Abstract
Background
The slowly-activating delayed rectifier current IKs contributes to repolarization of the cardiac action potential, and is composed of a pore-forming α-subunit, KCNQ1, and a modulatory β-subunit, KCNE1. Mutations in either subunit can cause long QT syndrome, a potentially fatal arrhythmic disorder. How KCNE1 controls the kinetics of IKs remains unresolved.
Methods and Results
We identified 2 adjacent mutations, S338F and F339S, in the KCNQ1 S6 domain in unrelated probands. The novel KCNQ1 S338F mutation segregated with prolonged QT interval and torsade de pointes; the second variant, F339S, was associated with fetal bradycardia and prolonged QT interval, but no other clinical events. S338F channels expressed in Xenopus oocytes had slightly increased peak conductance relative to wild type, with a more positive activation voltage. F339S channels conducted minimal current. Unexpectedly, S338F currents were abolished by co-expression with intact WT KCNE1 or its C-terminus (aa63-129), despite normal membrane trafficking and surface co-localization of KCNQ1 S338F and wt KCNE1. Structural modeling indicated that the S338F mutation specifically alters the interaction between the S6 domain of one KCNQ1 subunit and the S4-S5 linker of another, inhibiting voltage-induced movement synergistically with KCNE1 binding.
Conclusion
A novel KNCQ1 mutation specifically impaired channel function in the presence of KCNE1. Our structural model shows that this mutation effectively immobilizes voltage gating by an inhibitory interaction that is additive with that of KCNE1. Our findings illuminate a previously unreported mechanism for LQTS, and validate recent theoretical models of the closed state of the KCNQ1:KCNE1 complex.
Keywords: arrhythmia, ion channels, long QT syndrome, structural modeling, genetics
INTRODUCTION
Congenital long QT Syndrome (LQTS) is an inherited disorder carrying a high risk of lethal cardiac arrhythmias (1,2). Most cases of LQTS are caused by loss-of-function mutations in ion channels that regulate the repolarization phase of the cardiac action potential, including KCNQ1 (KVLQT1) and KCNE1 (Isk, MinK). These genes encode the α- and β- subunits of a heterotetrameric complex that produces the slowly activating delayed rectifier IKs (Kv 7.1), an outward potassium current important for the rapid phase (phase 3) of repolarization (3-5). Mutations in these same genes have been linked to short QT syndrome and familial atrial fibrillation (6). Although KCNQ1 by itself can form voltage-gated ion channels, the distinctive IKs current requires co-assembly with KCNE1. Interaction with KCNE1 greatly increases the voltage required for IKs channel opening, while increasing channel conductance, slowing the time course of activation and deactivation (3-5,7), and enabling adrenergic regulation (8). How KCNE1 interacts with KCNQ1 to produce these extensive functional alterations is incompletely understood (9,10).
In this report we analyze 2 mutations, KCNQ1 S338F and F339S, occupying adjacent sites within the S6 transmembrane domain. One of these (S338F) does not affect KCNQ1 channel function by itself, but causes a complete loss of conductance when co-assembled with KCNE1 or the C-terminal residues of KCNE1 that physically associate with KCNQ1 (10,11). The other mutant carries almost no current, and its voltage-dependent activation by KCNE1 is minimal and extremely right-shifted (12, 13). Structural modeling of the KCNQ1 homotetramer predicts that the variant phenylalanine at residue 338 in one subunit forms van der Waals attachments to the S4-S5 helical linker of a neighboring subunit, impeding channel opening. The combined inhibitory interaction of KCNE1 and the S338F mutation effectively immobilizes the S4-S5 linker, rendering the channel unable to open at physiologic voltages. Our findings elucidate a novel co-assembly-specific mechanism for LQTS, and provide high-resolution structural and mechanistic insights into KCNQ1:KCNE1 modulatory interaction and gating of IKs.
METHODS
A detailed Methods supplement is available online.
Subjects
Subjects of this study were referred after presenting with syncope and/or ventricular tachyarrhythmias with prolonged QT intervals, and family histories of unexplained or sudden cardiac deaths. Subjects were recruited and evaluated in accordance with a human subjects research protocol approved by the UM Institutional Review Board.
Mutation Identification
DNA was isolated using a commercial kit (Puregene DNA Isolation Kit, Gentra Systems/Qiagen, Minneapolis, MN), and individual exons from 5 genes (KCNQ1, KCNH2, SCN5A, KCNE1 and KCNE2) associated with LQTS were amplified with published primers (14, 15) using the polymerase chain reaction (PCR).
Electrophysiology
Stage V oocytes were prepared from mature Xenopus laevis follicular tissue (Nasco, Fort Atkinson, WI). Oocytes were injected with combinations of KCNQ1/KCNE1 cRNA, and current responses were analyzed under two-electrode voltage clamp using an OpusXpress 6000A Parallel Oocyte Voltage Clamp system running OPUSXPRESS 1.1 and CLAMPFIT 9.1 software (Molecular Devices).
Epitope tagging and membrane expression
We employed differential epitope-tagging to facilitate microscopic visualization and biotinylation of membrane-bound mutant and WT alpha and beta subunits. Details pertaining to these experimental procedures can be found in the online supplement.
