 23.2% of the isomiRs (471 out of 2026) had a modified seed sequence, due to seed-shifting caused by trimming or additions of nucleotides at the 5′-end, or due to nucleotide substitutions within the seed sequence itself (Supplementary Table S5).
The canonical miRNA was the most represented variant (i.e. contributing >51% of the total read counts) for the vast majority of retina-expressed miRNAs (n = 140) (Figure 2E, white bars). The contribution of different isomiRs to the total reads of a given miRNA was variable (Figure 2E). IsomiRs with a canonical seed contributed to the total miRNA expression with a similar trend to that of canonical miRNAs (Figure 2E, grey bars). Instead, in the majority of cases isomiRs with a modified seed sequence contributed little to the overall miRNA expression (Figure 2E, black bars). For 82.5% of individual miRNAs (n = 292), the expression of seed-modified isomiRs accounted for less than 10% of total read counts of the corresponding miRNA (Figure 2E; black bars). Nevertheless, it is noteworthy that for a limited set of miRNAs (7.3%, n = 26), the isomiRs with a modified seed sequence represented the most abundant variant in the human retina (Figure 2E; black bars).
We looked for independent evidence of expression and functional relevance for the isomiRs with a modified seed. We focused on the isomiRs of the 100 top-ranked miRNAs that have a modified seed and contribute >1% to the expression of the corresponding miRNA. The vast majority (96%) of these isomiRs were detected in other tissues according to the small RNAseq data available at the YM500v2 database (http://ngs.ym.edu.tw/ym500v2/index.php) (40), an integrated database for miRNA analysis that reports information on isomiRs identified in a number of human tissues. We also found evidence that 14.5% of the above seed-modified isomiRs were loaded onto the Argonaute (Ago) silencing complex, as determined by Ago PAR-CLIP experiments performed on human lymphoblastoid cell lines (41) (Supplementary Table S6).
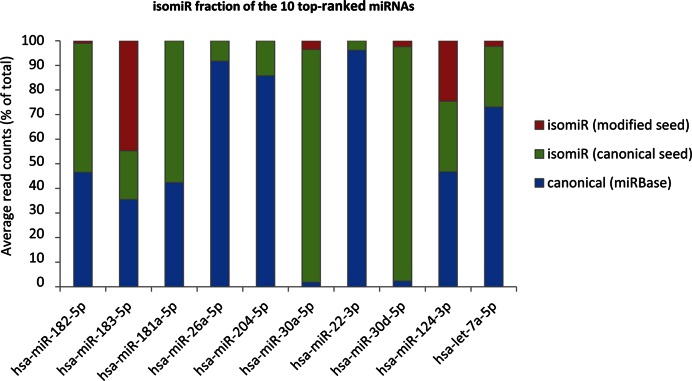
Two notable examples of isomiRs with a modified seed and significant expression in the human retina are represented by hsa-miR-183-5p and hsa-miR-124-3p. In the case of hsa-miR-183-5p, the isomiRs with a modified seed accounted on average for 45% of the total read counts across all samples (Figure 3 and Supplementary Table S3). The miR-183/96/182 cluster plays a key role in postnatal differentiation and synaptic connectivity of photoreceptors (34), and the relevant expression level of a hsa-miR-183-5p variant with a modified seed opens the possibility of a wider spectrum of target recognition in the human adult retina.
Figure 3.
IsomiR contribution to the expression of the most abundant miRNAs in the human retina. Bar plots showing the relative contribution to expression (shown as percentage of total read counts) for the ten most abundant miRNAs in the human retina. Blue area: canonical sequence (same sequence as the reference miRNA annotated in the miRBase); Green area: isomiRs bearing the same seed as the canonical miRNA; Red area: isomiRs with a modified seed sequence. The analysis was performed on isomiRs detected in all retina samples (n = 16).
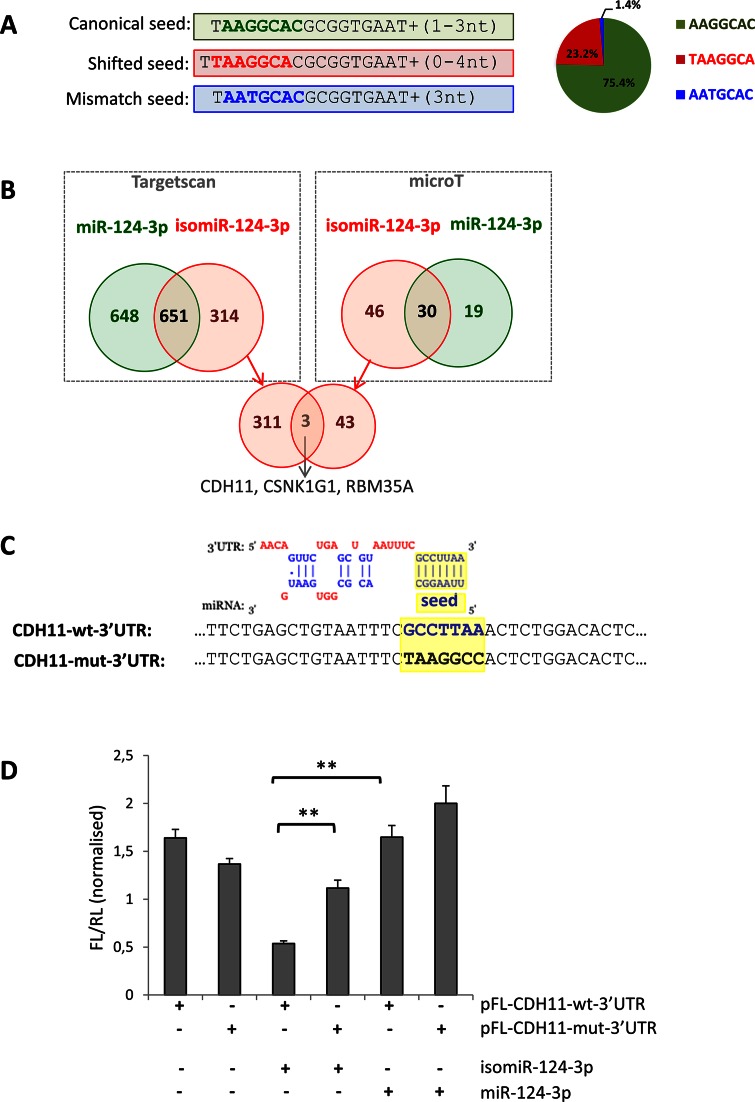
Concerning hsa-miR-124a-3p, an abundant neuronal-specific miRNA (18,23,42), the isomiRs with a modified seed accounted for one quarter of the total read counts (Figure 3 and Supplementary Table S7). Specifically, the reads of miR-124a-3p present three different seed sequences: (i) the canonical seed, which is present in 75.4% of cases, (ii) a modified seed due to seed-shifting (23.2% of reads) and (iii) a less represented seed (1.4%) bearing a base-substitution (Figure 4A and Supplementary Table S7). In silico prediction indicated that these variants may have distinct targeting properties (Figure 4B). According to TargetScan and microT software 33% and 61%, respectively, of the isomiR-124-3p targets (Figure 4B, red circles) are not predicted to be targeted by the canonical miR-124-3p (Figure 4B, green circles).
