Abstract
Objective
Epigenetic modifications contribute to the etiology of type 2 diabetes.
Method
We performed genome-wide methylome and transcriptome analysis in liver from severely obese men with or without type 2 diabetes and non-obese men to discover aberrant pathways underlying the development of insulin resistance. Results were validated by pyrosequencing.
Result
We identified hypomethylation of genes involved in hepatic glycolysis and insulin resistance, concomitant with increased mRNA expression and protein levels. Pyrosequencing revealed the CpG-site within ATF-motifs was hypomethylated in four of these genes in liver of severely obese non-diabetic and type 2 diabetic patients, suggesting epigenetic regulation of transcription by altered ATF-DNA binding.
Conclusion
Severely obese non-diabetic and type 2 diabetic patients have distinct alterations in the hepatic methylome and transcriptome, with hypomethylation of several genes controlling glucose metabolism within the ATF-motif regulatory site. Obesity appears to shift the epigenetic program of the liver towards increased glycolysis and lipogenesis, which may exacerbate the development of insulin resistance.
Keywords: Liver, Obesity, Type 2 Diabetes, Epigenetics, Lipid, Glucose
Highlights
-
•
Omic analysis in human liver reveals aberrant pathways in obesity and diabetes.
-
•
Genes regulating glycolysis and de novo lipogenesis were hypomethylated in obesity.
-
•
DNA methylation of ATF binding elements alters hepatic gene expression.
-
•
Obesity shifts the epigenome to increase lipid production and insulin resistance.
1. Introduction
Although the prevalence of type 2 diabetes is approaching epidemic proportions, the underlying pathophysiology is still not fully understood. The etiology of type 2 diabetes is complex and multifactorial, as it is influenced by both genetic predisposition [30] and lifestyle factors, such as diet and physical activity [14]. The liver is a key glucoregulatory organ involved in the maintenance of glucose homeostasis and the development of type 2 diabetes [17], [31]. In healthy individuals, insulin promotes hepatic glycogen synthesis and de novo lipogenesis, while inhibiting gluconeogenesis. However, in obese or type 2 diabetic patients, insulin fails to suppress hepatic glucose output, which leads to hyperglycemia via an upregulation of the gluconeogenic enzymes glucose-6-phosphatase and phosphoenolpyruvate carboxykinase [27]. The underlying molecular mechanism for these shifts in hepatic metabolism is still incompletely resolved and aberrant regulation of additional metabolic pathways are likely to be involved.
Epigenetic mechanisms such as DNA methylation integrate environmental factors and genetic susceptibility by modulating transcriptional regulation without changing the underlying DNA sequence. DNA methylation is an epigenetic mark that can change in response to environmental challenges to directly modify gene expression. DNA methylation can modify gene expression in several ways, for example by altering histone interactions, influencing transcription factor binding, and/or recruitment of methyl-binding proteins [16]. Dynamic DNA methylation often occurs distal to the transcription start site, with the position co-localizing with gene regulatory elements, particularly enhancers and transcription factor-binding sites [50]. Alterations in DNA methylation at differentially methylated sites or regions have been implicated in metabolic diseases such as obesity [24], [26], [48] and type 2 diabetes [5], [26], [36], [46], [47]. Since DNA methylation can lead to stable alterations in the transcriptional potential, epigenetic mechanisms may partly explain the rapidly increasing prevalence of type 2 diabetes [23]. Recent evidence suggests that DNA methylation of key metabolic genes in skeletal muscle is remodeled by interventions known to improve insulin sensitivity such as exercise [6], [36] or bariatric surgery [4]. Thus, changes in the epigenome may provide an underlying molecular mechanism for deleterious metabolic health outcomes associated with severe obesity or type 2 diabetes. Likewise, coordinated epigenetic changes may also improve metabolic health after therapeutic intervention.
Systematic studies of the DNA methylation landscape and the related transcriptome of metabolic tissues from obese and/or type 2 diabetic patients show that DNA methylation is altered in metabolic diseases [4], [24], [26], [32], [33], [35], [36], [40], [43], [46], [47]. For example DNA methylation at the HIF3A loci in blood cells is correlated with BMI in Caucasian adults [13]. Moreover, evidence from mouse models indicates that normally occurring variation in methylation levels contribute to clinically relevant hepatic traits [37]. Therefore, global epigenome and transcriptome analysis of human liver in various states of insulin resistance could offer valuable insight into regulatory mechanisms involved in the pathogenesis of type 2 diabetes.
To better understand the molecular mechanisms underlying the development of hepatic insulin resistance and type 2 diabetes, we performed a genome-wide methylome and transcriptome analysis of liver from age-matched non-obese metabolically healthy, obese non-diabetic and obese type 2 diabetic men. We found key genes involved in hepatic glycolysis and de novo lipogenesis were hypomethylated and activated in obese non-diabetic and obese type 2 diabetic patients compared to non-obese control subjects. Our results indicate the epigenetic landscape in liver is altered in obesity, concomitant with increased expression of genes involved in hepatic glycolysis and gluconeogenesis, as well as stearate biosynthesis. These genomic changes may contribute to the development of insulin resistance in obesity and type 2 diabetes.
2. Results
2.1. Obese and type 2 diabetic signature of the hepatic methylome and transcriptome
Liver biopsies were obtained from non-obese men during cholecystectomy and from obese non-diabetic and obese type 2 diabetic men during Roux-en Y gastric bypass surgery. Anthropometric and clinical parameters of the study cohort are presented (Table 1). Age was not different between the cohorts. Body weight, body mass index (BMI) and waist circumference did not differ between the obese non-diabetic and obese type 2 diabetic patients, but was increased compared to the non-obese participants. While plasma triglycerides, high density lipoprotein cholesterol (HDL) and low density lipoprotein cholesterol (LDL) levels were unchanged between the cohorts, fasting serum glucose, glycated hemoglobin (HbA1c) and homeostatic model assessment – insulin resistance (HOMA-IR) levels were increased in the obese type 2 diabetic patients compared to the non-obese controls and obese non-diabetic patients.
