Abstract
Ribosome synthesis employs a number of energy-consuming enzymes in both eukaryotes and prokaryotes. One such enzyme is the conserved circularly permuted GTPase Nug1 (nucleostemin in human). Nug1 is essential for 60S subunit assembly and nuclear export, but its role and time of action during maturation remained unclear. Based on in vitro enzymatic assays using the Chaetomium thermophilum (Ct) orthologue, we show that Nug1 exhibits a low intrinsic GTPase activity that is stimulated by potassium ions, rendering Nug1 a cation-dependent GTPase. In vivo we observe 60S biogenesis defects upon depletion of yeast Nug1 or expression of a Nug1 nucleotide-binding mutant. Most prominently, the RNA helicase Dbp10 was lost from early pre-60S particles, which suggested a physical interaction that could be reconstituted in vitro using CtNug1 and CtDbp10. In vivo rRNA–protein crosslinking revealed that Nug1 and Dbp10 bind at proximal and partially overlapping sites on the 60S pre-ribosome, most prominently to H89 that will constitute part of the peptidyl transferase center (PTC). The binding sites of Dbp10 are the same as those identified for the prokaryotic helicase DbpA bound to the 50S subunit. We suggest that Dbp10 and DbpA are performing a conserved role during PTC formation in all organisms.
INTRODUCTION
Ribosome biogenesis is a complex and highly dynamic process requiring the precise coordination of multiple processing, modification and assembly steps. In yeast, four rRNA species (18S, 5.8S, 25S and 5S rRNA) must assemble together with 79 ribosomal proteins (r-proteins) to form the small (40S) and the large (60S) subunits (1,2). This process occurs within a series of pre-ribosomal particles and requires the activity of a plethora of transiently associating biogenesis factors. In yeast, more than 200 ribosome biogenesis factors and 70 small nucleolar RNAs (snoRNAs) are involved in ribosome assembly, however, the exact function of most of the assembly factors remains elusive (3–5). Of the identified biogenesis factors, a small percentage is predicted or has been shown to display enzymatic activities, e.g. ATPase, GTPase, kinase or methyl-transferase activity (2).
Among the assembly factors that exhibit enzymatic activity is Nug1, an evolutionary conserved GTPase, found in all three domains of life that is required for the biogenesis of the large 60S subunit. Nug1 is a circularly permuted GTPase (cpGTPase) where the conserved G motifs have been reordered [(G5/DAR)-G4-G1-(G2)-G3]. Despite variation in the motif order, the three-dimensional structure of the G-domain is preserved as seen in the structures of the cpGTPases YlqF (B. subtilis) and YjeQ (Escherichia coli) (6,7). One distinguishing feature of cpGTPases is the presence of additional domains flanking the GTPase core. These are proposed to stabilize the permuted G-domain and are believed to propagate intra-molecular conformational changes (8). Correspondingly, the predicted domain architecture of Nug1 reveals the presence of N- and C-terminal domains flanking the central GTPase domain. The N-terminal domain is present only in eukaryotic orthologues and is rich in positively charged amino acids. This domain is essential for nucleolar targeting and association with pre-60S particles, but it has also been shown to exhibit rRNA binding activity (9). In contrast, the C-terminal domain is conserved from bacteria and archaea to eukaryotes (9,10), but little is known about its function. Previous studies have shown that yeast Nug1 can hydrolyse GTP in vitro (9). However, the Km (0.2 mM) and Kcat (0.11 min−1) calculated show that Nug1 displays an intrinsically low GTP hydrolysis activity.
In this study, we define a novel role for Nug1 in ribosome biogenesis. Mutant forms of Nug1, unable to bind nucleotide, were analyzed in vivo and found to display 60S biogenesis defects. Specifically, we show that the composition of early Ssf1 and Nsa1 pre-60S particles is altered in a Nug1 nucleotide-binding mutant or when Nug1 is depleted. One factor that clearly decreases in these particles is Dbp10, an RNA helicase, which is genetically linked to Nug1 (9). We show that Nug1 and Dbp10 bind adjacent to each other at a site on the 60S subunit that goes on to form the peptidyl-transferase center (PTC) in the mature ribosome. Together, our data indicate that Nug1 binding, but not its GTPase activity is required for the stable association of Dbp10 helicase with the pre-ribosome. We suggest that the Nug1 GTPase displays a function upon nucleotide binding that together with the helicase activity of Dbp10 are involved in the formation of the PTC.
MATERIALS AND METHODS
Yeast strains and genetic methods
All S. cerevisiae strains used in this study are listed in Supplementary Table S1 and, unless otherwise specified, are derivatives of W303 and DS1–2b. Preparation of media, yeast transformation and genetic manipulations were done according to standard procedures performed as previously described (11,12).
Plasmid constructs
All recombinant DNA techniques were performed according to standard procedures using E. coli DH5α for cloning and plasmid propagation. Site-directed mutagenesis was performed by overlap-extension PCR. All cloned DNA fragments generated by PCR amplification were verified by sequencing. Plasmids used in this study are listed in Supplementary Table S2. CtNug1 (accession number CTHT_0059920) and CtDbp10 (accession number CTHT_0033480) were amplified from a Chaetomium thermophilum cDNA library (13) and cloned into appropriate E. coli or yeast expression vectors.
Expression and purification of CtNug1 and CtDbp10
Full-length CtNug1 was cloned into the expression vector pET24a fused with a C-terminal 6xHis-tag. The CtNug1 expression vector was transformed to E. coli BL21 CodonPlus RIL strain (Stratagene), grown in LB media and induced with 1mM IPTG (30°C for 3 h). Harvested cell pellets were resuspended in lysis buffer (20 mM HEPES pH 8.0, 250 mM KCl, 10 mM NaCl, 5 mM MgCl2, 1 mM DTT and protease inhibitor). Lysis was performed using a high-pressure cavitation homogenizer (microfluidizer) and followed by centrifugation at 39 000 × g at 4°C for 20 min. The supernatant was incubated with 1 ml of pre-equilibrated slurry of SP-sepharose beads (Sigma) at 4°C for 1 h. Following extensive washing, CtNug1 protein was eluted from the SP-sepharose using lysis buffer supplemented with 600 mM KCl. The SP-sepharose eluate was then slowly diluted to a final KCl concentration of 400 mM. Pre-equilibrated Ni-NTA agarose slurry (0.5 ml) (Machery-Nagel) were added and incubated at 4°C for 1 h. After Ni-NTA binding, the beads were washed with lysis buffer and CtNug1 was eluted twice from the beads with 500 mM imidazole in lysis buffer (without DTT). The protein was further purified by size exclusion chromatography using Superdex 200 (GE Healthcare) on a ÄktaPurifier System (GE Healthcare), pre-equilibrated in lysis buffer. Purified CtNug1 was concentrated, flash-frozen in liquid nitrogen and stored at −80°C. The same purification scheme was followed for the CtNug1 G-domain mutants.
Full-length CtDbp10 was cloned into a yeast leu2d vector under the inducible GAL1–10 promoter, carrying an N-terminal pA-TEV-FLAG tag. Heterologous expression of C. thermophilum proteins in S. cerevisiae was carried out into DS1–2b cells. For galactose induction, cells were grown in 1L raffinose (SRC-) medium to an OD600 of 2 and then diluted to 2L with galactose medium (YPG) to induce expression. When the OD600 reached 4, cells were harvested and resuspended in lysis buffer (50 mM Tris-HCl pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.15% (v/v) Nonident P40, 2 mM CaCl2, 5% (v/v) glycerol and protease inhibitor mix. Lysis was performed with 0.5 mm glass beads using a ball mill (Fritsch Pulverisette), followed by centrifugation at 39 000 × g at 4°C for 20 min. The supernatant was incubated with 0.5 ml of pre-equilibrated IgG-Sepharose slurry (GE-Healthcare) at 4°C for 1 h. The IgG-Sepharose beads were washed with 10 column volumes of lysis buffer to remove any unbound protein. Bound CtDbp10 was subjected to TEV cleavage (1 mg/ml TEV) for 1.5 h at 16°C. The TEV-eluate was collected, added to 0.3 ml pre-equilibrated anti-FLAG slurry (Sigma) and incubated for 1 h at 4°C. After binding the anti-FLAG beads were washed with 10 column volumes of lysis buffer and elution was carried out with 0.6 ml of 2x FLAG peptide (Sigma) for 1 h at 4°C.