Figure 4.
Analysis of different targeting capabilities of the 5′ isomiR-124-3p. (A) The reads mapping to miR-124-3p present three different seed sequences; the canonical seed (bold green), a modified seed due to seed-shifting (bold red) and a seed bearing a base-substitution (bold blue). The pie-chart on the right shows their relative abundance. Detailed sequence information and average frequency is shown in Supplementary Table S7. (B) Venn diagrams showing the overlap of the predicted targets for the miR-124-3p (green) and the most abundant isomiR with a shifted seed (red). Predictions were performed using the microT (right box) and TargetScan (left box) software. The intersection of the Venn diagram (bottom of the panel) shows the three genes predicted in common by both software to be targeted only by the isomiR. (C) Top: alignment of the predicted binding of the seed-shifted isomiR-124-3p and the 3′ UTR of the CDH11 mRNA (as in microT output). Bottom: sequence alignment of the corresponding region in the vectors bearing the wild-type (CDH11-wt-3′UTR; bold blue) and mutated (CDH11-mut-3′UTR; bold black) predicted seed-binding site (highlighted in yellow). (D) Relative luciferase activity of pFL-CDH11-wt-3′UTR and pFL-CDH11-mut-3′UTR in HeLa cells co-transfected with mimics for miR-124–3p and the seed-shifted isomiR (isomiR-124-3p). Firefly luciferase values are normalized against the Renilla luciferase. Error bars are ±SEM of three independent experiments (performed in triplicate). **: P < 0.005.
To experimentally validate the latter prediction, we tested the ability of the seed-shifted isomiR to target the 3′ UTR of CDH11. Both TargetScan and microT predicted this gene to be an isomiR-specific target and, interestingly, CDH11 has a role in retinal differentiation (43). The wild-type 3′ UTR of CDH11 was cloned downstream a luciferase reporter, yielding the CDH11-wt-3′UTR vector. The predicted seed-binding sequence was mutated to yield the control construct CDH11-mut-3′UTR (Figure 4C). In HeLa cells, the luciferase activity of the CDH11-wt-3′UTR vector was significantly reduced upon co-transfection with a custom mimic for the most abundant seed-shifted variant, as opposed to co-transfection with the hsa-miR-124-3p mimic (Figure 4D). Luciferase activity levels were restored, when the predicted seed-binding sequence was mutated (CDH11-mut-3′UTR) (Figure 4D). Co-transfection of either construct with the hsa-miR-124-3p mimic did not affect luciferase activity (Figure 4D). This suggested the direct targeting of CDH11 by the isomiR and not by the canonical miR-124-3p, supporting the predicted differences in their targeting properties.
Identification of novel miRNAs in the human retina
It has been recently proposed that a significant number of tissue-specific miRNAs are yet to be discovered (44). To identify putative miRNAs originating from novel precursors in the human retina, we processed the reads obtained from the analysed samples using the miRDeep2 software (33). For each sample, putative novel miRNA precursors were identified by analysing the distribution of the sequencing reads and assessing its compatibility with miRNA biogenesis and precursor processing (33). We identified 51 putative novel miRNAs (named ‘nrm’, novel retina miRNA) deriving from 49 precursors that were predicted independently in at least half of the samples analysed (Table 2). We retained those with an expression level higher than 1/100 of the small nucleolar RNA (snoRNA) SNORD44 (44). These miRNAs were encoded in both intragenic (≈51% intronic and 12% exonic) and intergenic (37%) regions of the genome and one-third was found to overlap, partially or entirely, with repeated sequence elements (Table 2).
Table 2. Novel miRNA precursors identified in the human retina.
| Novel retina miRNA ID1 | Chr. | Coordinates | Strand | Detection2 | Mapped region | Overlapping gene | Overlap. repeats | Notes3 |
|---|---|---|---|---|---|---|---|---|
| nrm1 | 1 | 142850083–142850134 | - | 16 | intronic | PNAS-130 mRNA | - | - |
| nrm2 | 3 | 48357870–48357934 | + | 16 | intronic | SPINK8 | LINE | hsa-mir-2115 |
| nrm3 | 7 | 41697430–41697484 | + | 16 | intergenic | - | - | - |
| nrm4/5 | 8 | 65351159–65351217 | +/- | 16 | intergenic | - | - | - |
| nrm6 | 10 | 105362131–105362190 | + | 16 | intronic/exonic | NEURL, SH3PXD2A | - | - |
| nrm7 | 13 | 27259471–27259537 | + | 16 | intronic | WASF3 | MER3 | - |
| nrm8 | 19 | 57950479–57950542 | + | 16 | intronic | ZNF749 | LINE | - |
| nrm9 | 21 | 10205174–10205225 | + | 16 | intergenic | - | - | - |
| nrm10 | 22 | 31284149–31284211 | + | 16 | intronic | OSBP2 | - | - |
| nrm11/12 | 22 | 18190160–18190225 | +/- | 16 | intronic | BCL2L13 | - | - |
| nrm13 | 1 | 113424673–113424746 | - | 15 | intergenic | - | - | - |
| nrm14 | 3 | 129253485–129253531 | + | 15 | 3′ UTR | RHO | - | - |
| nrm15 | 10 | 135055629–135055715 | + | 15 | intergenic | - | - | - |