Table 1.
Clinical Parameters.
| Non-Obese Controls |
Obese Non-Diabetic |
Obese Type 2 Diabetic |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Transcriptomic analysis | Methylomic analysis | Complete cohort | Transcriptomic analysis | Methylomic analysis | Complete cohort | Transcriptomic analysis | Methylomic analysis | Complete cohort | |
| N (male) | 6 | 7 | 11 | 6 | 7 | 13 | 6 | 8 | 10 |
| Age [years] | 40.5 (36.7–49.0) ± 4.1 | 42.7 (36.2–59.1) ± 7.8 | 41 (34.0–59.1) ± 6.8 | 41.6 (32.5–50.5) ± 6.7 | 45.8 (36.4–59.8) ± 7.6 | 44 (32.5–59.8) ± 8.3 | 40.9 (21.4–62.1) ± 13.0 | 41.2 (21.4–62.1) ± 11.3 | 43 (21.4–62.1) ± 11.4 |
| Body weight [kg] | 83.3 (71.6–89.1) ± 7.2 | 80.8 (71.6–89.1) ± 6.8 | 82.3 (68.0–89.1) ± 7.3 | 120.0 (114.0–140.0) ± 9.26a | 123.6 (114.0–144.0) ± 12.0a | 136.9 (114.0–201.) ± 26.5a | 136.3 (104.0–154.0) ± 16.3a | 132.9 (104.0–154.0) ± 15.4a | 132.8 (104.0–154.0) ± 13.8a |
| BMI [kg/m2] | 26.1 (22.6–28.4) ± 2.0 | 25.4 (22.6–27.8) ± 1.7 | 25.7 (20.0–28.4) ± 2.4 | 38.9 (35.6–41.6) ± 2.1a | 38.6 (35.6–41.6) ± 2.0 | 42.1 (35.6–56.5) ± 6.3a | 40.9 (36.0–45.8) ±3.3a | 40.3 (36.0–45.8) ±3.1a | 40.6 (36.0–45.8) ±3.1a |
| Waist circumference [cm] | 91.5 (80.0–102.0) ± 7.4 | 90.6 (80.0–100.0) ± 6.6 | 92.0 (80.0–102.0) ± 7.2 | 126.3 (114.0–140.0) ± 8.7a | 125.7 (114.0–140.0) ± 8.7a | 126.9 (114.0–140.0) ± 8.7a | 137.5 (121.0–143.0) ± 7.6a | 133.5 (118.0–143.0) ± 9.7a | 133.5 (118.0–143.0) ± 9.7a |
| Fasting glucose [mmol/l] | 5.5 (4.9–5.9) ± 0.4 | 5.4 (4.8–5.9) ± 0.4 | 5.6 (4.8–6.3) ± 0.4 | 5.6 (5.3–5.9) ±0.2 | 5.5 (5.2–5.9) ±0.2 | 5.6 (5.1–6.1) ±0.3 | 8.4 (6.6–13.4) ± 2.3ab | 8.2 (6.6–13.4) ± 2.1ab | 8.4 (6.6–13.4) ± 2.0ab |
| HbA1c [%] | 4.6 (4.2–5.0) ± 0.2 | 4.6 (4.2–5.0) ± 0.2 | 4.6 (4.2–5.0) ± 0.2 | 4.7 (4.3–5.0) ± 0.3 | 4.7 (4.2–5.0) ± 0.3 | 4.7 (4.2–5.0) ± 0.3 | 6.0 (5.2–7.9) ± 0.8ab | 6.2 (5.2–7.9) ± 1.0a | 6.1 (5.2–7.9) ± 0.9ab |
| Fasting insulin [pmol/l] | 74.8 (40.0–189.0) ± 51.9 | 67.9 (27.0–189.0) ± 50.9 | 73.0 (27.0–189.0) ± 41.7 | 136.0 (72.5–183.9) ± 42.7a | 139.0 (72.5–199.7) ± 44.9 | 127.7 (72.5–199.7) ± 42.9a | 205.1 (151.6–299.6) ± 49.1a | 196.7 (146.5–299.6) ± 49.9a | 180.6 (115.0–299.6) ± 53.6a |
| HOMA-IR | 2.7 (1.5–6.9) ± 1.9 | 2.4 (0.9–6.9) ± 1.9 | 2.2 (0.9–6.9) ± 1.5 | 4.8 (2.6–6.4) ± 1.5 | 4.8 (2.6–6.6) ± 1.4 | 5.0 (2.6–6.8) ± 1.5a | 10.6 (8.0–13.0) ± 2.1ab | 10.2 (7.7–13.0) ± 2.2ab | 9.6 (7.5–13.0) ± 2.2ab |
| LDL [mmol/l] | 3.2 (1.8–3.9) ± 0.7 | 3.4 (1.8–4.7) ± 0.8 | 3.3 (1.8–4.7) ± 0.8 | 3.6 (2.3–4.4) ± 0.7 | 3.6 (2.3–4.4) ± 0.7 | 3.5 (2.3–4.4) ± 0.6 | 3.2 (1.7–4.9) ± 1.1 | 3.0 (1.3–4.9) ± 1.7 | 3.0 (1.3–4.9) ± 1.2 |
| HDL [mmol/l] | 1.2 (0.9–1.5) ± 0.2 | 1.1 (0.7–1.5) ± 0.3 | 1.1 (0.7–1.5) ± 0.3 | 1.1 (0.4–1.4) ± 0.3 | 1.2 (0.4–2.0) ± 0.4 | 1.2 (0.4–2.0) ± 0.4 | 0.8 (0.6–1.0) ± 0.2 | 1.2 (0.6–3.9) ± 1.0 | 1.2 (0.6–3.9) ± 1.0 |
| Plasma triglycerides [mmol/l] | 1.2 (0.6–2.2) ± 0.5 | 1.2 (0.6–2.2) ± 0.5 | 1.2 (0.6–2.2) ± 0.5 | 1.7 (1.2–2.6) ± 0.5 | 1.7 (0.9–2.6) ± 0.5 | 1.6 (0.9–2.6) ± 0.5 | 2.1 (1.2–3.3) ± 0.7 | 1.9 (1.2–3.3) ± 0.7 | 1.9 (1.2–3.3) ± 0.7 |
| Liver triglycerides [mM/mg] | 7.5 (2.0–16.9) ± 5.6 | 7.9 (2.0–16.9) ± 6.1 | 12.6 (2.0–43.1) ± 14.4 | 63.3 (12.4–95.6) ± 30.9a | 47.0 (12.4–95.6) ± 36.4c | 37.7 (6.0–95.6) ± 31.6 | 40.3 (6.1–88.3) ± 30.7 | 41.0 (6.1–88.3) ± 31.0 | 33.4 (6.1–88.3) ± 28.5 |
Data are mean (minimum–maximum) ± SD from the cohort used for transcriptomic and methylomic analysis and the entire cohort; Differences between groups were analyzed by one way ANOVA and Tukey's multiple comparisons test.
P < 0.05 versus non-obese controls in same cohort.
P < 0.05 versus obese non-diabetic patients in same cohort.
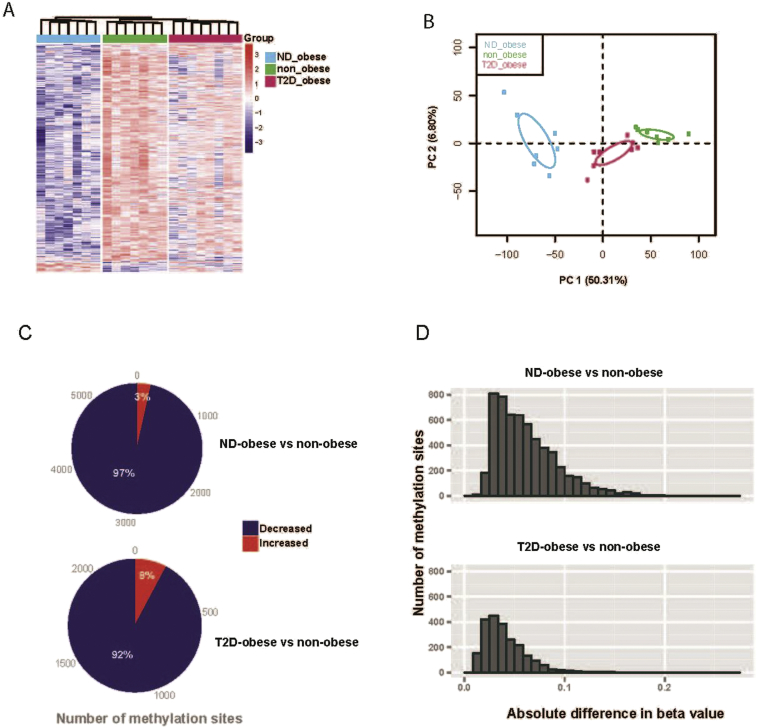
To identify novel regulatory mechanisms involved in the pathogenesis of obesity and type 2 diabetes, we determined the hepatic methylome of 7 non-obese metabolically healthy, 7 obese non-diabetic and 8 obese type 2 diabetic Caucasian men of similar age using the Illumina Infinium 450K Bead Chip. We also assessed the hepatic transcriptome using Affymetrix gene arrays of liver biopsies obtained from 18 individuals (6 from each group). Samples from 17 of the subjects were subjected to both the transcriptome and methylome analysis. Methylome data were filtered for detection and analyzed according to the Beta Mixture Quantile dilation (BMIQ) method [29]. Employing a multigroup comparison, 5834 CpG loci were identified to be differentially methylated among the three groups (FDR < 0.25). Hierarchical clustering and principal component analysis (PCA), based on the differentially methylated sites, revealed a clear separation of the three phenotypic groups, with the obese non-diabetic group being well-separated from the obese type 2 diabetic and non-obese group (Figure 1A,B). In total, 5682 CpG sites mapping to 3058 individual genes were differentially methylated in obese non-diabetic individuals versus non-obese controls (p < 0.05; Tables S1 and S2). Moreover, 2255 CpG sites mapping to 1388 individual genes were differentially methylated between obese type 2 diabetic individuals compared to non-obese controls (p < 0.05; Table Tables S1 and S2). In particular, the vast majority of CpG sites displayed decreased DNA methylation in obese non-diabetic and obese type 2 diabetic individuals, compared to non-obese controls (97 and 92%, respectively; Figure 1C). The distribution of the absolute differences in DNA-methylation between obese non-diabetic versus non-obese controls and obese type 2 diabetic individuals versus non-obese controls is shown in Figure 1D.
Figure 1.
The human methylome in obese non-diabetic, obese type 2 diabetic and non-obese individuals. (A) Hierarchical clustering for 5834 differentially methylated sites derived from multi-group comparison (FDR < 0.25). (B) Principal component analysis (PCA) of 5834 differentially methylated sites for the major two principal components (PC). The 95% confidence ellipses are shown for each group. (C) Pie chart indicating the number of sites that exhibit increased and decreased DNA methylation in liver from obese non-diabetic and obese type 2 diabetic individuals compared to non-obese controls. (D) Distribution of the absolute difference in DNA-methylation in liver from obese non-diabetic and obese type 2 diabetic individuals compared to non-obese controls.
