Abstract
Objectives
Chronic and high consumption of fat constitutes an environmental stress that leads to metabolic diseases. We hypothesized that high-fat diet (HFD) transgenerationally remodels the epigenome of spermatozoa and metabolism of the offspring.
Methods
F0-male rats fed either HFD or chow diet for 12 weeks were mated with chow-fed dams to generate F1 and F2 offspring. Motile spermatozoa were isolated from F0 and F1 breeders to determine DNA methylation and small non-coding RNA (sncRNA) expression pattern by deep sequencing.
Results
Newborn offspring of HFD-fed fathers had reduced body weight and pancreatic beta-cell mass. Adult female, but not male, offspring of HFD-fed fathers were glucose intolerant and resistant to HFD-induced weight gain. This phenotype was perpetuated in the F2 progeny, indicating transgenerational epigenetic inheritance. The epigenome of spermatozoa from HFD-fed F0 and their F1 male offspring showed common DNA methylation and small non-coding RNA expression signatures. Altered expression of sperm miRNA let-7c was passed down to metabolic tissues of the offspring, inducing a transcriptomic shift of the let-7c predicted targets.
Conclusion
Our results provide insight into mechanisms by which HFD transgenerationally reprograms the epigenome of sperm cells, thereby affecting metabolic tissues of offspring throughout two generations.
Keywords: Epigenetics, Obesity, Spermatozoa, DNA methylation, microRNA
Highlights
-
•
Body weight and glucose metabolism are altered in F1 and F2 offspring of F0-HFD fathers.
-
•
High-fat diet reprograms the epigenome of sperm cells.
-
•
Spermatozoa from F0-HFD fathers and F1 offspring share common epigenetic signatures.
-
•
Expression of let-7c is changed in sperm of founders and in the adipose tissue of the offspring.
1. Introduction
Obesity is a metabolic disorder caused by a chronic imbalance between energy intake and expenditure that increases the risk of developing type 2 diabetes, cardiovascular disease and malignancies. Epidemiological data showing that children of obese parents are at higher risk of developing metabolic disease later in life support the notion that a heritable component contributes to obesity and obesity-related traits [1]. Several lines of evidence reveal that the nutritional status of the parents can affect the metabolic phenotype of the offspring [2], [3], [4]. Although cultural factors could be involved in this phenomenon in humans, animal studies support the notion that gametes of obese parents contain an environmentally-induced message that influences glucose and energy metabolism.
Epigenetic inheritance describes transmission of a phenotype from one generation to the next that is not carried by the DNA code itself. By modulating the early stages of embryonic development, epigenetic information contained in the gamete has the potential to alter the phenotype of an offspring after birth [5], [6]. Epigenetic modifications such as DNA methylation and hydroxymethylation, histone modifications and non-coding RNA expression modulate the access of the transcription machinery to the chromatin, the repression of transposable elements or the regulation of mRNA targets, thereby regulating gene expression in time and space [7]. Most of the epigenetic marks are erased upon gametogenesis and fertilization to allow for de novo programming of the embryo, but specific epigenetic marks escape reprogramming and are potential carriers of environmentally-induced information to program phenotypes from one generation to the next.
Animal models of paternal epigenetic inheritance have been used to investigate the possible transfer of epigenetic information from one generation to next in order to exclude any confounding influence of gestational effects on somatic tissues during embryological development. Using these types of models, the nutritional status of the father has been reported to impair metabolism in the offspring, which strongly implicates that the spermatozoa carry information that is influenced by dietary factors [2], [8], [9]. However the nature and influence of the gametic epigenetic signature on metabolic features such as glucose metabolism and the predisposition towards developing obesity is unknown.
Here, we determined how paternal diet affected the epigenetic signature of spermatozoa and the metabolic function of the offspring over two generations. We provide evidence that a paternal high-fat diet induces a robust, sex specific disturbance in glucose metabolism and energy homeostasis within two following generations. We identified common altered DNA methylation signatures and small non-coding RNA expression profiles in the spermatozoa from F0 and F1 males, providing a mechanism for the propagation of metabolic dysfunction to the next generation. The predicted pathways affected by these epigenetic marks were perturbed in metabolic tissues of the offspring. Our results support the existence of transgenerational reprogramming of the gametic epigenome and inheritance of diet-induced metabolic dysfunction throughout two generations.
2. Material and methods
2.1. Animal care
Male and female Sprague–Dawley founder rats were obtained from Charles River Laboratories (Germany). At 4 weeks of age, F0 male breeders were fed either with a high-fat diet (HFD; TD.88137/TD.08811, 42/45% energy from fat, Harlan Laboratories, USA) or a control chow diet (R36-Lab For Lactamin, Sweden) for 12 weeks (Figure 1A). Water and food were provided ad libitum.
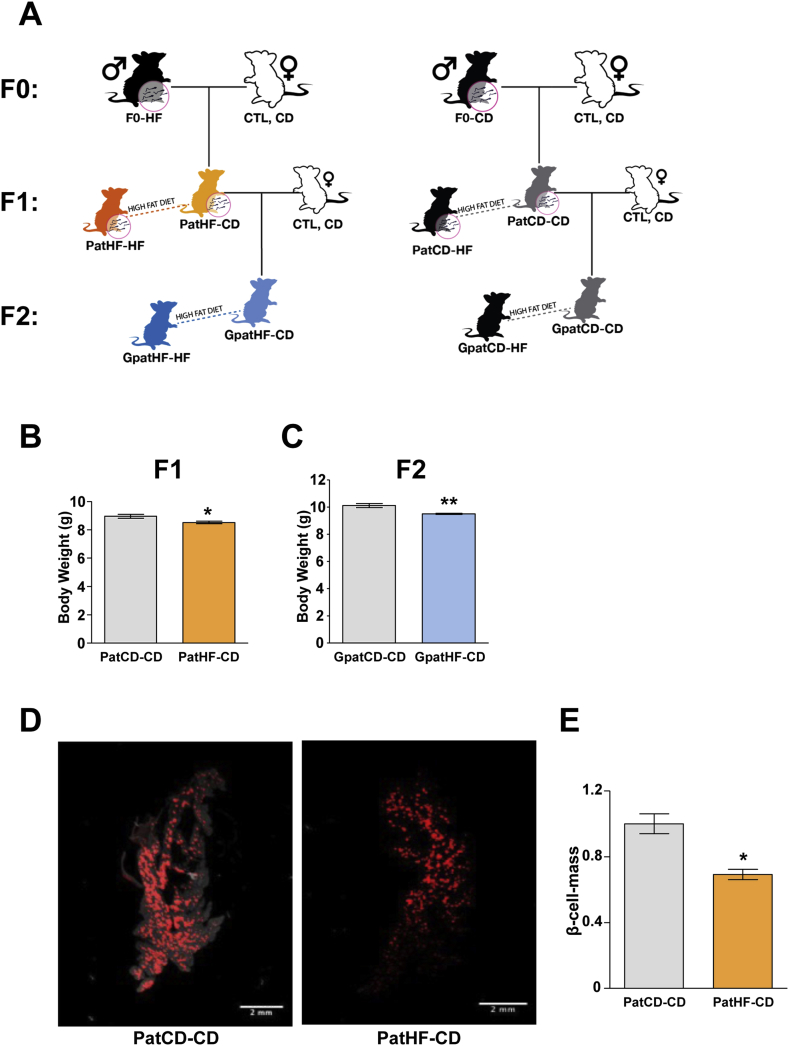
Figure 1.
Body weight and β-cell mass is altered in newborn offspring of HFD-fed founders. (A) Schematic diagram of study design: F0 male Sprague–Dawley rats fed a chow (F0-CD) or high-fat diet (F0-HF) for 12 weeks were mated with control females fed a chow diet (CTL, CD) to generate the F1 offspring. Twelve week-old chow-fed F1 male rats from F0-CD or F0-HF were mated with females from an independent line (also maintained on chow diet) to generate the F2 offspring. At 10 weeks of age, a sub-group of F1 and F2 female and male offspring was subjected to chow or high fat diet (HFD) for 12 weeks. Metabolic status of the offspring was assessed. (B) Body weight of 3-day-old F1-offspring (n = 190 and 181 from 16 litters of PatCD-CD and 14 litters of PatHF-CD, respectively). (C) Body weight of 3-day-old F2-offspring (n = 52 and 71 from 5 litters of GpatCD-CD and 6 litters of GpatHF-CD, respectively). Decreased pancreatic β-cell mass in female F1-offspring from HFD-fed fathers: (D) Representative images of Optical Projection Tomography (OPT) analysis of insulin-secreting cells in whole pancreas from 5-day-old female rats (E) quantification of OPT measures (n = 4 and 5 from 4 litters of PatCD-CD and 2 litters of PatHF-CD, respectively). Results are represented as mean ± SEM. Student's t-test: *p ≤ 0.05: PatHF-CD vs PatCD-CD; **p ≤ 0.05, GpatHF-CD vs GpatCD-CD. PatCD-CD: Paternal-Chow on Chow; PatHF-CD: Paternal-HFD on Chow; GpatCD-CD: Grandpaternal-Chow on Chow; GpatHF-CD: Grandpaternal-HFD on Chow.
To obtain the F1 offspring, one F0 male breeder was housed together with a 12 week-old female rat, with free access to chow diet from 7:00 to 18:00, for 8 consecutive days. Male F0 breeders returned to their respective cages with the original diet, while F0 female rats consumed only chow diet throughout mating, gestation and lactation. To assure that there were no differences between female breeders, body and tissue weight, as well as blood glucose levels were evaluated (Table T2). To control for postnatal nutrition, litter sizes were standardized to 12 pups at day-1 after birth. At day-3, all pups were weighed and at day-5 litter sizes were reduced to 8 pups (4 males and 4 females). The pups were weaned from mothers at 21 days of age, and male or female siblings were housed together and fed a chow diet. At week 10 of age, one of the F1 siblings was subjected to a HFD for 12 weeks, and another sibling was kept on chow diet (control group) (Figure 1A).
To generate F2 offspring, only chow-fed F1 male rats were mated with 12 week-old females from an independent line. During mating, one F1 male was housed with one female for 10 consecutive days. Exactly as for F1, the F2 offspring litter size was measured and pups were weaned to a chow diet at 21 days of age. At week 10, a subgroup of the F2 offspring was also subjected to a HFD treatment for 12 weeks (Figure 1A). Body weight and food intake of F0 male breeders and F1 and F2 offspring were monitored weekly by collecting and weighing offered and remaining food.