| nrm16 | 15 | 56657482–56657536 | - | 15 | intergenic | - | - | - |
| nrm17 | 22 | 37985238–37985300 | + | 15 | intergenic | - | - | - |
| nrm18 | 10 | 99635579–99635642 | - | 14 | intronic | CRTAC1 | - | mmu-mir-3085 |
| nrm19 | 8 | 56821959–56822008 | - | 13 | intronic | LYN | rRNA | - |
| nrm20 | 10 | 43966686–43966745 | + | 13 | intronic | ZNF487P | LTR | - |
| nrm21 | 12 | 87138854–87138913 | - | 13 | intronic | MGAT4C | - | mml-mir-3059 |
| nrm22 | 3 | 159000445–159000510 | - | 12 | intronic | SCHIP1 | LINE | hsa-mir-3919 |
| nrm23 | 1 | 46598160–46598217 | + | 11 | intronic; 5′ UTR | PIK3R3 | - | - |
| nrm24 | 1 | 18073682–18073757 | + | 11 | intergenic | - | MER81 | - |
| nrm25 | 2 | 71083968–71084025 | - | 11 | intergenic | - | LINE | - |
| nrm26 | 5 | 176941854–176941910 | - | 11 | intronic | DDX41 | - | - |
| nrm27 | 11 | 16812462–16812523 | - | 11 | intronic | PLEKHA7 | - | - |
| nrm28 | 11 | 100841607–100841663 | + | 11 | intronic | ARHGAP42 | LINE | - |
| nrm29 | 14 | 57273999–57274061 | - | 11 | intronic | OTX2 | - | - |
| nrm30 | 17 | 7738772–7738833 | - | 11 | intergenic | - | - | - |
| nrm31 | 19 | 58024385–58024430 | - | 11 | exonic | ZNF773 | LTR | - |
| nrm32 | X | 69674997–69675068 | + | 11 | intronic/exonic | DLG3 | low complexity | - |
| nrm33 | 1 | 109815217–109815283 | - | 10 | exonic | CELSR2 | - | - |
| nrm34 | 1 | 150207085–150207139 | + | 10 | intronic | ANP32E | - | - |
| nrm35 | 8 | 85320843–85320898 | + | 10 | intronic | RALYL | LTR | mml-mir-7198 |
| nrm36 | 11 | 112568119–112568195 | - | 10 | intergenic | - | - | - |
| nrm37 | 2 | 46575933–46576012 | + | 9 | intronic | EPAS1 | DNA | - |
| nrm38 | 17 | 31555853–31555923 | + | 9 | intronic | ASIC2 | - | - |
| nrm39 | 19 | 708865–708935 | - | 9 | intergenic | - | simple; low complexity | - |
| nrm40 | 19 | 50527860–50527911 | + | 9 | intronic | VRK3 | LINE | - |
| nrm41 | 10 | 73182013–73182067 | + | 8 | intronic | CDH23 | SINE | - |
| nrm42 | 11 | 1357505–1357575 | + | 8 | intergenic | - | low complexity | - |
| nrm43 | 11 | 76156409–76156473 | + | 8 | intronic | c11orf30 | - | - |
| nrm44 | 14 | 104005952–104006017 | + | 8 | intergenic | - | - | - |
| nrm45/46 | 14 | 31678113–31678198 | +/- | 8 | intergenic | - | - | - |
| nrm47 | 17 | 7210150–7210205 | - | 8 | intergenic | - | - | - |
| nrm48 | 17 | 65822070–65822138 | - | 8 | exonic | BPTF | low complexity | - |
| nrm49 | 17 | 79919292–79919363 | + | 8 | intergenic | - | low complexity | - |
| nrm50 | 19 | 5978316–5978373 | + | 8 | exonic | RANBP3 | - | - |
| nrm51 | 19 | 42837698–42837758 | + | 8 | intronic | MEGF8 | - | - |
1miRNAs analysed by qRT-PCR (Figure 5A) are shown in bold.
2Number of samples in which the novel miRNA was identified.
3miRNAs currently present in the miRBase v21.
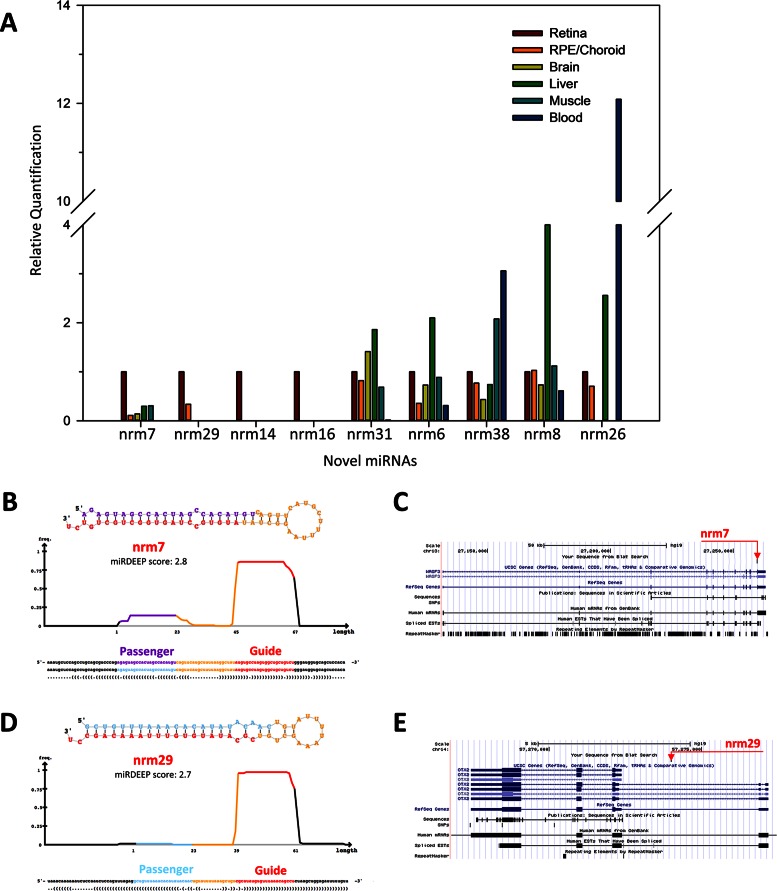
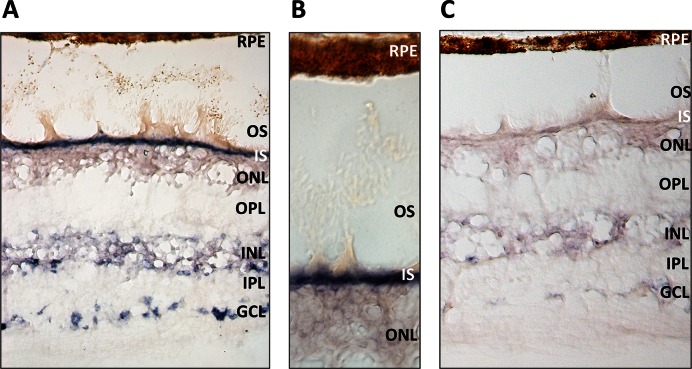
We aimed to validate the expression of ten predicted novel miRNAs (namely nrm 4, nrm6, nrm7, nrm8, nrm14, nrm16, nrm26, nrm29, nrm31 and nrm38) with an independent approach, a custom-designed Quantitative Reverse Transcription (qRT-)PCR assay. Nine of the analysed novel miRNAs were detected by qRT-PCR in the retina and, to a variable extent, in other human tissues (Figure 5A and Supplementary Figure S4). We further characterized two of them. Nrm7 derives from a putative precursor mapping within the last intron of the WASF3 gene (chr13: 27259470–27259537; hg19) (Figure 5B,C). WASF3 encodes a member of the Wiskott–Aldrich syndrome protein family. Its murine orthologue is expressed in the brain and retina during embryonic development (www.eurexpress.org) (45). The second miRNA, named nrm29, derives from a precursor mapping within the second intron of the longer OTX2 isoform (chr14: 57273990–57274069; hg19), (Figure 5D,E). OTX2 is a key transcription factor for the initial specification of the PR lineage and in the retina is expressed in cones, rods, bipolar cells and in the RPE (46). The qRT-PCR analysis indicated that both miRNAs were enriched in the retina, compared to the RPE/choroid as well as to the other human tissues analysed (Figure 5A and Supplementary Figure S4). This finding was consistent with the observation that reads mapping to these loci were barely detectable in the RPE/choroid small RNA-seq libraries (see below). To determine the spatial distribution of these two novel miRNAs in the human retina, we performed RNA in situ hybridization (ISH) experiments on sections of human eye using custom Locked Nucleic Acid (LNA)-modified DNA oligonucleotide probes. In our experimental conditions, we detected a specific signal over background for nrm7 (Figure 6), but not for nrm29. Nrm7 was specifically expressed in the area corresponding to the photoreceptor inner segments (Figure 6A,B). A weaker signal was observed in the Ganglion Cell Layer (GCL) as well as in the innermost part of the Inner Nuclear Layer (INL) (Figure 6A), while no staining was detectable in the RPE (Figure 6B).