To investigate whether obesity and/or type 2 diabetes is associated with alterations in global DNA methylation, and/or alterations in the methylation profile at specific genomic positions, the distribution of altered CpG sites was mapped according to defined CpG categories and in relation to the nearest gene. Although distinct changes in DNA methylation were associated with obesity and type 2 diabetes (Figure 1), global DNA methylation with regard to CpG categories was similar between the three cohorts (Figure S1A). Additionally, genomic distribution of the differentially methylated sites with regard to the nearest gene was similar between the cohorts (Figure S1B). In general, differential DNA-methylation was observed at specific loci, rather than spread over larger chromosomal areas (data not shown), as is often observed in various cancers [45].
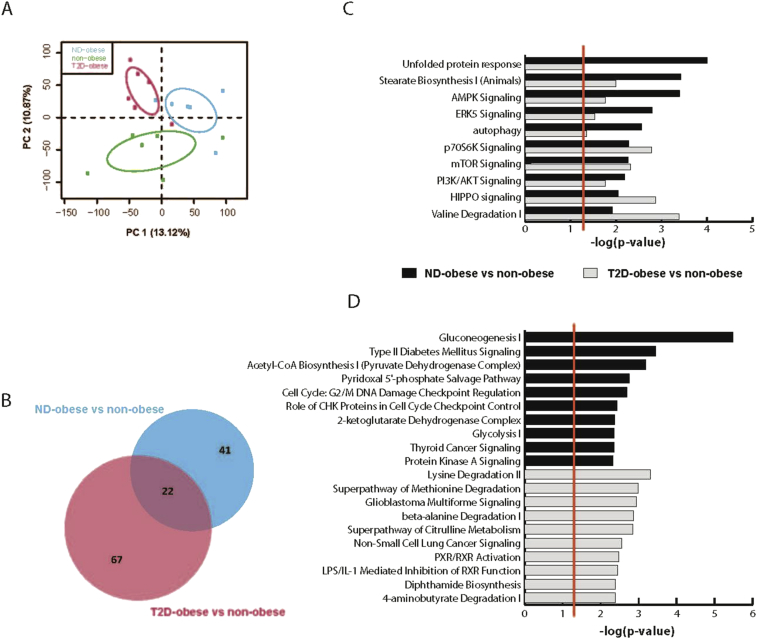
PCA analysis based on overall gene expression, separated the three groups into distinct clusters (Figure 2A). Analysis of differences in gene expression profiles between the groups revealed that 448 transcripts were differentially expressed in obese non-diabetic individuals versus non-obese controls (FDR ≤ 0.25; Table S3). Moreover, 1285 transcripts were differentially expressed in obese type 2 diabetic individuals versus non-obese controls (FDR ≤ 0.25; Table S3). Only 11 transcripts were identified as differentially expressed in obese non-diabetic individuals versus obese type 2 diabetic individuals.
Figure 2.
Canonical pathway analysis of changes in the hepatic transcriptome for obese non-diabetic and obese type 2 diabetic individuals compared to non-obese individuals. (A) PCA on the overall gene expression profile based on all annotated transcripts for two major principal components (PC). The 95% confidence ellipses are shown for each group. (B). Venn diagram of pathways changed in obese non-diabetic individuals compared to non-obese controls and obese type 2 diabetic individuals compared to non-obese controls. (C). The top 10 pathways changed in both obese type 2 diabetic individuals compared to non-obese controls and obese non-diabetic individuals compared to non-obese controls. (D) The top 10 pathways specifically changed in obese non-diabetic individuals compared to non-obese controls or obese type 2 diabetic individuals compared to non-obese controls, respectively. Log p-values are shown at the bottom of the C and D panels. The red lines in C and D indicate threshold significance for enrichment, P-value 0.05.
Canonical pathway analysis was employed to identify pathways in the liver that are altered in obesity and type 2 diabetes. Interestingly, there was substantial overlap between pathways altered in obese non-diabetic individuals compared to non-obese controls and pathways altered in obese type 2 diabetic individuals compared to non-obese controls (Figure 2B). For example, genes involved in stearate biosynthesis, AMPK signaling and PI3K/AKT signaling were affected in both obese non-diabetic, as well as obese type 2 diabetic individuals compared to non-obese controls (Figure 2C). Genes involved in gluconeogenesis and glycolysis were specifically altered in liver from obese non-diabetic individuals compared to non-obese controls (Figure 2D; upper panel). Moreover, transcripts involved in PXR/RXR activation were specifically altered in obese type 2 diabetic individuals compared to non-obese individuals (Figure 2D; lower panel).
2.2. Correlation of the obese and type 2 diabetic hepatic methylome and transcriptome
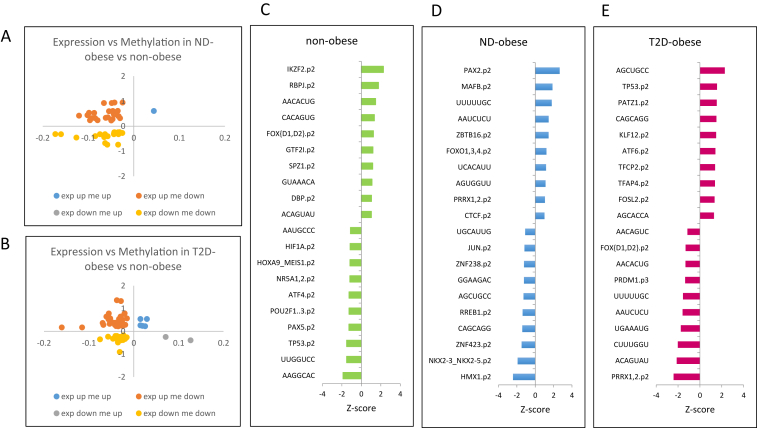
Figure 3A,B displays CpG sites with altered DNA-methylation and where the expression of the associated gene is changed for obese non-diabetic individuals versus non-obese controls and obese type 2 diabetic individuals versus non-obese controls, respectively. Overall, we identify 36 and 61 genes with significantly altered expression where one, or several, CpG sites display altered DNA-methylation. Among the genes with altered expression and DNA-methylation in obese type 2 diabetic individuals compared to non-obese controls are several genes belonging to the nerve growth factor signaling and Wnt signaling pathways. Specifically, six genes that are involved in the nerve growth factor signaling pathway display increased expression and decreased DNA-methylation in obese type 2 diabetic individuals compared to non-obese controls; PRKCE (protein kinase C, epsilon), ABR (active BCR-related), PDGFA (platelet-derived growth factor alpha polypeptide), ARHGEF16 Rho guanine nucleotide exchange factor (GEF) 16, ADCY6 (adenylate cyclase 6) and RPS6KA1 (ribosomal protein S6 kinase, 90 kDa, polypeptide 1). Activation of PRKCE has been implicated as a key switch leading to decreased insulin signaling in the pathogenesis of hepatic insulin resistance and type 2 diabetes, as well as NAFLD [9]. Three additional genes involved in the Wnt signaling pathway also showed increased expression and decreased DNA-methylation in obese type 2 diabetic individuals compared to non-obese controls; CTBP1 (C-terminal binding protein 1), CCND1 (Cyclin D1) and WNT11 (wingless-type MMTV integration site family, member 11). This analysis identifies several genes that are differently regulated in liver between obese type 2 diabetic individuals and obese non-diabetic individuals, respectively, and non-obese controls and provides a useful resource for future studies to interrogate gene regulatory responses in human liver, as well as the impact of metabolic disease.
Figure 3.
Top candidate genes and transcription factors altered in obesity and type 2 diabetes. Fold change gene expression (y-axis; log2 value) was plotted against the change in DNA methylation (x-axis; M-value) for non-diabetic obese individuals versus non-obese controls (A) and obese type 2 diabetic individuals versus non-obese controls (B). Top 10 transcription factors that are predicted by ISMARA to be activated (positive Z-values) or inactived (negative Z-values) in liver of (C) non-obese controls, (D) obese non-diabetic patients and (E) obese type 2 diabetic patients. See also Tables S5–S7.
We further identified differentially methylated CpG-sites that show an inverse change in mRNA expression of the associated gene (Table S2, column overlay). A summary of these sites is provided as Table S4. A detailed comparison between the liver methylome and the transcriptome revealed 23 genes, including 26 differentially methylated CpG sites, where decreased DNA-methylation is associated with increased mRNA expression in obese non-diabetic individuals compared to non-obese controls (Table S2 column negative correlation). In obese type 2 diabetic individuals compared to non-obese controls we observed a correlation between increased mRNA expression and decreased DNA-methylation for 33 genes, including 40 differentially methylated CpG sites (Table S2 column negative correlation). As the vast majority of CpG sites displayed decreased DNA methylation in the obese non-diabetic and obese type 2 diabetic individuals, the number of genes where increased DNA-methylation was associated with decreased mRNA expression is very low and only 2 genes, including 2 differentially methylated CpG sites, were observed, both in obese type 2 diabetic individuals compared to non-obese controls (Table S2 column negative correlation).