Molecular modeling
Structural models of WT and S338F/F339S mutants of KCNQ1 in the closed state were built using MODELLER software (16). We used as a template the closed-state structure of KCNQ1 modeled by Sanders and co-workers (17, 18). Atomic models were rendered using RIBBONS (19).
RESULTS
Clinical Subjects
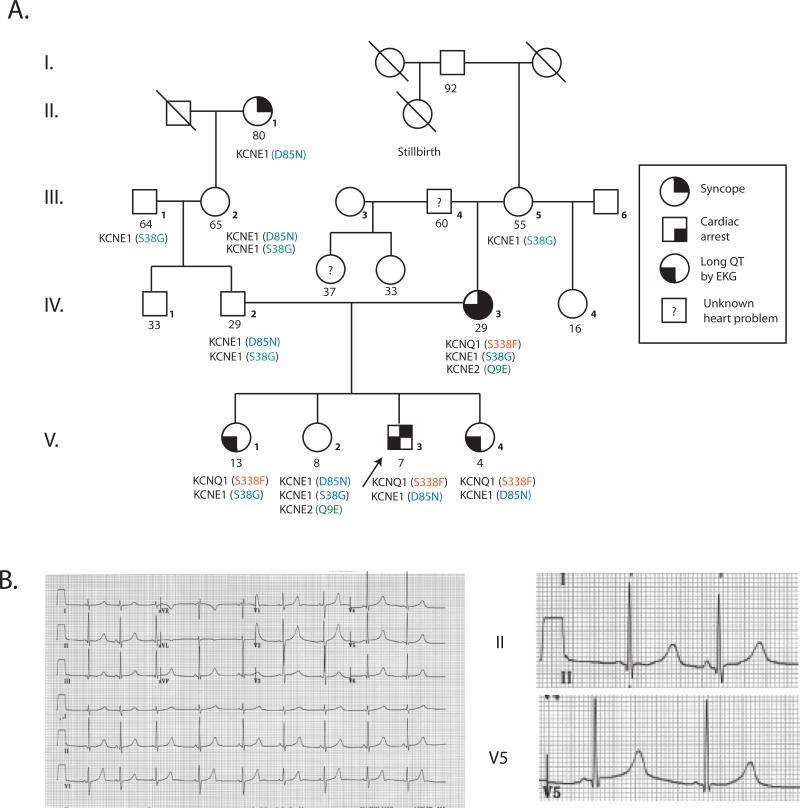
The first proband was a 7 year old Caucasian child referred for evaluation of syncope and LQTS. The proband's mother had LQTS and received an implantable cardioverter-defibrillator (AICD) at age 29 for recurrent syncope and cardiac arrest (Figure 1). At age 7, the proband experienced 3 episodes of loss of consciousness over a 6 month period. The first occurred while swimming at age 7, when he was noted to have fixed pupils, apnea, and cyanosis. He recovered spontaneously after brief chest compression; an EEG and CT scan of the brain were normal. A second episode occurred 2 months later while swimming; an EKG was reportedly normal. Four months later, he suddenly fell while running around the pool and lost consciousness for more than a minute. An EKG revealed sinus bradycardia at 58 bpm and a QT/QTc of 510/505 msec (Figure 1). He was treated with β--adrenergic blockade and at age 8 received an AICD.
Figure 1. Proband 1 pedigree and ECG.
A. Pedigree B. 12-lead electrocardiogram from proband 1 at 7 years of age.
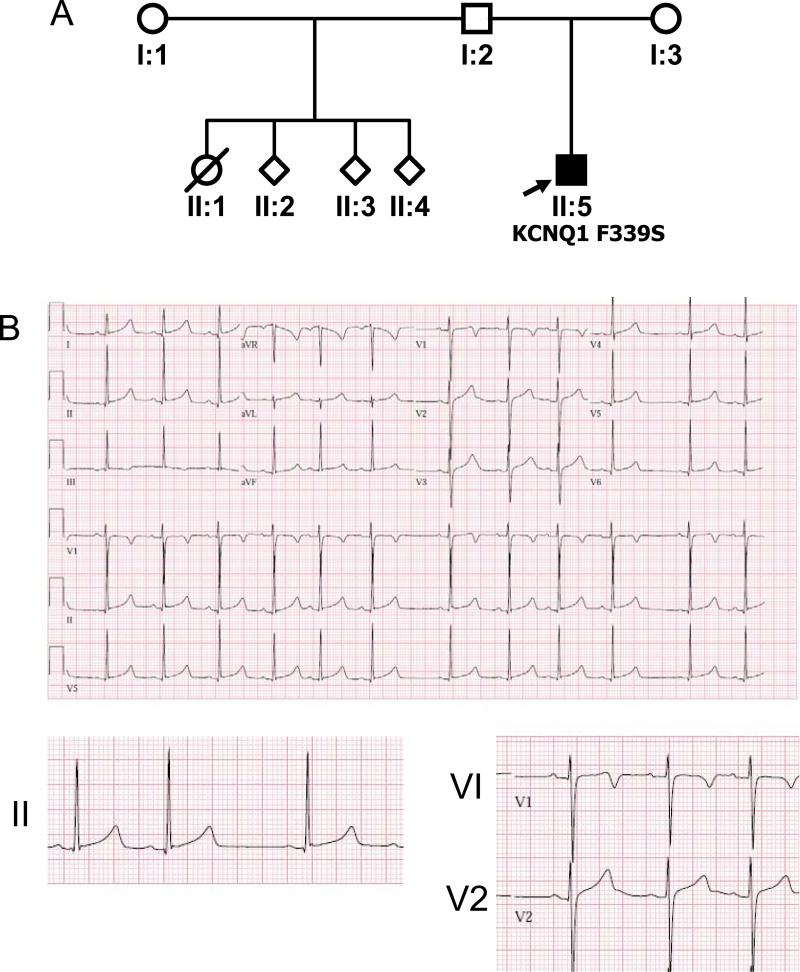
The second proband was a full-term infant with a strong paternal family history of premature sudden death (Figure 2A), who presented with fetal bradycardia (<110) and a prolonged QT interval during the first month of life (Figure 2B); his half-sister reportedly died suddenly in infancy, while crying. He was treated with beta-adrenergic blockade and is now 8 years old, with no complaints of palpitations, syncope or cardiac arrest, and recorded QTcs ranging from 440-500 msec.