Figure 5.
qRT-PCR analysis, predicted hairpin structure and genomic localization of putative novel retina miRNAs. (A) The expression of nine predicted novel miRNAs was validated by qRT-PCR on RNA from the human retina, RPE/choroid, brain, liver, muscle and blood. The retina was used as calibrator tissue. miRDeep output (predicted hairpin structure and associated score) (B, D) and genomic localization (C, E) of the putative novel miRNAs nrm7 and nrm29 that overlap with the transcriptional unit of WASF3 and OTX2, respectively.
Figure 6.
Expression analysis of the novel miRNA nrm7 by RNA in situ hybridization on sections of the adult human retina. (A) The novel miRNA nrm7 is expressed in different retina cell layers. (B) Close-up of the area corresponding to photoreceptor inner segments (IS) and outer segments (OS). (C) Hybridization with a scrambled probe, as negative control. Abbreviations: GCL, Ganglion Cell Layer; INL, Inner Nuclear Layer; IPL, Inner Plexiform Layer; ONL, Outer Nuclear Layer; OPL, Outer Plexiform Layer; RPE, Retinal Pigment Epithelium.
Analysis of small RNA expression in the human RPE/choroid
We carried out small RNA-Seq analysis on two human RPE/choroid samples (Supplementary Table S1). The RPE/choroid samples yielded 21 and 25 million reads (Supplementary Table S2). These data were analysed using the same pipeline described above for the retina. By using the same filtering criteria (i.e. at least 9 reads) we determined that 416 miRNAs are expressed in the RPE/choroid (Supplementary Table S8).
The RPE and the neural retina are contiguous tissues that physically interact. In particular, the apical processes of the RPE cells enwrap the photoreceptor outer segments. To some extent, carry- over of photoreceptor outer segments in the RPE/choroid preparations cannot be fully avoided. We therefore used the relative abundance of the reads mapping to the highly expressed, photoreceptor-specific miRNA cluster miR-183/96/182 (23) as an index to determine the extent of photoreceptor and outer segment carry over in the RPE/choroid samples. The mean expression of miR-182-5p, the most highly expressed member of the cluster, in the RPE/choroid samples was less than 1% (0.8%) of its expression in the neural retina (Supplementary Tables S3 and S8), indicating a very low level of carry-over contamination.
Since the data from the analysis of the human retina indicated that isomiRs can significantly contribute to the expression levels of a given miRNA, we characterized isomiR diversity in the human RPE/choroid using the mirAligner algorithm (Supplementary Table S9). We observed an overall similar representation of the various modifications across isomiRs, as seen for the retina samples, with the 3′-end variations, representing the most abundant class in terms of contribution to the total reads (Supplementary Figure S5). Variations at the 5′-end as well as nucleotide substitutions within the seed sequence were less frequent events (Supplementary Figure S5). The contribution of the seed-modifying isomiRs to the overall miRNA expression levels was limited with an overall relative frequency of <10% (black bars in Supplementary Figure S5 and Table S9), similar to what was observed for the retina.
We incorporated the reads corresponding to the isomiRs of each miRNA together with the ones that mapped on the canonical sequence and obtained the ranking of their relative abundance in the RPE/choroid (Table 3 and Supplementary Table S8). Following isomiR inclusion, we found that 419 miRNAs are expressed in the RPE/choroid with at least 9 read counts (Supplementary Table S8). Compared to previously published data (25), almost 40% of the miRNAs expressed in the human RPE/choroid were also detected in mouse (Supplementary Table S10). The miRNAs that were detected in both species accounted for more than 90% of the total reads (93.6% and 90.6% of the total reads in the human and mouse RPE/choroid, respectively) (Supplementary Table S10).
Table 3. The 40 most highly expressed miRNAs in the RPE/choroid.