We also investigated the correlation between changes in DNA-methylation and gene expression for the individual samples using Pearson correlation analysis. Supplementary Figure S2A shows the number of DNA-methylation sites with a negative or positive correlation to the expression of the associated gene that are equal or larger than a correlation coefficient r value of 0.6. Supplementary Figure S2B shows examples of DNA-methylation sites that are positively and negatively correlated to the expression of the associated gene. Finally, Supplementary Figure S2C shows the distribution of altered CpG sites with regard to defined CpG categories and in relation to the nearest gene.
2.3. Hypomethylation of ATF-motifs in liver from obese type 2 diabetic patients
To identify motifs and key transcription factors that drive the observed changes in hepatic gene expression between the obese non-diabetic, obese type 2-diabetic and non-obese cohorts, we performed an Integrated System for Motif Activity Response Analysis (ISMARA) [3]. This unbiased approach identified that distinct motifs and transcription factors are likely to be active in each of the cohorts, and this may explain the observed alterations in gene expression. While genes under the control of the transcription factor activating transcription factor 4 (ATF4) were predicted by the ISMARA analysis to be inactive in liver from the non-obese controls (Figure 3C), genes regulated by ATF6, a transcription factor belonging to the same family as ATF4, were predicted to be activated in obese type 2 diabetic patients (Figure 3E). Conversely, the activity of FOX[D1,D2] was predicted to be high in non-obese controls, and low in obese type 2 diabetic patients (Figure 3D,E). Furthermore, the ISMARA analysis suggests that PRRX1,2 target genes are inactive in obese type 2 diabetic patients, whereas target gene expression suggests high PRRX1,2 activity in obese non-diabetic patients (Figure 3D,E). Furthermore, we mapped DNA methylation and mRNA of the target genes from the top three active and top three inactive transcription factors in non-obese controls (Table S5), obese non-diabetic patients (Table S6), and obese type 2 diabetic patients (Table S7) using our genome-wide data. This approach identified the most likely active and inactive transcription factors in liver from obese non-diabetic and obese type 2 diabetic patients and serves as a resource for further studies to interrogate DNA methylation and gene expression of target genes.
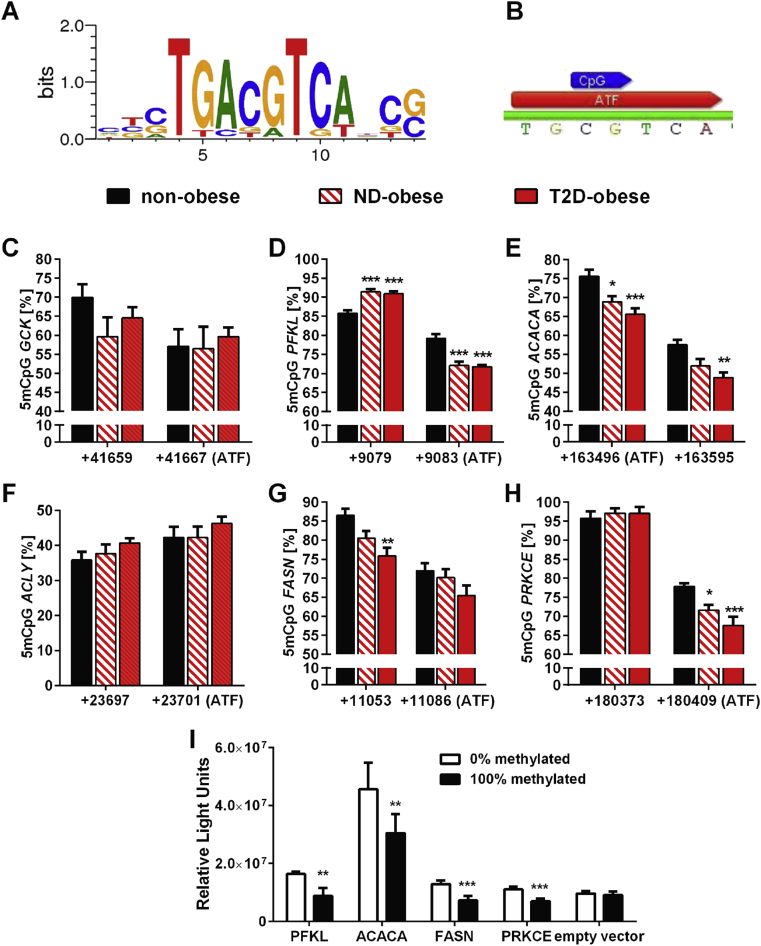
The ISMARA analysis implicates genes that are differently expressed between the non-obese controls and obese type 2 diabetic patients are likely under the transcriptional control of members of the ATF-family. Thus, we investigated methylation and activity of the ATF-family binding motif in human liver. The ATF-family shares a common recognition motif that contains a CpG-site (Figure 4A), which may play a role in the regulation of gene transcription by DNA methylation [10]. To test whether methylation of the CpG-site within this common ATF-motif is altered in obesity and/or type 2 diabetes, the hepatic methylation status of this ATF-CpG-site (TGCGTCA; Figure 4B) in candidate genes was quantified by pyrosequencing. Acetyl-CoA carboxylase alpha (ACACA) is a predicted target gene of ATF6 that was activated in liver from obese type 2 diabetic patients. ACACA transcription was also identified to be upregulated by the canonical pathway analysis (Figure 3). Therefore we searched for an ATF-family-motif within ACACA and other genes identified by the canonical pathway analysis belonging to “Type 2 diabetes signaling” and “Glycolysis/Gluconeogenesis” pathways. Several genes involved in hepatic glycolysis (GCK glucokinase and PFKL phosphofructokinase, liver), de novo lipogenesis (ACLY ATP citrate lyase, ACACA and FASN fatty acid synthase) and insulin signaling (PRKCE protein kinase Cε) contain an ATF-motif in their gene body. Thus, we selected these genes for in-depth analysis. Interestingly, the genome-wide DNA methylation assay identified at least one significantly hypomethylated CpG site in association with these target genes in obese non-diabetic and obese type 2 diabetic patients, compared to non-obese controls (Table S8).
Figure 4.
Hypomethylation of the common ATF-motif in liver. (A) Position weight matrix of the consensus sequence of the ATF binding motif (from Motifmap, www.motifmap.ics.uci.edu). (B) Consensus sequence of the ATF motif present in the gene body of GCK, PFKL, ACACA, ACLY, FASN and PRKCE was analyzed for CpG methylation by pyrosequencing. Methylation of the CpG-site covering an ATF binding motif and the next associated CpG site was measured in the (C) GCK, (D) PFKL, (E) ACACA, (F) ACLY (G) FASN and (H) PRKCE, respectively by pyrosequencing of bisulfite treated DNA in liver from non-obese controls (n = 11), obese non-diabetic patients (ND; n = 13) and obese type 2 diabetic patients (T2D; n = 11). Numbers on the x-axis represent the base count of the analyzed cytosines starting from the transcriptional start site of each gene. *p < 0.05 vs. non-obese controls; **p < 0.01 vs. non-obese controls; ***p < 0.001 vs. non-obese controls using one way ANOVA with Tukey's multiple comparisons test. (I) Luciferase assay for the specific region covering the common ATF motif in PFKL, ACACA, FASN and PRKCE that were pyrosequenced in B–H. **p < 0.01 vs. non-methylated; ***p < 0.001 vs. non-methylated using unpaired two-tailed Students t-test. See also Tables S8–S9.
Methylation of the common ATF-motif in GCK was unchanged (Figure 4C) in liver from obese non-diabetic and obese type 2 diabetic patients. Methylation of the ATF-motif in the gene body of PFKL (Figure 4D) and ACACA (Figure 4E) was reduced in obese non-diabetic and obese type 2 diabetic patients compared to non-obese controls. Conversely, ACLY methylation was unaltered (Figure 4F). ATF methylation in the FASN gene body tended to decrease, but this did not reach significance. However the proximal CpG-site to the ATF-motif in FASN (Figure 4G) and the ATF-motif in PRKCE (Figure 4H) was hypomethylated in obese non-diabetic and obese type 2 diabetic patients compared to non-obese controls.