All the rats were maintained under a temperature-controlled environment and 12:12-h light:dark cycle at the animal facility at the Department of Physiology and Pharmacology, Karolinska Institutet, Stockholm, Sweden. All the experimental procedures were approved by the animal ethical committee, Stockholm North, and conducted in accordance with regulations for protection of laboratory animals. All the experiments were repeated using two independent cohorts of F0 breeders and F1 and F2 offspring. Statistical analysis related to transgenerational effects was made considering one sibling per litter. If more than one sibling per litter was used, the number is stated in the figure legend.
2.2. Intraperitoneal glucose tolerance test (ipGTT)
The ipGTT was performed in the F0 male breeders and F1 and F2 offspring after 12 weeks under the assigned diet (control or HFD). Rats underwent an overnight fast (15 h) prior to the ipGTT. On the day of the experiment, the animals received an intraperitoneal injection of 2 g glucose/kg body weight. The blood collected from the tail was used to measure glucose levels at basal (0), 15, 30, 60, 90 and 120 min after glucose injection. Blood glucose levels were measured using Accu Check Advantage Glucometer (Roche) and insulin levels were measured using an ELISA kit (see details below).
2.3. Intraperitoneal insulin tolerance test (ipITT)
The ipITT was performed in the F0 male breeders after 12 weeks on the assigned diet. On the day of the experiment, food was removed from the cages 3 h prior to the ipITT. Insulin (Actrapid, Novo Nordisk; 1 U/kg) was administrated intraperitoneally. The blood collected from the tail was used to measure the blood glucose levels at basal (0), 10, 30, 45, 60 and 90 min after insulin injection. A different subset of animals underwent the glucose tolerance test or the insulin tolerance test to avoid stress response associated to injections and blood sampling.
2.4. Endpoint
Rats were subjected to food deprivation from 4 h prior to the termination. Animals were anesthetized with sodium pentobarbital (100 mg/kg, ip). Blood was collected and the plasma was stored at −20 °C until use. Tissues were harvested, snap-frozen in liquid nitrogen and stored at −80 °C until use.
2.5. Blood analysis
Blood insulin and leptin levels were measured using the ultrasensitive rat ELISA kits (#90060 and #90040, respectively, Crystal Chem Inc., USA). Blood triglycerides and free fatty acid levels were determined by colorimetric assays (#ab65336 and #ab65341, Abcam, UK). The assays were performed according to manufacturer's instructions.
2.6. Pancreas optical projection tomography (OPT)
The entire pancreas from 5 day-old F1 female offspring was fixed in 4% paraformaldehyde (PFA) for 2 h at 4 °C, followed by a subsequent dehydration in methanol 33%, 66% and 100% for 15 min at each step. To quench the autofluorescence, tissues were incubated for 24 h in a methanol:DMSO:H2O2 (2:1:3) solution, followed by two washes in methanol for 30 min. Samples were brought to −80 °C for 1 h and brought back to room temperature 3 times to ensure that antigens in the deeper parts of the tissue were rendered accessible. Samples were rehydrated in Tris-Buffered Saline and Tween 20 buffer (TBST) 33%, 66% and 100% for 15 min at each step, and blocked in 10% rat serum in TBST for 24 h. Samples were incubated for 48 h with primary insulin antibody, followed by 48 h incubation with secondary antibody. Finally, tissues were mounted in 1% low melting agarose and dehydrated in 100% methanol for 24 h. Samples were washed in BABB solution (1:2 Benzyle alcohol, Benzyle benzoate) for 24 h, scanned, visualized and quantified in a stereomicroscope equipped for epifluorescence fluorochrome (Alexa 488, Alexa 594, Cy 3).
2.7. Isolation of motile spermatozoa
The cauda epididymis was dissected from the anesthetized animal and punctured in a Petri dish containing sperm isolation buffer (Earle's Balanced Salt Solution, 25 mM Hepes, 48.5 mM bovine serum albumin) pre-warmed to 37 °C. Samples were transferred to a 14 ml round bottom tube, overlaid with isolation buffer and subjected to a swim-up assay. Samples were incubated at 37 °C at a 45-degree angle and the supernatant was harvested after 2 h.
2.8. Extraction of RNA and DNA from sperm cells
Extraction of DNA was performed using the kit Illustra™ Nucleon BACC 2 kit (#RPN8502; GE Healthcare, UK), modified for processing of sperm, according to the manufacturer's recommendations. Approximately 20 million motile mature sperm cells were used for RNA extraction using TRIzol® reagent (#15596-026; Invitrogen™, USA). RNA extraction was performed according to manufacturer's instructions.
2.9. Construction of sperm DNA methylation and sncRNA sequencing libraries
For the DNA methylation analysis, DNA fragments of 300–1000 bp obtained by sonication were enriched for methylation by Methyl Binding Domain (MBD)-capture (MethylMiner, Invitrogen). Captured methylated fractions were ligated to TruSeq (Illumina) sequencing adapters, and sequenced on a HiSeq Illumina platform. Total RNA was isolated by the TRIzol® method (Life Technologies). SncRNA sequencing libraries were prepared using the NEBNext® Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs), according to the manufacturer's instructions. Molecules of 20–50 nucleotides were separated by acrylamide gel electrophoresis, extracted, and sequenced on a HiSeq Illumina instrument.
2.10. Analysis of sequencing data
Analysis was performed in R using Bioconductor packages. Preprocessed reads of at least 15 nt were aligned to rat genome (rn4) with Bowtie [10]. Methylated DNA peaks were called by MACS2 peakcall [11]. Regions of differential methylation were identified using DiffBind. Rat mature miRNAs and precursor sequences were obtained from miRBase [12] version 20. piRNA sequences were downloaded from piRNABank [13]. Rat tRNAs were retrieved from UCSC genome browser [14]. tRFs were defined when more than 10% of all reads mapping to a particular sncRNA were identical to a tRNA and the overlapping sequence was used as a tRF. Before measuring differential expression, lowly expressed (less than one count per million reads) or inadequately represented (less than half of the samples) sncRNAs were excluded from the tests. Differential sncRNA expression was calculated by edgeR [15] with a p-value cut-off of 0.05 for miRNAs, and 0.05 or 0.01 for piRNAs and tRFs, in F0 and F1 respectively.
2.11. DNA methylation in WAT
DNA was extracted using the DNeasy blood & tissue kit (Qiagen; Hilden, Germany). Fragmented DNA obtained by sonication was enriched for methylation by (MBD)-capture or used as input control for qPCR analysis. Specific primers for each target are described in supplementary Table T7.
2.12. Transcriptomic and correlation analysis
Total RNA from white adipose tissue was isolated using the RNeasy Lipid Tissue kit (Qiagen). RNA quality was assured by the integrity of ribosomal subunits and lack of degradation by the Experion Automated Electrophoresis System (Bio-Rad Laboratories, CA, USA). Affymetrix whole-transcript expression analysis (GeneChip Rat Gene 1.1 ST) was used to measure transcripts differentially expressed between chow- and HFD-fed F2 females from F0-CD and F0-HF fathers (n = 6–7 per group; 1 sibling per litter). Analysis were performed at the Bioinformatics and Expression Analysis core facility (BEA) at Karolinska Institutet, Huddinge, Sweden, Data were normalized using the expression console plier method, followed by two-way ANOVA statistical analysis (p-values were corrected with the Benjamini and Hochberg method). Correlation analysis of predicted rat let-7c target genes found to be expressed in the gene array data was performed using R Project for statistical computing analysis. Gene ontology analysis of the genes inversely correlated with let-7c expression was performed using DAVID bioinformatics resources [16] (B–H corrected p-values were set to p < 0.06).
2.13. Quantitative real-time PCR
Total RNA from sperm, liver, EDL and gonadal white adipose tissue was isolated using TRIzol® reagent according to manufacturer's recommendations. For the miRNA expression analysis, cDNA was prepared using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA, USA), followed by a real-time PCR reaction using a pool of target-specific TaqMan probes (Table T7; Applied Biosystems).
For the gene expression analysis, cDNA was prepared using High Capacity cDNA Reverse Transcription Kit with random primers, followed by real-time PCR reaction using TaqMan Fast Universal PCR reagents and TaqMan target-specific probe assays (Applied Biosystems) (Table T7). All reactions were performed in duplicates and measured by Applied Biosystems StepOnePlus Real-Time PCR Systems.
2.14. Protein expression analysis
White adipose was homogenized in ice-cold buffer (10% glycerol, 5 mM sodium pyrophosphate, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 20 mM Tris (pH 7.8), 1% Triton X-100, 10 mM sodium fluoride, 1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 0.5 mM sodium vanadate, 1 mM benzamidine, 1 μM microcystin). The floating fatty layer was removed after centrifugation (12,000 g for 10 min) at 4 °C, and homogenates rotated end-over-end for 1 h at 4 °C. After centrifugation, protein abundance was measured in the supernatant (Pierce, Thermo Fisher Scientific Inc.). An equal amount of protein (25 μg) was separated by SDS-PAGE using Criterion XT Precast gels (Bio-Rad, Hercules, CA) and transferred to polyvinylidene difluoride membrane (PVDF) (Immobilon-P; Millipore, Billerica, MA). Protein content was determined using the following antibodies: AKT2 (#3063), IRS2 (#3089) and IR-β (#3025) from Cell Signaling Technology (Massachusetts, USA), and Caspase 9 (#ab2325) from Abcam (Cambridge, UK). Ponceau staining of the PVDF membranes was used for normalization and loading control.
3. Results
3.1. Paternal chronic high-fat diet consumption transgenerationally reprograms body weight and glucose metabolism of the offspring
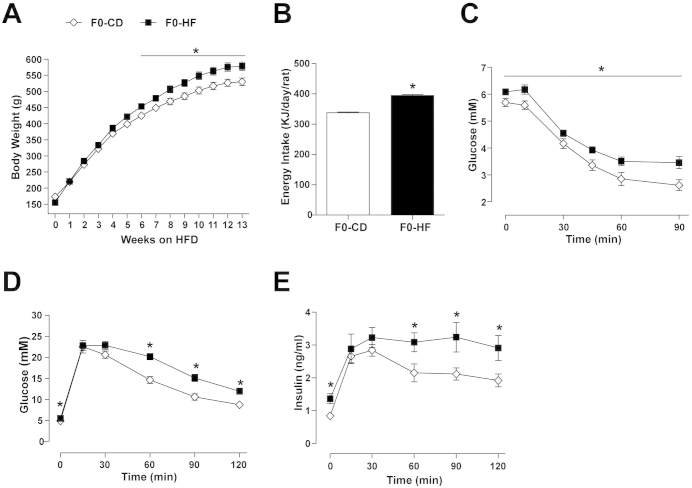
To identify gametic epigenetic signatures, we used an experimental design based on a rodent model of paternal inheritance of metabolic dysfunction [9], [17]. Male rats fed a HFD for 12 weeks (F0-HF) and exhibiting increased body weight and impaired glucose tolerance (Figure S1 and Table T1) were mated to chow-fed (control) dams (Table T2). A control group was maintained on chow diet before and during breeding (F0-CD). The metabolic phenotype of the offspring was characterized (study design depicted in Figure 1A). F1-offspring from founders fed a HFD (PatHF-CD) had decreased body weight at 3 days of age compared to F1-offspring from control rats (PatCD-CD) (Figure 1B). This phenotype was not associated with any change in litter size (data not shown), suggesting that the decreased body weight was not caused by nutritional competition in utero. A similar phenotype was observed in the F2 generation, where F2-offspring (GpatHF-CD) of F0 grandfathers fed a HFD showed reduced birth-weight compared to F2-offspring from control grandfathers (GpatCD-CD) (Figure 1C), indicating a transgenerational effect. As supported by previous observations using a similar model [9], we found beta-cell mass was decreased in F1 females from F0-HF fathers compared to controls, as revealed by optical tomography measurements of insulin-secreting cells in 5-day-old rats (Figure 1D,E).