| miRNA Name | Rank | Mean Expression Values (TMM) |
|---|---|---|
| hsa-miR-204-5p | 1 | 165078 |
| hsa-miR-143-3p | 2 | 106057 |
| hsa-miR-26a-5p | 3 | 124744 |
| hsa-miR-30a-5p | 4 | 22994 |
| hsa-miR-22-3p | 5 | 18928 |
| hsa-let-7a-5p | 6 | 18712 |
| hsa-miR-27b-3p | 7 | 18546 |
| hsa-miR-126-5p | 8 | 17939 |
| hsa-miR-181a-5p | 9 | 10261 |
| hsa-miR-211-5p | 10 | 9914 |
| hsa-miR-191-5p | 11 | 9534 |
| hsa-let-7f-5p | 12 | 9372 |
| hsa-miR-146a-5p | 13 | 8460 |
| hsa-miR-30d-5p | 14 | 7067 |
| hsa-miR-21-5p | 15 | 5927 |
| hsa-miR-125a-5p | 16 | 6203 |
| hsa-miR-125b-5p | 17 | 5881 |
| hsa-let-7c-5p | 18 | 4795 |
| hsa-miR-92a-3p | 19 | 5676 |
| hsa-miR-101-3p | 20 | 4587 |
| hsa-miR-126-3p | 21 | 3774 |
| hsa-miR-146b-5p | 22 | 3284 |
| hsa-miR-148a-3p | 23 | 3307 |
| hsa-miR-100-5p | 24 | 3140 |
| hsa-miR-26b-5p | 25 | 2974 |
| hsa-miR-486-5p | 26 | 6015 |
| hsa-miR-99b-5p | 27 | 3037 |
| hsa-miR-16-5p | 28 | 2768 |
| hsa-miR-182-5p | 29 | 2555 |
| hsa-let-7g-5p | 30 | 2406 |
| hsa-miR-99a-5p | 31 | 2462 |
| hsa-miR-186-5p | 32 | 2355 |
| hsa-miR-23b-3p | 33 | 2090 |
| hsa-miR-30e-5p | 34 | 2064 |
| hsa-let-7b-5p | 35 | 1939 |
| hsa-miR-151a-5p | 36 | 1642 |
| hsa-miR-10a-5p | 37 | 1671 |
| hsa-miR-27a-3p | 38 | 1487 |
| hsa-miR-127-3p | 39 | 1422 |
| hsa-miR-29a-3p | 40 | 1380 |
The 12 top-ranked miRNAs contributed about 75% of the total read counts. The most abundant miRNA in the RPE/choroid samples was miR-204-5p, which has already been shown in mouse to be strongly expressed in the RPE from early embryonic stages (23). Together with its paralogue miR-211, miR-204 accounted for almost 35% of the total reads in the RPE/choroid (Table 3), besides being also abundant in the human neural retina (Table 1).
We compared the data sets from the neural retina and RPE/choroid to identify miRNAs that were enriched in either tissue. We looked at the expression levels of the neural retina-expressed miRNAs in the RPE/choroid samples and vice versa. A list of selected miRNAs that show relative enrichment in either of these tissues is provided in Table 4.
Table 4. Selected miRNAs showing relative enrichment in either the retina (top panel) or RPE/choroid (bottom panel).
| miRNA | Retina rank | Retina mean1 | Ratio retina versus RPE/choroid2 | RPE/choroid rank | RPE/choroid mean1 | Ratio RPE/choroid versus retina2 |
|---|---|---|---|---|---|---|
| hsa-miR-182-5p | 1 | 310414 | 121.5 | 29 | 2555 | 0.0 |
| hsa-miR-183-5p | 2 | 177857 | 238.4 | 55 | 746 | 0.0 |
| hsa-miR-181a-5p | 3 | 104145 | 10.1 | 9 | 10261 | 0.1 |
| hsa-miR-124-3p | 8 | 18105 | 98.9 | 107 | 183 | 0.0 |
| hsa-miR-9-5p | 11 | 14355 | 16.8 | 49 | 855 | 0.1 |
| hsa-miR-192-5p | 17 | 9464 | 13.6 | 57 | 696 | 0.1 |
| hsa-miR-96-5p | 20 | 7776 | 79.6 | 129 | 98 | 0.0 |
| hsa-miR-181b-5p | 23 | 5317 | 10.9 | 66 | 490 | 0.1 |
| hsa-miR-181a-2-3p | 44 | 1696 | 13.1 | 111 | 130 | 0.1 |
| hsa-miR-181a-3p | 65 | 589 | 7.0 | 153 | 84 | 0.1 |
| hsa-miR-146a-5p | 138 | 86 | 0.0 | 13 | 8460 | 98.0 |
| hsa-miR-142-5p | 189 | 38 | 0.0 | 48 | 944 | 24.8 |
| hsa-miR-145-5p | 203 | 30 | 0.1 | 69 | 429 | 14.4 |
| hsa-miR-451a | 129 | 126 | 0.1 | 45 | 1731 | 13.7 |
| hsa-miR-126-3p | 82 | 354 | 0.1 | 21 | 3774 | 10.7 |
| hsa-miR-126-5p | 39 | 2075 | 0.1 | 8 | 17939 | 8.6 |
| hsa-miR-143-3p | 12 | 14389 | 0.1 | 2 | 106057 | 7.4 |
| hsa-miR-23a-3p | 116 | 119 | 0.2 | 58 | 697 | 5.8 |
| hsa-miR-27a-3p | 87 | 304 | 0.2 | 38 | 1487 | 4.9 |
| hsa-miR-204-5p | 5 | 37292 | 0.2 | 1 | 165078 | 4.4 |
| hsa-miR-211-5p | 35 | 2421 | 0.2 | 10 | 9914 | 4.1 |
1Mean expression values (TMM normalization).
2Bold fonts highlight relative enrichment in this tissue.
Finally, we determined the extent of similarity in isomiR representation in the two tissues by analysing the variants of miRNAs expressed in both the retina and RPE/choroid. More than half (54%) of the miRNA sequence variants, including isomiRs and reference miRNA sequences, were identified in both tissues, whereas 17% were retina-specific and 29% were detected only in the RPE/choroid (Supplementary Table S11). Despite the high number of tissue-specific isomiRs in the two contiguous tissues, these contributed little to the overall miRNA expression (1.3% and 2.4% of the total reads in the retina and RPE/choroid, respectively) (Supplementary Table S11). On the other hand, the common variants accounted for 98.7% and 97.6% of the total reads in the retina and RPE/choroid, respectively (Supplementary Table S11).
The miRetina database
To increase the usefulness of our analysis for the scientific community, we built the publicly available miRetina database (http://miretina.tigem.it). It allows case-insensitive queries by miRNA gene name (e.g. MIR182 or miR-182) or by genomic coordinates and provides graphical information on the genomic location by presenting an alignment of the reference miRNA in a Genome Browser window along with the respective isomiRs. Only sequence variants that were identified in all sixteen individuals were included in the database. The relative expression level of each miRNA variant in the retina is shown as relative frequency (RF) out of the total miRNA reads. For the 100 top-ranked miRNAs, we used the TargetScan custom tool to predict the targets of the isomiRs that bear a modified seed sequence and contribute >1% to the expression of the corresponding miRNA. We compared the list of predicted targets of each isomiR to that of the respective canonical miRNA and established three categories of predicted target genes: (i) those predicted to be targeted only by the isomiR, (ii) those targeted both by the isomiR and its respective reference miRNA and (iii) genes predicted to be targeted only by the reference miRNA. We provide the above gene lists in the database. An output example for miR-182 (MIR182) is presented in Supplementary Figure S6, along with a brief explanation of the colour code. The 51 predicted novel miRNAs can be visualized in the genome browser interface of the miRetina database. The user can download information on the predicted secondary structure of the hairpin as well as the alignment of reads across the predicted precursor, as provided by the miRDeep2 software (33). For each novel miRNA, the user can also retrieve the list of predicted target genes, obtained as described above, and download this information as a table with active hyperlinks to NCBI (Supplementary Figure S6).