GTPase assays with single-turnover reactions
The GTPase activity experiments were performed as previously described (14–17). CtNug1 wild-type and mutants were incubated with a final concentration of 0.1 μM GTP containing 750 nCi of [γ-32P]-labeled GTP in buffer containing 20 mM HEPES pH 8.0, 200 mM KCl, 5 mM MgCl2, 1 mM DTT for the indicated time at 37°C. For the different ion-dependent experiments 200 mM of NaCl, CsCl, NH4Cl or RbCl were used instead of KCl in the buffer. After the reaction, the hydrolyzed γ-phosphate was separated by thin layer chromatography, exposed overnight on a Phosphorimager screen (BAS-MS 2040 Fujifilm) and scanned with a FLA-7000 (Fujifilm). ImageJ and GraphPad PRISM software were used for quantification and analysis, respectively.
Fluorescence-based nucleotide binding assays
The nucleotide-binding assays were performed using the fluorescently labeled nucleotides mant-GTP [2′-/3′-O-(N’-methylanthraniloyl) guanosine-5′-O- triphosphate] (JenaBioscience)(17). Reactions of 100 μl were performed in 96 well-plates, with 1 μM of recombinant protein incubated with 0.5 μM of mant-nucleotides in buffer containing 20 mM HEPES pH 8.0, 200 mM KCl, 5 mM MgCl2, 1 mM DTT for 10 min at 30°C. The reaction mixture was then excited at 355 nm with a xenon lamp, and emission spectra were recorded between 385 and 600 nm with a 5-nm increment step using a Synergy 4 spectrophotometer (BioTek). All data were processed with Microsoft Office Excel and GraphPad PRISM.
CRAC analysis
The crosslinking and analysis of cDNA (CRAC) experiments were performed as described (18) using a 6xHis-TEV-ProtA tag either in the N- or C-terminal end of ScDbp10 and ScNug1, respectively. Cultures were grown in SDC medium to OD600 0.8 and cells were UV-irradiated in the Megatron UV chamber at a dose of 1.6 J cm−2 for 3 min and processed as described (18,19). The cDNAs originating from ScNug1 and ScDbp10 CRAC experiments were sequenced on the Illumina MiSeq system, according to manufacturer's procedures. Illumina sequencing data were aligned to the yeast genome using Novoalign (http://www.novocraft.com). Downstream analyses were performed using the pyCRAC tool suite (20) and the UCSF Chimera (21).
Miscellaneous
Further methods used in this study and previously described were TAP purifications of pre-60S particles (22) and sucrose gradient analysis to obtain ribosomal and polysomal profiles (23,24). RNA extractions, Northern analysis and primer extensions were performed as previously described (25,26). Probes used for Northern analysis and primer extension were as follows 003 (TGCTTACCTCTGGGCC), 020 (TGAGAAGGAAATGACGCT), 007(CTCCGC TTATTGATATGC), 004(CGGTTTTAATTGTCCTA), 008 (CATGGCTTAAT CTTTGAG), 017 (GCGTTGTTCATCGATGC) and 041 (CTACTCGGT CAGGCTC). Antibodies used for Western blotting in the following dilutions were anti-Nug1 (27) 1:1000; anti-Rsa4 (28) 1:10 000; anti-RpL35 (29) 1:35 000; anti-RpL3 (30) 1:5000; anti-Nsa2 (31) 1:10 000; anti-Nog1 (32) 1:30 000; anti-Yvh1 (27) 1:4000; anti-Mrt4 (33) 1:1000; anti-Rpp0 (34) 1:10; anti-Arc1 (35) 1:20 000; anti-Rpl12 (36) 1:10; anti-Tif6 (37) 1:10 000; anti-FLAG (monoclonal, Sigma, HRP-conjugated) 1:10 000; anti-CBP (Polyclonal, Open Biosystems) 1:70 000; goat-anti-mouse (BioRad, HRP-conjugated) 1:3000 and mouse-anti-rabbit (BioRad, HRP-conjugated) 1:3000. Page ruler unstained protein ladder (Thermo Scientific) was used as a protein marker; Brilliant Blue G-Colloidal Concentrate Electrophoresis Reagent (Sigma-Aldrich) was used for Coomassie stain, and 4–12% NuPAGE Bis-Tris gels (Novex) together with NuPAGE MOPS SDS running buffer (Invitrogen) were used for SDS–PAGE.
RESULTS
CtNug1 exhibits a low intrinsic GTPase activity in vitro that can be stimulated by potassium ions
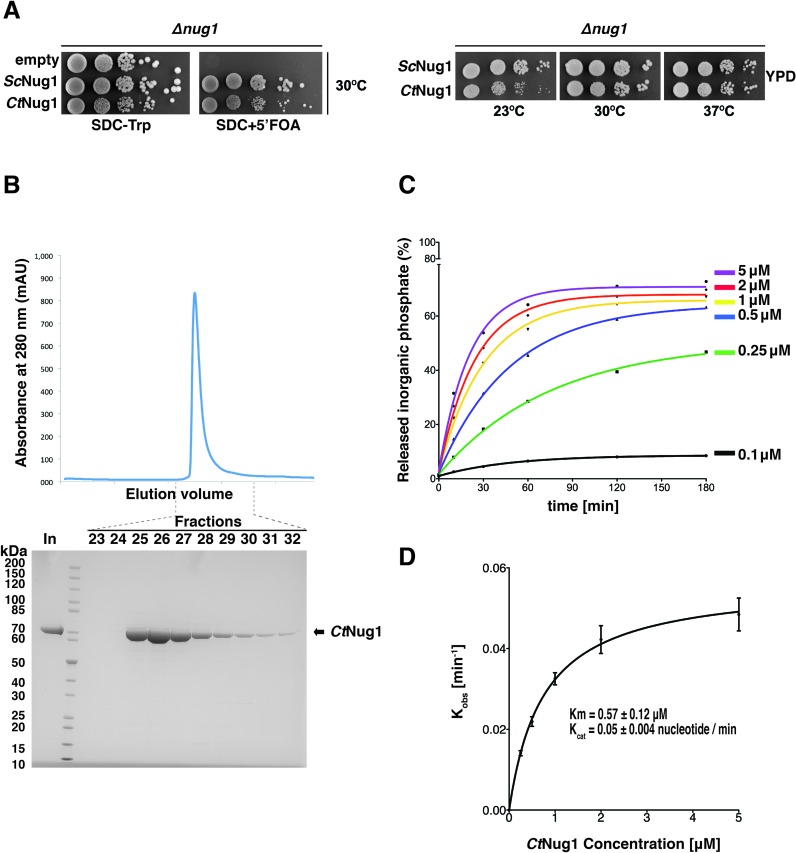
In order to elucidate Nug1's function in ribosome biogenesis, we sought to further characterise its GTPase activity by employing single turnover GTPase assays (14–17). As the yeast Nug1 protein could not be obtained in a high yield and soluble form when recombinantly expressed in E. coli, we used the Chaetomium thermophilum homologue of Nug1 (CtNug1) for the GTP binding and hydrolysis assays. Importantly, CtNug1 is functional in yeast, which was shown by expressing CtNug1 under the control of the endogenous yeast NUG1 promoter in a haploid nug1 null strain (Figure 1A). Apparently, CtNug1 complements the otherwise non-viable yeast nug1 disruption mutant very efficiently at 30°C and 37°C, similar to the endogenous yeast NUG1 (Figure 1A). It is only when propagated at lower temperatures (23°C) that a growth defect could be observed in the case of the thermophilic Nug1 protein in the mesophilic yeast.
Figure 1.