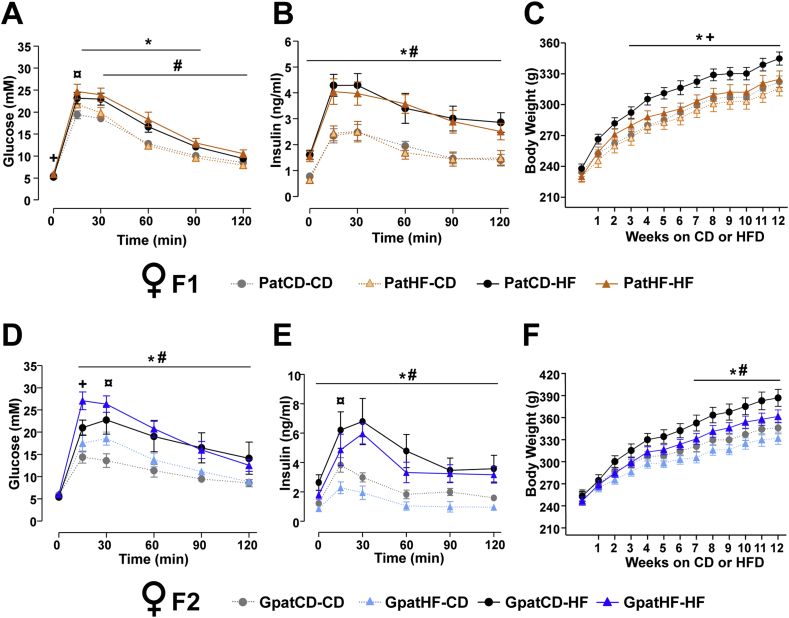
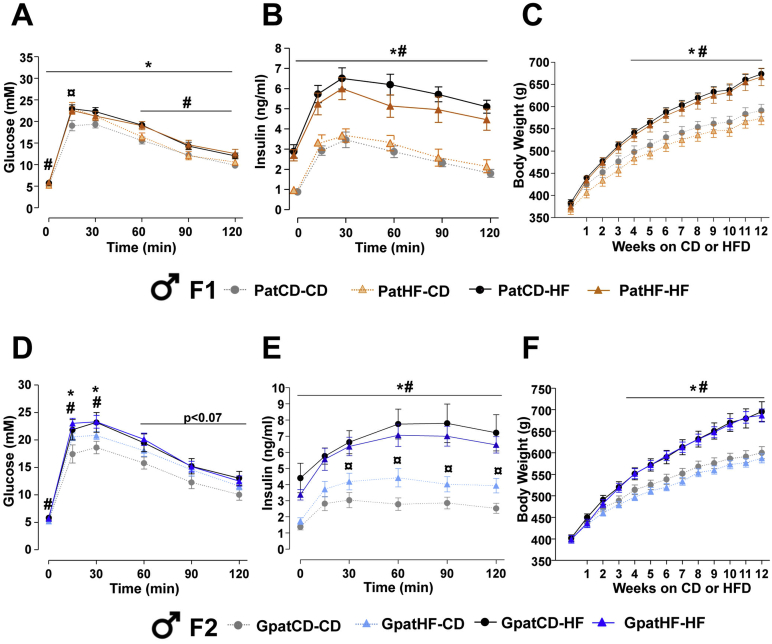
In adult life, F1 and F2 offspring were separated into two groups, one fed with HFD and another fed with control diet, for 12 weeks to assess the response to a metabolic challenge. Area under the curve for glucose levels during the glucose tolerance test was greater in both F1 and F2 offspring fed a HFD when compared offspring fed chow (F1: p < 0.0001 and F2: p < 0.001; data not shown). On control diet, F1 PatHF-CD and F2 GpatHF-CD females were moderately less glucose tolerant compared to offspring from control F0-CD rats (Figure 2A,D and Tables T3 and T4). Strikingly, when subjected to a HFD, F1 PatHF-HF and F2 GpatHF-HF females were strongly resistant to HFD-induced weight gain when compared to respective controls (Figure 2C, F), despite similar energy intake (data not shown). When fed with a HFD, F1 and F2 females from F0-HF had a 12–22% decrease in fat mass as compared to female offspring from F0-CD (Tables T3 and T4). However, only F2 females from F0-HF showed further impairments in glucose tolerance in response to HFD (Figure 2D,E), suggesting resistance to HFD-induced weight gain is not a causative factor for glucose intolerance in this model. Glucose intolerance was not associated with altered plasma insulin levels in F1 female rats (Figure 2B), while F2-females from grandfathers fed a HFD showed decreased insulin levels (Figure 2E).
Figure 2.
Impaired glucose tolerance and reduced body weight gain in response to HFD persists into two generations. (A–C) Metabolic profiling of F1 female offspring after 12 weeks of high fat feeding; blood (A) glucose and (B) insulin during an intraperitoneal glucose tolerance test (ipGTT) (n = 9–11 animals; 1 sibling per litter) and (C) body weight curve (n = 14–22 animals from 14 to 17 litters; used more than 1 sibling per litter). (D–F) Metabolic profiling of F2 female offspring after 12 weeks of high fat feeding: Blood (D) glucose and (E) insulin during an ipGTT (n = 5–6 litters; 1 sibling per litter) and (F) body weight curve (n = 10–13 animals from 5 to 6 litters; used more than one sibling per litter). Results are represented as mean ± SEM. Two-way ANOVA followed by Bonferroni post-hoc test: *p ≤ 0.05: PatCD-HF vs PatCD-CD or GpatCD-HF vs GpatCD-CD; ¤p ≤ 0.05: PatHF-CD vs PatCD-CD or GpatHF-CD vs GpatCD-CD; #p ≤ 0.05: PatHF-HF vs PatHF-CD or GpatHF-HF vs GpatHF-CD; +p ≤ 0.05: PatHF-HF vs PatCD-HF or GpatHF-HF vs GpatCD-HF. PatCD-CD: Paternal-Chow on Chow; PatHF-CD: Paternal-HFD on Chow; PatCD-HF: Paternal-Chow on HFD; PatHF-HF: Paternal-HFD on HFD; GpatCD-CD: Grandpaternal-Chow on Chow; GpatHF-CD: Grandpaternal-HFD on Chow; GpatCD-HF: Grandpaternal-Chow on HFD; GpatHF-HF: Grandpaternal-HFD on HFD.
In contrast, in F2 but not F1 male rats fed a chow diet, insulin levels were increased during the glucose tolerance test (Figure S2E). Also, F1 and F2 male offspring of F0-HF rats challenged with a HFD did not show major phenotypic differences as compared to respective controls (Figure S2 and Tables T5 and T6), suggesting transgenerational responses are sex-specific. Collectively, our results show that paternal high fat feeding is a strong inducer of transgenerational metabolic effects, mainly in female offspring.
3.2. High-fat diet reprograms the DNA methylation profile of spermatozoa
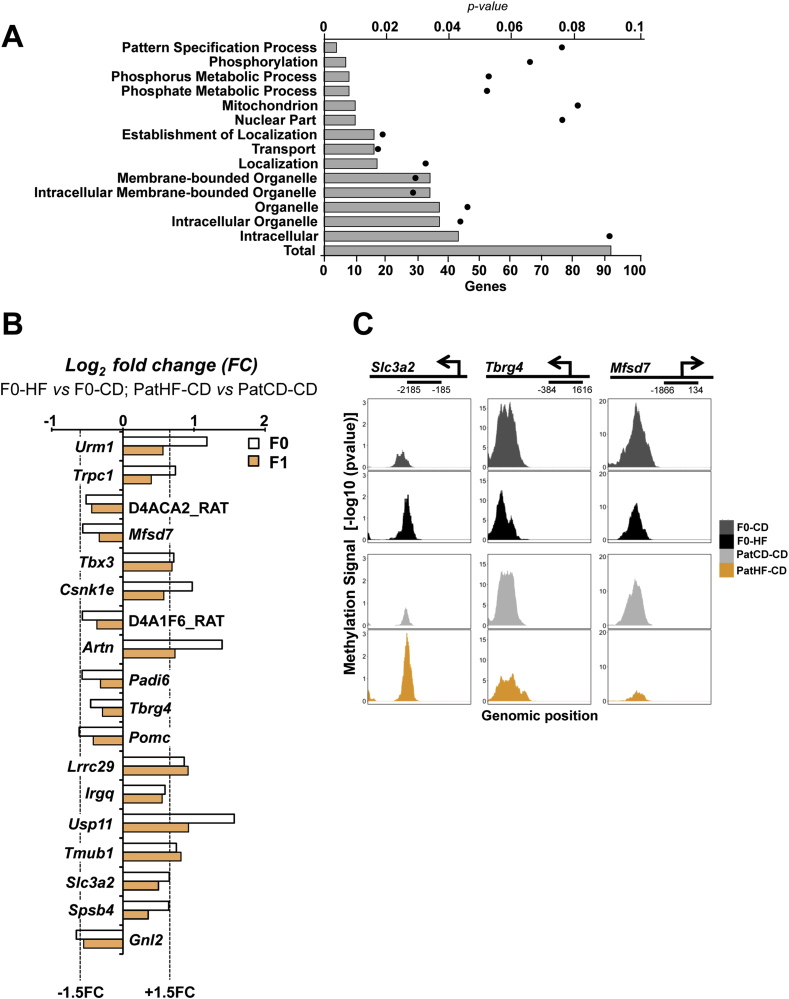
Spermatozoa from F0 and F1 males were collected from the cauda epididymis and purified using a swim-up assay [18]. In the ultra-pure fraction of motile spermatozoa, we first investigated the DNA methylation profile using methylated DNA capture followed by deep sequencing (MBD-Seq). We found numerous differentially methylated regions (DMR) in the spermatozoa of F0 and F1 rats fed a HFD as compared with spermatozoa from rats fed a control diet (Supplemental file SF1). Thus, HFD directly alters gametic DNA methylation. We identified that 92 genes related to the DMRs were commonly regulated by HFD in both F0 and F1 spermatozoa (Figure 3A and Supplemental file SF1). Gene ontology analysis of the nearest genes of the common DMRs showed enrichment of genes involved in cellular localization, transport, and metabolic processes (Figure 3A).