Figure 2. Proband 2 Pedigree and ECG.
A. Pedigree B. 12-lead electrocardiogram from proband 2.
Molecular Genetics
Proband 1 harbored a novel transition mutation in exon 7 of KCNQ1 that resulted in substitution of cytosine for thymine at position 1013 (NM 000218), causing a serine (TCT) to phenylalanine (TTT) switch at position 338 (S338F, Figure 3A). All relatives of Proband 1 with long QT intervals carried the same S338F mutation. Proband 1 was also heterozygous for a previously reported D85N (G253A) mutation in KCNE1, inherited from his asymptomatic father. Two S338F carrier siblings have had no documented arrhythmias or syncope to date.
Figure 3. Adjacent novel mutations in the KCNQ1 S6 transmembrane domain.
A. Partial KCNQ1 exon 7 DNA sequence from proband 1. B. Partial KCNQ1 exon 7 DNA sequence from proband 2.
In Proband 2, KCNQ1 sequencing revealed a T>C substitution at position 1016, resulting in replacement of a phenylalanine codon (TTC) with a serine codon (TCC) at position 339 (F339S, Figure 3B). This mutation was not identified in either parent. No other KCNQ1 or KCNE1 variants were found.
Protein Expression Studies
Voltage clamp experiments were performed in Xenopus oocytes on the two KCNQ1 α–subunit mutations (S338F and F339S), alone and in combination with wild-type (WT) and D85N-variant KCNE1 β-subunits. Background currents from endogenous oocyte α– and β-subunits were minimal (Figure 4A). Homomeric WT- and S338F-KCNQ1 conducted similar amounts of current, but the S338F current exhibited more positive activation voltage, slightly greater conductance at activation, and more rapid inactivation of the time-dependent outward current. S338F also lacked a rapidly deactivating tail current (Figure 4A). In contrast, homomeric F339S channels had extremely poor conductance at all voltages (Figure 4B).
Figure 4. KCNQ1 S338F mutation induces a dysfunctional interaction with KCNE1.
A. Homomeric currents generated by WT KCNQ1 and S338F and F339S variants. B. Addition of KCNE1 activates WT and suppresses mutant KCNQ1 currents. C-E. Current density–voltage relationships for KCNQ1 and IKs currents. F. KCNE1 C-terminal domain represses KCNQ1 S338F currents.
Mutant S338F exhibits defective activation by KCNE1
As expected, co-expression of KCNE1 with KCNQ1 generated a current exhibiting typical features of IKs, with a more positive voltage of activation, and current amplitudes (μA) and densities (μA/μF) that were approximately 40 times greater than that produced by WT KCNQ1 alone (Figures 4B, C). Channels consisting of KCNE1 alone conducted a small current (Figures 4B, C). Remarkably, channels formed by S338F-KCNQ1 with WT-KCNE1 exhibited severely depressed current amplitude and density compared with either protein alone (Figures 4B, 4D), and the resulting current did not display other typical kinetics of WT IKs (Figures 4B, C). Similarly, co-injection of KCNE1 and F339S produced channels that conducted significantly smaller time-dependent outward currents compared to either protein alone, with no observable tail currents (Figures 4B and E). Thus, both S338F and F339S mutations result in markedly abnormal interactions between KCNQ1 and its accessory subunit, producing an overall loss of function. A complete summary of current properties for all channel stoichiometries is provided in Supplementary Table 1.
Co-expression of a KCNE1 C-term fragment (aa63-129) previously shown to physically interact with the S6 domain of KCNQ1 (11) had a similar negative effect on S338F current amplitude (Figure 4F, n=19; Supplementary Table 1). Taken together, these data indicate a specific role for KCNE1:KCNQ1 contact in the manifestation of the KCNQ1 S338F defect.