CtNug1 is functionally interchangeable with ScNug1 and displays a low intrinsic GTPase activity. (A) In vivo complementation of the yeast Nug1 shuffle strain (nug1Δ, pURA3-NUG1) transformed with empty plasmid, TRP1 plasmids harboring yeast ScNUG1 or C. thermophilum CtNUG1. The yeast (Sc) and the thermophilic (Ct) Nug1 ORFs were introduced into the centromeric YCplac22 plasmid and expressed under the control of the native yeast NUG1 promoter (PNUG1). Cells were spotted in 10-fold serial dilutions on SDC-Trp (loading control) and SDC+5′ FOA plates (complementation) and grown at 30°C for 3 days (left panel). Growth analysis of the yeast nug1Δ strain complemented by Ycplac22-PNUG1::ScNUG1 or Ycplac22-PNUG1::CtNUG1. Transformants were spoted in 10-fold serial dilution on YPD plates and grown at the indicated temperatures for 2 days. (B) Purification of recombinantly expressed CtNug1 from E. coli. Upper panel: chromatogram from size-exclusion chromatography (Superdex200 10/300) of purified CtNug1. Y-axis, protein absorbance at 280 nm, expressed in absorbance units (mAU); X-axis, fractions from the gel filtration column. Lower panel: SDS-PAGE of the fractions from the size-exclusion chromatography. The numbers on top of the gel correspond to the gel-fitration fractions, and ‘In’ denotes the input of purified CtNug1. (C) GTPase activity of purified CtNug1 tested in single turnover experiments. Ratio of hydrolyzed phosphate to total GTP is plotted against time for each of the indicated CtNug1 concentrations using the highly purified CtNug1 (see B). (D) Characterization of CtNug1's Km and Kcat values based on single-turnover GTPase assays. For each of the curves obtained in the GTPase assays in (C), the observed rate constants (Kobs) were calculated and plotted against the different concentrations of CtNug1. Non-linear regression and the standard enzyme kinetics equations of GraphPad software were used to calculate the indicated Km and Kcat values.
When CtNug1 was recombinantly expressed and affinity-purified from E. coli, a high yield of pure, soluble protein could be obtained from the final gel filtration step (final concentration ∼20 mg/ml) (Figure 1B). The calculated molecular weight of CtNug1 from SEC-MALS analysis (data not shown) is consistent with Nug1 being a monomer. The purified CtNug1 was then tested in single turnover GTPase assays. The amount of phosphate released (Pi) following incubation of varying concentrations of CtNug1 protein with [γ-32P]-labeled GTP was used to calculate Km and Kcat values (Figure 1C). Accordingly, CtNug1 hydrolyzes GTP with a Km of 0.57 ± 0.12 μM and a Kcat of 0.05 ± 0.004 nucleotide/min (Figure 1D), which is comparable to what has been previously found for the yeast Nug1 (9). Thus, also Chaetomium thermophilum Nug1 has a low intrinsic GTPase activity, suggesting that eukaryotic Nug1 GTPases may have in general low GTP hydrolysis activity, which could be subject to regulation.
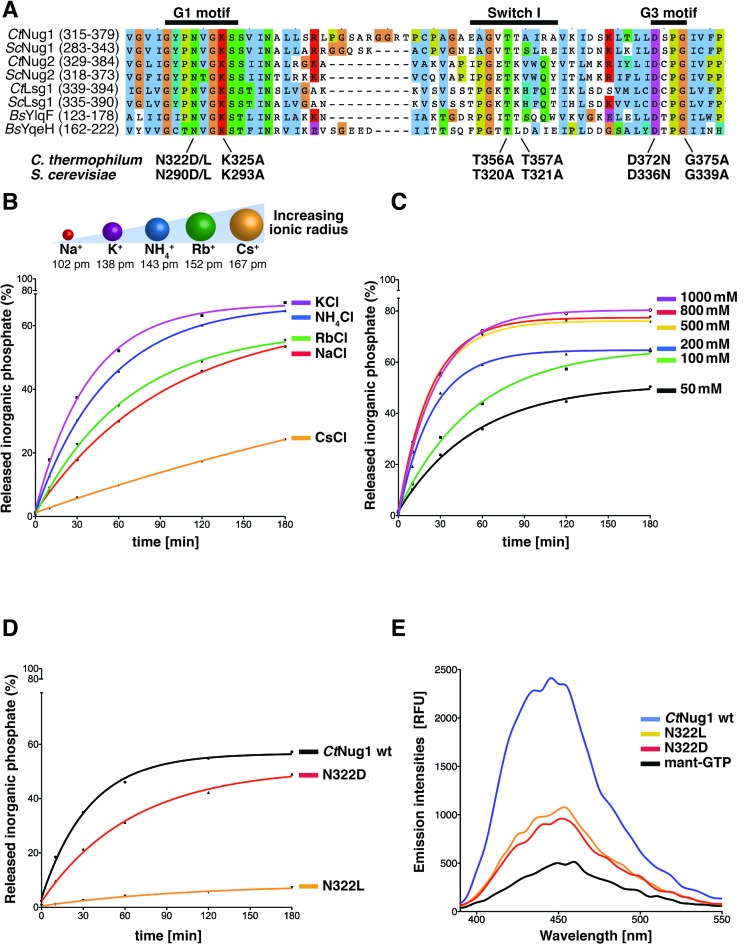
Recently, a new group of GTPases termed ‘cation-dependent’ (CD-GTPases) has been described (10). This sub-family exhibits enhanced GTPase activity in the presence of certain cations, in particular potassium ions. Furthermore, the crystal structures of members of these CD-GTPases (e.g. MnmE, FeoB) suggest that the radius of the cation is restricted, by the size of the enzymatic pocket (10,38–40). From multiple sequence alignment of Nug1 with these CD-GTPases, CtNug1 was found to contain the conserved residue within the G1 motif (N322) predicted to coordinate the cation (Figure 2A). To assess if the catalytic activity of Nug1 is affected by the presence of cations, GTPase assays were performed in the presence of cations of increasing ionic radii (Na+: 102 pm, K+: 138 pm, NH4+: 143 pm, Rb+: 152 pm, Cs+: 167 pm) (Figure 2B). Of the different ions tested, a maximal stimulation of CtNug1's GTPase activity was observed in the presence of potassium (K+) ions, whereas the lowest enzymatic activity was obtained for caesium (Cs+), the largest ion tested. Additionally, we tested whether increasing the concentration of potassium ions (50–1000 mM) could further stimulate the enzymatic activity of CtNug1 (Figure 2C). These experiments showed that the GTPase activity of CtNug1 increased as the concentration of potassium rose up to 500 mM KCl. However, above 500 mM of KCl no further stimulation on GTPase activity was observed.
Figure 2.
CtNug1 intrinsic low GTPase activity can be stimulated by potassium ions. (A) Multiple-sequence alignment of the G-domain of cation-dependent GTPases involved in ribosome biogenesis (eukaryotic Ct and Sc Nug1, Nug2 and Lsg1, aligned with the prokaryotic YlqF and YqeH). G1 [GxxNxGKS] (where the conserved asparagine residue N322 in CtNug1 is responsible for the cation binding), Switch I and G3 [DxxG] motifs are labeled. The conserved residues mutagenized in these motifs in CtNug1 and ScNug1 are indicated. (B–D) GTPase activity of CtNug1 tested in single turnover experiments under different conditions (B) in the presence of cations with increasing ionic radius, (C) with increasing concentrations of KCl in the reaction and (D) when CtNug1 wild-type (wt) was compared to potassium coordination mutants N322D and N322L. The concentration of potassium used is 300 mM. The cartoon in (B) depicts the ionic radii and the indicated numbers corresponding to coordination number VI. (E) Fluorescence-based nucleotide binding assay. Mant-GTP was mixed with purified CtNug1 wild-type or mutants (N322D, N322L) and the change in the emission spectra between the indicated proteins were measured in relative fluorescence units (RFUs). All in vitro assays were performed at least twice with highly reproducible data sets.
To confirm that the enhanced activity seen for CtNug1 was due to cation stimulation, the conserved asparagine in the G1 motif [GxxNxGKS] predicted to be involved in K+ coordination was mutated (N322D and N322L, referred thereafter as K+-loop mutants). As anticipated, the K+-loop mutants exhibited decreased GTPase activity, particularly for the Nug1 N322L mutant, when compared to wild-type protein (Figure 2D). Additionally, while both mutants were found to bind guanine nucleotide, the binding was decreased when compared to the wild-type CtNug1 (Figure 2E). Together, these in vitro studies show that CtNug1 displays an intrinsically low GTPase activity that can be stimulated by potassium ions.