We believe that this user-friendly, searchable set-up will provide a practical and valuable resource for future miRNA genomics or functional studies of the human retina.
DISCUSSION
Using high-throughput sequencing technologies, we report a systematic and comprehensive profiling of miRNA expression in the human retina at an unprecedented resolution. Thanks to the high coverage of our sequencing experiment, we were able to detect miRNAs and sequence variants that were lowly expressed. Moreover, the parallel analysis of retinae from a large number of individuals (n = 16) allowed us to establish a reliable consensus ranking and a detailed isomiR catalogue, overcoming possible inter-individual variability in miRNA expression levels and nucleotide sequences. We did not find any evident clustering of the analysed retinae based on miRNA expression according to age, gender, post-mortem time and associated morbidity of donors (Supplementary Figure S1). A larger number of samples may be required to assess the potential impact of these factors on miRNA profiles.
Our data demonstrated that almost a fifth of known human miRNAs are expressed in the neural retina. MiRNA quantification indicated a wide range of expression levels, with a limited set of miRNAs (n = 20) representing the most abundant portion of the retina miRNome. As expected, this set comprized miRNAs (e.g. miR-182-5p, miR-183-5p, miR-96-5p, miR-124-3p, miR-9-5p) with a well-established role in retina development and function in vertebrates (14,17,34,42,47). We also found miRNAs, such as miR-192-5p (Table 4), that, based on our results, are highly enriched in the retina compared to the RPE/choroid, and whose functional role in the retina has not been addressed thus far. On the other hand, a number of miRNAs with a previously reported role in retinal development and function in vertebrates, such as miR-216a and miR-23a (48,49), ranked at much lower positions in our list. The latter observation indicated that miRNAs with relatively low expression levels can be endowed with a tissue-specific physiological role, highlighting the importance of a high-coverage for the study of human tissues.
Analysis of miRNA sequence diversity indicated that a vast range of variants are expressed in the neural retina, bearing different and often composite sequence modifications. Over 90% of the retina-expressed miRNAs presented isomiRs. This high percentage refers to sequence variants consistently detected in all sixteen samples, making a significant contribution of errors generated during library preparation or sequencing very unlikely. Nevertheless, it should be added that library preparation can introduce a bias in miRNA representation (50,51). In some cases, a given isomiR was remarkably more abundant compared to the corresponding canonical mature sequence deposited in the miRBase. However, for mature miRNA sequences the term ‘canonical’ should be considered with caution as it mainly corresponds, in the miRBase, to the expression data obtained from the first tissue and/or species in which a particular miRNA was identified. Our data confirmed that analyses exclusively based on the detection of canonical sequences may present serious limitations when studying miRNA expression in specific tissues.
Both the number of individual variants and their contribution to miRNA expression indicated that 3′-modified isomiRs are the predominant category, consistent with the model that heterogeneity at the 5′ is expected to have a major impact on mRNA targeting (39). More than a fifth of the isomiRs had variations at the 5′-end or nucleotide substitutions in positions 2–8, which result in a modified seed sequence. For these isomiRs, the contribution to the overall miRNA expression levels was lower (less than 5% of total reads). Taking into account the different types of sequence alteration detected, the lower frequency and contribution to expression of seed-modifying isomiRs imply that for a significant proportion of miRNAs, seed-modified variants are likely to be restricted by functional constraints (35,39). For instance, we did not identify seed-modified isomiRs of miR-204, suggesting that in a trend of great sequence variability, some modifications may be under a strong evolutionary pressure. We have recently demonstrated that a heterozygous mutation (n.37C > T) in the seed region of the human miR-204 is responsible for an autosomal dominant condition of retinal dystrophy and bilateral coloboma (21).
For two of the ten most abundantly expressed miRNAs in the retina (miR-183-5p and miR-124a-3p), the seed-modified isomiRs represented a substantial proportion of the total reads. The genes that were predicted to be targeted by the isomiR-183-5p (and not by the respective canonical miRNA) had binding sites also for miR-182 and miR-96 (members of the miR-183/96/182 cluster), giving reasons to believe that their combinatorial action might mask the effect of isomiR-183-5p targeting (52). We therefore focused our attention on miR-124-3p. The potential significance of the predicted targeting differences between the isomiR and the canonical miRNA was validated by luciferase assay. Our data showed that CDH11, a gene involved in retinal differentiation (43), is post-transcriptionally regulated by isomiR-124-3p, but not by the canonical sequence. This result indicated that a single nucleotide addition at the 5′-end conferred to an isomiR of miR-124-3p the specific ability to target a gene with a known tissue-specific role in the retina. Our observations indicate that the specific targeting properties of seed-modifying isomiRs, especially those expressed at significant levels, deserves further systematic investigation because it may lead to the discovery of a much broader range of miRNA action in the retina as well as in other tissues.
The present study also revealed 51 novel retinal miRNAs, corroborating the recent proposition that a number of tissue-specific miRNAs are yet to be discovered (44). The newly predicted miRNAs were identified across several human retina samples (i.e. in at least half of the ones analysed) and, to the best of our knowledge, more than two thirds are novel also considering the very recent report by Londin and collaborators (44). This suggested that the predicted novel miRNAs may not have been previously discovered due to their strong tissue- or species-specificity. We did not find compelling evidence of conservation of these miRNAs in mouse, most likely because almost all human novel miRNAs are thought to be primate lineage-specific (44). Unfortunately, information on miRNA expression in the retina of non-human primates is scarce. We cannot fully exclude that some of the putative novel miRNAs may represent degradation products from genes expressed at high levels in the retina (see for instance nrm14 that is located within the 3′ UTR of the highly expressed Rhodopsin transcript). However, the size homogeneity of the reads, their relatively invariant 5′ end and coherent alignment across the hairpin (shown in the miRDeep output in the miRetina database), as well as the experimental validation of the expression of nine of them (including nrm14; Figure 5A and Supplementary Figure S4) further supports our prediction.
RNA in situ hybridization indicated that the novel miRNA nrm7 was expressed primarily at the area corresponding to the photoreceptor inner segments, supporting the specificity of the miRNA profiles described in this study. Although further investigation is needed to define the targeting properties of the predicted novel miRNAs, it is reasonable to speculate that at least a fraction of the novel miRNAs described herein are involved in modulating relevant processes for retina development and function in humans.