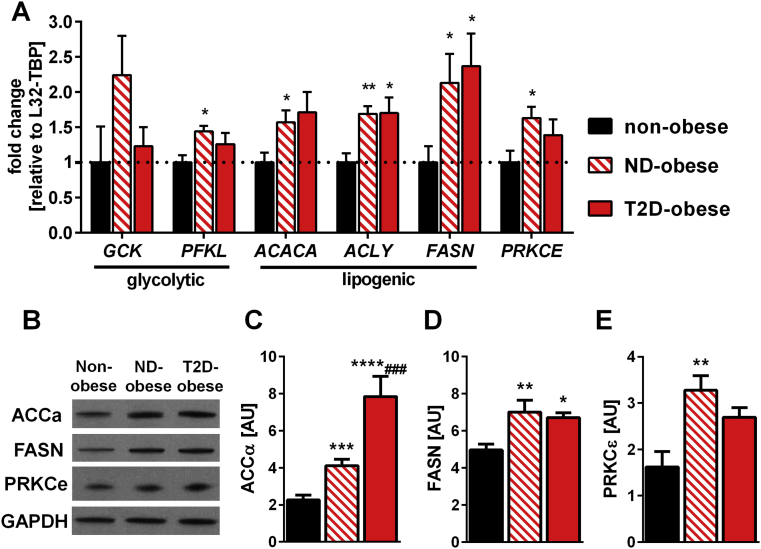
mRNA expression and protein abundance of these genes was assessed (Figure 5). RT-qPCR showed that GCK expression was unchanged between the cohorts, consistent with unchanged DNA methylation at the ATF-motif, while PFKL mRNA expression was increased in obese non-diabetic patients versus non-obese controls (Figure 5A). However, total liver glycogen content was unaltered between the cohorts (Fig. S3). mRNA expression of the lipogenic genes ACLY, ACACA and FASN was increased in obese non-diabetic and obese type 2-diabetic patients (Figure 5A). Consistent with the increased mRNA expression, protein abundance of ACCα/β (Figure 5B and Figure 5C) and FASN (Figure 5B and Figure 5D) was increased in obese non-diabetic and obese type 2 diabetic patients. PRKCE mRNA expression (Figure 5A) and protein content (Figure 5B and Figure 5E) was increased only in obese non-diabetic patients. Notably, the pattern of DNA methylation at the ATF-family-motif in non-diabetic and type 2 diabetic obese patients inversely mirrored mRNA expression, except for ACLY. Thus, our results suggest DNA methylation of the ATF-binding motif regulates the expression of these genes.
Figure 5.
mRNA expression and protein abundance of glycolytic and lipogenic genes in liver. A) Gene expression of glycolytic and lipogenic genes containing the common ATF-motif in liver of non-obese controls (n = 9), obese non-diabetic patients (ND-obese, n = 12) and obese type 2 diabetic patients (T2D-obese n = 8) was measured by TaqMan qPCR. Data are normalized to the geometric mean of RPL32 and TBP gene expression and calculated as fold-change relative to the non-obese controls. (B) Western Blot analysis of liver abundance of FASN, ACC and GAPDH. A representative blot each group is shown and bar graphs show quantified data for liver (C) ACCα, (D) FASN and (E) PRCKε from non-obese controls (n = 10), obese non-diabetic patients (n = 7) and obese type 2 diabetic patients (n = 5). Data are mean ± SEM analyzed using one-way ANOVA with Tukey's multiple comparisons test. *p < 0.05 vs. non-obese; **p < 0.01 vs. non-obese controls; ***p < 0.0001 vs. non-obese controls; ****p < 0.00001 vs. non-obese controls; ###p < 0.0001 vs. ND-obese. See also Figs. S3 and S5.
We performed luciferase reporter assays to determine whether DNA methylation of the ATF-binding motif within PFKL, ACACA, FASN and PRCKE regulate mRNA expression of each respective gene. Gene sequences of PFKL, ACACA, FASN and PRCKE that were analyzed by pyrosequencing above (Figure 4) were cloned into a CpG-free vector containing a promoter and a luciferase gene. Methylation of the ATF-motif decreased luciferase activity for all four candidate genes (Figure 4I). ATF6 silencing altered expression of PFKL, FASN, ACLY, ACACA and PRKCE in Huh7 cells studied under basal or tunicamycin-stimulated condition (Fig. S4).
Methylation of the ATF-motif within PFKL and ACACA correlated with body weight, BMI and waist circumference (Table S9), whereas that of FASN, PRKCE, PFKL and ACACA correlated with blood glucose, HbA1c, insulin and HOMA-IR. Methylation of the ATF-motif within ACLY, which was not altered between cohorts, did not correlate with the clinical characteristics of the participants (Table S9).
To further interrogate our results, we compared our results in human liver against the ENCODE ChIP-SEQ data available for MBD4 (HepG2 cells). When all differentially methylated sites identified in human liver in this study were overlaid with the MBD4 ChIP SEQ data, we identified 35 DNA methylation sites with 31 unique genes in obese non-diabetic patients versus non-obese controls, 19 DNA methylation sites with 15 unique genes in obese type 2-diabetic versus non-obese patients, and 12 DNA methylation sites with 11 unique genes were found in obese non-diabetic versus obese type 2 diabetic patients (Table S10). Nevertheless, this overlay analysis did not reveal binding of MBD4 to the ATF-motifs studied in Figure 4.
3. Discussion
We provide a genome-wide cartography of the liver methylome and transcriptome in age-matched healthy, obese non-diabetic, and obese type 2 diabetic Caucasian men. Unlike other studies [1], [15], [19], [33], [49], liver biopsies were also collected from non-obese otherwise healthy men undergoing elective gallbladder surgery. These individuals did not suffer from non-alcoholic fatty liver disease (NAFLD) or other malign conditions such as cancer, which is known to alter DNA methylation [45]. The genomic maps presented here for healthy versus metabolic disease states constitute a resource that will facilitate translational approaches to identify and validate rational intervention strategies to improve hepatic insulin sensitivity in obesity or type 2 diabetes. Consistent with an earlier study [35], we report that the majority of DNA-methylation sites modified in liver from obese type 2 diabetic individuals show hypomethylation. Moreover, the expression of PRKCE may undergo regulation by DNA methylation. This finding is of particular interest given that Protein Kinase C (PKC) epsilon activation is implicated in hepatic insulin resistance [41], [42].
Using unbiased ISMARA and gene ontology approaches, we predicted differential activation of the ATF-family and identified expression changes of ATF-target genes in liver from obese non-diabetic and obese type 2 diabetic patients. Using a pyrosequencing approach, we validated that the ATF motif present in metabolic genes is hypomethylated in liver from obese non-diabetic and obese type 2 diabetic patients compared to non-obese controls. Overexpression of ATF6 in zebra fish leads to alcohol-induced hepatic steatosis via activation of fatty acid synthase [20], highlighting the metabolic role of this transcription factor. Somewhat surprisingly, ATF6 silencing in cell cultures increased expression of the investigated genes. While our results support a role for ATF in the regulation of these genes, the unexpected direction of this expression change could be related to the nature of the in vitro studies, in particular given that in vitro modulation of ATF expression does not always reflect the direction of gene expression change observed in vivo [20]. Nevertheless, we report that DNA methylation of ATF binding elements decreases expression of glycolytic and lipogenic target genes in vitro (Figure 4I). Thus, the methylation state of ATF binding sites may regulate glycolytic and lipogenic genes, providing a possible mechanism for an epigenomic regulation of glucose and lipid metabolism in liver from obese non-diabetic and obese type 2 diabetic patients. This hypothesis is supported by the finding that the CpG-site within the ATF3-motif is a critical recognition site, which is unmethylated in HCT116 colorectal cancer cells when ATF3 is bound [10].
The gluconeogenic genes glucose-6-phosphatase and phosphoenolpyruvate carboxykinase are upregulated in liver of obese humans and rodents [17], [27]. Here we report that DNA methylation of these two genes was unaltered (data not shown), while glucose-6-phosphatase mRNA was down-regulated in obese type 2 diabetic individuals compared to non-obese controls (Tabel S2). Moreover, we report that transcription of the glycolytic gene PFKL was increased in conjunction with DNA hypomethylation, implicating increased glycolysis with obesity. In addition, we found genes involved in stearate synthesis, a pathway downstream of glycolysis, were hypomethylated and the transcripts were simultaneously increased in liver from obese patients. Increased lipogenesis may be an adaptive response in obesity to cope with the increased hepatic glucose load since excess glucose not utilized by the liver for energy production is converted to lipids. However, hepatic lipid production also impairs insulin signaling, thus increased lipogenesis is likely to promote insulin resistance [25]. Moreover, insulin directly stimulates hepatic de novo lipogenesis [18], [38] even in states of hepatic insulin resistance [11]. We also found altered DNA methylation of lipid and cholesterol metabolism pathways in liver from obese non-diabetic and obese type 2 diabetic patients. In particular, lipogenic genes ACLY, ACACA and FASN were hypomethylated in obese non-diabetic and obese type 2 diabetic patients. Thus, an upregulation of glycolysis facilitated by hypomethylation of the ATF-binding motif may fuel de novo lipogenesis. Taken together the ATF-associated hypomethylation of these lipogenic genes may disrupt hepatic insulin signaling and contribute to development of insulin resistance in obesity.