Dominant-negative effect of KCNQ1 heterotetramers
As is shown in figure 5A, homomeric S338F had greater peak current density than WT KCNQ1, but a more positive activation voltage, while homomeric F339S current density was extremely low (Figure 5A). Co-expression of S338F with WT KCNQ1 at a 1:1 ratio generated a current density that was greatly reduced compared with either protein expressed separately (Figures 5B and 5C), suggesting the formation of heterotetramers with reduced cooperativity or stability, and/or inability to interact with endogenous oocyte KCNE1-like proteins.
Figure 5. Dominant negative effects of KCNQ1 S338F and F339s mutations.
A. Current density-voltage relationships for WT, mutant and mixed α-subunits. B. Effect of KCNE1 co-expression on WT and mutant KCNQ1 current density. C. Inhibitory effect of KCNQ1 mutants on KCNE1-generated currents. D. Dominant negative effect of S338F on WT KCNQ1/IKs current. E. Differential effect of KCNE1 D85N variant on WT and S338F mutant KCNQ1/IKs currents.
Dominant negative effect of KCNQ1 mutants on activation by KCNE1
To confirm the dominant negative effect of the S338F mutation, we expressed both WT and mutant α–subunits at varying ratios in the presence of KCNE1 (Figure 5D). When S338F and WT KCNQ1 were co-expressed at approximately equal stoichiometry, almost no activation by KCNE1 was observed, suggesting that the depressive effect of the S338F mutation is dominant.
Functional interaction of KCNE1 D85N mutant with KCNQ1 WT and S338F mutants
In addition to the S338F mutation, proband 1 also had a rare variant in KCNE1, D85N (rs1805128, MAF = 0.8 to 2.5%), previously associated with drug-induced LQTS (20-23). Accordingly, we analyzed channels containing various combinations of WT and mutant α- and β-subunits. WT KCNE1 and D85N (Figure 5E) generated identical, small currents in the absence of co-expressed α-subunits (<1 μA/μF at 20mV), likely through interaction with endogenous Xenopus α-subunits. However, the D85 variant activated WT KCNQ1 channels with reduced efficacy compared to WT KCNE1. S338F had identical inhibitory interactions with both WT and variant KCNE1 (Figure 5E).
S338F mutant trafficks normally in the presence of KCNE1
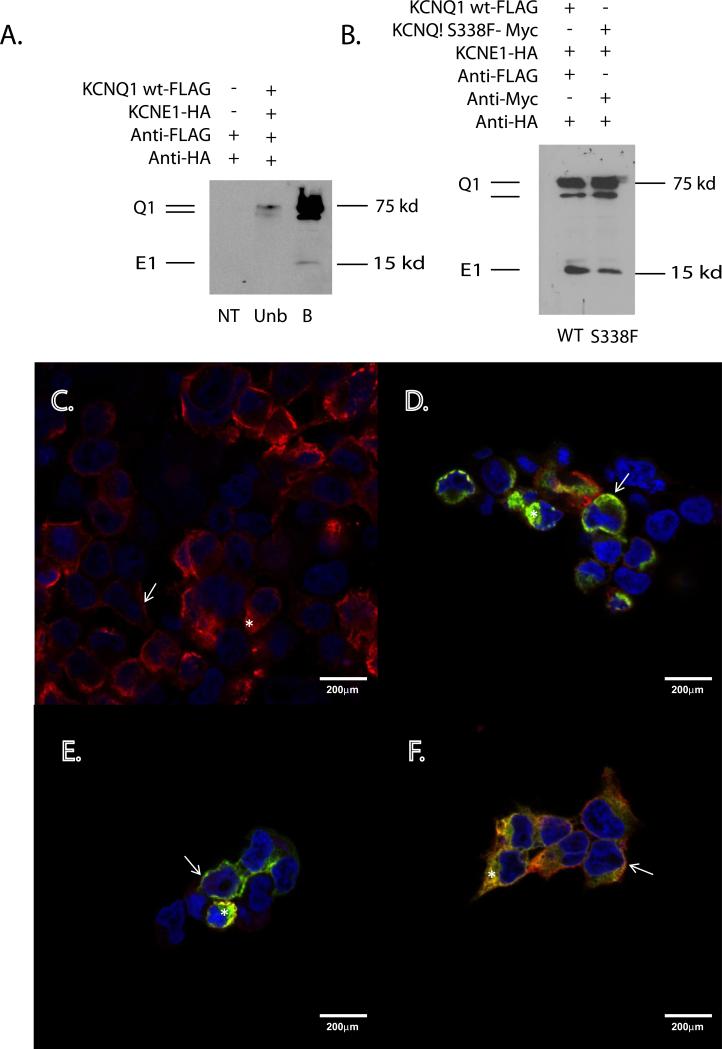
To explore the possibility of trafficking defects, cell surface biotinylation experiments were performed in HEK293 cells transfected with combinations of FLAG-tagged WT-KCNQ1, Myc-tagged S338F-KCNQ1, and HA-tagged KCNE1 (see Supplemental Methods). As expected, WT Q1:E1 channels appeared as bands at 75 and 100kD (KCNQ1), and 15kD (KCNE1), which were enriched at the membrane surface (Figure 6A). Equivalent membrane enrichment was seen for S338F Q1:E1 channels (Figure 6B), arguing against a KCNE1-induced trafficking defect. Confirming these findings, immunocytochemistry demonstrated co-localization of all 3 proteins at the cell surface, separately and in combination (Figure 6 C-F). WT KCNQ1 was observed at the cell surface when co-expressed with KCNE1 (Figure 6C), with some intracellular staining reflecting high channel overexpression as noted elsewhere (24). KCNQ1 S338F similarly co-localized with KCNE1 at the plasma membrane, both in the absence (Figure 6D) and presence (Figure 6EF) of WT KCNQ1. These results additionally exclude the possibility that the dominant negative effect of the S338F mutant on WT channels results from a trafficking defect arising in heteromeric channels.