Figure 3.
High-fat diet alters the DNA methylation signature of spermatozoa. (A) Main biological functions obtained by DAVID ontology analysis of genes commonly differentially methylated in the sperm of F0 (HF vs CD) and F1 (PatHF-HF vs PatCD-HF). Bars represent the number of genes involved in each of the functional clusters that were differentially methylated and black dots indicate p-values. Total is the total number of genes near or overlapping with common differentially methylated regions. (B) Differentially methylated regions common in sperm of F0 and F1 indicates a direct gametic transgenerational reprogramming. Data are represented as log2 of the fold change in F0 (HF vs CD) or F1 (PatHF-CD vs PatCD-CD) founders. (C) Representative figure of genomic views of differentially methylated regions near transcription start sites (−/+1000 bp relative to the TSS) in sperm from F0-HF and PatHF-CD as compared to their respective controls. Data is represented as –log10(p value) of methylation signal and was generated using MACS2 bdgcmp with the ‘-m ppois’ parameter set (F0, n = 7 per group; F1-Chow, n = 8–11 litters; F1-HFD, 5–7 litters). F0-CD: F0 on chow diet; F0-HF: F0 on HFD; PatCD-CD: Paternal-Chow on Chow; PatHF-CD: Paternal-HFD on Chow; PatCD-HF: Paternal-Chow on HFD; PatHF-HF: Paternal-HFD on HFD.
To identify potential DNA methylation marks associated with transgenerational effects, we next compared the DNA methylation profile of the spermatozoa from F0 founders and their F1 offspring on control diet. We identified 18 regions that were differentially methylated in both F0-HF and their F1 (PatHF-CD) offspring in comparison to the respective controls (Figure 3B and supplemental file SF1). For instance, we identified DMRs located at proximity to the transcription start site of Slc3a2 (solute carrier family 3 (amino acid transporter heavy chain), member 2), Tbrg4 (transforming growth factor beta regulator 4) and Mfsd7 (major facilitator superfamily domain containing 7) (Figure 3C). We found Slc3a2 and Tbrg4 were hypermethylated, while Mfsd7 was hypomethylated in the sperm of F0-HF and F1 PatHF-CD when compared to their controls.
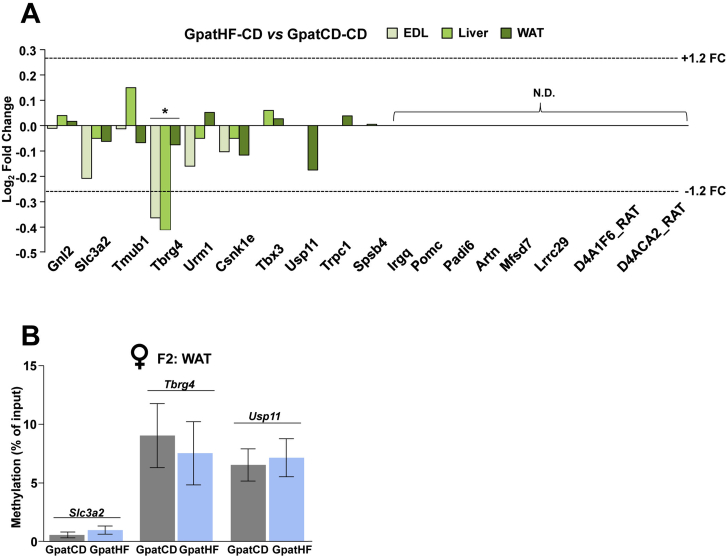
Transcriptomic analysis of liver, Extensor Digitorum Longus (EDL) muscle and white adipose tissue (WAT), corresponding to the nearest genes of the 18 sperm DMRs common to F0-HF and their F1 male offspring showed that the expression of only Slc3a2, Tbrg4 and Usp11 (Ubiquitin Specific Peptidase 11) was altered in somatic tissues of F2 offspring (Figure S3A). For these genes, the analysis of DNA methylation in WAT of F2 females from F0-HF showed no difference in mRNA expression compared to controls (Figure S3B). Collectively, these findings suggest a modest link, if any, between differential methylation of parental sperm and the altered methylation and gene expression signature in somatic tissues of adult offspring.
3.3. Small non-coding RNA expression is remodeled in spermatozoa of F0 and F1 founders
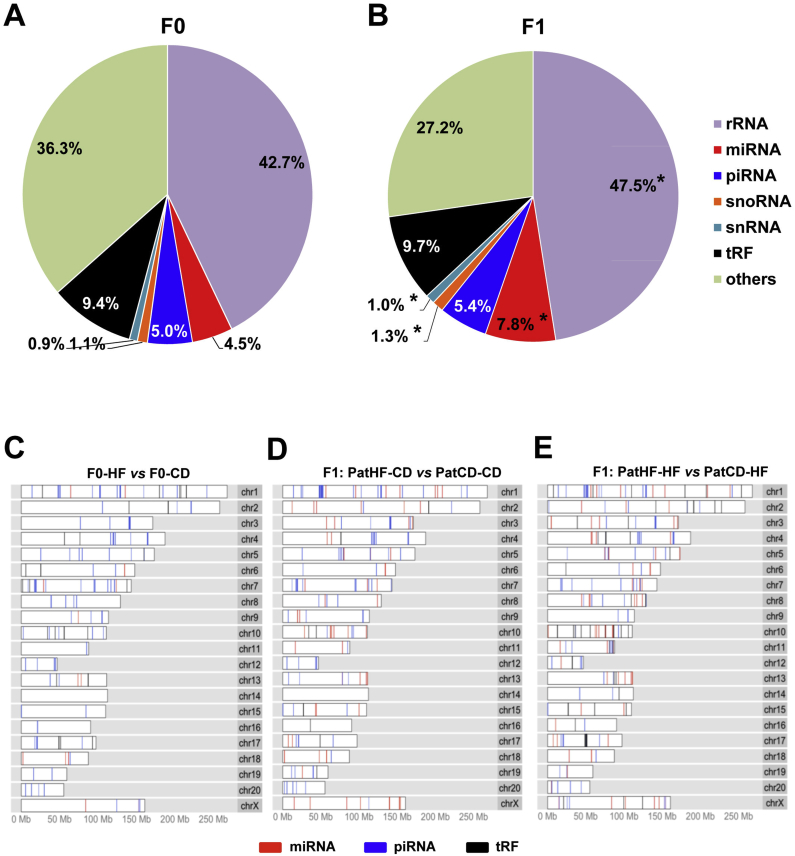
Small non-coding RNAs (sncRNAs) can carry epigenetic information through their potential to be inherited in dividing cells. Expression of sncRNAs was determined by small RNA-Seq. Similar to what is found in mouse and human [5], [19] we found that rat spermatozoa contain an important variety of small RNA subtypes, notably ribosomal RNA (rRNA), microRNAs (miRNA), PIWI-interacting RNAs (piRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA) and tRNA-derived fragments (tRFs) (Figures S4A and S4B).
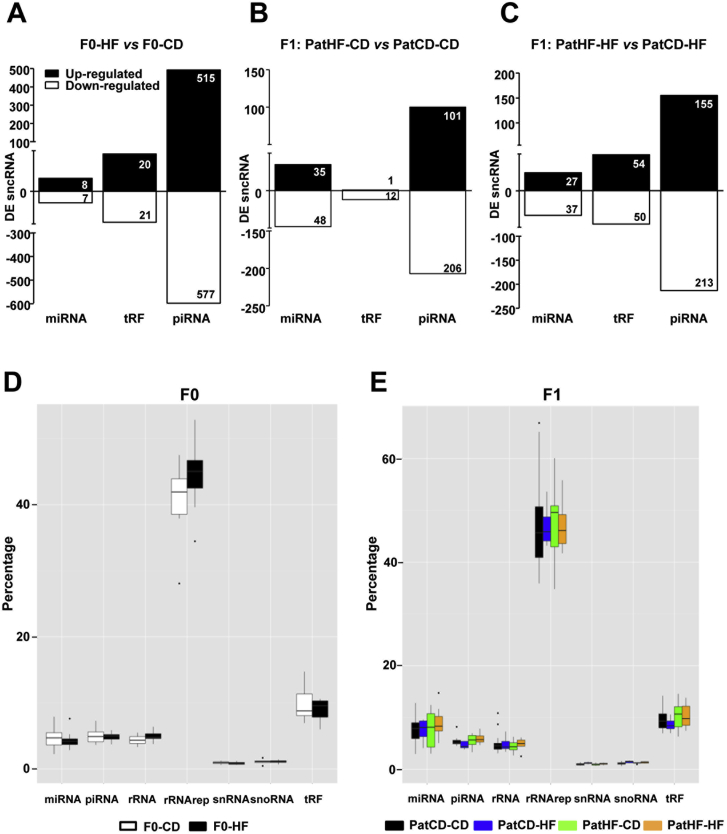
We next compared the expression profiles within each sncRNA subtype and identified several miRNAs, piRNAs and tRFs differentially expressed as a result of HFD in F0 founders (Figure S5A and Supplemental files SF2, SF3 and SF4). We found 15 miRNAs, 41 tRFs and 1092 piRNAs were differentially expressed in sperm of F0-HD vs F0-CD (Figure S5A). Similarly, sperm from F1 offspring of F0-HF fathers showed several differentially expressed sncRNAs as compared to F1 from F0-CD (Figures S5B and S5C, Supplemental files SF2, SF3 and SF4). Diet itself was without effect on the overall proportion of each respective sperm sncRNA category in F0 or F1 rats, showing that differential expression was not driven by an overrepresentation of a respective sncRNA subtype (Figures S5D and S5E). Differentially expressed sncRNAs presented equivalent distribution across chromosomes in both generations, indicating no preferable genomic hot spot for sncRNA expression changes (Figures S4C, S4D and S4E). Interestingly, when analyzing the proportions of each sncRNA subtype in sperm from all F0 compared to all F1 rats (irrespective of diet), we found increased proportions of snRNA, snoRNA, miRNA and rRNA, while tRF and piRNA were significantly unchanged (Figure S4A and S4B). In particular, we found that sperm cells from F1 present 1.8 fold more pri-miRNA than F0 sperm. Moreover, we found that counts per million reads were increased for 280 pri-miRNAs and decreased 60 pri-miRNAs in F1 compared to F0 (data not shown). Collectively, these results suggest that miRNA biogenesis is differently regulated between generations.