Activation of PRKCε is implicated in hepatic insulin resistance in NAFLD [1], [21], [25]. Here we provide evidence for PRKCε activation in liver from obese non-diabetic and obese type 2 diabetic patients, which may contribute to the development of hepatic insulin resistance and type 2 diabetes. We propose that site specific DNA hypomethylation promotes pathological activation of glycolysis and subsequently de novo lipogenesis via increased stearate synthesis, which may be facilitated by ATF transcription factors activation and increased gene transcription (Figure 6). Transcription factors belonging to the ATF-family are stress-inducible and responsive to metabolic dysregulation. For example, pancreatic ATF3 activation is involved in glucose homeostasis after partial pancreatectomy or streptozotocin treatment [2]. Furthermore, peroxisome proliferator-activator receptor (PPAR) activators can induce ATF3 activation providing additional support for a role for ATF3 in regulation of metabolism [34].
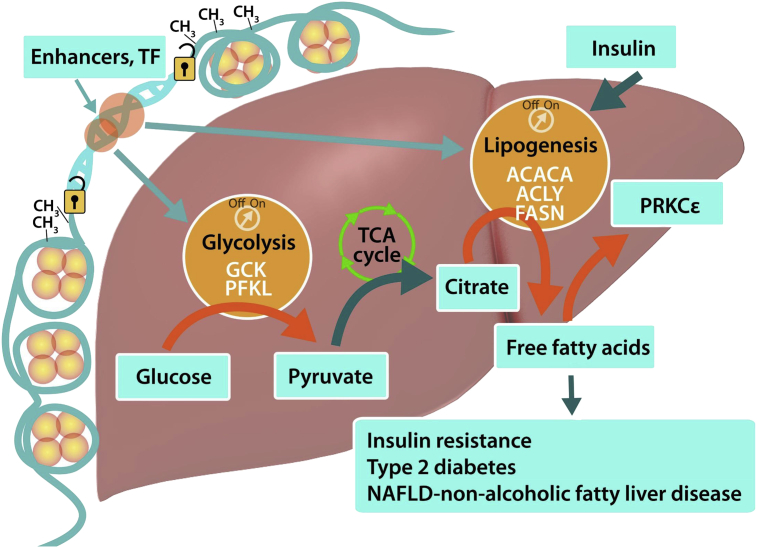
Figure 6.
Increased hepatic glycolysis and de novo lipogenesis is associated with DNA hypomethylation. Hypomethylation of the ATF-binding motif of the obese liver “unlocks” the glycolytic and lipogenic pathways. An overabundance of pyruvate from glycolysis is not used for ATP production in the TCA cycle, but is instead converted to fatty acids. These in turn activates protein kinase Cε, which is also locked into an “on position” by hypomethylation. Thus, the obese liver is programmed by a feed forward loop to become insulin resistant, possibly contributing to the development of type 2 diabetes and fatty liver. Orange arrows represent activation of glycolysis and lipogenesis that were studied. Turquoise arrows represent hypothesis.
Distinct hepatic epigenetic signatures characterize patients with NAFLD [1], [33]. In patients undergoing liver biopsy for suspected NAFLD, or intraoperatively for assessment of liver histology, obesity accelerates epigenetic aging of liver by 3.3 years for each 10 BMI units [20]. Our results in obese non-diabetic and obese type 2 diabetic patients provide additional insight into mechanisms by which epigenetic modifications reprogram the liver towards insulin resistance. Gene-specific alterations in DNA methylation can be induced in cultured cells exposed to high levels of lipids or inflammatory cytokines [5], Hence, DNA methylation may be altered in response to obesity-induced metabolic changes. Although we are unable to determine whether alterations in DNA methylation are a cause or consequence of obesity or type 2 diabetes, similarities between DNA methylation profiles in obese non-diabetic and obese type 2 diabetic patients were observed. Our finding of normal HbA1c and lower HOMA-IR in the obese non-diabetic versus obese type 2 diabetic patients support the notion that obesity alters DNA methylation before or at an early stage in the development of type 2 diabetes. Moreover, as the data by necessity are from a cross-sectional analysis from human subjects undergoing surgery, we are unable to establish a direct causation between altered methylation and gene expression, and it remains possible that the relationship is bidirectional. Notably, obesity-alterations in DNA methylation in whole blood, skeletal muscle, adipose tissue and liver can be partly reversed by gastric bypass [1], [4], [22], [32], indicating DNA methylation is a dynamic process.
Overall, the difference in hepatic DNA methylation between both obese patient groups and the non-obese control subjects were significant, but the changes were modest, consistent with previous findings that environmental influences, including metabolic diseases, alter DNA methylation in a site-specific and subtle manner [1], [36], [47], [50]. Conversely, in cancer, DNA methylation of larger genomic areas is robustly modified, and differences between cohorts more evident [45]. The modest sample size in our study may account for the fact that only subtle changes in gene expression and DNA methylation were noted between groups. Nevertheless, the glycolytic and lipogenic target genes selected for more thorough analysis were also validated by qPCR, pyrosequencing, and protein abundance, providing secondary validation that the changes in expression and DNA methylation impact specific gene function. We also provide evidence of methylation-dependent function of the ATF-motif and ATF6 expression, highlighting a possible mechanism for the regulation of specific target genes. Thus, metabolic diseases induce modest, but clinically relevant changes in the epigenome, consistent with the epigenetic signature for enhanced hepatic de novo lipogenesis observed in independent cohort of obese patients [1].
In conclusion, mRNA expression of targets along the glycolytic pathway are increased in liver of obese people, concomitant with DNA hypomethylation of their respective genes at an important ATF-motif regulatory site. Constitutive activation of theses pathways is associated with PRKCE activation and hypomethylation, which may lead to the development of hepatic insulin resistance and hepatic steatosis. This hepatic epigenetic signature contributes to the understanding of the underlying etiology of metabolic dysfunction in obesity and type 2 diabetes and identifies new targets for the development of therapeutic strategies to combat insulin resistance.
4. Experimental procedures
4.1. Patients
This study was conducted according to the principles described in the Declaration of Helsinki. The regional ethics committee at Karolinska Insitutet approved the study. All participants provided informed written consent. Liver biopsies (100–200 mg) were collected from 11 non-obese men during cholecystectomy and from 13 obese non-diabetic and 11 obese type 2 diabetic men during Roux-en Y gastric bypass surgery, providing a total cohort of 35 subjects. Biopsies were flash-frozen and stored in liquid nitrogen until analysis. Clinical parameters of the study cohort are presented (Table 1).
4.2. Genome-wide DNA methylation analysis
Genomic DNA was extracted with the DNeasy kit (Qiagen, Hilden, Germany) and assays were performed according to the manufacturer's protocol. The hepatic methylome was analyzed using the Illumina Infinium human methylation 450K beadchip arrays (Illumina, San Diego, CA, USA) from 22 randomly chosen subjects out of the entire cohort (7 non-obese, 7 obese non-diabetic and 8 obese type 2 diabetic). Each array includes 485,577 probes, targeting 21,231 (99%) RefSeq genes and 125401 (96%) CpG island regions [7], [8]. The Illumina 450K array contains two different assays, Infinium I and Infinium II. The methylation values acquired from these two assays show different distributions [12]. To correct for this bias, several data normalization methods were proposed [12], [28], [44]. The BMIQ method was recently shown to outperform other methods [29]. Thus, we applied the BMIQ method in the present study. All samples were filtered for detection p-value <0.001, adjusted for color bias and normalized using the Quantile normalization method and the lumi package, followed by the BMIQ normalization method in R Bioconductor (http://www.bioconductor.org). Cross reactive probes and probes including SNPs with minor allele frequency (MAF) > 5% according to Illumina were removed leaving 427,550 methylation sites for further analysis. The Beta-value, which is the ratio of the methylated probe intensity versus the unmethylated probe intensity and the M-value, which is the log2 ratio of the intensity of the methylated probe versus the unmethylated probe values, were generated. The M value was used for further analysis and significant differences in methylation were identified after multi-group comparison with FDR <0.25. Differentially methylated sites between 2 groups were identified by t-test (p < 0.05). Qlucore Omics Explorer (Qlucore AB, Lund, Sweden) was used to calculate multi-group comparison and to generate heat maps.