Figure 6. KCNQ1 S338F-KCNE1 channels traffic normally to plasma membrane.
A. Biotinylation identifies membrane-localized WT KCNQ1-KCNE1 channels. NT = nontransfected, WT = KCNQ1 WT-FLAG, Unb = unbound and B = bound fraction of cell lysates. B. Equivalent membrane localization of WT and S338F mutant KCNQ1 in the presence of KCNE1. (Left lane) Biotinylated fraction of WT KCNQ1-FLAG and KCNE1-HA-transfected HEK293 cells. (Right lane) Biotinylated fraction of KCNQ1- S338F-MYC and KCNE1-HA-transfected HEK293 cells. C-F. Membrane co-localization of WT and S338F mutant KCNQ1 in the presence of KCNE1. C. Membrane localization of WT KCNQ1-FLAG in the presence of WT KCNE1-HA. Arrows= Fluorescent signal at the cell surface. Star = signal within the intracellular compartment. The nucleus has been counterstained with DAPI (blue). D. Membrane co-localization of KCNQ1 S338F and KCNE1. Green = S338F KCNQ1-MYC; red = WT KCNE1-HA. Co-localization (yellow) can be seen at the cell surface (arrow) and within the interior of the cell (star). E. Membrane co-localization of KCNQ1 S338F and KCNE1 in the presence of WT KCNQ1. Red = anti-HA (KCNE1). Green = anti-MYC (S338F KCNQ1). F. Membrane co-localization of KCNQ1 S338F and KCNQ1 WT in the presence of KCNE1. Triple-transfected cells as in E were imaged for S338F (MYC, green) and WT KCNQ1 (FLAG, red). Again note signal co-localization at the cell membrane (arrow) and to a lesser extent within the cell interior (star).
Structural modeling of KCNQ1 WT and S338F mutant interactions with KCNE1
We used MODELLER software to build closed-state structures of the WT and S338F/F339S variants of KCNQ1 channels, assuming a tetramer with a fourfold axis of symmetry (Figure 7A); each monomer contains an N-terminal voltage sensor (VS) domain and a C-terminal pore forming domain (17). The pore domain of each monomer crosses over its neighbor in an orthogonal fashion, creating the central ion-conducting channel. Because the extracellular opening of the pore always remains open, gating of the cytosolic vestibule is key to controlling ion flow (Figure 7B).
Figure 7. Structural modeling of KCNQ1 mutant channels. A and B. Model of WT KCNQ1 channel in the closed state.
A. Bottom view, as visualized along an axis perpendicular to the plane of the cell membrane from the cytosolic side. Dashed circles and connecting arrows show direction of rotation of the VS domain during channel opening. Red dot denotes the central pore. B. Side view, in the plane of the cell membrane. Blue, green, cyan and purple: individual monomers. Gray: the S4-S5 helical linker connecting the VS domain (helices S1-S4) to the pore forming domain (helices S5/P/S6) within each monomer. Red arrow: path of potassium ions. C and D. Impact of the S338F variant on KCNQ1 channel structure. In C and D, the interface of two monomers (cyan and green) is highlighted within the tetrameric KCNQ1. Grey: S4-S5 helical linker. C. Model of KCNQ1 WT in the closed state. D. Model of KCNQ1 S338F in the closed state. Red: sidechain moieties of residues S338/F338 in the S6 helix are shown interacting through van der Waals forces with residues W248/L251 residues in the S4-S5 helical linker of an adjacent monomer. E and F. Impact of the F339S variant on KCNQ1 channel structure. In these models, only one monomer is shown (blue) so as to highlight inter-helical interactions within each monomer. Gray: S4-S5 helical linker. Red: sidechain moieties of F339/S339 and T265/I268 involved in intramolecular van der Waals contacts between S5 and S6 helices. E. Model of KCNQE1 WT in the closed state. F. Model of KCNQ1 F339S in the closed state.