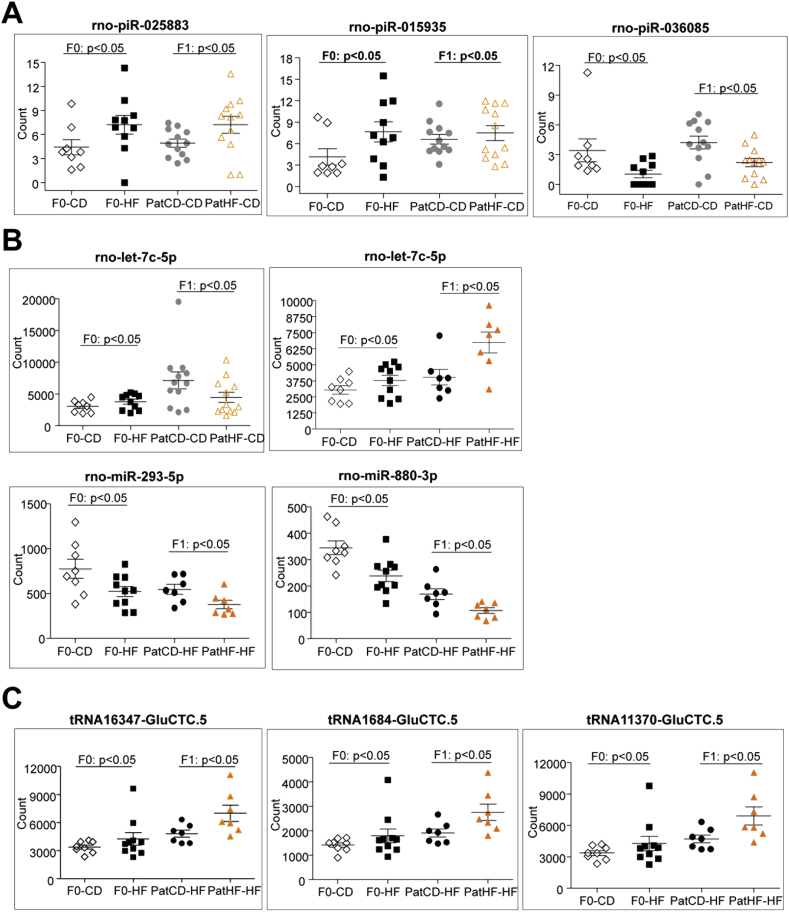
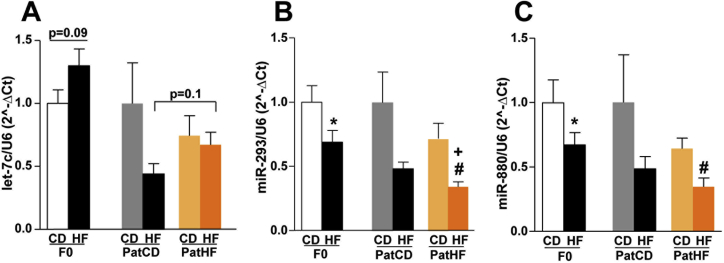
To identify the sncRNA molecules potentially carrying transgenerational effects, we searched for sncRNAs commonly regulated in the sperm of F0 founders fed a HFD and their F1 offspring on chow diet. We found three piRNAs (piR-025883, piR-015935, piR-036085), and the miRNA let-7c differentially expressed in the sperm of F0 fathers fed a HFD and their F1 sons, when compared to their respective controls (Figure 4A,B).
Figure 4.
Small non-coding RNA expression in sperm of F0 and F1 rats shows transgenerational programming. (A) sncRNA molecules differentially expressed in sperm of high fat diet fed F0 founders (F0-HF vs F0-CD) and their F1 offspring (PatHF-CD vs PatCD-CD). Effect of HFD and/or paternal-HFD on the expression of (B) miRNAs in sperm of F0 and F1 rats and (C) tRNA fragments (tRF). F0: n = 8–10 and F1: n = 7–12. Y-axis represents normalized counts per total number of reads of each specific sncRNA category (counts per million). Values are represented as mean ± SEM. F0-CD: F0 on chow diet; F0-HF: F0 on HFD; PatCD-CD: Paternal-Chow on Chow; PatHF-CD: Paternal-HFD on Chow; PatCD-HF: Paternal-Chow on HFD; PatHF-HF: Paternal-HFD on HFD.
A subset of sncRNA was changed under the combined effect of HFD and paternal inheritance; the expression of several tRFs and miRNA species was altered in both F0 fathers under HFD and in their F1 sons only when fed a HFD (PatHF-HF vs PatCD-HF). In these rats, we found that two miRNAs, miR-293-5p and miR-880-3p, were downregulated whereas let-7c-5p, a subset of GLU-CTC.5 tRFs and the tRF GLU-TTC.5-1087 were upregulated in both F0 and F1 generations (Figure 4B, C and S7). Collectively, these results indicate that the expression of sncRNA in sperm is altered in rats transmitting metabolic dysfunction to their offspring.
Analysis of the putative regulatory regions of individual miRNAs and tRFs reported in Figure 4 could not establish a link between sncRNA expression and DNA methylation, suggesting that expression of miRNA and tRF is not under the direct control of DNA methylation (data not shown).
3.4. Expression of let-7c is altered in metabolic tissues of the offspring
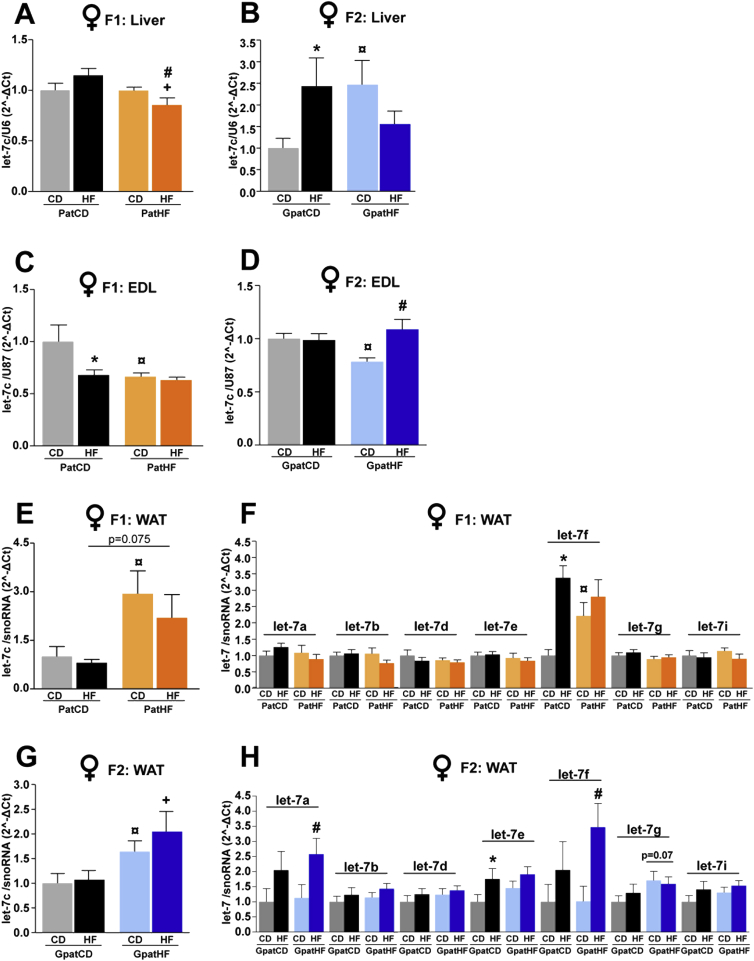
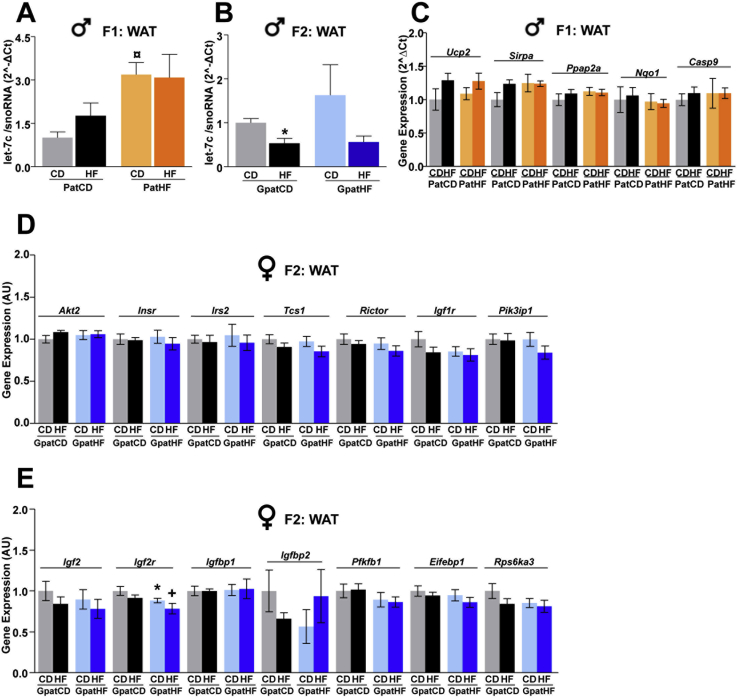
We investigated the functional significance of the differentially regulated miRNAs identified in sperm of F0 and F1 founders and determined the role of these miRNAs in orchestrating the metabolic phenotype of their F1 and F2 female offspring by measuring the expression of miR-293-5p, miR-880-3p and let-7c in liver, gonadal white adipose tissue (WAT) and EDL muscle. Among these three miRNAs, only let-7c was detected in somatic tissues of adult offspring. In liver, let-7c expression was decreased in response to HFD in F1 and F2 offspring of F0-HF fathers but not in offspring of F0-CD fathers (Figure 5A,B). Conversely, let-7c expression was altered in EDL muscle from F1 and F2 females from F0-HF when compared to control offspring (Figure 5C,D). HFD treatment decreased let-7c expression only in EDL of F1 females from F0-CD (Figure 5C). Remarkably, in WAT, let-7c expression was further elevated in F1 and F2 female offspring of F0-HF fathers, independently of their diet (Figure 5E,G). Let-7c expression was also increased in WAT of F1 male offspring from HFD-fed fathers (Figure S6A), but unaltered in male F2 offspring (Figure S6B). Expression analysis of known let-7 family members in WAT from F1 and F2 did not reveal any consistent change in the expression pattern of other let-7 family members between F1 and F2 (Figure 5F,H). For instance, unlike let-7c, let-7f expression was upregulated by HFD in F1, but not in F2 offspring from PatCD (Figure 5F,G). This further suggests that amongst the let-7 family members, let-7c has the greatest potential to contribute to the phenotype of the offspring.