Mutations in the GTPase domain of CtNug1 impair nucleotide binding or hydrolysis
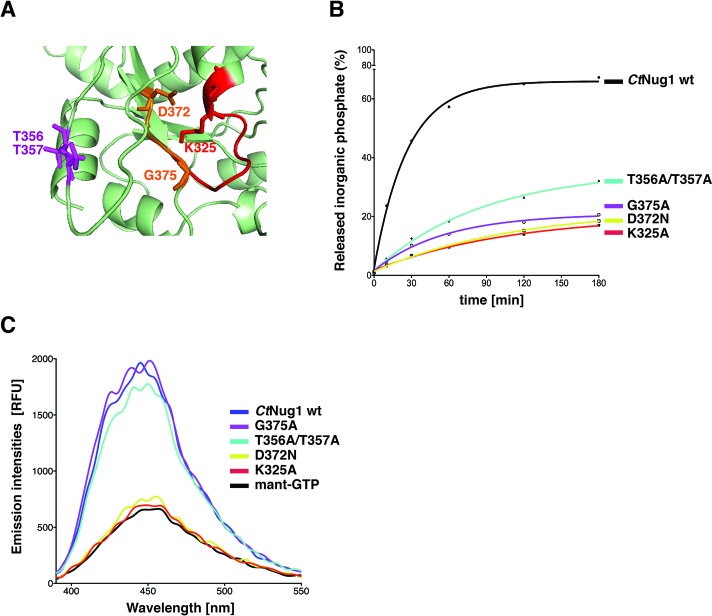
To further characterize Nug1's nucleotide binding and hydrolysis activity, we generated a series of point mutations within each of the conserved motifs (G1 to G3) present in the CtNug1 GTPase domain (Figures 2A and 3A). The residues mutated in the G1 (K325A) or G3 motif (D372N) are predicted to inhibit nucleotide binding, while those generated in either the G2 (T356A/T357A) or the G3 motif (G375A) were designed to affect only GTP hydrolysis. Subsequent GTPase assays, as described above, revealed that each of the Nug1 mutants showed a decrease in GTP hydrolysis activity, when compared to wild-type CtNug1 (Figure 3B). Interestingly, the mutants predicted to be involved in nucleotide binding (K325A and D372N) exhibited the greatest decrease in activity. Furthermore, fluorescence-based nucleotide-binding assays (Figure 3C) demonstrated that the Nug1 K325A and D372N mutants are indeed inhibited in nucleotide binding, as their emission spectra were similar to background (fluorescent-analog alone). In contrast, the Nug1 G375A and T356A/T357A mutants predicted to be only defective in GTP hydrolysis indeed could bind mant-GTP at the same levels as the wild-type CtNug1 (Figure 3C). Thus, the Nug1 mutants generated, allowed us to effectively distinguish between defects in GTP binding versus hydrolysis.
Figure 3.
GTPase-domain mutants of CtNug1 are impaired in nucleotide binding or hydrolysis. (A) A magnified view of the CtNug1 GTPase-domain model, based upon the crystal structure of the B. subtilis YlqF (PDB ID: 1PUJ) homologue using the Phyre2 software (58). The conserved residues that are mutagenized in G1 (red), Switch I (purple) and G3 (orange) are shown. (B) GTPase activity of recombinantly purified CtNug1 wild-type and representative G-domain mutants, as tested in single turnover assays. (C) Fluorescence-based nucleotide binding assay. Mant-GTP was mixed with purified CtNug1 wild-type and the indicated G-domain mutants.
Nug1 nucleotide-binding mutant causes defects in 60S subunit maturation
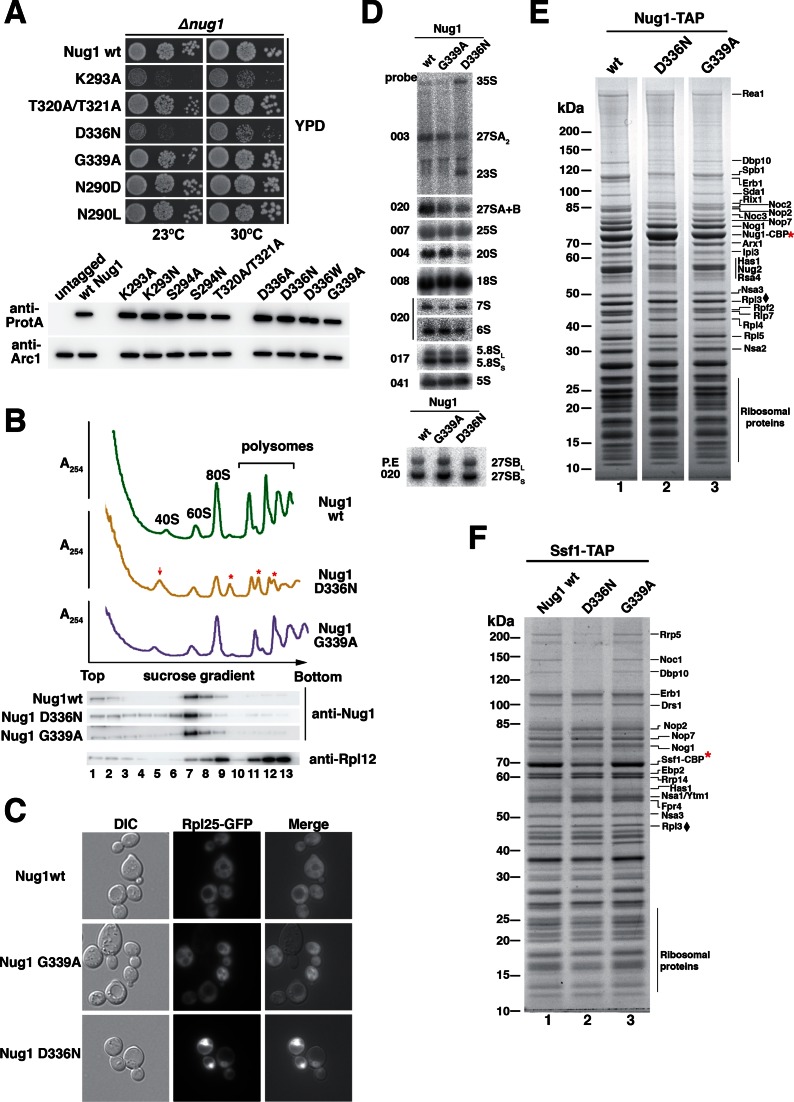
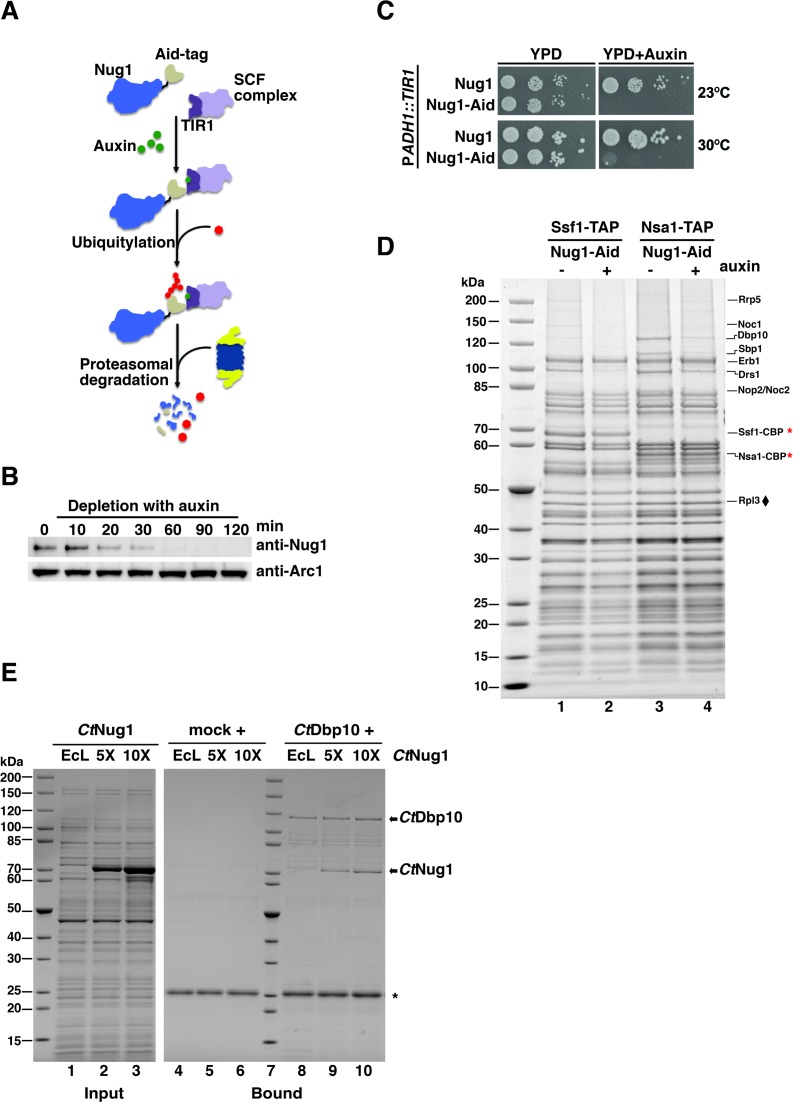
To gain in vivo insight into the role played by Nug1 nucleotide binding versus GTP hydrolysis, the orthologous yeast mutants (Figure 2A) were generated based on the data obtained with CtNug1 and tested for complementation using the Nug1 shuffle strain (Figure 4A). While all yeast Nug1 mutants (nug1K283A, nug1T320A/T321A, nug1D336N, nug1G339A, nug1N290L and nug1N290D) could complement the non-viable nug1Δ strain, their growth behaviour differed (Figure 4A). The Nug1 constructs carrying mutations shown in vitro to affect nucleotide binding (K293A, D336N) exhibited substantially slower growth when compared to the wild-type Nug1. In contrast, growth appeared to be unaffected in mutants defective in GTP hydrolysis (T320A/T321A, G339A) or in the K+-loop mutants (N290D and N290L). These differences in growth were not due to either altered levels of expression or protein instability as assessed by Western blotting of whole cell lysates (Figure 4A; lower panel). This suggests that the growth defects seen in the nucleotide-binding Nug1 mutants are due to functional differences from the wild-type protein.