Finally, we determined the miRNome of the human RPE/choroid. In comparison with the neural retina, the RPE/choroid exhibited notably distinct miRNA enrichment patterns that may account for the compositional and functional differences of the two adjacent tissues. By analysing the expression profiles of the most abundant miRNAs in the retina and RPE/choroid, we identified three main groups. The first consists of miRNAs expressed at high levels in both tissues (e.g. miR-26a and miR-22) for which we assume a housekeeping role, also based on previous data (22,53). The second group comprises miRNAs (e.g. miR-204/miR-211 and miR-181a) abundantly expressed in both tissues, yet without evidence of a ubiquitous expression in other tissues (23). This group of miRNAs may be important for vision by controlling differentiation and function of several ocular tissues, including the RPE, as previously shown for miR-204 (15,54–56). The third group, represented by miRNAs enriched either in the retina or in RPE/choroid, includes the most attractive candidates for exerting a tissue-specific functional role, such as miR-143-3p, miR-126-5p and miR-146a-5p in the case of the RPE and choroid (57,58).
In conclusion, our data represent the first thorough investigation of the complexity of the human retina miRNome at nucleotide resolution. The novel resource described herein constitutes a valuable reference for scientists interested in retinal and miRNA biology. To facilitate these studies, we built an easily searchable database where both predicted novel miRNAs and sequence variants with their relative frequencies are shown in relation to their genomic location. In a long-term perspective, our study provides extended information that will ultimately improve the understanding of the contribution of miRNAs to the pathophysiology of the human retina.
DATA AVAILABILITY
The data sets supporting the results of this article are included in the Supplementary Data. The raw data sets of the sequencing experiments described herein are available from the ArrayExpress repository (http://www.ebi.ac.uk/arrayexpress/) under the accession number E-MTAB-3792.
Supplementary Material
Acknowledgments
The authors are grateful to Mohit Parekh from the Eye Bank of Venice for the collection of human retina and RPE/choroid samples. The authors would also like to thank Manuela Dionisi and the Next Generation Sequencing Facility (TIGEM).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for this work and for open access charge: Italian Telethon Foundation [TGM11SB2 to S.B., TGM11SB1 to D.d.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ameres S.L., Zamore P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 2.Plasterk R.H. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 3.Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 4.Wienholds E., Plasterk R.H. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 5.Couzin J. MicroRNAs make big impression in disease after disease. Science. 2008;319:1782–1784. doi: 10.1126/science.319.5871.1782. [DOI] [PubMed] [Google Scholar]
- 6.Meola N., Gennarino V.A., Banfi S. microRNAs and genetic diseases. PathoGenetics. 2009;2:7. doi: 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iorio M.V., Croce C.M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiland M., Gao X.H., Zhou L., Mi Q.S. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 9.Sundermeier T.R., Palczewski K. The physiological impact of microRNA gene regulation in the retina. Cell. Mol. Life Sci. 2012;69:2739–2750. doi: 10.1007/s00018-012-0976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damiani D., Alexander J.J., O'Rourke J.R., McManus M., Jadhav A.P., Cepko C.L., Hauswirth W.W., Harfe B.D., Strettoi E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J. Neurosci. 2008;28:4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis N., Mor E., Ashery-Padan R. Roles for Dicer1 in the patterning and differentiation of the optic cup neuroepithelium. Development. 2011;138:127–138. doi: 10.1242/dev.053637. [DOI] [PubMed] [Google Scholar]
- 12.Georgi S.A., Reh T.A. Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J. Neurosci. 2010;30:4048–4061. doi: 10.1523/JNEUROSCI.4982-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinter R., Hindges R. Perturbations of microRNA function in mouse dicer mutants produce retinal defects and lead to aberrant axon pathfinding at the optic chiasm. PLoS One. 2010;5:e10021. doi: 10.1371/journal.pone.0010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Busskamp V., Krol J., Nelidova D., Daum J., Szikra T., Tsuda B., Juttner J., Farrow K., Scherf B.G., Alvarez C.P., et al. miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron. 2014;83:586–600. doi: 10.1016/j.neuron.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Conte I., Carrella S., Avellino R., Karali M., Marco-Ferreres R., Bovolenta P., Banfi S. miR-204 is required for lens and retinal development via Meis2 targeting. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15491–15496. doi: 10.1073/pnas.0914785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Z.B., Hirokawa G., Gui L., Takahashi R., Osakada F., Hiura Y., Takahashi M., Yasuhara O., Iwai N. Targeted deletion of miR-182, an abundant retinal microRNA. Mol. Vis. 2009;15:523–533. [PMC free article] [PubMed] [Google Scholar]
- 17.Krol J., Busskamp V., Markiewicz I., Stadler M.B., Ribi S., Richter J., Duebel J., Bicker S., Fehling H.J., Schubeler D., et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 18.Sanuki R., Onishi A., Koike C., Muramatsu R., Watanabe S., Muranishi Y., Irie S., Uneo S., Koyasu T., Matsui R., et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat. Neurosci. 2011;14:1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 19.Walker J.C., Harland R.M. microRNA-24a is required to repress apoptosis in the developing neural retina. Genes Dev. 2009;23:1046–1051. doi: 10.1101/gad.1777709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Q., Sun W., Okano K., Chen Y., Zhang N., Maeda T., Palczewski K. Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J. Biol. Chem. 2011;286:31749–31760. doi: 10.1074/jbc.M111.259028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conte I., Hadfield K.D., Barbato S., Carrella S., Pizzo M., Bhat R.S., Carissimo A., Karali M., Porter L.F., Urquhart J., et al. MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E3236–E3245. doi: 10.1073/pnas.1401464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karali M., Peluso I., Gennarino V.A., Bilio M., Verde R., Lago G., Dolle P., Banfi S. miRNeye: a microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karali M., Peluso I., Marigo V., Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest. Ophthalmol. Vis. Sci. 2007;48:509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- 24.Loscher C.J., Hokamp K., Kenna P.F., Ivens A.C., Humphries P., Palfi A., Farrar G.J. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soundara Pandi S.P., Chen M., Guduric-Fuchs J., Xu H., Simpson D.A. Extremely complex populations of small RNAs in the mouse retina and RPE/choroid. Invest. Ophthalmol. Vis. Sci. 2013;54:8140–8151. doi: 10.1167/iovs.13-12631. [DOI] [PubMed] [Google Scholar]