Figure 5.
Let-7c expression is altered in metabolic tissues of offspring. Let-7c expression in liver of (A) F1 (n = 11–12 from 11 to 12 litters), and (B) F2 (n = 6–7 litters; 1 sibling per litter); Extensor Digitorum Longus (EDL) muscle of (C) F1 (n = 5 litters; 1 sibling per litter) and (D) F2 (n = 5 litters; 1 sibling per litter); white adipose tissue (WAT) of (E) F1 (n = 8–10 from 6 to 7 litters; used more than 1 sibling per litter) or (G) F2 (n = 8–13 from 6 to 7 litters; used more than 1 sibling per litter). Expression of all let-7 family members in WAT of (F) F1 and (H) F2 female offspring. U6, U87 or snoRNA were used as internal controls depending on the tissue. Two-way ANOVA followed by Bonferroni post-hoc test: *p ≤ 0.05: PatCD-HF vs PatCD-CD or GpatCD-HF vs GpatCD-CD; ¤p ≤ 0.05: PatHF-CD vs PatCD-CD or GpatHF-CD vs GpatCD-CD; #p ≤ 0.05: PatHF-HF vs PatHF-CD or GpatHF-HF vs GpatHF-CD; +p ≤ 0.05: PatHF-HF vs PatCD-HF or GpatHF-HF vs GpatCD-HF. Results are represented as mean ± SEM. PatCD-CD: Paternal-Chow on Chow; PatHF-CD: Paternal-HFD on Chow; PatCD-HF: Paternal-Chow on HFD; PatHF-HF: Paternal-HFD on HFD; GpatCD-CD: Grandpaternal-Chow on Chow; GpatHF-CD: Grandpaternal-HFD on Chow; GpatCD-HF: Grandpaternal-Chow on HFD; GpatHF-HF: Grandpaternal-HFD on HFD.
3.5. Let-7c regulates the expression of metabolic genes in adipose tissue
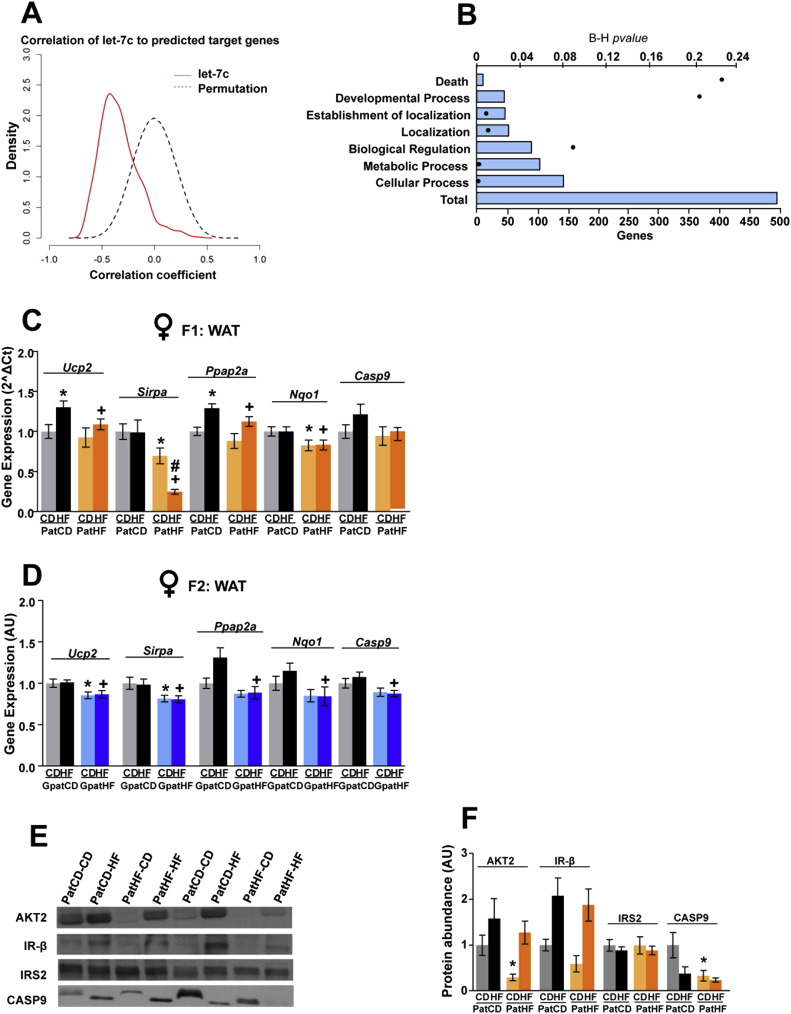
The let-7 miRNA family influences glucose metabolism and insulin sensitivity by regulating the expression of target genes involved in the control of lipid and glucose metabolism, and in the insulin signaling pathway [20], [21]. Transcriptomic analysis of white adipose tissue from F2-female offspring revealed an inversed relationship between let-7c expression and expression of the vast majority of predicted let-7c target genes in rats (Figure 6A). Gene ontology analysis of the targets having an expression negatively correlated to let-7c expression revealed specific enrichment for the GO terms Localization, Cellular process and Metabolic process (Figure 6B). Additionally, we evaluated the expression of several previously reported predicted targets of the broad let-7 family [21]. This transcriptomic analysis showed that the mRNA expression of genes in the insulin/Igf signaling pathway, such as Akt2, Insr and Irs2, was unchanged, whereas Igf2r was decreased in WAT from F2 females from F0-HF (Figures S6D and S6E). However, other predicted target genes specific to let-7c in rats involved in lipid metabolism, such as Ucp2 (Uncoupling protein 2) and Ppap2a (phosphatic acid phosphase 2a), were downregulated in WAT of both F1 and F2-female offspring of F0-HF fathers (Figure 6C,D). Considering that miRNAs can regulate the expression of their targets at the protein level [22], [23], we quantified the protein content of let-7c predicted targets in WAT of F1-female offspring. We found that the protein abundance of AKT2 and Caspase 9 was reduced in WAT of PatHF-CD females when compared to PatCD-CD (Figure 6E,F) and showed a weak correlation (R = 0.38; p = 0.16 and R = 0.44; p = 0.11) with increased let-7c expression (Figure 5E). IR-β expression was reduced by 40% and 10% in PatHF-CD and PatHF-HF, respectively, when compared to controls (Figure 6E,F). Let-7c expression was also upregulated in WAT of F1 male offspring from F0-HF (Figure S6A); however, the mRNA expression of let-7c predicted target genes was not altered (Figure S6C).
Figure 6.
Let-7c expression in WAT of female offspring shows inverse relationship with predicted target genes. (A) Inverse relationship between expression of let-7c and its predicted target genes in white adipose tissue (WAT) of F2 female offspring. Dotted black curve represents the permutation analysis of let-7c expression correlation to target genes. Red curve represents the correlation coefficients of let-7c to predicted target gene. Expression of let-7c predicted target genes was obtained by Affymetrix transcriptomic analysis. (B) Gene ontology analysis of let-7c predicted target genes with an inverse relationship to let-7c expression in WAT of F2-females (performed with DAVID Bioinformatics Resources). Bars represent the number of genes involved in each of the functional clusters and black dots indicate the p values. Total is the total number of genes found to have a significant inverse relationship to let-7c expression. (C) Quantitative PCR validation of let-7c predicted target genes inversely correlated to let-7c expression in WAT of F1 females (n = 10–12 per group; 1 sibling per litter). Geometric mean of the housekeeping genes RPLP0 and β-actin was used for normalization. Values are represented as mean ± SEM (D) Representation of let-7c predicted target genes inversely correlated to let-7c expression in transcriptomic analysis of F2 offspring WAT (n = 6–7 per group; 1 sibling per litter). (E) Representative blots of protein abundance of AKT2, IR-β, IRS2 and Caspase 9 (CASP9) in WAT of F1-female. (F) Protein abundance of AKT2, IR-β, IRS2 and CASP9. Ponceau staining of the membranes was used for data normalization. Results are represented as mean ± SEM. Two-way ANOVA followed by Bonferroni post-hoc test: *p ≤ 0.05: PatCD-HF vs PatCD-CD; ¤p ≤ 0.05: GpatCD-HF vs GpatCD-CD; +p ≤ 0.05: PatHF-HF vs PatCD-HF or GpatHF-HF vs GpatCD-HF. PatCD-CD: Paternal-Chow on Chow; PatHF-CD: Paternal-HFD on Chow; GpatCD-CD: Grandpaternal-Chow on Chow; GpatHF-CD: Grandpaternal-HFD on Chow; GpatCD-HF: Grandpaternal-Chow on HFD; GpatHF-HF: Grandpaternal-HFD on HFD. B–H: Benjamini and Hochberg corrected p-values. AU: arbitrary units.
Together, these data indicate that upregulation of let-7c in WAT of the offspring is associated with changes in expression of predicted let-7c targets at the mRNA or protein level and suggest that let-7c contributes to the changes in the WAT phenotype in response to HFD feeding in founders.
4. Discussion
Here we provide evidence that high-fat diet reprograms the epigenome of rat sperm cells. We demonstrate that high fat diet alters the expression of the miRNA let-7c in the sperm of F0 rats and their F1 offspring. Moreover, differential expression of miRNA let-7c concurs in metabolic tissues of the offspring, altering the expression of its predicted target genes in adipose tissue. These discoveries support the role of spermatozoa in carrying transgenerational epigenetic inheritance.
We used an animal model of paternal inheritance to investigate the role of high-fat diet on the epigenetic reprogramming of gametes. Emerging evidence provides molecular support that transfer of non-genetic information induced by dietary factors, and carried by the gametes, such as changes in the DNA methylation profile [9], [17], histone modification [2] and miRNA expression [17] may influence the metabolic phenotype of the offspring. However, it is possible that other signals, not carried by the gamete itself, may transfer environmentally-induced signals to the offspring. For instance, an abnormal metabolic and hormonal milieu can affect the seminal fluid composition, which, in turn, could alter embryonic and placental development [24]. Additionally, the contribution of simple genetic factors to the paternal effects has been proposed [25], [26], [27], but this putative phenomenon, implying a selective pressure to triage specific spermatozoal genomes that will fertilize the egg, lacks mechanistic foundation. Our identification of similar epigenetic signatures in sperm from both F0 and F1 founders of metabolically disturbed offspring strongly underscores a role of the gametic epigenome in the transmission of transgenerational effects.