Importantly, the VS and pore domains are connected by a short S4-S5 helical linker that is critical for transmitting voltage-induced conformational changes in the VS to opening of the pore. At the cytosolic gate, the S6 C-terminus hinges outward, enforced by a proline at residue 343, creating an opening for ions (17, 25). Our model predicts that in the closed state (Figure 7A), the S6 C hinge is forced to straighten by steric hindrance applied downward by the S4-S5 helical linker. In this conformation, the interwoven S6 helices close off the cytosolic vestibule of the pore. During membrane depolarization, the VS domains undergo a synchronized rotation about an axis perpendicular to the membrane plane (17, 25) (see rotational arrow in Figure 7A). This movement pulls the S4-S5 helical linker away from S6, allowing the S6 helix to bend at its hinge point at residue P343 and splay outward from the central axis of the channel. This bending results in the opening of the cytosolic pore to K+ ions (Figure 7B).
Both S338 and F339 residues are located within the functionally critical S6 helix and lie just one helical turn above the hinge at residue P343. Structural modeling of the tetrameric channel revealed that S338 in one monomer lies close to residues W248 and L251 within the S4-S5 linker of the neighboring monomer (Figure 7C and Supplementary File 2). Thus, substitution of a small polar serine with a more bulky and apolar phenylalanine at this position will likely result in the formation of a tight network of van der Waals contacts among these residues (Figure 7D and Supplementary File 3). In particular, overlapping of electron clouds of the benzyl ring of F338 with those of the indole moiety of W248 and the aliphatic sidechain of L152 would be highly energetically favorable. The result is to create a new stable interaction between the S4-S5 linker of one monomer and the S6 helix of another that does not exist in the WT tetramer. Such an interaction would increase the energy required to displace the S4-S5 linker away from S6, and channel opening would thus require a higher membrane potential.
Structural modeling of KCNQ1 WT and F339S mutant interactions
As described above, opening of the inner channel pore involves bending and outward splaying of the S6 helices at the hinge centered at position P343. During depolarization, rotation of the VS domains (Figure 7A) applies torque to the S5 helix that is transmitted to S6 just above the hinge, promoting its outward bending and opening of the channel. Transmission of this torque requires a strong network of van der Waals adhesive contacts between the anti-parallel S5 and S6 helices (Figure 7A). One strong adhesive contact is formed by the sandwiching of the benzyl ring of F339 in S6 with the aliphatic sidechains of T265 and I268 in S5 (Figure 7E and Supplementary File 2). Our model shows that substitution of the bulky apolar phenylalanine with a small polar serine in the F339S mutant would disrupt this contact, uncouple the S6 C-terminus from voltage-dependent opening forces (Figure 7F and Supplementary File 4), and irreversibly inactivate the KCNQ1 channel.
DISCUSSION
Our observations identify a novel KCNQ1 mutation that qualitatively alters the effect of its accessory subunit KCNE1 from a positive modulation to a negative one, thereby profoundly depressing the current density of IKs over a wide range of voltages. In the absence of KCNE1, homomeric S338F forms functional channels exhibiting a positive shift in voltage-dependent activation, reminiscent of the rightward shift in voltage dependence of channel activation that occurs when KCNQ1 complexes with KCNE1 (26). Our model shows that the S338F substitution causes this rightward shift by creating a new “sticky” intermolecular bond between S6 and the S4-S5 linker that impedes voltage-induced movement of the linker away from the pore. Since this movement is required for pore opening, as shown by molecular dynamics simulations (17, 27, 28) (see Supplementary Files 2 and 3), greater energy is required to open the channel, in excellent agreement with our electrophysiologic data.
Our model further explains the catastrophic impact of adding KCNE1 to S338F homotetramers. Cysteine mapping and mutagenesis have identified multiple points of contact between KCNQ1 and KCNE1, notably between the E1 transmembrane domain and the Q1 S6 domain (including residues S338 and F339), between the E1 transmembrane domain and Q1 S1 domain of a different subunit, and between the E1 extracellular domain and S1-S2 linker of Q1 (10, 26, 27, 29). Importantly, a recent structural model suggests that KCNE1 contacts the S4-S5 helix from the side opposite to that pressing on the S6 helix. This contact impedes S4-S5 linker movement, and thereby increases the voltage required for pore opening (17, 30). The “sandwiching” of the S4-S5 linker between the new sticky interior phenylalanine and the exterior KCNE1 C-terminus effectively immobilizes the S4-S5 helical linker, rendering the channel unable to open.
Another interesting observation is that heterotetramers of WT and S338F channels conduct much less current than homotetramers of either protein. Again, our model predicts that the mutant phenylalanine of one monomer interacts with residues of a neighboring subunit, so that WT KCNQ1 gating or interaction with endogenous oocyte KCNE-like proteins could be affected by the presence of one or more S338F subunits within the same complex. Alternatively, the presence of the large substitution at aa338 could impair the 4-sided symmetry of the heterotetrameric pore structure.