4.3. Genome-wide mRNA expression and RT-qPCR analysis
Total RNA was extracted using the miRNeasy kit (Qiagen, Hilden, Germany) and RNA integrity was validated with the Bioanalyzer (Aglient, Santa Clara, CA, USA). Global gene expression profiling of liver was performed for 18 subjects studied in the methylome analysis (6 non-obese, 6 obese non-diabetic and 6 obese type 2 diabetic) using the Human Gene ST 1.1 array (Affymetrix, Santa Clara, CA, USA). Data was pre-processed using Plier (in Expression Console). The Robust Multi-array Average (RMA) normalization method was used to normalize all data sets. The Bioconductor Limma package [39] was used to define differentially expressed genes (FDR < 0.25). The Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Inc., Redwood City, CA, USA) tool was used to perform pathway analysis. Expression of selected target genes was validated in 8 non-obese, 12 obese non-diabetic and 8 obese type 2 diabetic subjects (reduced sample size due to RNA degradation in some samples) by TaqMan (Life Technologies, Carlsbad, CA, USA) qPCR and normalized to the geometric mean of RPL32 and TBP expression. cDNA synthesis was performed with the Superscript Vilo cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA).
4.4. The Integrated System for Motif Activity Response Analysis (ISMARA)
ISMARA was performed using the genome-wide mRNA expression data measured by Affymetrix array (see above) using methods for automated processing and modeling of the data as described earlier [3].
4.5. Pyrosequencing
Genomic DNA from all subjects (11 non-obese, 13 obese non-diabetic and 11 obese type 2 diabetic) was treated with sodium bisulfite using the EpiTect bisulfite conversion kit (Qiagen, Hilden, Germany). PCR and sequencing primers (Table S9) were designed using the PyroMark Assay Design software (Qiagen, Hilden, Germany). PCR was performed using the PyroMark PCR Kit (Qiagen, Hilden, Germany). PCR products were consequently analyzed using the PyroMark Q24 Advanced machine (Qiagen, Hilden, Germany).
4.6. Luciferase assay
The CpG-free plasmid containing a universal promoter and the Lucia gene was obtained from InvivoGen (Cat. Code pCpGfree-promoter-Lucia). PCR products that covered the common AFT-motif sequence that was pyrosequenced as described above were generated by PCR amplification using human genomic DNA and PCR primers indicated in Table S11. Empty pCpGfree-promoter-Lucia and PCR products were double-digested with NsiI and BamHI (New England Biolabs, Ipswich, MA, USA) and gel purified. Digested PCR fragments were inserted into the digested empty plasmid using the quick ligation kit (New England Biolabs, Ipswich, MA, USA). PIR1 competent E. coli (Life Technologies, Carlsbad, CA, USA) were transformed with ligated DNA to amplify the constructs. After purification, plasmids with inserts (as validated by Sanger-sequencing), as well as the empty plasmid were methylated with Sss1 and SAM (New England Biolabs, Ipswich, MA, USA) for 6 h or mock-methylated (addition of SAM without SssI). HEK293 cells were subsequently transfected with methylated or mock-methylated plasmid DNA using lipofectamine2000 (Life Technologies, Carlsbad, CA, USA). Luminescence was measured 24 h after transfection in 6 replicates per gene and condition.
4.7. Western Blot analysis
Liver tissues (25 mg) were homogenized in cold lysis buffer to extract total proteins [20 mM Tris·HCl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mM β-glycerolphosphate, 2.5 mM sodium pyrophosphate, 1 mM NaF, and 1 mM Na3VO4, pH 7.4, with protease inhibitors (Roche Diagnostics, Mannheim, Germany)]. Homogenates were subjected to 1 h of rotation at 4 °C and centrifugation at 12,000g for 15 min at 4 °C. Protein concentration was measured using the Pierce BCA Protein assay kit (Thermo Scientific, Waltham, MA). Total proteins (30–40 μg) were separated by SDS-PAGE on 4–10% Tris–HCl separation gels and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Immunoblot analysis was performed using the following antibodies: Acetyl-CoA Carboxylase (Cell Signaling, Cat.No #3662), FASN (Sigma, Cat.No #F9554), PRKCε (BD transduction laboratories, Cat.No #610086) and GAPDH (FL-335) (Santa Cruz, CatNo. #sc-25778). Blots were quantified using Quantity One software (BioRad, Hercules, CA, USA). Due to unsatisfactory protein quality of some samples protein abundance from only 9 non-obese, 7 obese non-diabetic and 5 obese type 2 diabetic T2D-subjects could be quantified (bar graphs).
4.8. Hepatic glycogen content
Hepatic glycogen content was measured in liver tissue from all subjects (n = 35). Liver tissue (10 mg) was homogenized in PBS and glycogen was determined using a glycogen assay kit (Abcam, Cambridge, UK).
4.9. Hepatic triglyceride content
Liver tissue (∼10 mg) was homogenized in PBS. Total triglycerides were quantified using the Hitachi Triglyceride/Glycerol kit from Roche (Cat. No 12146029216).
4.10. ATF6 silencing
Huh7 cell were cultured in high glucose DMEM (31966, Life Technologies) and transfected with ATF6 or scrambled control silencer select siRNAs (s223544 and 4390843, respectively, Life Technologies) utilizing the RNAiMAX tranfection reagent. Following transfection (48 h), cells were treated for 6 h with tunicamycin (10 μM) (Sigma Aldrich, St. Louis, MO, USA) or DMSO control. Cells were lysed in Trizol followed by high capacity cDNA synthesis and Taqman qPCR expression analysis.
4.11. Statistics
Gene expression, protein content and pyrosequencing results are presented as mean ± SEM. Clinical parameters are presented as mean (minimum–maximum) ± SD. These data were tested for normal distribution using the Skewness and Kurtosis test in SPSS Statistics Software 21.0. When a normal distribution was observed, the statistical difference between non-obese controls, obese non-diabetic patients and obese type 2 diabetic patients was determined by one-way ANOVA with Tukey's multiple comparisons test. Statistical analysis of the genome-wide data from the gene array and Infinium 450K methylation array is described above.
4.12. Genome-wide data deposition
The data are deposited in the Gene Expression Omnibus (GEO) database with the accession number GSE65058.
Acknowledgments
The authors thank Dr. Carsten Daub from SciLifeLab, Science for Life Laboratory, Solna, Sweden and Rasmus Sjögren, Karolinska Institutet, for their advice regarding the genome-wide data analysis. H.K. is supported by an EMBO long-term fellowship. This work was supported by grants from the Thurings Stiftelse, The Tore Nilson Stiftelse, The Strategic Diabetes Program at Karolinska Institutet, Cardiovascular Research Program at Karolinska Institutet, European Research Council Ideas Program (ICEBERG, ERC-2008-AdG23285), Swedish Research Council (2011-3550), Swedish Diabetes Foundation (DIA2012-082), Swedish Foundation for Strategic Research (SRL10-0027), and Stockholm County Council (2012-0086) supported this research. Gene expression profiling and DNA-methylation assays were performed at the Bioinformatics and Expression Analysis core facility (www.bea.ki.se). The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent Research Center at the University of Copenhagen partially funded by an unrestricted donation from the Foundation (www.metabol.ku.dk).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2015.12.004.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Supplementary Figure 1. Genomic distribution of DNA methylation. Differentially methylated CpG sites (DMSs) (q < 0.25; 5,834 sites) were mapped in relation to genomic distribution. (A) Genomic distribution of DMSs in relation to defined CpG categories. (B) Genomic distribution of DMSs in relation to the nearest gene. This figure is related to Figure 1.
Supplementary Figure 2. Pairwise comparison of methylation and expression data. Pearson correlation of changes in DNA-methylation and gene expression for individual samples. Number of DNA-methylation sites where the absolute negative or positive correlation coefficient r value to the expression of the associated gene is equal or larger than 0.6 (A). Examples of DNA-methylation sites that are positively and negatively correlated to the expression of the associated gene (B). Distribution of altered CpG sites with regard to defined CpG categories and in relation to the nearest gene (C).
Supplementary Figure 3. Hepatic glycogen content. Glycogen content was measured in liver from non-obese controls (n = 8), obese non-diabetic (ND-obese) individuals (n = 9) and obese type 2 diabetic (T2D-obese) individuals (n = 8) using a Glycogen Assay Kit from Abcam. Liver glycogen content is expressed per gram (g) wet weight. Data are mean ± SEM. This figure is related to Figure 5.
Supplementary Figure 4. ATF6 gene silencing. Huh7 cells were transfected with siRNA against ATF6 or a scrambled sequence and incubated in the absence or presence of tunicamycin. Target gene expression was measured 24 h after ATF6 gene silencing. This figure is related to Figure 4.
Supplementary Figure 5. Immunoblots for protein quantification in liver. Immunoblots were performed using liver protein extract for ACC (A), FASN (B) and PKCe (C). The entire blots are shown here, while only three representative samples are shown in Figure 5 of the manuscript. (D) Ponceau staining of the membranes for FASN, ACC and PKCe used for quantification (Figure 5C–D). This figure is related to Figure 5.