We report that 18 loci are differentially methylated in sperm from F0 rats fed a high-fat diet and their F1 offspring, suggesting a role for gametic DNA methylation in transgenerational inheritance of metabolic dysfunction. However, when we tested the DNA methylation in F2 offspring adipose tissue in 3 regions corresponding to DMRs altered in sperm, we found that none of these regions showed differential methylation, despite altered gene expression. This suggests that differential gametic methylation is not carried on or, alternatively, is not persistent in somatic tissues of offspring. Similarly, other research groups found a discrepancy between sperm DNA methylation and expression of the respective genes in metabolic tissue [2], [28]. For example, the paternal effects induced by in utero caloric restriction were associated with alterations in sperm DNA of founders, but the same DMRs were not detected in the somatic tissues of the offspring [28]. Thus, altered sperm DNA methylation may not constitute a program that solely functions in differentiated somatic tissues but may instead be a mechanism modulating the early phases of embryological development with a potential to influence phenotype later in life.
We used a “swim-up” purification method to sort highly pure motile spermatozoa from epithelial cells, which represent a contaminant that introduces artifacts into spermatozoal RNA preparations [18], and sequenced the small RNA content of ultrapure rat sperm cells. We found a substantial number of sequences corresponding to ribosomal rRNA (40–50% of mapped sequences). Although their role in gene interference remains elusive, the high percentage of rRNA in rat sperm does not suggest non-functional RNA degradation products. Rather, the short tRNA-derived fragments detected (9–10% of mapped sequences) could be considered as tRNA byproducts. However, a role of tRFs in the regulation of fundamental biological processes such as regulation of the cell cycle has been proposed [29], potentially involving the regulation of the silencing activity of other classes of small RNAs [30]. We also report a substantial amount of novel sequences that could not be matched to the current small RNA databases (25–40%). These sequences matched poorly with exonic (<1% of sequences) or intronic (<7% of sequences) sequences, supporting the notion that they are not simple degradation products from mRNAs. The fourth most abundant class of small RNA we detected in rat spermatozoa was piRNA, which is specifically enriched in germ line cells [31], compared to somatic tissues. Moreover, piRNAs are classically described to be involved in defending the hypomethylated gametic genome from the rearrangement of transposable elements and in maintaining transposon silencing [32]. In our data set, the differentially expressed piRNAs represents a small number of sequencing counts compared to miRNAs, which could suggest moderate biological relevance.
Several groups have demonstrated the potential of microinjected sperm RNA to both influence embryonic development and mediate transgenerational effects [5], [6], [33]. We provide evidence that HFD transgenerationally reprograms the small RNA content of sperm cells. Our analysis of small RNA species specifically dysregulated in sperm from both F0 and F1 founders of offspring with metabolic dysfunction retrieved three small RNAs including miRNA let-7c. A role of miRNAs in embryonic development is suggested by experiments in mice whereby microinjection of miR-124 into fertilized eggs reduced body weight of the newborn [34]. Our data suggest a functional role of miRNAs, especially let-7c, in mediating transgenerational inheritance of metabolic dysfunction.
The blunted response to HFD-induced weight gain in female F1 and F2 offspring from HFD-fed F0 fathers implies that the plasticity of the adipose tissue was affected. Of interest, adipose tissue showed the tightest association between expression of let-7c in sperm and a specific metabolic tissue in the next generation. Moreover, the transcriptomic analysis of metabolic tissues showed that the strongest link between let-7c expression and expression of its predicted targets occurred in adipose tissue of female offspring. Collectively, our results support a specific role of gametic let-7c expression to functionally reprogram adipose tissue in the offspring.
The miRNA let-7c is a member of the let-7 miRNA family and has been implicated in the regulation of glucose homeostasis, insulin sensitivity and the predisposition to Type 2 diabetes [21], [35]. Maturation of the let-7 family of miRNAs involves the RNA-binding proteins Lin28a and Lin28b [36], [37]. Overexpression of let-7 family members in mice reduces body size and body weight, due to decreased fat mass, and impairs glucose tolerance [35]. Conversely, inhibition of let-7 prevents obesity-induced glucose intolerance and restores insulin signaling in skeletal muscle and liver by upregulating the expression of let-7 targets, such as the insulin receptor and the insulin receptor substrate 2 [35]. In Lin28a/b transgenic mice, suppression of let-7 activates multiple let-7 targets involved in insulin-PI3K-mTOR signaling [21]. Of interest, the repression of let-7c maturation by Lin28a/b deregulates genes involved in cell proliferation and metabolism [21], [38]. We were unable to detect differential mRNA expression of insulin-PI3K signaling-related genes previously shown to be regulated by expression of the broad let-7 family [21], whereas more pronounced changes were observed at the protein level, suggesting that let-7c could regulate these predicted targets by inhibiting translation. Of potential let-7c targets in rats, is the Ppap2a gene that encodes a lipid phosphatic phosphatase enzyme involved in the dephosphorylation of lipid phosphate esters such as phophatidic acid (PA) and lysophosphatidic acid (LPA), two important intermediates of intracellular lipid metabolism and signaling pathways [39]. Let-7c expression was increased in WAT of F1 and F2 offspring from HFD-fed fathers and grandfathers, while expression of its potential target Ppap2a was reduced. This may contribute to the altered body fat composition only observed in F1 and F2 females from the F0-HF lineage. Strikingly, while let-7c expression was dysregulated in sperm from F0 and F1 of paternal HFD lineage, let-7c expression in sperm was upregulated in F0 and downregulated in F1. Given that sperm let-7c is upregulated in response to HFD in F0 and F1 rats, the downregulation of let-7c in sperm from F1 offspring of HFD founders suggests that the regulation of sperm let-7c in these animals is caused by factors other than those inducing an upregulation of let-7c. The effect of HFD itself, however, was identical across generations, as sperm from F1 offspring fed a HFD showed a similar let-7c expression pattern to the F0-HFD group. Collectively, our findings show that sperm let-7c expression is consistently reprogrammed by dietary factors and implicate a role of let-7c in the metabolic phenotype of the offspring.
We induced paternally inherited metabolic dysfunction using approaches described earlier [9], [17] and report that paternal HFD was associated with a decreased pancreatic beta-cell mass in F1 offspring. Here, we found decreased beta-cell mass in newborn female offspring. However, insulin levels were normal in adult offspring, indicating a normal beta-cell function. These data support the notion that the metabolic phenotype in offspring is driven by altered peripheral glucose metabolism, possibly in WAT, rather than by alterations in insulin secretion. We also observed sex-specific differences in the metabolic phenotypes of offspring from HFD-fed fathers. Thus, our results suggest sexual dimorphism in adipose tissue plasticity. Of note, in mice, in a similar model of HFD-induced transgenerational inheritance [17], adipose tissue mass was increased in female, but not male-offspring of HFD fathers. Collectively, these studies provide evidence that adipose tissue is a target for HFD-induced transgenerational reprogramming. Furthermore, these studies reveal that response to paternal obesity robustly targets sex-specific differences, the pancreatic beta-cells and adipose tissue in female offspring. The evolutionary nature of these transgenerational effects is unclear, given that in all cases the metabolic responses in offspring are maladaptive [2], [8], [9], [17], [40]. Maladaptive responses such as a worsening of metabolism in response to environmental insults experienced by the previous generation could partly explain why the prevalence of obesity and associated comorbidities is increasing at exponential, rather than linear rate worldwide. This concept is supported by observations that the transmission of epigenetically driven phenotypes induced by dietary factors is accumulated from one generation to the next [41].
5. Conclusions
Our results provide insight into mechanisms by which high-fat diet reprograms the epigenome of sperm cells and affects the metabolic phenotype of the offspring throughout several generations. Furthermore, we provide evidence that altered miRNA let-7c expression in sperm concurs in the adipose tissue of the offspring, leading to a transcriptomic shift in let-7c predicted targets. In conclusion, reprogramming of sperm let-7c expression may constitute a mechanism by which high-fat diet can alter the metabolism of the offspring.
Author contributions
This study was designed by TDCB, JRZ and RB. Data were acquired by TDCB, PA, JM, JM, MR, LV and SG; analyzed by TDCB, LRI, PA, MR, ID, RS, SG, AK, JRZ and RB; and interpreted by TDCB, LRI, PA, SV, AK, JRZ and RB. The manuscript was written by TDCB, JRZ and RB; and reviewed and approved by all authors.
Acknowledgment
This study was funded by the Novo Nordisk Foundation, Swedish Research Council (#2009-1068 and #2011-3550), European Research Council (233285), Swedish Diabetes Foundation (DIA2012-082), Swedish Foundation for Strategic Research (#SRL10-0027), Fredrik and Ingrid Thurings Foundation (#4-3202/2013), Erik and Edith Fernströms, Karolinska Institutet Fonder (#2013p3re39037) and the Strategic Diabetes Research Program at Karolinska Institutet. The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent Research Center at the University of Copenhagen partially funded by an unrestricted donation from the Novo Nordisk Foundation (www.metabol.ku.dk).
We thank Ann-Marie Pettersson and Dr. Kui Qian for technical assistance and Dr. Marie Björnholm for support with animal experiments. We thank Dan Holmberg for access to the Optical Projection Tomography instruments.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2015.12.002
Conflict of interest
The authors have no conflict of interest to declare.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Supplementary Figure S1.
Metabolic phenotype of F0-male rats after 12 weeks on HFD. (A) Body weight curve (n = 19 (CD) and 20 (HFD)). (B) Energy Intake (n = 20/19). (C) Blood glucose levels during an intraperitoneal insulin tolerance test (ipITT) (n = 9/10). Blood (D) glucose and (E) insulin during the glucose tolerance test (ipGTT), n = 8). Results are represented as mean ± SEM. Student's t-test: *p ≤ 0.05, F0-HF vs F0-CD. N-numbers are indicated for CD and HF, respectively.
Supplementary Figure S2.