Figure 4.
Nug1 nucleotide-binding mutant causes defects in 60S subunit biogenesis. (A) Growth behavior of Nug1 GTPase mutants were tested in a nug1Δ background complemented by wild-type NUG1 or mutant nug1 K293A, T320A/T321A, D336N, G339A, N290D or N290L, all carried on a plasmid (YCplac22, CEN3, TRP1). Ten-fold serial dilutions of the indicated strains were spotted on YPD plates for 2 days at the indicated temperatures. Lower panel: whole cell lysates were prepared from exponentially growing cells for each of the indicated mutants. Samples were analyzed on SDS-PAGE and the protein levels of the Nug1 wild-type or mutants were determined by Western blotting using antibodies against the C-terminal TAP-tag (anti-protA). The Arc1 Western blot served as loading control and untagged Nug1 as negative control. (B) Top: Ribosome and polysome analysis of Nug1 wild-type and representative nucleotide-binding (D336N) and hydrolysis (G339A) mutants. Whole cell lysates were analyzed by sucrose gradient centrifugation. A254nm profiles of the fractions collected are depicted. The red arrow denotes the increase of the 40S subunit and the red asterisks the half-mers. Bottom: Western blot of the gradient fractions using antibodies against Nug1 and Rpl12. (C) Rpl25-GFP localization in Nug1 wild-type and representative nucleotide-binding (D336N) and hydrolysis (G339A) mutants. (D) Northern hybridization and primer extension analysis (lower panel) of RNA extracted from Nug1 WT, nucleotide-binding (D336N) and Nug1 hydrolysis (G339A) mutants. Oligonucleotides used for hybridization or primer extension (P.E) are shown on the left of gel panels (E) Affinity-purification of Nug1 wild-type and nug1 D336N or G339A mutants. (F) Affinity-purified Ssf1 pre-ribosomes in the presence of Nug1 wild-type or G-domain mutants (D336N, G339A). For (C) and (D) final eluates were analyzed by SDS-PAGE and Coomassie staining. The indicated bands were identified by mass spectrometry and/or by comparison with characterized TAP pre-ribosomal particles. Red asterisks (*) denote the baits. Black diamond (♦) corresponds to Rpl3, which was used as a loading control.
To assess the impact of representative Nug1 nucleotide-binding (D336N) and GTP hydrolysis (G339A) mutants on ribosome biogenesis, ribosomal subunits and polysomes were analyzed by sucrose density gradient centrifugation (Figure 4B). When compared to the wild-type Nug1, the nucleotide-binding mutant (D336N) exhibited a substantial decrease in 60S subunits and a corresponding increase in free 40S subunits. This together with the appearance of ‘halfmers’ supported the idea that the nug1 D336N mutant causes a defect in 60S subunit assembly. This is not observed for the catalytic nug1 G339A mutant, which shows normal growth and unaffected 60S subunit assembly (Figure 4A and B). Further, western analysis of the gradient fractions showed that Nug1 D336N and G339A proteins were efficiently assembled into pre-60S subunits (Figure 4B, lower panel). However, some of Nug1 D336N mutant protein could also be seen in lower molecular weight fractions, suggesting that it may be less stably bound to pre-60S particles.
To further analyze the apparent 60S subunit assembly defect caused by the Nug1 D336N mutant, we performed the Rpl25-GFP reporter assay to detect a defect in nuclear export of the large subunit (Figure 4C). Consistent with a delay in the maturation of the 60S subunit, the Rpl25-GFP reporter was found to accumulate in the nucleus in the Nug1 nucleotide binding mutant (D336N), but not the hydrolysis (G339A) mutant. No defect in the export of the small subunit was seen in the D336N mutant (data not shown). Next we compared the pre-rRNA processing intermediates in the Nug1 WT, nucleotide binding and hydrolysis mutants (Figure 4D). Consistent with the idea that the nucleotide binding mutant of Nug1, but not the hydrolysis mutant, showed a delay in ribosome assembly, we see a clear accumulation of the 35S pre-rRNA, (the earliest of the rRNA precursors) combined with a decrease in the 27SA2 species and a corresponding appearance of the 23S rRNA. This suggests a delay in the processing of site A2, consistent with the idea that rRNA processing is generally slowed in this nug1 mutant.
Previous studies have shown that Nug1 associates with a broad range of nuclear pre-60S particles, from the early nucleolar Ssf1 to the later nucleoplasmic Arx1 particles (41). To assess the composition of Nug1 wild-type and mutant pre-ribosomal particles, tandem affinity purifications (TAP) were performed and the co-precipitating proteins were analysed (Figure 4E). Both wild-type and mutant Nug1 bait proteins (either nucleotide binding or hydrolysis defective) were efficiently affinity-purified from yeast whole cell lysates and revealed no drastic alteration in composition of associated assembly factors (Figure 4E, lanes 1–3). However, in the case of the pre-60S particles derived from the nug1 D336N mutant (defective in GTP binding), a slight reduction in some of the early assembly factors including Dbp10 could be observed (Figure 4E, lane 2).
Pre-60S particles isolated from the nug1 nucleotide-binding and nug1 degron mutants are specifically decreased in the early assembly factor Dbp10
To verify that early assembly factors are absent from pre-60S particles when carrying Nug1 defective in GTP binding, we sought to affinity-purify the assembly factor Ssf1 known to precipitate an early pre-60S particle (42). Indeed, several early 60S factors (e.g. Rrp5, Noc1 and Dbp10) but not intermediate assembly factors (e.g. Nog1, Nop7, Nsa3) were largely absent from the Ssf1-TAP purification derived from the Nug1 D336N nucleotide binding mutant (Figure 4F, lane 2). In contrast, the affinity-purified Ssf1-TAP particle from wild-type Nug1 or the GTP hydrolysis mutant Nug1 (G339A) exhibited a similar co-enrichment of both types of factors (Figure 4F, lanes 1 and 3). Notably, Ssf1-TAP affinity-purified from a nug1 degron strain (43), which showed a rapid depletion of Nug1, (Figure 5A–C), also exhibited such a decrease of early 60S assembly factors (Figure 5D, lane 2). To demonstrate such a decrease of early 60S assembly factors with another bait protein, Nsa1-TAP was affinity-purified from the nug1 degron mutant. In this case, the assembly factor and RNA helicase Dbp10 decreased the most dramatically, when compared to the non-depleted strain (Figure 5D, lanes 3 and 4). Together, these data suggest that the nucleotide binding ability of Nug1 or the absence of Nug1 affects the stable association of a subset of early 60S biogenesis factors, in particular Dbp10, with pre-60S particles.