The adjacent F339S mutation from Proband 2 results in a non-functional channel. It, like the S338F mutant, exerted dominant-negative effects on WT channel function, providing strong support for the concept that it is translated and assembled into complexes with WT KCNQ1. Although we cannot exclude a co-assembly-induced trafficking defect, our structural model predicts that the F339S mutation disrupts a critical strong contact between S5 and S6 (Figure 7F and Supplementary File 4) that is highly likely to result in misfolding, and consequently either irreversible inactivation of the channel or failure to form functional channels at the surface. Our functional and structural data are consistent with and expand upon those of Yang et al., who previously identified the F339S mutation and localized it to a critical interhelical position in S6 (13). It is intriguing to speculate that the lack of clinical events in proband 2 is related to the presence of a non-functioning channel, as opposed to the dysfunctional channel in the family of Proband 1.
The apparent lack of impact of co-existing rare KCNE1 and KCNE2 variants in the family of Proband 1 is noteworthy, although the small number of individuals prevents conclusive interpretation (21, 23, 31). The D85N variant lies near a KCNQ1-interacting D76 residue (11), and has been associated with greater QT interval length (+10.5 msec) (32, 33). Both KCNE1 D85N and KCNE2 Q9E variants have been implicated in drug-induced LQTS and torsade de pointes (20-23), and D85N is frequently identified as a second variant in LQTS patients with other mutations (34). In our studies, presence of the D85N variant had no effect, positive or negative, on S338F channel conductance, possibly because inhibitory interaction between KCNE1 and KCNQ1 involves other contacts, or because the negative effect of S338F conceals the more subtle effects of KCNE1 D85N on IKs.
One potential limitation of these studies is the presence of competing effects of endogenous KCNQ1-like and KCNE1-like channels previously identified in X. laevis oocytes. However, the background currents contributed by these channels were minimal throughout these experiments, and it is unlikely that such currents masked important differences between the wt and mutant channels, or had mutation-specific impact on the exogenous channels.
The novel insight provided by our study is the identification of a mutation that causes abnormal bonding between alpha-subunit amino acids within the tetramer, the effect of which is magnified by the stabilizing effect of KCNE1 binding. Our findings further reinforce the importance for channel gating and kinetics of direct contacts between positions S338 and F339 in Q1 with E1 transmembrane residues, as reported by others (26, 35), and additionally provide a new view of steric interactions among subunits of the homotetramer and accessory subunits that affect the energy requirements for gating.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Lijing You for her technical support, and to Dr. Charles Luetje for providing access to the OpusXpress system. The authors also thank Dr. Peter Larsson for his reading and comments on the manuscript.
SOURCES OF FUNDING
Major support for this work was provided by the Florida Heart Research Institute (to N.H.B. and R.J.M.), the National Institutes of Health Grants R01-HL71094 (to N.H.B.) and R01-GM083897 (to A.F.), the American Heart Association Florida/Puerto Rico Affiliate (to L.B.-R.), and the Fondation Leducq (Trans-Atlantic Network of Excellence Grant, “Preventing Sudden Cardiac Death” 05-CVD-01, to R.J.M. and N.H.B.). Dr. Hoosien is supported by an NIH T32 Training Grant in Cardiovascular Signaling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
N.B.: Supplemental File 1 contains a list of key terms and their definitions.
DISCLOSURES: The authors have no conflicts of interest to report.
REFERENCES
- 1.Modell SM, Lehmann MH. The long QT syndrome family of cardiac ion channelopathies: a HuGE review. Genet Med. 2006;8:143–155. doi: 10.1097/01.gim.0000204468.85308.86. [DOI] [PubMed] [Google Scholar]
- 2.Moss AJ, Shimizu W, Wilde AA, et al. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation. 2007;115:2481–2489. doi: 10.1161/CIRCULATIONAHA.106.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanguinetti MC, Curran ME, Zou A, et al. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 4.Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 5.Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev. 2005;85:1205–1253. doi: 10.1152/physrev.00002.2005. [DOI] [PubMed] [Google Scholar]
- 6.Chen YH, Xu SJ, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 7.Osteen JD, Sampson KJ, Kass RS. The cardiac IKs channel, complex indeed. Proc Natl Acad Sci USA. 2010;107:18751–18752. doi: 10.1073/pnas.1014150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marx SO, Kurokawa J, Reiken S, et al. Requirement of a macromolecular signaling complex for - adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]