Glucose tolerance is impaired, while HFD-induced weight gain is not, in male F1 and F2 offspring. (A–C) Metabolic phenotyping of F1 male offspring after 12 weeks of high fat feeding; blood (A) glucose and (B) insulin during an intraperitoneal glucose tolerance test (ipGTT) (n = 11–14 animals and litters) and (C) body weight curve (n = 18–22 animals from 15 to 16 litters; used more than 1 sibling per litter). (D–F) Metabolic phenotyping of F2 male offspring after 12 weeks of high fat diet feeding. Blood (D) glucose and (E) insulin during an ipGTT (n = 10–11 litters; 1 sibling per litter) and (F) body weight curve (n = 18–24 animals from 10 to 13 litters; used more than 1 sibling per litter). Results are represented as mean ± SEM. Two-way ANOVA followed by Bonferroni post-hoc test: *p ≤ 0.05: PatCD-HF vs PatCD-CD or GpatCD-HF vs GpatCD-CD; ¤p ≤ 0.05: PatHF-CD vs PatCD-CD or GpatHF-CD vs GpatCD-CD; #p ≤ 0.05: PatHF-HF vs PatHF-CD or GpatHF-HF vs GpatHF-CD; +p ≤ 0.05: PatHF-HF vs PatCD-HF or GpatHF-HF vs GpatCD-HF; p < 0.07: GpatHF-HF vs GpatHF-CD or GpatCD-HF vs GpatCD-CD. PatCD-CD: Paternal-Chow on Chow; PatHF-CD: Paternal-HFD on Chow; PatCD-HF: Paternal-Chow on HFD; PatHF-HF: Paternal-HFD on HFD; GpatCD-CD: Grandpaternal-Chow on Chow; GpatHF-CD: Grandpaternal-HFD on Chow; GpatCD-HF: Grandpaternal-Chow on HFD; GpatHF-HF: Grandpaternal-HFD on HFD.
Supplementary Figure S3.
Transcriptomic analysis and DNA methylation in metabolic tissues of offspring shows modest association to sperm DNA methylation. (A) Gene expression representation in Extensor Digitorum Longus (EDL) muscle, liver and white adipose tissue (WAT) of the F2-female offspring. Represented targets are the nearest genes of the 18 sperm DMRs common to F0 and F1 offspring. Data are represented as log2 of the fold change of GpatHF-CD vs GpatCD-CD F2-females. (B) Target-specific DNA methylation in WAT of F2 females. DNA methylation is represented as percentage of methylated DNA relative to input (total) DNA of each target measured by quantitative PCR after MBD-capture (n = 7–9). Student's t-test: *p < 0.05: GpatHF-CD vs GpatCD-CD. GpatCD-CD: Grandpaternal-Chow on Chow; GPatHF-CD: Grandpaternal-HFD on Chow. N.D.: Not Detected. FC: Fold Change.
Supplementary Figure S4.
SncRNA expression pattern in sperm from F0 and F1 rats. Distribution of different small non-coding RNAs (sncRNAs) categories in sperm of (A) F0 (n = 8–10) and (B) F1 (n = 7 per group) rats. (C–E) Chromosomal distribution (sperm karyogram) of differentially expressed sncRNA: (C) F0-HF vs F0-CD; (D) F1 (PatHF-CD vs PatCD-CD) and (E) F1 (PatHF-HF vs PatCD-HF). F0-CD: F0 on chow diet; F0-HF: F0 on HFD; PatCD-CD: Paternal-Chow on Chow; PatHF-CD: Paternal-HFD on Chow; PatCD-HF: Paternal-Chow on HFD; PatHF-HF: Paternal-HFD on HFD. Student's t-test: *p ≤ 0.05 (F1 vs F0). F0-CD: F0 on chow diet; F0-HF: F0 on HFD; PatCD-CD: Paternal-Chow on Chow; PatHF-CD: Paternal-HFD on Chow; PatCD-HF: Paternal-Chow on HFD; PatHF-HF: Paternal-HFD on HFD.
Supplementary Figure S5.
Gametic small non-coding RNA expression is altered by HFD and paternal diet. (A–C) Number of differentially expressed sncRNAs (miRNAs, tRFs and piRNAs) in the sperm of (A) F0-HF vs F0-CD (n = 8–10), (B) F1 offspring of F0-HF (PatHF-CD) vs F1 of F0-CD (PatCD-CD) fed a chow diet or (C) a high fat diet (PatHF-HF vs PatCD-HF) (n = 7 per group; 1 sibling per litter). Sperm sncRNA expression pattern in percentage per group in (D) F0 and (E) F1. F0-CD: F0 on chow diet; F0-HF: F0 on HFD; PatCD-CD: Paternal-Chow on Chow; PatHF-CD: Paternal-HFD on Chow; PatCD-HF: Paternal-Chow on HFD; PatHF-HF: Paternal-HFD on HFD. DE: Differentially Expressed. rRNA (rRNA annotation from Ensemble) and rRNArep (rRNA annotation from repeatmask2).
Supplementary Figure S6.
Expression of let-7c in WAT of male offspring and mRNA expression of predicted targets. Expression of let-7c in WAT of male offspring (A) F1 and (B) F2. The snoRNA was used as internal control. (C) Quantitative PCR validation of let-7c predicted target genes in WAT of F1 males (n = 6 per group; 1 sibling per litter). Geometric mean of the housekeeping genes RPLP0 and β-actin was used for normalization. Transcriptomic analysis in WAT of F2-female offspring (D) Known let-7 target genes in the insulin/Igf signaling pathway and (E) Insulin/Igf signaling predicted target genes of let-7c in rats (n = 6–7). Values are represented as mean ± SEM. *p ≤ 0.05: PatCD-HF vs PatCD-CD; ¤p ≤ 0.05: PatHF-CD vs PatCD-CD; +p ≤ 0.05: PatHF-HF vs PatCD-HF.
Supplementary Figure S7.
Validation of miRNA expression in sperm of F0 and F1 offspring. miRNA expression measured by quantitative PCR using TaqMan specific probes for (A) let-7c; (B) miR-293 and (C) miR-880. U6 was used as internal control. Values are represented as mean ± SEM. Student's t-test or two-way ANOVA followed by Bonferroni post-hoc test: *p ≤ 0.05: F0-HF vs F0-CD; +p ≤ 0.05: PatHF-HF vs PatCD-HF; #p ≤ 0.05: PatHF-HF vs PatHF-CD. F0 (n = 13–16); F1 (n = 7–12).
References
- 1.Lake J.K., Power C., Cole T.J. Child to adult body mass index in the 1958 British birth cohort: associations with parental obesity. Archives of Disease in Childhood. 1997;77:376–381. doi: 10.1136/adc.77.5.376. [DOI] [PubMed] [Google Scholar]
- 2.Carone B.R., Fauquier L., Habib N., Shea J.M., Hart C.E., Li R. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jimenez-Chillaron J.C., Isganaitis E., Charalambous M., Gesta S., Pentinat-Pelegrin T., Faucette R.R. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes. 2009;58:460–468. doi: 10.2337/db08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pembrey M.E., Bygren L.O., Kaati G., Edvinsson S., Northstone K., Sjostrom M. Sex-specific, male-line transgenerational responses in humans. European Journal of Human Genetics. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 5.Gapp K., Jawaid A., Sarkies P., Bohacek J., Pelczar P., Prados J. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nature Neuroscience. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner K.D., Wagner N., Ghanbarian H., Grandjean V., Gounon P., Cuzin F. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Developmental Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Ghildiyal M., Zamore P.D. Small silencing RNAs: an expanding universe. Nature Reviews Genetics. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson L.M., Riffle L., Wilson R., Travlos G.S., Lubomirski M.S., Alvord W.G. Preconceptional fasting of fathers alters serum glucose in offspring of mice. Nutrition. 2006;22:327–331. doi: 10.1016/j.nut.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Ng S.F., Lin R.C., Laybutt D.R., Barres R., Owens J.A., Morris M.J. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 10.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E. Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sai Lakshmi S., Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Research. 2008;36:D173–D177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D. The UCSC table browser data retrieval tool. Nucleic Acids Research. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao X., Sherman B.T., Huang da W., Stephens R., Baseler M.W., Lane H.C. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fullston T., Ohlsson Teague E.M., Palmer N.O., DeBlasio M.J., Mitchell M., Corbett M. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB Journal. 2013;27:4226–4243. doi: 10.1096/fj.12-224048. [DOI] [PubMed] [Google Scholar]
- 18.Mortimer D. Sperm preparation methods. Journal of Andrology. 2000;21:357–366. [PubMed] [Google Scholar]
- 19.Krawetz S.A., Kruger A., Lalancette C., Tagett R., Anton E., Draghici S. A survey of small RNAs in human sperm. Human Reproduction. 2011;26:3401–3412. doi: 10.1093/humrep/der329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papaioannou G., Inloes J.B., Nakamura Y., Paltrinieri E., Kobayashi T. let-7 and miR-140 microRNAs coordinately regulate skeletal development. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E3291–E3300. doi: 10.1073/pnas.1302797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H., Shyh-Chang N., Segre A.V., Shinoda G., Shah S.P., Einhorn W.S. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 24.Robertson S.A. GM-CSF regulation of embryo development and pregnancy. Cytokine & Growth Factor Reviews. 2007;18:287–298. doi: 10.1016/j.cytogfr.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 25.Rando O.J. Daddy issues: paternal effects on phenotype. Cell. 2012;151:702–708. doi: 10.1016/j.cell.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waterland R.A. Epigenetic mechanisms affecting regulation of energy balance: many questions, few answers. Annual Review of Nutrition. 2014;34:337–355. doi: 10.1146/annurev-nutr-071813-105315. [DOI] [PubMed] [Google Scholar]
- 27.Nelson V.R., Heaney J.D., Tesar P.J., Davidson N.O., Nadeau J.H. Transgenerational epigenetic effects of the Apobec1 cytidine deaminase deficiency on testicular germ cell tumor susceptibility and embryonic viability. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2766–E2773. doi: 10.1073/pnas.1207169109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radford E.J., Ito M., Shi H., Corish J.A., Yamazawa K., Isganaitis E. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014;345:1255903. doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y.S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes & Development. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A.Z., Kay M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siomi M.C., Sato K., Pezic D., Aravin A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nature Review Molecular Cell Biology. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 32.O'Donnell K.A., Boeke J.D. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rassoulzadegan M., Grandjean V., Gounon P., Vincent S., Gillot I., Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 34.Grandjean V., Gounon P., Wagner N., Martin L., Wagner K.D., Bernex F. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- 35.Frost R.J., Olson E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman M.A., Thomson J.M., Hammond S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E.E., Nitsch R. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature Cell Biology. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 38.Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E.G., Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes & Development. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris A.J., Smyth S.S. Lipid phosphate phosphatases: more than one way to put the brakes on LPA signaling? Journal of Lipid Research. 2014;55:2195–2197. doi: 10.1194/jlr.C054957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braunschweig M., Jagannathan V., Gutzwiller A., Bee G. Investigations on transgenerational epigenetic response down the male line in F2 pigs. PLoS One. 2012;7:e30583. doi: 10.1371/journal.pone.0030583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waterland R.A., Travisano M., Tahiliani K.G. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. FASEB Journal. 2007;21:3380–3385. doi: 10.1096/fj.07-8229com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.