Figure 5.
Nug1 depletion inhibits cell growth and causes defects in 60S subunit maturation. (A) The auxin-inducible degron system targets proteins for proteasomal degradation. Auxin (indole-3-acetic acid; IAA) induces degradation by mediating the interaction of the Aid-degron (fused to the protein target) with the substrate recognition domain of the TIR1 (F-box protein, auxin receptor). TIR is part of the SCF complex (E3 ubiquitin ligase) and leads to ubiquitination of the target and finally proteasomal degradation. (B and C) Depletion of Nug1 results in growth inhibition. Nug1 was genomically tagged at the C-terminal end with the Aid-tag. The ubiquitin E3 ligase TIR1 was genomically integrated and expressed under the constitutive ADH1 promoter (PADH1). (B) Cells expressing the Nug1-Aid was treated with 0.5 mM auxin and samples were taken at different time points (t = 0, 10, 20, 30, 60, 90 and 120 min). Whole-cell lysates were analyzed by SDS-PAGE followed by Western blotting using an anti-Nug1 antibody. The Arc1 Western blot served as loading control. (C) Growth analysis of yeast cells expressing Aid-tagged or untagged Nug1 in the ADH1::TIR1 background. Cells were spoted in 10-fold serial dilutions on YPD plates with or without 0.5 mM auxin and incubated at 23°C and 30°C for 1 day. (D) Ssf1-TAP and Nsa1-TAP pre-ribosomes were affinity-purified from yeast cells expressing the fusion Nug1-Aid protein following treatment with auxin. TAP eluates were analyzed by SDS-PAGE followed by Coomassie staining. Rpl3 (♦) was used as a loading control. Bait proteins are marked with a star (*). (E) Binding assays of CtNug1 and CtDbp10. FLAG-CtDbp10 was immobilized on anti-FLAG beads and full-length CtNug1 was added in 5- or 10-fold excess in the presence of E. coli lysate (EcL) to compete for unspecific binding. The bound material was eluted with loading buffer and analyzed by SDS-PAGE followed by Coomassie staining. As a negative control (mock), CtNug1 was incubated with anti-FLAG beads. The bands corresponding to CtDbp10 and CtNug1 are indicated with arrows and the IgG light chain is labeled by *.
Nug1 physically interacts with the DEAD-box RNA helicase Dbp10 and binds at proximal rRNA sites on the pre-60S ribosome
The observed decrease in levels of Dbp10 in pre-ribosomal particles derived from the different nug1 mutants, together with the previous observation that Dbp10 interacts genetically with Nug1 (9), prompted us to test for a physical interaction between these two assembly factors. To assess this, in vitro binding assays were performed using Chaetomium thermophilum orthologs CtNug1 and CtDbp10, due to enhanced solubility when compared to the yeast proteins. Affinity-purified CtDbp10 was immobilized on beads and incubated with increasing amounts of purified CtNug1 in the presence of competitor E. coli lysate (Figure 5E). This reconstitution experiment revealed that CtNug1 specifically bound to immobilized CtDbp10, suggesting complex formation between these two assembly factors.
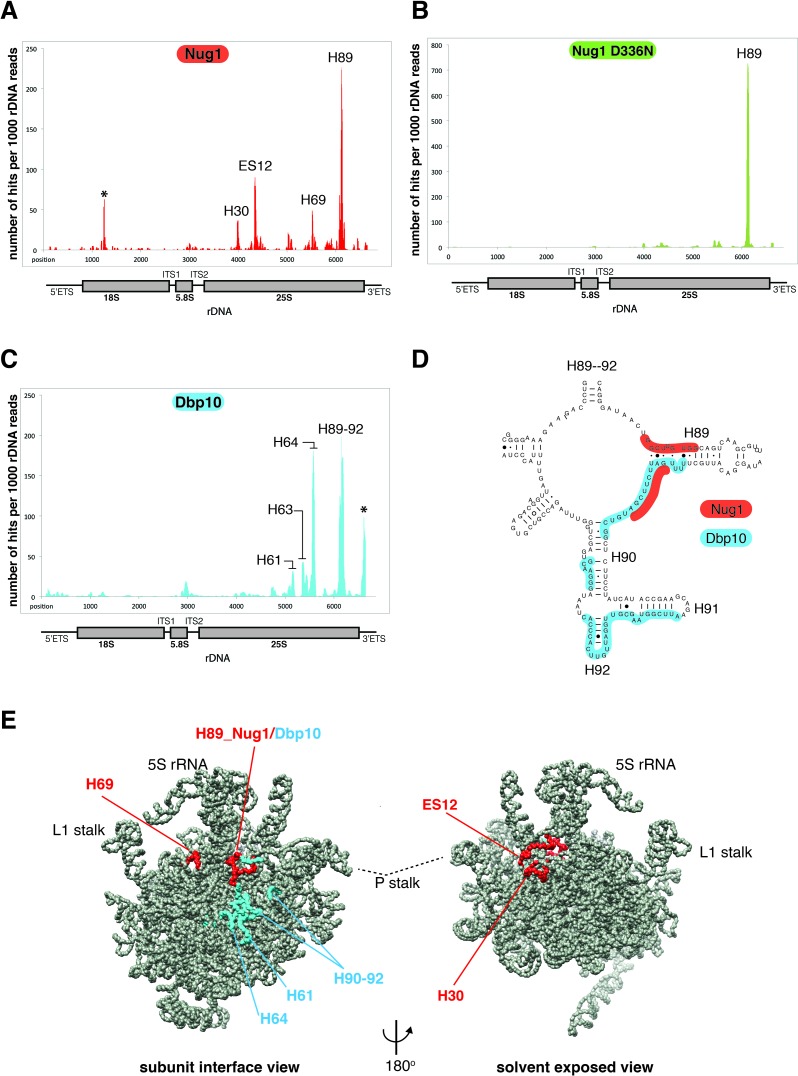
We next asked if Nug1 and Dbp10 bind on the pre-ribosome at positions consistent with their direct physical interaction. We therefore employed the UV CRAC technique, to identify the rRNA interaction sites of Nug1 and Dbp10. Both Nug1 and Dbp10 were efficiently crosslinked to RNA (Figure 6A and C). The major Nug1 crosslink corresponds to the base of H89 within the 25S rRNA, which is located on the subunit interface of the 60S subunit. H89 is functionally important in the mature ribosome as it forms part of the PTC. Another site of crosslink corresponds to H69, which is part of the A and P sites in the mature ribosome, and both sites H89 and H69 are close to each other, when the 3D volume of the Arx1 pre-60S particle (41) was used as a model (Figure 6E). Additional Nug1 crosslink hits were found at H30 and ES12, which are neighboring RNAs on the 60S solvent side, in close vicinity to the 5S rRNA (Figure 6A and E). When CRAC analysis was performed with the nug1 D336N mutant, Nug1 was still efficiently crosslinked, but now only to helix 89 and the other CRAC sites were lost (Figure 6B; see also Discussion).
Figure 6.