References
- 1.Ahrens M., Ammerpohl O., von Schönfels W., Kolarova J., Bens S., Itzel T. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metabolism. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Allen-Jennings A.E., Hartman M.G., Kociba G.J., Hai T. The roles of ATF3 in glucose homeostasis. A transgenic mouse model with liver dysfunction and defects in endocrine pancreas. Journal of Biological Chemistry. 2001;276:29507–29514. doi: 10.1074/jbc.M100986200. [DOI] [PubMed] [Google Scholar]
- 3.Balwierz P.J., Pachkov M., Arnold P., Gruber A.J., Zavolan M., van Nimwegen E. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Research. 2014;24:869–884. doi: 10.1101/gr.169508.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barres R., Kirchner H., Rasmussen M., Yan J., Kantor F.R., Krook A. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Reports. 2013;3:1020–1027. doi: 10.1016/j.celrep.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Barres R., Osler M.E., Yan J., Rune A., Fritz T., Caidahl K. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metabolism. 2009;10:189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Barres R., Yan J., Egan B., Treebak J.T., Rasmussen M., Fritz T. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metabolism. 2012;15:405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Bibikova M., Le J., Barnes B., Saedinia-Melnyk S., Zhou L., Shen R. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 9.Birkenfeld A.L., Shulman G.I. Nonalcoholic fatty liver disease, hepatic insulin resistance, and type 2 diabetes. Hepatology. 2014;59:713–723. doi: 10.1002/hep.26672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blattler A., Farnham P.J. Cross-talk between site-specific transcription factors and DNA methylation states. Journal of Biological Chemistry. 2013;288:34287–34294. doi: 10.1074/jbc.R113.512517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown M.S., Goldstein J.L. Selective versus total insulin resistance: a pathogenic paradox. Cell Metabolism. 2008;7:95–96. doi: 10.1016/j.cmet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Dedeurwaerder S., Defrance M., Calonne E., Denis H., Sotiriou C., Fuks F. Evaluation of the Infinium methylation 450K technology. Epigenomics. 2011;3:771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- 13.Dick K.J., Nelson C.P., Tsaprouni L., Sandling J.K., Aissi D., Wahl S. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 14.Egan B., Zierath J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metabolism. 2013;17:162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Eissing L., Scherer T., Todter K., Knippschild U., Greve J.W., Buurman W.A. De novo lipogenesis in human fat and liver is linked to ChREBP-beta and metabolic health. Nature Communications. 2013;4:1528. doi: 10.1038/ncomms2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng S., Jacobsen S.E., Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher S.J., Kahn C.R. Insulin signaling is required for insulin's direct and indirect action on hepatic glucose production. Journal of Clinical Investigation. 2003;111:463–468. doi: 10.1172/JCI16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foretz M., Guichard C., Ferre P., Foufelle F. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagg S., Skogsberg J., Lundstrom J., Noori P., Nilsson R., Zhong H. Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: the Stockholm atherosclerosis gene expression (STAGE) study. PLoS Genetics. 2009;5:e1000754. doi: 10.1371/journal.pgen.1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howarth D.L., Lindtner C., Vacaru A.M., Sachidanandam R., Tsedensodnom O., Vasilkova T. Activating transcription factor 6 is necessary and sufficient for alcoholic fatty liver disease in zebrafish. PLoS Genetics. 2014;10:e1004335. doi: 10.1371/journal.pgen.1004335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jornayvaz F.R., Shulman G.I. Diacylglycerol activation of protein kinase Cepsilon and hepatic insulin resistance. Cell Metabolism. 2012;15:574–584. doi: 10.1016/j.cmet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchner H., Nylen C., Laber S., Barres R., Yan J., Krook A. Altered promoter methylation of PDK4, IL1 B, IL6, and TNF after Roux-en Y gastric bypass. Surgery for Obesity and Related Diseases. 2014;10:671–678. doi: 10.1016/j.soard.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Kirchner H., Osler M.E., Krook A., Zierath J.R. Epigenetic flexibility in metabolic regulation: disease cause and prevention? Trends in Cell Biology. 2013;23:203–209. doi: 10.1016/j.tcb.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Kuehnen P., Mischke M., Wiegand S., Sers C., Horsthemke B., Lau S. An alu element-associated hypermethylation variant of the POMC gene is associated with childhood obesity. PLoS Genetics. 2012;8:e1002543. doi: 10.1371/journal.pgen.1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumashiro N., Erion D.M., Zhang D., Kahn M., Beddow S.A., Chu X. Cellular mechanism of insulin resistance in nonalcoholic fatty liver disease. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16381–16385. doi: 10.1073/pnas.1113359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C.C., Young P.E., Maloney C.A., Eaton S.A., Cowley M.J., Buckland M.E. Maternal obesity and diabetes induces latent metabolic defects and widespread epigenetic changes in isogenic mice. Epigenetics. 2013;8:602–611. doi: 10.4161/epi.24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magnusson I., Rothman D.L., Katz L.D., Shulman R.G., Shulman G.I. Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. Journal of Clinical Investigation. 1992;90:1323–1327. doi: 10.1172/JCI115997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maksimovic J., Gordon L., Oshlack A. SWAN: subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biology. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marabita F., Almgren M., Lindholm M.E., Ruhrmann S., Fagerstrom-Billai F., Jagodic M. An evaluation of analysis pipelines for DNA methylation profiling using the Illumina HumanMethylation450 BeadChip platform. Epigenetics. 2013;8:333–346. doi: 10.4161/epi.24008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarthy M.I. Genomics, type 2 diabetes, and obesity. The New England Journal of Medicine. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 31.Michael M.D., Kulkarni R.N., Postic C., Previs S.F., Shulman G.I., Magnuson M.A. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Molecular Cell. 2000;6:87–97. [PubMed] [Google Scholar]
- 32.Multhaup M.L., Seldin M.M., Jaffe A.E., Lei X., Kirchner H., Mondal P. Mouse-human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell Metabolism. 2015;21:138–149. doi: 10.1016/j.cmet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy S.K., Yang H., Moylan C.A., Pang H., Dellinger A., Abdelmalek M.F. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nawa T., Nawa M.T., Cai Y., Zhang C., Uchimura I., Narumi S. Repression of TNF-alpha-induced E-selectin expression by PPAR activators: involvement of transcriptional repressor LRF-1/ATF3. Biochemical and Biophysical Research Communications. 2000;275:406–411. doi: 10.1006/bbrc.2000.3332. [DOI] [PubMed] [Google Scholar]
- 35.Nilsson E., Matte A., Perfilyev A., de Mello V.D., Kakela P., Pihlajamaki J. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. Journal of Clinical Endocrinology and Metabolism. 2015;100:E1491–E1501. doi: 10.1210/jc.2015-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nitert M.D., Dayeh T., Volkov P., Elgzyri T., Hall E., Nilsson E. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61:3322–3332. doi: 10.2337/db11-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orozco L.D., Morselli M., Rubbi L., Guo W., Go J., Shi H. Epigenome-wide association of liver methylation patterns and complex metabolic traits in mice. Cell Metabolism. 2015;21:905–917. doi: 10.1016/j.cmet.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Postic C., Dentin R., Denechaud P.D., Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annual Review of Nutrition. 2007;27:179–192. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- 39.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronn T., Volkov P., Davegardh C., Dayeh T., Hall E., Olsson A.H. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genetics. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samuel V.T., Liu Z.X., Qu X., Elder B.D., Bilz S., Befroy D. Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. Journal of Biological Chemistry. 2004;279:32345–32353. doi: 10.1074/jbc.M313478200. [DOI] [PubMed] [Google Scholar]
- 42.Samuel V.T., Liu Z.X., Wang A., Beddow S.A., Geisler J.G., Kahn M. Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. Journal of Clinical Investigation. 2007;117:739–745. doi: 10.1172/JCI30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sookoian S., Rosselli M.S., Gemma C., Burgueno A.L., Fernandez Gianotti T., Castano G.O. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor gamma coactivator 1alpha promoter. Hepatology. 2010;52:1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 44.Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timp W., Feinberg A.P. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nature Reviews Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toperoff G., Aran D., Kark J.D., Rosenberg M., Dubnikov T., Nissan B. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Human Molecular Genetics. 2012;21:371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volkmar M., Dedeurwaerder S., Cunha D.A., Ndlovu M.N., Defrance M., Deplus R. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO Journal. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu X., Su S., Barnes V.A., De Miguel C., Pollock J., Ownby D. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics. 2013;8:522–533. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Y., Ping J., Chen H., Jiao L., Zheng S., Han Z.G. A comparative analysis of liver transcriptome suggests divergent liver function among human, mouse and rat. Genomics. 2010;96:281–289. doi: 10.1016/j.ygeno.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Ziller M.J., Gu H., Muller F., Donaghey J., Tsai L.T., Kohlbacher O. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.