- 26.Aherne A., Kennan A., Kenna P.F., McNally N., Lloyd D.G., Alberts I.L., Kiang A.S., Humphries M.M., Ayuso C., Engel P.C., et al. On the molecular pathology of neurodegeneration in IMPDH1-based retinitis pigmentosa. Hum. Mol. Genet. 2004;13:641–650. doi: 10.1093/hmg/ddh061. [DOI] [PubMed] [Google Scholar]
- 27.Parekh M., Ferrari S., Di Iorio E., Barbaro V., Camposampiero D., Karali M., Ponzin D., Salvalaio G. A simplified technique for in situ excision of cornea and evisceration of retinal tissue from human ocular globe. J. Vis. Exp. 2012:e3765. doi: 10.3791/3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pantano L., Estivill X., Marti E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010;38:e34. doi: 10.1093/nar/gkp1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maragkakis M., Reczko M., Simossis V.A., Alexiou P., Papadopoulos G.L., Dalamagas T., Giannopoulos G., Goumas G., Koukis E., Kourtis K., et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 33.Friedlander M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lumayag S., Haldin C.E., Corbett N.J., Wahlin K.J., Cowan C., Turturro S., Larsen P.E., Kovacs B., Witmer P.D., Valle D., et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E507–E516. doi: 10.1073/pnas.1212655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cammaerts S., Strazisar M., De Rijk P., Del Favero J. Genetic variants in microRNA genes: impact on microRNA expression, function, and disease. Front. Genet. 2015;6:186. doi: 10.3389/fgene.2015.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morin R.D., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neilsen C.T., Goodall G.J., Bracken C.P. IsomiRs–the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012;28:544–549. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Burroughs A.M., Ando Y., de Hoon M.J., Tomaru Y., Nishibu T., Ukekawa R., Funakoshi T., Kurokawa T., Suzuki H., Hayashizaki Y., et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan G.C., Chan E., Molnar A., Sarkar R., Alexieva D., Isa I.M., Robinson S., Zhang S., Ellis P., Langford C.F., et al. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014;42:9424–9435. doi: 10.1093/nar/gku656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng W.C., Chung I.F., Tsai C.F., Huang T.S., Chen C.Y., Wang S.C., Chang T.Y., Sun H.J., Chao J.Y., Cheng C.C., et al. YM500v2: a small RNA sequencing (smRNA-seq) database for human cancer miRNome research. Nucleic Acids Res. 2015;43:D862–D867. doi: 10.1093/nar/gku1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loher P., Londin E.R., Rigoutsos I. IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies. Oncotarget. 2014;5:8790–8802. doi: 10.18632/oncotarget.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu K., Liu Y., Mo W., Qiu R., Wang X., Wu J.Y., He R. MiR-124 regulates early neurogenesis in the optic vesicle and forebrain, targeting NeuroD1. Nucleic Acids Res. 2011;39:2869–2879. doi: 10.1093/nar/gkq904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clendenon S.G., Sarmah S., Shah B., Liu Q., Marrs J.A. Zebrafish cadherin-11 participates in retinal differentiation and retinotectal axon projection during visual system development. Dev. Dyn. 2012;241:442–454. doi: 10.1002/dvdy.23729. [DOI] [PubMed] [Google Scholar]
- 44.Londin E., Loher P., Telonis A.G., Quann K., Clark P., Jing Y., Hatzimichael E., Kirino Y., Honda S., Lally M., et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E1106–E1115. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., Magen A., Canidio E., Pagani M., Peluso I., et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishida A., Furukawa A., Koike C., Tano Y., Aizawa S., Matsuo I., Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 47.La Torre A., Georgi S., Reh T.A. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E2362–E2370. doi: 10.1073/pnas.1301837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olena A.F., Rao M.B., Thatcher E.J., Wu S.Y., Patton J.G. miR-216a regulates snx5, a novel notch signaling pathway component, during zebrafish retinal development. Dev. Biol. 2015;400:72–81. doi: 10.1016/j.ydbio.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rasheed V.A., Sreekanth S., Dhanesh S.B., Divya M.S., Divya T.S., Akhila P.K., Subashini C., Chandrika Sivakumar K., Das A.V., James J. Developmental wave of Brn3b expression leading to RGC fate specification is synergistically maintained by miR-23a and miR-374. Dev. Neurobiol. 2014;74:1155–1171. doi: 10.1002/dneu.22191. [DOI] [PubMed] [Google Scholar]
- 50.Sorefan K., Pais H., Hall A.E., Kozomara A., Griffiths-Jones S., Moulton V., Dalmay T. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence. 2012;3:4. doi: 10.1186/1758-907X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hafner M., Renwick N., Brown M., Mihailovic A., Holoch D., Lin C., Pena J.T., Nusbaum J.D., Morozov P., Ludwig J., et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dambal S., Shah M., Mihelich B., Nonn L. The microRNA-183 cluster: the family that plays together stays together. Nucleic Acids Res. 2015;43:7173–7188. doi: 10.1093/nar/gkv703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddy A.M., Zheng Y., Jagadeeswaran G., Macmil S.L., Graham W.B., Roe B.A., Desilva U., Zhang W., Sunkar R. Cloning, characterization and expression analysis of porcine microRNAs. BMC Genomics. 2009;10:65. doi: 10.1186/1471-2164-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adijanto J., Castorino J.J., Wang Z.X., Maminishkis A., Grunwald G.B., Philp N.J. Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J. Biol. Chem. 2012;287:20491–20503. doi: 10.1074/jbc.M112.354761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Avellino R., Carrella S., Pirozzi M., Risolino M., Salierno F.G., Franco P., Stoppelli P., Verde P., Banfi S., Conte I. miR-204 targeting of Ankrd13A controls both mesenchymal neural crest and lens cell migration. PLoS One. 2013;8:e61099. doi: 10.1371/journal.pone.0061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang F.E., Zhang C., Maminishkis A., Dong L., Zhi C., Li R., Zhao J., Majerciak V., Gaur A.B., Chen S., et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kutty R.K., Nagineni C.N., Samuel W., Vijayasarathy C., Jaworski C., Duncan T., Cameron J.E., Flemington E.K., Hooks J.J., Redmond T.M. Differential regulation of microRNA-146a and microRNA-146b-5p in human retinal pigment epithelial cells by interleukin-1beta, tumor necrosis factor-alpha, and interferon-gamma. Mol. Vis. 2013;19:737–750. [PMC free article] [PubMed] [Google Scholar]
- 58.Wang S., Aurora A.B., Johnson B.A., Qi X., McAnally J., Hill J.A., Richardson J.A., Bassel-Duby R., Olson E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the results of this article are included in the Supplementary Data. The raw data sets of the sequencing experiments described herein are available from the ArrayExpress repository (http://www.ebi.ac.uk/arrayexpress/) under the accession number E-MTAB-3792.