Nug1 and Dbp10 bind at proximal sites on the intersubunit face of the 60S subunit. CRAC analysis of (A) Nug1, (B) Nug1 D336N mutant and (C) Dbp10. (A–C) Illumina-Miseq sequencing results were aligned to the yeast 35S rDNA and plotted. The histogram shows the position and distribution of crosslink sites of Nug1 wild-type (red), Nug1 D336N mutant (green) and Dbp10 (blue) on the 35S rRNA. Position of mature rRNA sequences and spacers are indicated below the x-axis. The y-axis displays number of times each nucleotide was mapped (hits) per 1000 rDNA reads. The location of the peaks in the secondary structure of the rRNA is indicated with helix (H) numbers. The asterisks indicate frequently observed contaminants. Each experiment was performed at least twice, and representative results are shown. (D) Overlapping interaction region of Nug1 and Dbp10 on helix 89. Binding sites of Nug1 (red) and Dbp10 (blue) are shown on the secondary structure map of rRNA helices 89–92 (http://www.rna.ccbb.utexas.edu/). (E) Mapping of the Nug1 (red) and Dbp10 (blue) crosslink sites on the cryo-EM structure of the early Arx1 pre-60S particle.
Dbp10 shows CRAC sites at H64 and H89–92, which when mapped into the 3D volume of the pre-60S subunit are found very close to each other at a distinct area on the intersubunit face (Figure 6C, D and E). Comparison of the Dbp10 crosslink sites with those of Nug1 shows that the sites of interaction are very close and at one region, around the base of helix 89, partially overlap (Figure 6B and D). This finding further strengthens the idea of direct interaction between these two assembly factors, occupying neighboring sites. However, the partially overlapping binding pattern suggests that Dbp10 and Nug1 may also act sequentially at helix 89.
DISCUSSION
This study revealed that Nug1 is a cation-dependent potassium-selective GTPase and that the conserved asparagine in the G1 motif [GxxNxGKS] coordinates the cation. Furthermore, stimulation of CtNug1's activity was observed when increasing concentrations of potassium ions were used. This is in agreement with the increased enzymatic activities observed when similar experiments were performed with the bacterial CD-GTPases FeoB and MnmE (10,39,44). Despite the stimulatory role of potassium ions seen in vitro, the intracellular concentration of potassium in yeast is around 200–300 mM (45). This concentration is lower than required for maximal stimulation (500 mM) of CtNug1 in vitro, and thus raises the question of whether all CtNug1 molecules bind potassium ions in vivo. Hence, we postulate that the presence of potassium is not a strict requirement for GTPase hydrolysis, but rather an additional in vivo ‘co-factor’ to achieve a catalysis-competent state. Interestingly, for some CD-GTPases the maximum activity is achieved not only by the presence of cations, but also by additional mechanisms, including dimerization for MnmE and dynamin (40,46), or binding to the ribosome for RbgA and YqeH (38,47). Similar mechanisms could be envisaged for Nug1 activity.
To date, nine GTPases have been implicated in ribosome assembly (2). While a comprehensive analysis of different G-domain mutants has yet to be performed, it appears clear that the function of some GTPases relies upon the ability to hydrolyse GTP, e.g. Efl1 and Nug2 (17,48). Unexpectedly, nucleotide binding, but not GTP-hydrolysis appears to be of key importance to Nug1's in vivo function, since the Nug1 D336N is impaired in cell growth and shows a strong defect in 60S subunit maturation. This is likely caused by the fact that in the Nug1 null (following depletion) or nucleotide-binding mutant early assembly factors are less efficiently associated with the pre-ribosomal particles. One rationale for this observation is that upon GTP binding conformational changes within the G-domain of Nug1 take place. These conformational changes could then be transmitted to the flanking N- or C-terminal domains and could thus affect Nug1's association with the pre-ribosome and interaction with other biogenesis factors (e.g. Dbp10). Indeed, the additional domains flanking the GTPase core in cpGTPase have been proposed to propagate intramolecular conformational changes upon GTP binding or hydrolysis (8). Interestingly, the N-terminus of Nug1 has been shown to mediate binding to the pre-ribosome (9). Thus, if the conformation or accessibility of the N-terminus is altered, then Nug1's binding to pre-ribosomes could consequently be affected. This may explain why in the Nug1 D336N mutant, RNA crosslinking to the ES12-H30 region was lost. It is tempting to speculate that these two distant Nug1 crosslink sites on the solvent side of the pre-ribosome correspond to a site of contact of the N-domain of Nug1. The N-domain of Nug1 is predicted to fold into a long flexible α-helical structure, which can be envisaged to extend over a long distance from the main binding site of Nug1 on the subunit joining side over the top of the ribosome to the solvent side. Indeed, uncharacterized extra density in the Arx1 pre-60S particle can be seen to bridge between the interface and solvent side and pass under the unrotated 5S RNP (41). In this way Nug1 could sense conformational changes on the pre-60S ribosome over large distances and could transmit this information to distant sites or other assembly factors. Whilst all our data support a role of Nug1 in the early steps of 60S biogenesis, we cannot exclude the possibility of Nug1 playing additional roles at later points during 60S maturation.
The major crosslink site of Nug1 was at the base of H89, which is part of the PTC, an area that undergoes conformational changes during pre-60S maturation (41). Interestingly, Dbp10 crosslink sites were also mapped to this area and are in close proximity to Nug1. Unexpectedly, both of these proteins have a partial overlap on the base of H89, but we postulate they bind to H89 at distinct time points during 60S maturation. Dbp10 is an essential ribosome biogenesis factor in yeast and is predicted to be an ATP-dependent RNA helicase. Additionally, it displays a synthetic lethal interaction with Nug1 (9), and has previously been implicated in the processing of 27SB pre-rRNA (49). Indeed we observed a delay in the processing of early pre-rRNAs in the Nug1 D336N mutant, similar to what is seen following longer times of depletion of Dbp10 (49). In both cases this phenotype could be explained by the sequestration of biogenesis factors on stalled pre-ribosomes.
Interestingly, the bacterial helicase DbpA has been shown to bind to H92 of the 23S rRNA via its C-terminal domain (CTD) (50–52). It is suggested that anchoring the CTD of DbpA allows the targeting of its catalytic N-terminal ATPase domain to nearby rRNA regions. Footprinting studies in the presence of AMppNp (a non-hydrolyzable ATP analog) showed that the catalytic domain of DbpA binds and acts upon the H89 region of rRNA (53). The fact that the yeast Dbp10 binds at the same position (H92 and H89) in the 25S rRNA as the prokaryotic DbpA does in the 23S rRNA, suggests that they could be functional homologues. Thus, an analogous model where Dbp10 plays a role in the unwinding of H89, allowing other assembly factors to bind, is an attractive possibility. In yeast, Nug1 would be a potential candidate, as we have now shown that it also binds to H89. This raises the question of whether Nug1 influences the ATPase/helicase activity of Dbp10. Interestingly, the GTPase Snu114 regulates unwinding of U4/U6 and therefore spliceosome dynamics by affecting Brr2 RNA helicase activity (54). In future, it would be interesting to identify whether Nug1 and Dbp10 display a similar relationship.
Notably, two methyl-transferases, Spb1 and Nop2, can be found on earlier pre-60S particles (17). Both function at the PTC, where Nop2 modifies C2870 in helix H89 and Spb1 modifies G2922 in Helix 92 (55,56). However, the precise timing of association and action of these two factors remains unclear. Spb1 has been reported to modify one of the 27S pre-rRNA species (55) and Nop2, considered a 27SB factor, is required for the association of most other assembly factors required for 27SB processing, with the notable exception of Dbp10 (57). This suggests that the helicase activity of Dbp10 might be employed at a distinct step during the structural maturation of the PTC and would allow access to Spb1 and Nop2 for base modification. Interestingly, upon depletion of Nug1 and purification of the Nsa1 particle, association of both Spb1 and Nop2 decreases when compared to particles from a wild-type control (Figure 5D). Thus, the Nug1 GTPase may mediate crosstalk between early assembly factors on the pre-60S ribosome so that they can be correctly positioned at the PTC.