- 9.Nakajo K, Ulbrich MH, Kubo Y, Isacoff EY. Stoichiometry of the KCNQ1-KCNE1 ion channel complex. Proc Natl Acad Sci USA. 2010;107(44):18862–7. doi: 10.1073/pnas.1010354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung DY, Chan PJ, Bankston JR, et al. Location of KCNE1 relative to KCNQ1 in the IKs potassium channel by disulfide cross-linking of substituted cysteines. Proc Natl Acad Sci U S A. 2009;106(3):743–8. doi: 10.1073/pnas.0811897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Zheng R, Melman YF, McDonald TV. Functional interactions between KCNE1 C-terminus and the KCNQ1 channel. PLoS One. 2009;4:5143. doi: 10.1371/journal.pone.0005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung SK, MacCormick JM, McCulley CH, et al. Long QT and Brugada syndrome gene mutations in New Zealand. Heart Rhythm. 2007;4(10):1306–1314. doi: 10.1016/j.hrthm.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 13.Yang T, Chung SK, Zhang W, et al. Biophysical properties of 9 KCNQ1 mutations associated with long-QT syndrome. Circ Arrhythm Electrophysiol. 2009;2(4):417–426. doi: 10.1161/CIRCEP.109.850149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Splawski I, Shen J, Timothy KW, Vincent GM, Lehmann MH, Keating MT. Genomic structure of three long QT syndrome genes: KVLQT1, HERG, and KCNE1. Genomics. 1998;51(1):86–97. doi: 10.1006/geno.1998.5361. [DOI] [PubMed] [Google Scholar]
- 15.Syrris P, Murray A, Carter ND, McKenna WM, Jeffery S. Mutation detection in long QT syndrome: a comprehensive set of primers and PCR conditions. J Med Genet. 2001;38(10):705–10. doi: 10.1136/jmg.38.10.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 17.Smith JA, Vanoye CG, George AL, Jr, Meiler J, Sanders CR. Structural models for the KCNQ1 voltage-gated potassium channel. Biochemistry. 2007;46:14141–14152. doi: 10.1021/bi701597s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 19.Carson M. Ribbons 2.0. J. Appl. Crystallogr. 1991;24:958–961. [Google Scholar]
- 20.Paulussen AD, Gilissen RA, Armstrong M, et al. Genetic variations of KCNQ1, KCNH2, SCN5A, KCNE1, and KCNE2 in drug-induced long QT syndrome patients. J Mol Med. 2004;82:182–188. doi: 10.1007/s00109-003-0522-z. [DOI] [PubMed] [Google Scholar]
- 21.Salisbury B, Judson RS, et al. AB47-4: The single nucleotide polymorphism D85NKCNE1 is associated with both congenital and drug-induced Long QT. Abstract. Heart Rhythm. 2006;3:S98. [Google Scholar]
- 22.Kääb S, Crawford DC, Sinner MF, et al. A Large Candidate Gene Survey Identifies the KCNE1 D85N Polymorphism as a Possible Modulator of Drug-Induced Torsades de Pointes. Circ Cardiovasc Genet. 2011 Nov 18; doi: 10.1161/CIRCGENETICS.111.960930. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishio Y, Makiyama T, Itoh H, et al. D85N, a KCNE1 polymorphism, is a disease-causing gene variant in Long QT syndrome. J Am Coll Cardiol. 2009;54:812–819. doi: 10.1016/j.jacc.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Grunnet M, Jespersen T, Rasmussen HB, et al. KCNE4 is an inhibitory subunit to the KCNQ1 channel. J Physiol. 2002;542:119–30. doi: 10.1113/jphysiol.2002.017301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang C, Tian C, Sönnichsen FD, et al. Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panaghie G, Tai KK, Abbott GW. Interaction of KCNE subunits with the KCNQ1 K+ channel pore. J Physiol. 2006;570:455–467. doi: 10.1113/jphysiol.2005.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melman YF, Um SY, Krumerman A, Kagan A, McDonald TV. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron. 2004;42:927–937. doi: 10.1016/j.neuron.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Jensen MØ, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–33. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Jiang M, Hsu KL, Zhang M, Tseng GN. KCNQ1 and KCNE1 in the IKs channel complex make state-dependent contacts in their extracellular domains. J Gen Physiol. 2008;131:283–91. doi: 10.1085/jgp.200809976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Horn WD, Vanoye CG, Sanders CR. Working model for the structural basis for KCNE1 modulation of the KCNQ1 potassium channel. Curr Opin Struct Biol. 2011;21(2):283–91. doi: 10.1016/j.sbi.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ackerman MJ, Tester DJ, Jones GS, Will ML, Burrow CR, Curran ME. Ethnic differences in cardiac potassium channel variants: implications for genetic susceptibility to sudden cardiac death and genetic testing for congenital long QT syndrome. Mayo Clin Proc. 2003;78:1479–1487. doi: 10.4065/78.12.1479. [DOI] [PubMed] [Google Scholar]
- 32.Gouas L, Nicaud V, Berthet M, et al. Association of KCNQ1, KCNE1, KCNH2 and SCN5A polymorphisms with QTc interval length in a healthy population. Eur J Hum Genet. 2005;13:1213–1222. doi: 10.1038/sj.ejhg.5201489. [DOI] [PubMed] [Google Scholar]
- 33.Marjamaa A, Newton-Cheh C, Porthan K, et al. Common candidate gene variants are associated with QT interval duration in the general population. J Intern Med. 2009;265:448–458. doi: 10.1111/j.1365-2796.2008.02026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westenskow P, Splawski I, Timothy KW, Keating MT, Sanguinetti MC. Compound mutations: a common cause of severe long-QT syndrome. Circulation. 2004;109:1834–1841. doi: 10.1161/01.CIR.0000125524.34234.13. [DOI] [PubMed] [Google Scholar]
- 35.Strutz-Seebohm N, Pusch M, Wolf S, Stoll R, Tapken D, Gerwert K, et al. Structural basis of slow activation gating in the cardiac I Ks channel complex. Cell Physiol Biochem. 2011;27:443–52. doi: 10.1159/000329965. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.