Supplementary Material
Acknowledgments
We would like to thank David Tollervey and Sander Granneman (University of Edinburgh) for carrying out initial low throughput CRAC analysis on Nug1. We would also like to thank David Ibberson at the Cell Networks Deep sequencing core facility for performing MiSeq sequencing and the mass spec facility (BZH, Heidelberg) of J Lechner for performing all mass-spec analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [BA2316/1–4, HU363/10–5 to E.H]. Funding for open access charge: Lab grant money from the DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Woolford J.L., Jr, Baserga S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kressler D., Hurt E., Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Henras A.K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lafontaine D.L., Tollervey D. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2001;2:514–520. doi: 10.1038/35080045. [DOI] [PubMed] [Google Scholar]
- 5.Staley J.P., Woolford J.L., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levdikov V.M., Blagova E.V., Brannigan J.A., Cladiere L., Antson A.A., Isupov M.N., Seror S.J., Wilkinson A.J. The crystal structure of YloQ, a circularly permuted GTPase essential for Bacillus subtilis viability. J. Mol. Biol. 2004;340:767–782. doi: 10.1016/j.jmb.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 7.Shin D.H., Lou Y., Jancarik J., Yokota H., Kim R., Kim S.H. Crystal structure of YjeQ from Thermotoga maritima contains a circularly permuted GTPase domain. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13198–13203. doi: 10.1073/pnas.0405202101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anand B., Verma S.K., Prakash B. Structural stabilization of GTP-binding domains in circularly permuted GTPases: implications for RNA binding. Nucleic Acids Res. 2006;34:2196–2205. doi: 10.1093/nar/gkl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassler J., Kallas M., Hurt E. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60 S pre-ribosomal particles. J. Biol. Chem. 2006;281:24737–24744. doi: 10.1074/jbc.M604261200. [DOI] [PubMed] [Google Scholar]
- 10.Ash M.R., Maher M.J., Mitchell Guss J., Jormakka M. The cation-dependent G-proteins: in a class of their own. FEBS Lett. 2012;586:2218–2224. doi: 10.1016/j.febslet.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 11.Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 12.Longtine M.S., McKenzie A. 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 13.Amlacher S., Sarges P., Flemming D., van Noort V., Kunze R., Devos D.P., Arumugam M., Bork P., Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 14.Peluso P., Shan S.O., Nock S., Herschlag D., Walter P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 15.Ferreira-Cerca S., Kiburu I., Thomson E., LaRonde N., Hurt E. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res. 2014;42:8635–8647. doi: 10.1093/nar/gku542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira-Cerca S., Sagar V., Schafer T., Diop M., Wesseling A.M., Lu H., Chai E., Hurt E., LaRonde-LeBlanc N. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol. 2012;19:1316–1323. doi: 10.1038/nsmb.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuo Y., Granneman S., Thoms M., Manikas R.G., Tollervey D., Hurt E. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature. 2014;505:112–116. doi: 10.1038/nature12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granneman S., Kudla G., Petfalski E., Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granneman S., Petfalski E., Tollervey D. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011;30:4006–4019. doi: 10.1038/emboj.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webb S., Hector R.D., Kudla G., Granneman S. PAR-CLIP data indicate that Nrd1-Nab3-dependent transcription termination regulates expression of hundreds of protein coding genes in yeast. Genome Biol. 2014;15:R8. doi: 10.1186/gb-2014-15-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 22.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 23.Kressler D., delaCruz J., Rojo M., Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 25.Beltrame M., Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tollervey D., Mattaj I.W. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987;6:469–476. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altvater M., Chang Y., Melnik A., Occhipinti L., Schutz S., Rothenbusch U., Picotti P., Panse V.G. Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export. Mol. Syst. Biol. 2012;8:628. doi: 10.1038/msb.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de la Cruz J., Sanz-Martinez E., Remacha M. The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits. Nucleic Acids Res. 2005;33:5728–5739. doi: 10.1093/nar/gki887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frey S., Pool M., Seedorf M. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 2001;276:15905–15912. doi: 10.1074/jbc.M009430200. [DOI] [PubMed] [Google Scholar]
- 30.Vilardell J., Warner J.R. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lebreton A., Saveanu C., Decourty L., Jacquier A., Fromont-Racine M. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J. Biol. Chem. 2006;281:27099–27108. doi: 10.1074/jbc.M602199200. [DOI] [PubMed] [Google Scholar]
- 32.Saveanu C., Namane A., Gleizes P.E., Lebreton A., Rousselle J.C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez-Mateos M., Abia D., Garcia-Gomez J.J., Morreale A., de la Cruz J., Santos C., Remacha M., Ballesta J.P. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res. 2009;37:3514–3521. doi: 10.1093/nar/gkp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Mateos M., Garcia-Gomez J.J., Francisco-Velilla R., Remacha M., de la Cruz J., Ballesta J.P. Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:7519–7532. doi: 10.1093/nar/gkp806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simos G., Segref A., Fasiolo F., Hellmuth K., Shevchenko A., Mann M., Hurt E.C. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 36.Vilella M.D., Remacha M., Ortiz B.L., Mendez E., Ballesta J.P.G. Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal-antibodies - proteins L44/L45 and L44’ have different functional roles. Eur. J. Biochem. 1991;196:407–414. doi: 10.1111/j.1432-1033.1991.tb15831.x. [DOI] [PubMed] [Google Scholar]
- 37.Senger B., Lafontaine D.L., Graindorge J.S., Gadal O., Camasses A., Sanni A., Garnier J.M., Breitenbach M., Hurt E., Fasiolo F. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 38.Achila D., Gulati M., Jain N., Britton R.A. Biochemical characterization of ribosome assembly GTPase RbgA in Bacillus subtilis. J. Biol. Chem. 2012;287:8417–8423. doi: 10.1074/jbc.M111.331322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ash M.R., Guilfoyle A., Clarke R.J., Guss J.M., Maher M.J., Jormakka M. Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B. J. Biol. Chem. 2010;285:14594–14602. doi: 10.1074/jbc.M110.111914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scrima A., Wittinghofer A. Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J. 2006;25:2940–2951. doi: 10.1038/sj.emboj.7601171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leidig C., Thoms M., Holdermann I., Bradatsch B., Berninghausen O., Bange G., Sinning I., Hurt E., Beckmann R. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat. Commun. 2014;5:3491. doi: 10.1038/ncomms4491. [DOI] [PubMed] [Google Scholar]
- 42.Fatica A., Cronshaw A.D., Dlakic M., Tollervey D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- 43.Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 44.Yamanaka K., Hwang J., Inouye M. Characterization of GTPase activity of TrmE, a member of a novel GTPase superfamily, from Thermotoga maritima. J. Bacteriol. 2000;182:7078–7082. doi: 10.1128/jb.182.24.7078-7082.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arino J., Ramos J., Sychrova H. Alkali metal cation transport and homeostasis in yeasts. Microbiol. Mol. Biol. Rev. 2010;74:95–120. doi: 10.1128/MMBR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chappie J.S., Acharya S., Leonard M., Schmid S.L., Dyda F. G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature. 2010;465:435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolanczyk M., Pech M., Zemojtel T., Yamamoto H., Mikula I., Calvaruso M.A., van den Brand M., Richter R., Fischer B., Ritz A., et al. NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol. Biol. Cell. 2011;22:1–11. doi: 10.1091/mbc.E10-07-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finch A.J., Hilcenko C., Basse N., Drynan L.F., Goyenechea B., Menne T.F., Gonzalez Fernandez A., Simpson P., D'Santos C.S., Arends M.J., et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25:917–929. doi: 10.1101/gad.623011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burger F., Daugeron M.C., Linder P. Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res. 2000;28:2315–2323. doi: 10.1093/nar/28.12.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicol S.M., Fuller-Pace F.V. The ‘DEAD box’ protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11681–11685. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diges C.M., Uhlenbeck O.C. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diges C.M., Uhlenbeck O.C. Escherichia coli DbpA is a 3′ –> 5′ RNA helicase. Biochemistry. 2005;44:7903–7911. doi: 10.1021/bi050033x. [DOI] [PubMed] [Google Scholar]
- 53.Karginov F.V., Uhlenbeck O.C. Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res. 2004;32:3028–3032. doi: 10.1093/nar/gkh640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Small E.C., Leggett S.R., Winans A.A., Staley J.P. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lapeyre B., Purushothaman S.K. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell. 2004;16:663–669. doi: 10.1016/j.molcel.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 56.Sharma S., Yang J., Watzinger P., Kotter P., Entian K.D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41:9062–9076. doi: 10.1093/nar/gkt679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Talkish J., Zhang J., Jakovljevic J., Horsey E.W., Woolford J.L., Jr Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:8646–8661. doi: 10.1093/nar/gks609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelley L.A., Sternberg M.J. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.